

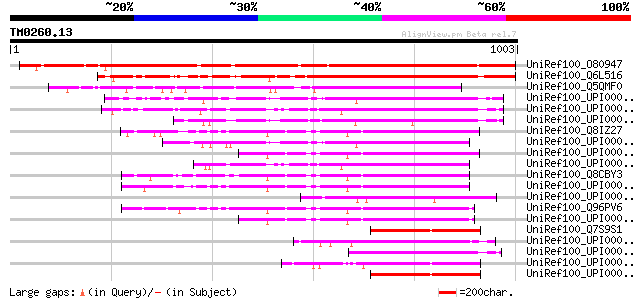
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0260.13
(1003 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80947 Hypothetical protein At2g39340 [Arabidopsis tha... 1018 0.0
UniRef100_Q6L516 Putative GANP protein [Oryza sativa] 681 0.0
UniRef100_Q5QMF0 Putative SAC3/GANP family protein [Oryza sativa] 628 e-178
UniRef100_UPI0000361091 UPI0000361091 UniRef100 entry 272 4e-71
UniRef100_UPI0000247721 UPI0000247721 UniRef100 entry 265 7e-69
UniRef100_UPI000027CB3C UPI000027CB3C UniRef100 entry 261 1e-67
UniRef100_Q8IZ27 Leukocyte receptor cluster (LRC) member 8 [Homo... 257 1e-66
UniRef100_UPI0000361090 UPI0000361090 UniRef100 entry 255 5e-66
UniRef100_UPI000036C0A0 UPI000036C0A0 UniRef100 entry 249 2e-64
UniRef100_UPI0000436B5C UPI0000436B5C UniRef100 entry 246 2e-63
UniRef100_Q8CBY3 Mus musculus adult male diencephalon cDNA, RIKE... 242 5e-62
UniRef100_UPI00001CE789 UPI00001CE789 UniRef100 entry 240 1e-61
UniRef100_UPI00003C2180 UPI00003C2180 UniRef100 entry 238 5e-61
UniRef100_Q96PV6 KIAA1932 protein [Homo sapiens] 236 3e-60
UniRef100_UPI000036C09F UPI000036C09F UniRef100 entry 234 1e-59
UniRef100_Q7S9S1 Hypothetical protein [Neurospora crassa] 218 1e-54
UniRef100_UPI000042C24F UPI000042C24F UniRef100 entry 215 6e-54
UniRef100_UPI0000248E94 UPI0000248E94 UniRef100 entry 214 1e-53
UniRef100_UPI000021B5A6 UPI000021B5A6 UniRef100 entry 208 6e-52
UniRef100_UPI000023CC0B UPI000023CC0B UniRef100 entry 207 2e-51
>UniRef100_O80947 Hypothetical protein At2g39340 [Arabidopsis thaliana]
Length = 989
Score = 1018 bits (2632), Expect = 0.0
Identities = 555/1013 (54%), Positives = 693/1013 (67%), Gaps = 58/1013 (5%)
Query: 20 ENRH-FDPGQNQPPSYAPTTPGSQAVSWAVQS----STANGGYSNPTYQYDQHPQPPGRS 74
+NR+ D Q Q SY +T GS++ W S + NG YSN Y HPQP G +
Sbjct: 2 QNRYGVDGSQTQKYSYQYST-GSESAPWTGHSVENQAVENGNYSNSNYY---HPQPTGPA 57
Query: 75 IQDGQNVSSVAGN--SSNLGTANVPQDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPN 132
+ Q + + SS GTANV QDY+ YT Y +SS+P+ Y + GY+ YY+ YQQQP+
Sbjct: 58 TGNVQEIPNTVSFTISSTSGTANVAQDYSGYTPYQTSSDPHNYSNTGYSNYYSGYQQQPS 117
Query: 133 QTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQTTGVYQN----- 187
Q+Y QPVGAYQNTGAP QP+SSFQN GSYAG+ +YS TYYNPA DYQT G YQ+
Sbjct: 118 QSYPQPVGAYQNTGAP-QPLSSFQNPGSYAGTPSYSGTYYNPA--DYQTAGGYQSTNYNN 174
Query: 188 -------STGYANQAPPWNNGSYSSYASHPHTNYTPDS---NSSGAATTSVQYQQQQQNP 237
ST Y+NQ P N G+Y+ Y S+P+ NYTPD+ +SS ATT + QQ
Sbjct: 175 QTAGSYPSTNYSNQTPASNQGNYTDYTSNPYQNYTPDAANTHSSTIATTPPVHYQQNYQQ 234
Query: 238 WADYYNQTEVSCAPGTENLSVTSSSTFGG--PIPAATSGYATPNSQPQQSYPPYWRQDSS 295
W +YY+QTEV CAPGTE LS ++S + P+P TS NSQP SY WR ++
Sbjct: 235 WTEYYSQTEVPCAPGTEKLSTPTTSAYSQSFPVPGVTSEMPASNSQPAPSYVQPWRPETD 294
Query: 296 ASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPIQPNYQSPLDLKSSYDN-FQDQQKT 354
+S PS QP A S +D YW H A + Q H P Q NYQSPL+ K Y+ FQ Q+
Sbjct: 295 SSHPPSQQPGAAVSTSNDTYWMHQAPSLQAHHPVPPQNNYQSPLETKPLYETPFQGHQRA 354
Query: 355 VSSQGTNLYLP-PPPPLPPPPQQVVSTPIQSAPSLDAKRVSKMQIPTNPRIASNLTYGQP 413
Q N PL P Q+AP +D++RVSK+QIPTNPRIASNL G
Sbjct: 355 TYPQEMNSQSSFHQAPLG------YRQPTQTAPLVDSQRVSKVQIPTNPRIASNLPSGFT 408
Query: 414 KIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAANSILKPGMFPKSLRGYVERALTRCK 473
K++KDS+ + A PAY++VS+PK D ++ PG FPKSLRG+VERA RCK
Sbjct: 409 KMDKDSTAASAAQAPAYVSVSMPKP------KDHTTAMSDPGTFPKSLRGFVERAFARCK 462
Query: 474 DDKQMKACQAVMKEMITKATADGTLCTRNWDMEPLFPMPDSDVVNKDSV---LSSTHKRS 530
DDK+ ++C+ +++++ KA D TL TR+WD EPL + ++V N +S LSS +S
Sbjct: 463 DDKEKESCEVALRKIVKKAKEDNTLYTRDWDTEPLSTVTTTNVTNSESSSAQLSSLQNKS 522
Query: 531 P-RRSKSRWEPLPEEKPVDNPVSISNDTVKYSAWVPNVKDRKVVMENKESKEDDLRNTKF 589
P RR KSRWEPL E KP P S + VK+ W ++ K ++ ES + T F
Sbjct: 523 PTRRPKSRWEPLVEGKPFVKPASTFSSAVKFGVWNHQNENNK---KSSESFQKVDAATGF 579
Query: 590 SPFI--QRTSSKAPQRPFKKQRLTDASIAHENGDASSGSDKEQSLTAYYSAAMAFSDTPE 647
P Q ++ K+ QRP K+QR + + + +ASS SDK+ LT YYS+AMA + + E
Sbjct: 580 KPTYSGQNSAKKSFQRPVKRQRFSGGAATAIDDEASSDSDKD--LTPYYSSAMALAGSAE 637
Query: 648 EKKRRENRSKRFDLGQGHRTENNHFRRKNAGAGNLYSRRASALVLSKSFEDGVSKAVEDI 707
EKKRR++RSKRF+ QGH N+ + KNA GNL+SRRA+AL LSK F++ S+AVEDI
Sbjct: 638 EKKRRDSRSKRFEKIQGHSRGNDLTKPKNANVGNLHSRRATALRLSKVFDESGSRAVEDI 697
Query: 708 DWDALTVKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQNSQKNYLYKCDQLKSI 767
DWDALTVKG CQEIEKRYLRLTSAPDPATVRPE+VLEKAL+MVQ+SQKNYL+KCDQLKSI
Sbjct: 698 DWDALTVKGTCQEIEKRYLRLTSAPDPATVRPEDVLEKALIMVQDSQKNYLFKCDQLKSI 757
Query: 768 RQDLTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYN 827
