

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0262.10
(145 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
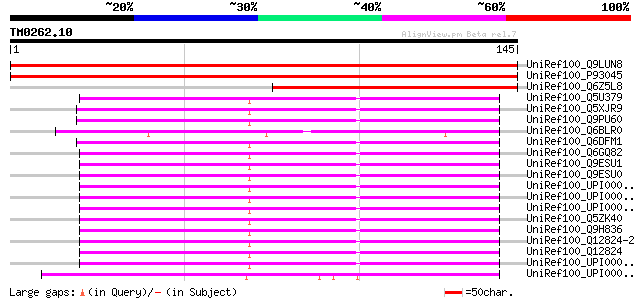
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LUN8 SNF5, transcription regulatory protein homolog ... 235 1e-61
UniRef100_P93045 SNF5 homolog BSH [Arabidopsis thaliana] 232 1e-60
UniRef100_Q6Z5L8 Putative SNF5 homolog BSH [Oryza sativa] 94 8e-19
UniRef100_Q5U379 Zgc:92517 [Brachydanio rerio] 78 5e-14
UniRef100_Q5XJR9 SWI/SNF-related matrix associated protein [Brac... 77 6e-14
UniRef100_Q9PU60 SWI/SNF-related matrix associated protein [Brac... 77 6e-14
UniRef100_Q6BLR0 Debaryomyces hansenii chromosome F of strain CB... 77 6e-14
UniRef100_Q6DFM1 SWI/SNF related, matrix associated, actin depen... 77 1e-13
UniRef100_Q6GQ82 MGC80271 protein [Xenopus laevis] 76 1e-13
UniRef100_Q9ESU1 Integrase interactor 1 [Mus musculus] 76 1e-13
UniRef100_Q9ESU0 Integrase interactor [Mus musculus] 76 1e-13
UniRef100_UPI000036C861 UPI000036C861 UniRef100 entry 75 3e-13
UniRef100_UPI00003AB0EC UPI00003AB0EC UniRef100 entry 75 3e-13
UniRef100_UPI00003AB0EB UPI00003AB0EB UniRef100 entry 75 3e-13
UniRef100_Q5ZK40 Hypothetical protein [Gallus gallus] 75 3e-13
UniRef100_Q9H836 Hypothetical protein FLJ13963 [Homo sapiens] 75 3e-13
UniRef100_Q12824-2 Splice isoform B of Q12824 [Homo sapiens] 75 3e-13
UniRef100_Q12824 SWI/SNF related, matrix associated, actin depen... 75 3e-13
UniRef100_UPI0000361E77 UPI0000361E77 UniRef100 entry 74 5e-13
UniRef100_UPI00002365A3 UPI00002365A3 UniRef100 entry 74 9e-13
>UniRef100_Q9LUN8 SNF5, transcription regulatory protein homolog BSH [Arabidopsis
thaliana]
Length = 240
Score = 235 bits (600), Expect = 1e-61
Identities = 112/145 (77%), Positives = 127/145 (87%)
Query: 1 MKTPASVFYRNPVKFRMPTAENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTV 60
MK S ++ PVKFRMPTAENLVPIRLDI+ + +RYKDAFTWNPSDPD+EV +FA+RTV
Sbjct: 1 MKGLVSTGWKGPVKFRMPTAENLVPIRLDIQFEGQRYKDAFTWNPSDPDNEVVIFAKRTV 60
Query: 61 KDLKLPPPFVTQIAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDL 120
KDLKLP FVTQIAQSI+SQL++FR+YEGQDMY GEKIIPIKLDL VNH L+KDQFLWDL
Sbjct: 61 KDLKLPYAFVTQIAQSIQSQLSDFRAYEGQDMYTGEKIIPIKLDLRVNHTLIKDQFLWDL 120
Query: 121 NNFESDLEELARIFCKDMGIEDPEV 145
NNFESD EE AR CKD+G+EDPEV
Sbjct: 121 NNFESDPEEFARTLCKDLGVEDPEV 145
>UniRef100_P93045 SNF5 homolog BSH [Arabidopsis thaliana]
Length = 240
Score = 232 bits (592), Expect = 1e-60
Identities = 111/145 (76%), Positives = 126/145 (86%)
Query: 1 MKTPASVFYRNPVKFRMPTAENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTV 60
MK S ++ PVKFRMPTAENLVPIRLDI+ + +RYKDAFTWNPSDPD+EV +FA+RTV
Sbjct: 1 MKGLVSTGWKGPVKFRMPTAENLVPIRLDIQFEGQRYKDAFTWNPSDPDNEVVIFAKRTV 60
Query: 61 KDLKLPPPFVTQIAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDL 120
KDLKLP FVTQIAQSI+SQL++FR+YEGQDMY GEKIIPIKLDL VN L+KDQFLWDL
Sbjct: 61 KDLKLPYAFVTQIAQSIQSQLSDFRAYEGQDMYTGEKIIPIKLDLRVNQTLIKDQFLWDL 120
Query: 121 NNFESDLEELARIFCKDMGIEDPEV 145
NNFESD EE AR CKD+G+EDPEV
Sbjct: 121 NNFESDPEEFARTLCKDLGVEDPEV 145
>UniRef100_Q6Z5L8 Putative SNF5 homolog BSH [Oryza sativa]
Length = 171
Score = 93.6 bits (231), Expect = 8e-19
Identities = 40/70 (57%), Positives = 54/70 (77%)
Query: 76 SIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFC 135
SI+ QL EFRSYEG++M EKI+P+K+DL +N+ +++DQFLWD+ N +SD EE AR C
Sbjct: 4 SIQGQLTEFRSYEGEEMQIREKIVPLKIDLRINNTVIRDQFLWDIGNLDSDPEEFARTLC 63
Query: 136 KDMGIEDPEV 145
D+ I DPEV