RQDLTVQRI N LT KVYETHAR A+EAGDL EYNQC SQL LYA+G+EG +EFAAY+
Sbjct: 758 RQDLTVQRIHNHLTAKVYETHARLALEAGDLPEYNQCLSQLKTLYAEGVEGCSLEFAAYS 817
Query: 828 LLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLN 887
LL + +HSNNNR+LLSSM+RLS+E KKDEAV+HAL+VRAAVTSGNYV FFRLYK APN+N
Sbjct: 818 LLYITLHSNNNRELLSSMSRLSEEDKKDEAVRHALSVRAAVTSGNYVMFFRLYKTAPNMN 877
Query: 888 TCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVANGGSDEKDTEAFEECL 947
+CLMDLYVEKMRYKAV+ M RS RPT+PVSYI VLGF + A+ G+DEK+T+ E+CL
Sbjct: 878 SCLMDLYVEKMRYKAVNFMSRSCRPTIPVSYIVQVLGF--TGAASEGTDEKETDGMEDCL 935
Query: 948 EWLKAHGASIMTDNNKDILVDTKVSSSSLFMPEPEDAVAHGDANLAVNDFLAK 1000
EWLK HGA+I+TD+N D+L+DTK +S+SLFMPEPEDAVAHGD NL VNDF +
Sbjct: 936 EWLKTHGANIITDSNGDMLLDTKATSTSLFMPEPEDAVAHGDRNLDVNDFFTR 988
>UniRef100_Q6L516 Putative GANP protein [Oryza sativa]
Length = 836
Score = 681 bits (1756), Expect = 0.0
Identities = 401/838 (47%), Positives = 511/838 (60%), Gaps = 94/838 (11%)
Query: 175 APADYQTTGVYQNSTGYANQAPPWNNGSYS-----SYASHPHTNYTPDSNSSGAATTSVQ 229
AP + YQNST WN GS+ SY ++P +N +S T S
Sbjct: 80 APGYGTSNSYYQNST--------WNGGSFENNYAQSYQNYPSSNTNTVQHSISVPTNSFS 131
Query: 230 YQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYPPY 289
YQQQ N W YYN T P V +S++ I TS Y+ P+ QP
Sbjct: 132 YQQQY-NQWPYYYNHT----VPNPAGDPVGNSNS----IVNTTSSYSYPSIQPPPPGTTS 182
Query: 290 WRQDSSASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPIQPNYQSPLDLKSSYDNFQ 349
W+ +SS+S P Q A+ G D Y N Q H P+ N +
Sbjct: 183 WKSNSSSSIAPPIQ-ASGGPGPQDQYIN------QAHA--PVLENQYAG----------- 222
Query: 350 DQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTPIQSAPSLDAKRVSKMQIPTNPRIASNLT 409
Q + + N Y P P V P+Q + D + T PRIA +
Sbjct: 223 --QVAGNPRSQNHYASQTPACPQ--STVNLNPVQQSNHGDQQN-------TVPRIAPGFS 271
Query: 410 YGQPKIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAANSILKPGMFPKSLRGYVERAL 469
PK EK + + KPAY++VS+ K NDA + P SL Y R L
Sbjct: 272 MVIPKSEKKILGADLSKKPAYVSVSMVK-------NDARS-------LPFSLHNYATRNL 317
Query: 470 TRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPLFPM-------PDSDVVNKDSV 522
CKD+ Q ACQ++++E+ A ADGTL T+NWD EPL P+ P++ N S
Sbjct: 318 NCCKDEAQKAACQSMIEEIKNSAIADGTLLTKNWDTEPLLPLVQNVATIPETSSANNSSP 377
Query: 523 LSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSAWVPNVKDRKVVMENKESKED 582
ST + RR KSRWEP+ EEK D + V + +N+ S
Sbjct: 378 SLST-STNRRRQKSRWEPVVEEKVTDKVEPVKG-------LVNGTTHNNLEAKNRMSNNW 429
Query: 583 DLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAHENGDASSGSDKEQSLTAYYSAAMAF 642
D R KF T++K QRP KKQ+++ S +NG+ASS SDKEQ LT YY++A A
Sbjct: 430 DSR--KFFQSHHATANKVSQRPAKKQKISSYSDQMQNGNASSDSDKEQDLTKYYASATAL 487
Query: 643 SDTPEEKKRRENRSKRFDLGQGHRTENNHFRRKNAGAGNLYSRRASALVLSKSFEDGVSK 702
+++PEEKKRRE+RSKRF+ Q +++ + N+++RRA + +L++S EDG +
Sbjct: 488 ANSPEEKKRREHRSKRFEKNQNSSSKSRNSAASKDVMANIHARRAVSALLARSCEDGTTL 547
Query: 703 AVEDIDWDALTVKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQNSQKNYLYKCD 762
AVED+DWDALTVKG CQEIEKRYLRLTSAPDPATVRPE VLEKAL MV+ SQKNYLYKCD
Sbjct: 548 AVEDMDWDALTVKGTCQEIEKRYLRLTSAPDPATVRPEHVLEKALSMVETSQKNYLYKCD 607
Query: 763 QLKSIRQDLTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYME 822
QLKSIRQDLTVQRI+N+LTVKVYETHAR AM+AGDL EYNQCQSQL LYA+GI+G Y E
Sbjct: 608 QLKSIRQDLTVQRIQNELTVKVYETHARLAMQAGDLPEYNQCQSQLKRLYAEGIKGCYFE 667
Query: 823 FAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKE 882
F+AYNLLCV++HSNN RDLLSS+ RLS +AK+DEAVKHALAV +AV+SGNYV FF+LYK+
Sbjct: 668 FSAYNLLCVMLHSNNKRDLLSSLARLSKQAKQDEAVKHALAVHSAVSSGNYVLFFKLYKQ 727
Query: 883 APNLNTCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVANGGSDEKDTEA 942
APNLN+CLMDLYVE+MR++AV CM +SYRPT+PV Y++ +LGFS D+EA
Sbjct: 728 APNLNSCLMDLYVERMRFEAVKCMSKSYRPTIPVGYVAQILGFS----------RIDSEA 777
Query: 943 FEECLEWLKAHGASIMTDNNKDILVDTKVSSSSLFMPEPEDAVAHGDANLAVNDFLAK 1000
EEC WLKAHGA + DN++D+ +DTK S+++L+MPEPE+AVAHGDA+LAVNDFLA+
Sbjct: 778 SEECEMWLKAHGAILSIDNSRDLQLDTKASTTTLYMPEPENAVAHGDASLAVNDFLAR 835
Score = 35.4 bits (80), Expect = 8.7
Identities = 44/168 (26%), Positives = 61/168 (36%), Gaps = 26/168 (15%)
Query: 19 FENRHFDPGQNQPPSYAPTTPGSQAVSWAVQSSTANGGYSNPTYQYDQH-PQPPGRSIQD 77
FEN + QN P S T S +S S + Y+ Y Y+ P P G +
Sbjct: 100 FENNYAQSYQNYPSSNTNTVQHS--ISVPTNSFSYQQQYNQWPYYYNHTVPNPAGDPV-- 155
Query: 78 GQNVSSVAGNSSNLGTANVPQDYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQ---PNQT 134
GNS+++ N Y SYPS P G T + +N P Q
Sbjct: 156 --------GNSNSI--VNTTSSY----SYPSIQPP----PPGTTSWKSNSSSSIAPPIQA 197
Query: 135 YSQPVGAYQNTGAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQTT 182
P Q + P+ Q G AG+ + Y + PA Q+T
Sbjct: 198 SGGPGPQDQYINQAHAPVLENQYAGQVAGNPRSQNHYASQTPACPQST 245
>UniRef100_Q5QMF0 Putative SAC3/GANP family protein [Oryza sativa]
Length = 893
Score = 628 bits (1620), Expect = e-178
Identities = 392/878 (44%), Positives = 520/878 (58%), Gaps = 93/878 (10%)
Query: 77 DGQNVSSVAGNSSNL--GTANVPQDYNAYTSYPSSSNPY------GYGSAGYTGYYNNYQ 128
D Q SV G++ N+ G +V Q T S PY GY + Y YY N
Sbjct: 42 DPQRDVSVPGSTENVTSGATHVVQSAMGITGATDSYAPYSNSVQPGYNAPQYPNYYYNCP 101
Query: 129 QQPNQTYSQPVGAYQNTGAPYQPISSFQNTGSYAGSANYSSTYYNPAPADYQTTGVYQNS 188
Q N++ Q G Q++GA YQP++SFQN+GSY G S+TYYN +QT Y S
Sbjct: 102 QSTNESSVQQ-GVDQSSGAAYQPLTSFQNSGSYVGPT--SNTYYNAGA--HQTAPGYATS 156
Query: 189 TGYANQAPPWNNGSYSSYASHPHTNYTPDSNSSGAATTSVQ----YQQQQQNPWADYYNQ 244
Y Q+ W GS + +YTP ++ +++S+ + QQ N W YY+Q
Sbjct: 157 NNYY-QSNSWTGGSSGDNHVQSYQSYTPSDTNAAQSSSSLPNNSYHYHQQYNQWPYYYDQ 215
Query: 245 TEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYPP--YWRQDSSASAVPS- 301
+ AP + +V SS + +SGY P++QP PP WR D+ A+AVP
Sbjct: 216 S----APSSGGPAVAVSSVSDANTASVSSGYVYPSTQPP---PPGTTWRSDAGATAVPPP 268
Query: 302 -------FQPAAIN-SGDHDGYWNH---------GAQTSQIHQTNPIQPNYQSPL-DLKS 343
FQ +N + GY N G Q ++Q P P +Q+ +L
Sbjct: 269 QAPGTPVFQNQHVNQAAGPPGYQNQYVNQAPGTPGFQNQYVNQA-PAVPGFQNQYANLAP 327
Query: 344 SYD------------NFQDQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTPIQSAPSLDAK 391
+Y N DQQK QG + + Q Q + + DA
Sbjct: 328 TYQPGTTYYSQLPLSNQADQQKASRWQGPISNVSSVNHVSESSQPT----FQGSATSDAL 383
Query: 392 RVSKMQIPTNPRIASNLTYGQPKIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAANSI 451
RV+K+QIPTNPRIA L PK+EK + +++ KPAY++V++ K+ K + + +
Sbjct: 384 RVNKIQIPTNPRIAPTLPMAMPKVEKRNLEADSSKKPAYVSVAVQKNDVKAAQD--GHEA 441
Query: 452 LKPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPLFPM 511
+ G P SLR YV R ++RCKDD Q A Q ++KE+ITKATADG L T+NWD+EPL P+
Sbjct: 442 VTQGSIPVSLRTYVGRNVSRCKDDAQRSAVQNILKEIITKATADGILHTKNWDIEPLVPL 501
Query: 512 PDS----DVVNKDSVLS----STHKRSP-RRSKSRWEPLPEEKPVDNPVSISNDTVKYSA 562
P++ ++ + LS ST +RSP RR+KSRWEP+ EEK + IS ++ K +
Sbjct: 502 PENITSTNLTSSAKDLSPFSFSTSRRSPSRRAKSRWEPVVEEKVANKVELISKESAKTNT 561
Query: 563 WVPNVKDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAP-----QRPFKKQRLTDASIAH 617
+ + E+ + R+ F+Q S +AP QRP KK+R+ S
Sbjct: 562 Y-----------NSSETTKRAGRSWDIGKFLQ--SRQAPLSQYNQRPSKKKRIGGNSSLT 608
Query: 618 ENGDASSGSDKEQSLTAYYSAAMAFSDTPEEKKRRENRSKRFDLGQGHRTENNHFRRKNA 677
ENG+ SS SDKEQ LT YY+ A+ +++PEEKKRRE+RSKRF+ QG + + +
Sbjct: 609 ENGNVSSDSDKEQDLTKYYANAITIANSPEEKKRREHRSKRFERSQGAASSKSRSSVPDK 668
Query: 678 -GAGNLYSRRASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKRYLRLTSAPDPAT 736
G N Y+RR+ ++LS+S D VS AVED+DWDALTVKG CQEIEKRYLRLTSAPDPAT
Sbjct: 669 DGTSNTYARRSMPMLLSRSNGDDVSFAVEDLDWDALTVKGTCQEIEKRYLRLTSAPDPAT 728
Query: 737 VRPEEVLEKALLMVQNSQKNYLYKCDQLKSIRQDLTVQRIRNQLTVKVYETHARFAMEAG 796
VRPE+VLEKAL MV+ SQKNYLYKCDQLKSIRQDLTVQRI+N+LTV VYETHAR A+++G
Sbjct: 729 VRPEDVLEKALHMVETSQKNYLYKCDQLKSIRQDLTVQRIQNELTVMVYETHARLALQSG 788
Query: 797 DLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDE 856
DL E+NQCQSQL LYA+GI+G + EF+AYNLLCV++HSNN RDLLSSM L EAK+D
Sbjct: 789 DLPEFNQCQSQLKRLYAEGIKGCHFEFSAYNLLCVMLHSNNKRDLLSSMASLPKEAKQDR 848
Query: 857 AVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLY 894
VKHALAV +AV+SGNYV FF+LYK AP+LN+CLM Y
Sbjct: 849 TVKHALAVHSAVSSGNYVLFFKLYKTAPDLNSCLMGKY 886
>UniRef100_UPI0000361091 UPI0000361091 UniRef100 entry
Length = 762
Score = 272 bits (695), Expect = 4e-71
Identities = 235/824 (28%), Positives = 361/824 (43%), Gaps = 145/824 (17%)
Query: 187 NSTGYANQAPPWNNGSYSSYA--SHPHTNYTPDSNSSGAATTSVQYQQQQQNPWADYYNQ 244
+S Q PW + Y ++P+ Y P TS Y Q P
Sbjct: 48 DSAALQQQYYPWYQQAQQHYPGYTYPYNYYYP-------MATSPTYSPQPPPP------- 93
Query: 245 TEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYPPYWRQDSSASAVPSFQP 304
P VTS P+P +T P+ Q S+PP SS++++
Sbjct: 94 ----ATPQPPQSPVTSEQP---PLPPST---IPPSPNSQYSHPPSQNTYSSSNSM----- 138
Query: 305 AAINSGDHDGYWNHGAQTSQIHQTNPIQPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYL 364
A GD Q+H + +PNY +SSY Q + SSQ + +L
Sbjct: 139 -AYPPGDRSQL--------QVHNHSQGRPNYGQGFQAQSSY------QPSQSSQQSGQFL 183
Query: 365 PPPPPLPPPPQQVVSTPI----QSAPSLDAK-RVSK--MQIPTNPRIASNLTYGQPKIEK 417
P Q + + AP K + K +Q+ +P + G+
Sbjct: 184 QGPGRGESSKNQKQGQQLWHRMKQAPGTSMKFNIPKRPLQVQCSPVSEQSFAPGE----- 238
Query: 418 DSSTTNTALKPAYLAVSLPKSTEKISSNDAANSILKPGMFPKSLRGYVERALTRCKDDKQ 477
N A PA A + +A + +P +P++++ YV+R T C+ ++
Sbjct: 239 QPPGNNPASSPALAAATA-----------SAGAQTRPQDWPQAMKEYVQRCFTACETEED 287