Sbjct: 64 DDLNITDPEV 73
Score = 41.6 bits (96), Expect = 0.004
Identities = 25/71 (35%), Positives = 36/71 (50%), Gaps = 1/71 (1%)
Query: 17 MPTAENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFV-TQIAQ 75
M E +VP+++D+ I+ +D F W+ + D + FAR DL + P V IA
Sbjct: 20 MQIREKIVPLKIDLRINNTVIRDQFLWDIGNLDSDPEEFARTLCDDLNITDPEVGPAIAV 79
Query: 76 SIESQLAEFRS 86
SI QL E S
Sbjct: 80 SIREQLYEIAS 90
>UniRef100_Q5U379 Zgc:92517 [Brachydanio rerio]
Length = 373
Score = 77.8 bits (190), Expect = 5e-14
Identities = 45/121 (37%), Positives = 71/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +FA DL L P FV IA +I
Sbjct: 172 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFAEILCDDLDLSPLTFVPAIASAIRQ 231
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + D +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 232 QIESYPTDSILDEQMDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 290
Query: 140 I 140
+
Sbjct: 291 L 291
Score = 35.8 bits (81), Expect = 0.20
Identities = 21/59 (35%), Positives = 31/59 (51%)
Query: 24 VPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIESQLA 82
V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI QL+
Sbjct: 250 VIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRGQLS 308
>UniRef100_Q5XJR9 SWI/SNF-related matrix associated protein [Brachydanio rerio]
Length = 370
Score = 77.4 bits (189), Expect = 6e-14
Identities = 45/122 (36%), Positives = 71/122 (57%), Gaps = 2/122 (1%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIE 78
AE LVPIRLD+EID ++ +DAFTWN ++ +FA DL L P FV IA +I
Sbjct: 164 AEVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFAEILCDDLDLNPLTFVPAIASAIR 223
Query: 79 SQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDM 138
Q+ + + + +++I IKL++ V ++ + DQF WD++ E+ E A C ++
Sbjct: 224 QQIESYPTDSILEEQTDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPESFALKLCSEL 282
Query: 139 GI 140
G+
Sbjct: 283 GL 284
Score = 35.8 bits (81), Expect = 0.20
Identities = 21/59 (35%), Positives = 31/59 (51%)
Query: 24 VPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIESQLA 82
V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI QL+
Sbjct: 243 VIIKLNIHVGNISLVDQFEWDMSEKENSPESFALKLCSELGLGGEFVTTIAYSIRGQLS 301
>UniRef100_Q9PU60 SWI/SNF-related matrix associated protein [Brachydanio rerio]
Length = 239
Score = 77.4 bits (189), Expect = 6e-14
Identities = 45/122 (36%), Positives = 71/122 (57%), Gaps = 2/122 (1%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIE 78
AE LVPIRLD+EID ++ +DAFTWN ++ +FA DL L P FV IA +I
Sbjct: 33 AEVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFAEILCDDLDLNPLTFVPAIASAIR 92
Query: 79 SQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDM 138
Q+ + + + +++I IKL++ V ++ + DQF WD++ E+ E A C ++
Sbjct: 93 QQIESYPTDSILEEQTDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPESFALKLCSEL 151
Query: 139 GI 140
G+
Sbjct: 152 GL 153
Score = 35.8 bits (81), Expect = 0.20
Identities = 21/59 (35%), Positives = 31/59 (51%)
Query: 24 VPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIESQLA 82
V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI QL+
Sbjct: 112 VIIKLNIHVGNISLVDQFEWDMSEKENSPESFALKLCSELGLGGEFVTTIAYSIRGQLS 170
>UniRef100_Q6BLR0 Debaryomyces hansenii chromosome F of strain CBS767 of Debaryomyces
hansenii [Debaryomyces hansenii]
Length = 577
Score = 77.4 bits (189), Expect = 6e-14
Identities = 46/131 (35%), Positives = 72/131 (54%), Gaps = 6/131 (4%)