Query: 478 MKACQAVMKEMITKATADGTLCTRNWDMEPLFPMPDSDVVNKDSVLSSTHKRSPRRSKSR 537
+ V+KE++ DG+ T +W EPL PD V +SR
Sbjct: 288 KDRTEKVLKEILQDRLKDGSAYTIDWTREPL---PDLKV-----------------KQSR 327
Query: 538 WEPLPEEKPVDNPVSISNDTVKYSAWVPN---VKDRKVVMENKESKEDDLRNTKFSPFIQ 594
WE +P ++ D VS T ++ + + + + R++ SP
Sbjct: 328 WEAVPFQRASDGSVSRGGKTTSSASRARGGHRLGHYRNIFSQRSPSSSSSRSSSRSP--- 384
Query: 595 RTSSKAPQRPFKKQRLTDASIAHENGDASSGSDKEQSLTAYYSAAMAFSDTPEEKKRREN 654
+ S + R QR H D SGS + S+++ ++ + R +
Sbjct: 385 -SPSNSRYRDHSNQR-------HRRSD--SGSQSDSSISSDLRPQLSRRNQRGGPGRGND 434
Query: 655 RSK-RFDLGQGHRTENNHFRRKNAGAGNLY------------------SRRASALVLSKS 695
R + R D G+G + N RRK AG ++ R LVLS +
Sbjct: 435 RERGRGDRGRGRGGKANRGRRKGGTAGFVFHDPNKEAKKQKRAARFQKQLRTEPLVLSVN 494
Query: 696 FEDGVSKAVEDIDWDALTVKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQ---N 752
+ D E + W+ + + G CQ+I K YLRLT APDP+TVRP VL K+L +V+
Sbjct: 495 YLDLPDGTQEGLSWEDVPIVGTCQDITKPYLRLTCAPDPSTVRPVHVLRKSLQVVKAHWK 554
Query: 753 SQKNYLYKCDQLKSIRQDLTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALY 812
+ ++Y Y C+Q+KSIRQDLTVQ IR + TV+VYE HAR A+E GD E+NQCQ+QL +LY
Sbjct: 555 TNQDYPYACEQMKSIRQDLTVQGIRTEFTVEVYECHARIALEKGDHEEFNQCQTQLKSLY 614
Query: 813 ADGIEGSYMEFAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGN 872
D + EF AY LL I + N DL + + L+ E + D V HALA+RAA GN
Sbjct: 615 KDVPSENIGEFTAYRLLYYIF-TRNTGDLTTELVYLTPELRADACVSHALALRAAWALGN 673
Query: 873 YVAFFRLYKEAPNLNTCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVAN 932
Y FF+LY EAP + + L+D +VE+ R A+ M +++RP +PV Y +LGF+
Sbjct: 674 YHRFFKLYLEAPRMASYLIDKFVERERKIALRAMVKTFRPDLPVQYAQSLLGFT------ 727
Query: 933 GGSDEKDTEAFEECLEWLKAHGASIMTDNNKDILVDTKVSSSSL 976
+ + C+ +L G S+ + +D K S+++L
Sbjct: 728 ---------SLDSCMAFLT--GLSVTFTPSDPSKIDCKASTAAL 760
>UniRef100_UPI0000247721 UPI0000247721 UniRef100 entry
Length = 767
Score = 265 bits (676), Expect = 7e-69
Identities = 240/838 (28%), Positives = 363/838 (42%), Gaps = 116/838 (13%)
Query: 183 GVYQNSTGYANQAPPWNNG-------SYSSYASHPHTNYTPDSNSSGAATTSVQYQQQQQ 235
G + G ++ P W S S ++ P TN T + +S QQQ
Sbjct: 1 GNNSSGEGPVHENPEWEKARQALASISKSQNSTKPSTNQTTQQVT--VTDSSAMQQQQYY 58
Query: 236 NPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIPAATSGYATPNSQPQQSYPPYWRQDSS 295
W NQ + + P N + P+P S P P + P + +S
Sbjct: 59 QQWYQQ-NQQQYAGYPYPYN--------YYYPMPPDQSSNYPPQPPPPATPQPPPQSPTS 109
Query: 296 ASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPIQPNYQSPLDLKSSYDNFQDQQKTV 355
+ P P I + Y +QT + N + P SP D + Q
Sbjct: 110 SDQPPPPPPPPIPPQPNAQYPRPPSQTP--YSPNGVLP--YSPTDSNPMMRGYNPGQIRP 165
Query: 356 SSQG--TNLYLPPPPPLPPPPQ---------------------QVVSTPIQSAPSLDAKR 392
QG T +Y P P P Q + ++ AP A
Sbjct: 166 GYQGYQTPVYQPHQQQAPAPGLYGGAGRVDAKKPNNSNMNKGGQQLWQRMKQAPGTGAV- 224
Query: 393 VSKMQIPTNPRIASNLTYGQPKIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAANSIL 452
K IP P + SN + P + TA P+ ST SS +
Sbjct: 225 --KFNIPKRPYVMSNQNF-PPGEQGPGLAPPTATNPS------SGSTSATSSASTGGAQT 275
Query: 453 KPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPLFPMP 512
+P +P++++ YV+R T C+ ++ + ++K+++ DG+ T +W+ EPL P
Sbjct: 276 RPQDWPQAMKEYVQRCFTACESEEDKDRTEKMLKDLLQSRLQDGSAYTIDWNREPL---P 332
Query: 513 DSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSAWVPNVKDRKV 572
D VL ++ K P SRWE +P+ +S N + A R
Sbjct: 333 D--------VLFNSLKPKP----SRWEVMPQ-------LSQENSIGRGGATRGRGGARLG 373
Query: 573 VMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAH---ENGDASSGSDKE 629
N S+ ++ S R+SS++P P + R D S H + G G+++E
Sbjct: 374 NYRNVFSQRSPSSSSSGS---SRSSSRSPS-PHSRHR--DRSRHHRRQQKGGRGRGAERE 427
Query: 630 QSLTAYYSAAMAFSDTPEEKKRREN-------RSKRFDLGQGHRTENNHFRRKNAGAGNL 682
+ A ++ E+ + K + G N ++++ A
Sbjct: 428 RGRGAAGERGRGRGKANRGRRNMEDLASGVGKKRKGGNAGLDFHDPNREAKKQSRAARFH 487
Query: 683 YSRRASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKRYLRLTSAPDPATVRPEEV 742
R LVL+ + D + E + WD + G CQ+I K YLRLT APDP+TVRP V
Sbjct: 488 TKLRTEPLVLNINVFDLPNGTQEGLSWDDCPIVGTCQDITKNYLRLTCAPDPSTVRPVPV 547
Query: 743 LEKALLMVQ---NSQKNYLYKCDQLKSIRQDLTVQRIRNQLTVKVYETHARFAMEAGDLS 799
L K+L+ V+ S ++Y+Y C+Q+KSIRQDLTVQ +R TV+VYETHAR A+E GD
Sbjct: 548 LRKSLIAVKAHWKSNQDYVYACEQMKSIRQDLTVQGVRTDFTVEVYETHARIALEKGDHE 607
Query: 800 EYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVK 859
E+NQCQ+QL ALY D + EF AY L+ I + N+ DL + + L+ E + D V
Sbjct: 608 EFNQCQTQLKALYKDCPSDNVGEFTAYRLIYYIF-TKNSGDLTTELVYLTTELRADPCVA 666
Query: 860 HALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYI 919
HAL +R A GN+ FFRLY++AP + L+D +VE+ R A+ + +S+RP+V V Y+
Sbjct: 667 HALELRTAWALGNFHRFFRLYQKAPRMAAYLIDKFVERERNIALRAILKSFRPSVSVEYV 726
Query: 920 SHVLGFSTSAVANGGSDEKDTEAFEECLEWLKAHGASIM-TDNNKDILVDTKVSSSSL 976
L F + CL +L G S +D +K +D KVSS+SL
Sbjct: 727 QSSLAFPD---------------LDTCLAFLTGLGISFTPSDPSK---IDCKVSSASL 766
>UniRef100_UPI000027CB3C UPI000027CB3C UniRef100 entry
Length = 758
Score = 261 bits (666), Expect = 1e-67
Identities = 207/694 (29%), Positives = 318/694 (44%), Gaps = 107/694 (15%)
Query: 324 QIHQTNPIQPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTPI- 382
Q H +PNY +SSY Q + +SQ +L P Q +
Sbjct: 128 QAHNHGQGRPNYGQSFQGQSSY------QPSQTSQQPGQFLQGPGRGEGNKNQKQGQQLW 181
Query: 383 ---QSAPSLDAKRVS----KMQIPTNPRIASNLTYG-QPKIEKDSSTTNTALKPAYLAVS 434
+ AP + + + +Q+ +P + + G QP +S+ TA PA
Sbjct: 182 HRMKQAPGTSSVKFNIPKRPLQVQCSPVSEQSFSPGEQPSANNPTSSPVTATAPA----- 236
Query: 435 LPKSTEKISSNDAANSILKPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATA 494
+A + +P +P++++ YV+R T C+ ++ + V+KE++
Sbjct: 237 ------------SAGAQTRPQDWPQAMKEYVQRCFTACETEEDKDRTEKVLKEILQDRLK 284
Query: 495 DGTLCTRNWDMEPLFPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSIS 554
DG+ T +W EPL PD V +SRWE +P ++ D VS
Sbjct: 285 DGSAYTIDWTREPL---PDLKV-----------------KQSRWEAVPAQRASDGSVSRG 324
Query: 555 NDTVKYSAWVP---NVKDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLT 611
T S+ + + + + R++ SP TSS+ R ++ + +
Sbjct: 325 GKTAASSSRGRAGHRLGHYRNIFSQRSPSSSPSRSSSRSP--SPTSSRYRDRNNQRHKRS 382
Query: 612 DASIAHENGDASSGSDKEQSLTAYYSAAMAFSDTPEEKKRREN-RSKRFDLGQGHRTENN 670
D+ D+S SD L+ + E+ R E R + +G R +
Sbjct: 383 DSG---SQSDSSISSDLRPQLSRRNRRGGSGRGNDRERARGERGRGRGGKANRGRRNTDE 439
Query: 671 HFRRKNAGAGNL-------------------YSRRASALVLSKSFEDGVSKAVEDIDWDA 711
+++ G L R+ LVLS + D E I W+
Sbjct: 440 SGKKRKGGTAGLDFHDPNKEAKKQKRAARFQKQLRSEPLVLSVNDLDLTDGTQEGISWED 499
Query: 712 LTVKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQ---NSQKNYLYKCDQLKSIR 768
+ + G CQ+I K YLRLT APDP+TVRP VL+K+L +V+ S ++YLY C+Q+KSIR
Sbjct: 500 VPIVGTCQDITKHYLRLTCAPDPSTVRPVLVLKKSLQVVKAHWKSNQDYLYACEQMKSIR 559
Query: 769 QDLTVQRIRNQLTVKVYETHARFAMEA------GDLSEYNQCQSQLTALYADGIEGSYME 822
QDLTVQ IR + TV+VYE HAR A+E GD E+NQCQ+QL ALY D + E
Sbjct: 560 QDLTVQGIRTEFTVEVYECHARVALEKLDVCCQGDHEEFNQCQTQLKALYKDVPSENIGE 619
Query: 823 FAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKE 882
F AY LL I + N DL + + L+ E + D+ V HALA+RAA GNY FF+LY E
Sbjct: 620 FTAYRLLYYIF-TRNTGDLTTELVYLTPELRADDCVAHALALRAAWALGNYHRFFKLYLE 678
Query: 883 APNLNTCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVANGGSDEKDTEA 942
AP + + L+D +VE+ R A+ M +++RP +PV Y VLGFS +
Sbjct: 679 APRMASYLIDKFVERERKIALRAMVKTFRPDLPVQYAQSVLGFS---------------S 723
Query: 943 FEECLEWLKAHGASIMTDNNKDILVDTKVSSSSL 976
+ C+ +L G + + + I D K S+++L
Sbjct: 724 LDSCVAFLTGLGVTFTSSDPSKI--DCKASTAAL 755
>UniRef100_Q8IZ27 Leukocyte receptor cluster (LRC) member 8 [Homo sapiens]
Length = 800
Score = 257 bits (657), Expect = 1e-66
Identities = 221/763 (28%), Positives = 336/763 (43%), Gaps = 108/763 (14%)
Query: 219 NSSGAATTSVQY---------QQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIP 269
+SS S QY QQQQ W YN P + ++ ++G P
Sbjct: 61 SSSNGPVASAQYVSQAEASALQQQQYYQWYQQYNYAY----PYSYYYPMSMYQSYGSPSQ 116
Query: 270 ---AATSGYATPN--SQPQQ----SYPPYWRQDSSAS------AVPSFQPAAINSGDHDG 314
A + G ATP S PQ + PP D S S +PS QP ++ H
Sbjct: 117 YGMAGSYGSATPQQPSAPQHQGTLNQPPVPGMDESMSYQAPPQQLPSAQPPQPSNPPHGA 176
Query: 315 YW-NHGAQTSQIHQTNPIQPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPPLPPP 373
+ N G Q T Q T + G + Y P P
Sbjct: 177 HTLNSGPQPGTAPATQHSQAG-----------------PATGQAYGPHTYTEPAKP---K 216
Query: 374 PQQVVSTPIQSAPSLDAKRVSKMQIPTNPRIASNLTYGQPKIEKDSSTTNTALKPAYLAV 433
Q + ++ AP K I P + ++G ++ ++ P
Sbjct: 217 KGQQLWNRMKPAPGTGGL---KFNIQKRPFAVTTQSFGS-----NAEGQHSGFGPQ---- 264
Query: 434 SLPKSTEKISSNDAANSIL-KPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKA 492
P+ + S + A ++ KP +P+ ++ YVER T C+ ++ + ++KE++
Sbjct: 265 PNPEKVQNHSGSSARGNLSGKPDDWPQDMKEYVERCFTACESEEDKDRTEKLLKEVLQAR 324
Query: 493 TADGTLCTRNWDMEPL-----FPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPV 547
DG+ T +W EPL P+ +S + SS H S +R P ++
Sbjct: 325 LQDGSAYTIDWSREPLPGLTREPVAESPKKKRWEAASSLHPPRGAGSATRGGGAPSQR-- 382
Query: 548 DNPVSISNDTVKYSAWVPNVKDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKK 607
P + + +++ + +N S D R+ R+SS++P R F++
Sbjct: 383 GTPGAGGAGRARGNSFTKFGNRNVFMKDNSSSSSTDSRS--------RSSSRSPTRHFRR 434
Query: 608 QRLTDASIAHENGDASSGSDKEQSLTAYYSAAMAFSDTPEEKKR---RENRSKRFDLGQG 664
S +H + D+S ++ + R R R KR DL
Sbjct: 435 ------SDSHSDSDSSYSGNECHPVGRRNPPPKGRGGRGAHMDRGRGRAQRGKRHDLAPT 488