Query: 14 KFRMPTAENLVPIRLDIEIDAKRYK--DAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVT 71
+ R + LVPIRL+ + + ++K D F W+ ++ + F ++D K PP
Sbjct: 216 RLRPSETQQLVPIRLEFDQERDKFKLRDTFLWDLNEDVLPLENFVALLIEDYKFIPPHHA 275
Query: 72 Q-IAQSIESQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNF-ESDLEE 129
Q I SI Q+ +F + D GE +PIK+D+ +N+ + DQF WD+ NF ESD E+
Sbjct: 276 QTILASIREQIRDF--HRKPDKVMGELRVPIKIDVTINNTQLVDQFEWDILNFGESDPED 333
Query: 130 LARIFCKDMGI 140
ARI C +M +
Sbjct: 334 FARIMCDEMNL 344
Score = 42.0 bits (97), Expect = 0.003
Identities = 32/102 (31%), Positives = 46/102 (44%), Gaps = 20/102 (19%)
Query: 8 FYRNPVKFRMPTAENLVPIRLDIEIDAKRYKDAFTWN-----PSDPDHEVGVFARRTVKD 62
F+R P K E VPI++D+ I+ + D F W+ SDP+ FAR +
Sbjct: 289 FHRKPDKVM---GELRVPIKIDVTINNTQLVDQFEWDILNFGESDPED----FARIMCDE 341
Query: 63 LKLPPPFVTQIAQSIESQLAEFR--------SYEGQDMYAGE 96
+ LP F T IA +I Q F S++G +Y E
Sbjct: 342 MNLPGEFTTAIAHTIREQTQLFHKSLFLVGYSFDGSPVYEDE 383
>UniRef100_Q6DFM1 SWI/SNF related, matrix associated, actin dependent regulator of
chromatin, subfamily b, member 1 [Xenopus tropicalis]
Length = 378
Score = 76.6 bits (187), Expect = 1e-13
Identities = 44/122 (36%), Positives = 73/122 (59%), Gaps = 2/122 (1%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIE 78
+E LVPIRLD+EID ++ +DAFTWN ++ +FA DL L P FV IA +I
Sbjct: 176 SEVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFAEILCDDLDLNPLAFVPAIASAIR 235
Query: 79 SQLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDM 138
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++
Sbjct: 236 QQIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSEL 294
Query: 139 GI 140
G+
Sbjct: 295 GL 296
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 251 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 310
Query: 80 QLA 82
QL+
Sbjct: 311 QLS 313
>UniRef100_Q6GQ82 MGC80271 protein [Xenopus laevis]
Length = 378
Score = 76.3 bits (186), Expect = 1e-13
Identities = 44/121 (36%), Positives = 72/121 (59%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +FA DL L P FV IA +I
Sbjct: 177 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFAEILCDDLDLNPLAFVPAIASAIRQ 236
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 237 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 295
Query: 140 I 140
+
Sbjct: 296 L 296
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 251 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 310
Query: 80 QLA 82
QL+
Sbjct: 311 QLS 313
>UniRef100_Q9ESU1 Integrase interactor 1 [Mus musculus]
Length = 385
Score = 76.3 bits (186), Expect = 1e-13
Identities = 43/121 (35%), Positives = 73/121 (59%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ + +F+ DL L P FV IA +I
Sbjct: 184 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTLEMFSEILCDDLDLNPLTFVPAIASAIRQ 243
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 244 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 302
Query: 140 I 140
+
Sbjct: 303 L 303
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 258 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 317
Query: 80 QLA 82
QL+
Sbjct: 318 QLS 320
>UniRef100_Q9ESU0 Integrase interactor [Mus musculus]
Length = 376
Score = 76.3 bits (186), Expect = 1e-13
Identities = 43/121 (35%), Positives = 73/121 (59%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ + +F+ DL L P FV IA +I
Sbjct: 175 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTLEMFSEILCDDLDLNPLTFVPAIASAIRQ 234