Query: 665 HRT------------ENNHFRRKNAGA---GNLYSRRASALVLSKSFEDGVSKAVEDIDW 709
R+ E ++K A G+ R LVL S + + D DW
Sbjct: 489 KRSRKKMAALECEDPERELKKQKRAARFQHGHSRRLRLEPLVLQMS---SLESSGADPDW 545
Query: 710 DALTVKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQ---NSQKNYLYKCDQLKS 766
L + G C +I K YLRLT APDP+TVRP VL+K+L MV+ +++Y + C+Q+KS
Sbjct: 546 QELQIVGTCPDITKHYLRLTCAPDPSTVRPVAVLKKSLCMVKCHWKEKQDYAFACEQMKS 605
Query: 767 IRQDLTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAY 826
IRQDLTVQ IR + TV+VYETHAR A+E GD E+NQCQ+QL +LYA+ + G+ EF AY
Sbjct: 606 IRQDLTVQGIRTEFTVEVYETHARIALEKGDHEEFNQCQTQLKSLYAENLPGNVGEFTAY 665
Query: 827 NLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNL 886
+L I + N+ D+ + + L+ E K D V HALA+R A GNY FFRLY AP +
Sbjct: 666 RILYYIF-TKNSGDITTELAYLTRELKADPCVAHALALRTAWALGNYHRFFRLYCHAPCM 724
Query: 887 NTCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSA 929
+ L+D + ++ R A+ M +++RP +PVSY+ L F A
Sbjct: 725 SGYLVDKFADRERKVALKAMIKTFRPALPVSYLQAELAFEGEA 767
Score = 35.4 bits (80), Expect = 8.7
Identities = 45/200 (22%), Positives = 71/200 (35%), Gaps = 22/200 (11%)
Query: 42 QAVSWAVQSSTANGGYSNPTYQYDQHPQPP---GRSIQDGQNVSSVAGNSSNLGTANVPQ 98
++ W+ Q S G + H P R + S AG S+ ++N P
Sbjct: 9 RSTDWSSQYSMVAGAGRENGMETPMHENPEWEKARQALASISKSGAAGGSAK-SSSNGPV 67
Query: 99 DYNAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAPYQPISSFQNT 158
A Y S + Y +Y Y +Y P+ YQ+ G+P S +
Sbjct: 68 ---ASAQYVSQAEASALQQQQYYQWYQQYNYAYPYSYYYPMSMYQSYGSP----SQYGMA 120
Query: 159 GSYAGSANYSSTYYNPAPADYQTT----GVYQNSTGYANQAPPWN--NGSYSSYASHPHT 212
GSY S+T P+ +Q T V + QAPP + ++ PH
Sbjct: 121 GSYG-----SATPQQPSAPQHQGTLNQPPVPGMDESMSYQAPPQQLPSAQPPQPSNPPHG 175
Query: 213 NYTPDSNSSGAATTSVQYQQ 232
+T +S + Q+ Q
Sbjct: 176 AHTLNSGPQPGTAPATQHSQ 195
>UniRef100_UPI0000361090 UPI0000361090 UniRef100 entry
Length = 679
Score = 255 bits (651), Expect = 5e-66
Identities = 203/673 (30%), Positives = 304/673 (45%), Gaps = 104/673 (15%)
Query: 302 FQPAAINSGDHDGYWNHGAQTSQIHQTNPIQP-NYQSPLDLKSSYDNFQDQQKTVSSQGT 360
FQ A +S + Q +Q H P NY P+ +DQ T S Q
Sbjct: 42 FQTAMTDSAALQQQYYPWYQQAQQHYPGYTYPYNYYYPMATPPPVPGMEDQSPTYSPQ-- 99
Query: 361 NLYLPPPPPLPPPPQQVVST-----PIQSAPSLDAKRVSK----------MQIPTNPRIA 405
PPPP P PPQ V++ P + P + S +Q P +
Sbjct: 100 ----PPPPATPQPPQSPVTSEQPPLPPSTIPPSPNSQYSHPPSQNTYSQFLQGPGRGESS 155
Query: 406 SNLTYGQPKIEKDSSTTNTALK------PAYLAVSL----------------PKSTEKIS 443
N GQ + T++K P + S P S+ ++
Sbjct: 156 KNQKQGQQLWHRMKQAPGTSMKFNIPKRPLQVQCSPVSEQSFAPGEQPPGNNPASSPALA 215
Query: 444 SNDA-ANSILKPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRN 502
+ A A + +P +P++++ YV+R T C+ ++ + V+KE++ DG+ T +
Sbjct: 216 AATASAGAQTRPQDWPQAMKEYVQRCFTACETEEDKDRTEKVLKEILQDRLKDGSAYTID 275
Query: 503 WDMEPLFPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSA 562
W EPL PD V +SRWE +P ++ D VS T ++
Sbjct: 276 WTREPL---PDLKV-----------------KQSRWEAVPFQRASDGSVSRGGKTTSSAS 315
Query: 563 WVPN---VKDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAHEN 619
+ + + + R++ SP + S + R QR H
Sbjct: 316 RARGGHRLGHYRNIFSQRSPSSSSSRSSSRSP----SPSNSRYRDHSNQR-------HRR 364
Query: 620 GDASSGSDKEQSLTAYYSAAMAFSDTPEEKKRRENRSK-RFDLGQGHRTENNHFRRKNAG 678
D SGS + S+++ ++ + R +R + R D G+G + N RRK
Sbjct: 365 SD--SGSQSDSSISSDLRPQLSRRNQRGGPGRGNDRERGRGDRGRGRGGKANRGRRKGGT 422
Query: 679 AGNLY------------------SRRASALVLSKSFEDGVSKAVEDIDWDALTVKGACQE 720
AG ++ R LVLS ++ D E + W+ + + G CQ+
Sbjct: 423 AGFVFHDPNKEAKKQKRAARFQKQLRTEPLVLSVNYLDLPDGTQEGLSWEDVPIVGTCQD 482
Query: 721 IEKRYLRLTSAPDPATVRPEEVLEKALLMVQ---NSQKNYLYKCDQLKSIRQDLTVQRIR 777
I K YLRLT APDP+TVRP VL K+L +V+ + ++Y Y C+Q+KSIRQDLTVQ IR
Sbjct: 483 ITKPYLRLTCAPDPSTVRPVHVLRKSLQVVKAHWKTNQDYPYACEQMKSIRQDLTVQGIR 542
Query: 778 NQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNN 837
+ TV+VYE HAR A+E GD E+NQCQ+QL +LY D + EF AY LL I + N
Sbjct: 543 TEFTVEVYECHARIALEKGDHEEFNQCQTQLKSLYKDVPSENIGEFTAYRLLYYIF-TRN 601
Query: 838 NRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEK 897
DL + + L+ E + D V HALA+RAA GNY FF+LY EAP + + L+D +VE+
Sbjct: 602 TGDLTTELVYLTPELRADACVSHALALRAAWALGNYHRFFKLYLEAPRMASYLIDKFVER 661
Query: 898 MRYKAVSCMCRSY 910
R A+ M ++Y
Sbjct: 662 ERKIALRAMVKTY 674
>UniRef100_UPI000036C0A0 UPI000036C0A0 UniRef100 entry
Length = 527
Score = 249 bits (637), Expect = 2e-64
Identities = 167/503 (33%), Positives = 253/503 (50%), Gaps = 46/503 (9%)
Query: 453 KPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPL---- 508
KP +P+ ++ YVER T C+ ++ + ++KE++ DG+ T +W EPL
Sbjct: 12 KPDDWPQDMKEYVERCFTACESEEDKDRTEKLLKEVLQARLQDGSAYTIDWSREPLPGLT 71