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 235 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 293
Query: 140 I 140
+
Sbjct: 294 L 294
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 249 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 308
Query: 80 QLA 82
QL+
Sbjct: 309 QLS 311
>UniRef100_UPI000036C861 UPI000036C861 UniRef100 entry
Length = 219
Score = 75.1 bits (183), Expect = 3e-13
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 18 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 77
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 78 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 136
Query: 140 I 140
+
Sbjct: 137 L 137
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 92 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 151
Query: 80 QLA 82
QL+
Sbjct: 152 QLS 154
>UniRef100_UPI00003AB0EC UPI00003AB0EC UniRef100 entry
Length = 376
Score = 75.1 bits (183), Expect = 3e-13
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 175 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 234
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 235 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 293
Query: 140 I 140
+
Sbjct: 294 L 294
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 249 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 308
Query: 80 QLA 82
QL+
Sbjct: 309 QLS 311
>UniRef100_UPI00003AB0EB UPI00003AB0EB UniRef100 entry
Length = 386
Score = 75.1 bits (183), Expect = 3e-13
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 185 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 244
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 245 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 303
Query: 140 I 140
+
Sbjct: 304 L 304
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 259 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 318
Query: 80 QLA 82
QL+
Sbjct: 319 QLS 321
>UniRef100_Q5ZK40 Hypothetical protein [Gallus gallus]
Length = 386
Score = 75.1 bits (183), Expect = 3e-13
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 185 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 244
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 245 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 303
Query: 140 I 140
+
Sbjct: 304 L 304
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 259 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 318
Query: 80 QLA 82
QL+
Sbjct: 319 QLS 321
>UniRef100_Q9H836 Hypothetical protein FLJ13963 [Homo sapiens]
Length = 394
Score = 75.1 bits (183), Expect = 3e-13
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 193 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 252
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 253 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 311
Query: 140 I 140
+
Sbjct: 312 L 312
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 267 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 326
Query: 80 QLA 82
QL+
Sbjct: 327 QLS 329
>UniRef100_Q12824-2 Splice isoform B of Q12824 [Homo sapiens]
Length = 376
Score = 75.1 bits (183), Expect = 3e-13
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 175 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 234
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 235 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 293
Query: 140 I 140
+
Sbjct: 294 L 294
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 249 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 308
Query: 80 QLA 82
QL+
Sbjct: 309 QLS 311
>UniRef100_Q12824 SWI/SNF related, matrix associated, actin dependent regulator of
chromatin subfamily B member 1 [Homo sapiens]
Length = 385
Score = 75.1 bits (183), Expect = 3e-13
Identities = 43/121 (35%), Positives = 72/121 (58%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +DAFTWN ++ +F+ DL L P FV IA +I
Sbjct: 184 EVLVPIRLDMEIDGQKLRDAFTWNMNEKLMTPEMFSEILCDDLDLNPLTFVPAIASAIRQ 243
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + + + +++I IKL++ V ++ + DQF WD++ E+ E+ A C ++G
Sbjct: 244 QIESYPTDSILEDQSDQRVI-IKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELG 302
Query: 140 I 140
+
Sbjct: 303 L 303
Score = 36.2 bits (82), Expect = 0.16
Identities = 21/63 (33%), Positives = 33/63 (52%)
Query: 20 AENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIES 79
++ V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI
Sbjct: 258 SDQRVIIKLNIHVGNISLVDQFEWDMSEKENSPEKFALKLCSELGLGGEFVTTIAYSIRG 317
Query: 80 QLA 82
QL+
Sbjct: 318 QLS 320
>UniRef100_UPI0000361E77 UPI0000361E77 UniRef100 entry
Length = 236
Score = 74.3 bits (181), Expect = 5e-13
Identities = 43/121 (35%), Positives = 68/121 (55%), Gaps = 2/121 (1%)
Query: 21 ENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPP-PFVTQIAQSIES 79
E LVPIRLD+EID ++ +D FTWN ++ +FA DL L P FV IA +I
Sbjct: 17 EALVPIRLDMEIDGQKLRDTFTWNMNEKLMTPEMFAEILCDDLDLSPLAFVPAIASAIRQ 76
Query: 80 QLAEFRSYEGQDMYAGEKIIPIKLDLCVNHMLVKDQFLWDLNNFESDLEELARIFCKDMG 139
Q+ + + +++I IKL++ V ++ + DQF WD++ E+ E A C ++G
Sbjct: 77 QIESYPMDTILEEQTDQRVI-IKLNIHVGNISLMDQFEWDMSERENSPESFALKLCSELG 135
Query: 140 I 140
+
Sbjct: 136 L 136
Score = 35.8 bits (81), Expect = 0.20
Identities = 21/59 (35%), Positives = 31/59 (51%)
Query: 24 VPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIESQLA 82
V I+L+I + D F W+ S+ ++ FA + +L L FVT IA SI QL+
Sbjct: 95 VIIKLNIHVGNISLMDQFEWDMSERENSPESFALKLCSELGLGGEFVTTIAYSIRGQLS 153
>UniRef100_UPI00002365A3 UPI00002365A3 UniRef100 entry
Length = 798
Score = 73.6 bits (179), Expect = 9e-13
Identities = 48/147 (32%), Positives = 76/147 (51%), Gaps = 16/147 (10%)
Query: 10 RNPVKFRMPTAENLVPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLP--- 66
R +K + E+LVPIRLDI+ + + +D FTWN D +FA + V+DL LP
Sbjct: 223 RKDMKTQSEQLEHLVPIRLDIDWEKVKIRDTFTWNLHDRVVSPDLFAEKLVEDLGLPLES 282
Query: 67 -PPFVTQIAQSIESQLAEFRSY-----EGQD------MYAGEKI-IPIKLDLCVNHMLVK 113
P V I+QSI+ Q+ +F + E D Y +++ I +KL++ + +
Sbjct: 283 CGPLVRMISQSIQEQICDFYPHVHIEEEALDPHLPYSAYKNDELRIVVKLNITIGQHTLV 342
Query: 114 DQFLWDLNNFESDLEELARIFCKDMGI 140
DQF WD+N+ + EE A D+ +
Sbjct: 343 DQFEWDINDPHNSPEEFAARMTTDLSL 369
Score = 39.7 bits (91), Expect = 0.014
Identities = 21/57 (36%), Positives = 28/57 (48%)
Query: 24 VPIRLDIEIDAKRYKDAFTWNPSDPDHEVGVFARRTVKDLKLPPPFVTQIAQSIESQ 80
+ ++L+I I D F W+ +DP + FA R DL L F T IA SI Q
Sbjct: 328 IVVKLNITIGQHTLVDQFEWDINDPHNSPEEFAARMTTDLSLSGEFTTAIAHSIREQ 384
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 254,683,772
Number of Sequences: 2790947
Number of extensions: 9853867
Number of successful extensions: 19397
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 19174
Number of HSP's gapped (non-prelim): 145
length of query: 145
length of database: 848,049,833
effective HSP length: 121
effective length of query: 24
effective length of database: 510,345,246
effective search space: 12248285904
effective search space used: 12248285904
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0262.10