Query: 509 -FPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSAWVPNV 567
P+ +S + SS H S +R P ++ P + + +++
Sbjct: 72 REPVAESPKKKRWEAASSLHPPRGAGSATRGGGAPSQR--GTPGAGGAGRARGNSFTKFG 129
Query: 568 KDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAHENGDASSGSD 627
+ +N S D R+ R+SS++P R F++ S +H + D+S +
Sbjct: 130 NRNVFMKDNSSSSSTDSRS--------RSSSRSPTRHFRR------SDSHSDSDSSYSGN 175
Query: 628 KEQSLTAYYSAAMAFSDTPEEKKR---RENRSKRFDLGQGHRT------------ENNHF 672
+ + R R R KR DL R+ E
Sbjct: 176 ECHPVGRRNPPPKGRGGRGAHMDRGRGRAQRGKRHDLAPTKRSRKKMAALECEDPERELK 235
Query: 673 RRKNAGA---GNLYSRRASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKRYLRLT 729
++K A G+ R LVL S + + D DW L + G C +I K YLRLT
Sbjct: 236 KQKRAARFQHGHSRRLRLEPLVLQMS---SLESSGADPDWQELQIVGTCPDITKHYLRLT 292
Query: 730 SAPDPATVRPEEVLEKALLMVQ---NSQKNYLYKCDQLKSIRQDLTVQRIRNQLTVKVYE 786
APDP+TVRP VL+K+L MV+ +++Y + C+Q+KSIRQDLTVQ IR + TV+VYE
Sbjct: 293 CAPDPSTVRPVAVLKKSLCMVKCHWKEKQDYAFACEQMKSIRQDLTVQGIRTEFTVEVYE 352
Query: 787 THARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDLLSSMT 846
THAR A+E GD E+NQCQ+QL +LYA+ + G+ EF AY +L I + N+ D+ + +
Sbjct: 353 THARIALEKGDHEEFNQCQTQLKSLYAENLPGNVGEFTAYRILYYIF-TKNSGDITTELA 411
Query: 847 RLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEKMRYKAVSCM 906
L+ E K D V HALA+R A GNY FFRLY AP ++ L+D + ++ R A+ M
Sbjct: 412 YLTRELKADPCVAHALALRTAWALGNYHRFFRLYCHAPCMSGYLVDKFADRERKVALKAM 471
Query: 907 CRSYRPTVPVSYISHVLGFSTSA 929
+++RP +PVSY+ L F A
Sbjct: 472 IKTFRPALPVSYLQAELAFEGEA 494
>UniRef100_UPI0000436B5C UPI0000436B5C UniRef100 entry
Length = 674
Score = 246 bits (629), Expect = 2e-63
Identities = 183/587 (31%), Positives = 277/587 (47%), Gaps = 78/587 (13%)
Query: 365 PPPPPLPPPPQQVVSTPIQSAP-----SLDAKRVS------------------------K 395
PPPPP+PP P P P +DAK+ + K
Sbjct: 124 PPPPPIPPQPNAQYPRPPSQTPYSPNGRVDAKKPNNSNMNKGGQQLWQRMKQAPGTGAVK 183
Query: 396 MQIPTNPRIASNLTYGQPKIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAANSILKPG 455
IP P + SN + P + TA P+ ST SS + +P
Sbjct: 184 FNIPKRPYVMSNQNF-PPGEQGPGLAPPTATNPS------SGSTSATSSASTGGAQTRPQ 236
Query: 456 MFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPLFPMPDSD 515
+P++++ YV+R T C+ ++ + ++K+++ DG+ T +W+ EPL PD
Sbjct: 237 DWPQAMKEYVQRCFTACESEEDKDRTEKMLKDLLQSRLQDGSAYTIDWNREPL---PD-- 291
Query: 516 VVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSAWVPNVKDRKVVME 575
VL ++ K P SRWE +P+ +S N + A R
Sbjct: 292 ------VLFNSLKPKP----SRWEVMPQ-------LSQENSIGRGGATRGRGGARLGNYR 334
Query: 576 NKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAH---ENGDASSGSDKEQSL 632
N S+ ++ S R+SS++P P + R D S H + G G+++E+
Sbjct: 335 NVFSQRSPSSSSSGS---SRSSSRSPS-PHSRHR--DRSRHHRRQQKGGRGRGAERERGR 388
Query: 633 TAYYSAAMAFSDTPEEKKRREN-------RSKRFDLGQGHRTENNHFRRKNAGAGNLYSR 685
A ++ E+ + K + G N ++++ A
Sbjct: 389 GAAGERGRGRGKANRGRRNMEDLASGVGKKRKGGNAGLDFHDPNREAKKQSRAARFHTKL 448
Query: 686 RASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKRYLRLTSAPDPATVRPEEVLEK 745
R LVL+ + D + E + WD + G CQ+I K YLRLT APDP+TVRP VL K
Sbjct: 449 RTEPLVLNINVFDLPNGTQEGLSWDDCPIVGTCQDITKNYLRLTCAPDPSTVRPVPVLRK 508
Query: 746 ALLMVQ---NSQKNYLYKCDQLKSIRQDLTVQRIRNQLTVKVYETHARFAMEAGDLSEYN 802
+L+ V+ S ++Y+Y C+Q+KSIRQDLTVQ +R TV+VYETHAR A+E GD E+N
Sbjct: 509 SLIAVKAHWKSNQDYVYACEQMKSIRQDLTVQGVRTDFTVEVYETHARIALEKGDHEEFN 568
Query: 803 QCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHAL 862
QCQ+QL ALY D + EF AY L+ I + N+ DL + + L+ E + D V HAL
Sbjct: 569 QCQTQLKALYKDCPSDNVGEFTAYRLIYYIF-TKNSGDLTTELVYLTTELRADPCVAHAL 627
Query: 863 AVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEKMRYKAVSCMCRS 909
+R A GN+ FFRLY++AP + L+D +VE+ R A+ + +S
Sbjct: 628 ELRTAWALGNFHRFFRLYQKAPRMAAYLIDKFVERERNIALRAILKS 674
>UniRef100_Q8CBY3 Mus musculus adult male diencephalon cDNA, RIKEN full-length
enriched library, clone:9330175N02 product:hypothetical
SAC3/GANP family containing protein, full insert
sequence [Mus musculus]
Length = 785
Score = 242 bits (617), Expect = 5e-62
Identities = 210/728 (28%), Positives = 322/728 (43%), Gaps = 89/728 (12%)
Query: 221 SGAATTSVQYQQQQQNPWADYYNQTEVSCAPGTENLSVTSSSTFGGPIP---AATSGYAT 277
S A +++Q QQQQ W YN P + ++ ++G P A++ G AT
Sbjct: 73 SQAEASALQQQQQQYYQWYQQYNYAY----PYSYYYPMSMYQSYGSPSQYGMASSYGSAT 128
Query: 278 ------PNSQPQQSYPPYWRQDSSASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPI 331
P Q + PP D S + S Q HG + +N
Sbjct: 129 AQQPSAPQHQGTLNQPPVPGMDESMAYQASPQQLPAAQPPQPSNSQHGTHSL----SNGP 184
Query: 332 QPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTPIQSAPSLDAK 391
QP S + Q T + G + Y P P Q + T ++ AP
Sbjct: 185 QPG------TAPSTQHSQAGAPTGQAYGPHSY---SEPAKPKKGQQLWTRMKPAPGTGGL 235
Query: 392 RVSKMQIP---TNPRIASNLTYGQPKIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAA 448
+ + + P T+ +SN ++ NT + S P +S
Sbjct: 236 KFNIQKRPFAVTSQSFSSNSEGQHSSFGPQPNSENTQNR------SGPSGRGNLSG---- 285
Query: 449 NSILKPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPL 508
KP +P+ ++ YVER T C+ ++ + ++KE++ DG+ T +W EPL
Sbjct: 286 ----KPDDWPQDMKEYVERCFTACESEEDKDRTEKLLKEVLQARLQDGSAYTIDWSREPL 341
Query: 509 -----FPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSAW 563
P+ +S + SS H S +R ++ P + + S++
Sbjct: 342 PGLTREPVAESPKKKRWEAPSSLHPSRGAGSVTRGGGAQSQR--GTPGAGGAGRARGSSF 399
Query: 564 VPNVKDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAHENGDAS 623
+ +N S D R+ R+SS++P R F++ S +H + D+S
Sbjct: 400 TKFGNRNVFMKDNSSSSSTDSRS--------RSSSRSPTRHFRR------SDSHSDSDSS 445
Query: 624 SGSDKEQSLTAYYSAAMAFSDTPEEKKR---RENRSKRFDLGQGHRT------------E 668
++ + R R R KR DL R+ E
Sbjct: 446 YSGNECHPVGRRNPPPKGRGGRGAHMDRGRGRAQRGKRHDLAPTKRSRKKMAALECEDPE 505
Query: 669 NNHFRRKNAGA---GNLYSRRASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKRY 725
++K A G+ R LVL S + + D DW L + G C +I K Y
Sbjct: 506 RELKKQKRAARFQHGHSRRLRLEPLVLQMS---NLESSGADPDWQELQIVGTCPDITKHY 562
Query: 726 LRLTSAPDPATVRPEEVLEKALLMVQN---SQKNYLYKCDQLKSIRQDLTVQRIRNQLTV 782
LRLT APDP+TVRP VL+K+L MV++ +++Y + C+Q+KSIRQDLTVQ IR + TV
Sbjct: 563 LRLTCAPDPSTVRPVAVLKKSLCMVKSHWKEKQDYAFACEQMKSIRQDLTVQGIRTEFTV 622
Query: 783 KVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDLL 842
+VYETHAR A+E GD E+NQCQ+QL +LYA+ + G+ EF AY +L I + N+ D+
Sbjct: 623 EVYETHARIALEKGDHEEFNQCQTQLKSLYAENLAGNVGEFTAYRILYYIF-TKNSGDIT 681
Query: 843 SSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEKMRYKA 902
+ + L+ E K D V HALA+RAA GNY FFRLY AP ++ L+D + ++ R A
Sbjct: 682 TELAYLTREMKADPCVAHALALRAAWALGNYHRFFRLYCHAPCMSGYLVDKFADRERKAA 741
Query: 903 VSCMCRSY 910
+ M ++Y
Sbjct: 742 LKAMIKTY 749
Score = 41.2 bits (95), Expect = 0.16
Identities = 58/266 (21%), Positives = 92/266 (33%), Gaps = 23/266 (8%)
Query: 42 QAVSWAVQSSTANGGYSNPTYQYDQHPQPPGRSIQDG-QNVSSVAGNSSNLGTANVPQDY 100
+A W+ Q S G + H P + ++S SS+ +++ P
Sbjct: 9 RAADWSSQYSMVTGNSRENGMETPMHENPEWEKARQALASISKAGATSSSKASSSGPVAS 68
Query: 101 NAYTSYPSSSNPYGYGSAGYTGYYNNYQQQPNQTYSQPVGAYQNTGAP--YQPISSFQN- 157
Y S +S Y +Y Y +Y P+ YQ+ G+P Y SS+ +
Sbjct: 69 AQYVSQAEAS-ALQQQQQQYYQWYQQYNYAYPYSYYYPMSMYQSYGSPSQYGMASSYGSA 127
Query: 158 TGSYAGSANYSSTYYNPAPADYQTTGVYQNSTGY--ANQAPPWNNGSYSSYA----SHPH 211
T + + T P + YQ S A Q P +N + +++ P
Sbjct: 128 TAQQPSAPQHQGTLNQPPVPGMDESMAYQASPQQLPAAQPPQPSNSQHGTHSLSNGPQPG 187
Query: 212 TNYTPDSNSSGAATTSVQYQQQQQNPWADYYNQ---TEVSCAPGTENL---------SVT 259
T + + +GA T P Q T + APGT L +VT
Sbjct: 188 TAPSTQHSQAGAPTGQAYGPHSYSEPAKPKKGQQLWTRMKPAPGTGGLKFNIQKRPFAVT 247
Query: 260 SSSTFGGPIPAATSGYATPNSQPQQS 285
S S +S PNS+ Q+
Sbjct: 248 SQSFSSNSEGQHSSFGPQPNSENTQN 273
>UniRef100_UPI00001CE789 UPI00001CE789 UniRef100 entry
Length = 930
Score = 240 bits (613), Expect = 1e-61
Identities = 206/728 (28%), Positives = 322/728 (43%), Gaps = 89/728 (12%)
Query: 221 SGAATTSVQYQQQQQNPWADYYNQTEVSCA--PGTENLSVTSSSTFG---------GPIP 269
S A +++Q QQQQ W YN P + S +S S +G P
Sbjct: 210 SQAEASALQQQQQQYYQWYQQYNYAYPYSYYYPMSMYQSYSSPSQYGMASSYGSATAQQP 269
Query: 270 AATSGYATPNSQPQQSYPPYWRQDSSASAVPSFQPAAINSGDHDGY-WNHGAQTSQIHQT 328
+A T N P +S +P+ QP ++ H + N+G Q+ T
Sbjct: 270 SAPQHQGTLNQPPVPGMDESMAYQASPQQLPAAQPPQPSNPQHGTHSLNNGPQSG----T 325
Query: 329 NPIQPNYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTPIQSAPSL 388
P + Q+ +Y G + Y P P Q + T ++ AP
Sbjct: 326 APTTQHNQAGAPTGQTY-------------GPHSY---SEPAKPKKGQQLWTRMKPAPGT 369
Query: 389 DAKRVSKMQIPTNPRIASNLTYGQPKIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAA 448
K I P +N ++ +S ++ P + K+ +
Sbjct: 370 GGL---KFNIQKRPFAVTNQSFSS-----NSEGQHSGFGPQ---PNSEKTQNHSGPSGRG 418
Query: 449 NSILKPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPL 508
N KP +P+ ++ YVER T C+ ++ + ++KE++ DG+ T +W EPL
Sbjct: 419 NLSGKPDDWPQDMKEYVERCFTACESEEDKDRTEKLLKEVLQARLQDGSAYTIDWSREPL 478
Query: 509 -----FPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSAW 563
P+ +S + SS H S +R ++ P + + S++
Sbjct: 479 PGLTREPVAESPKKKRWEAPSSLHPSRGAGSVTRGGGAQSQR--GTPGAGGAGRARGSSF 536
Query: 564 VPNVKDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAHENGDAS 623
+ +N S D R+ R+SS++P R F++ S +H + D+S
Sbjct: 537 AKFGNRNVFMKDNSSSSSTDSRS--------RSSSRSPTRHFRR------SDSHSDSDSS 582
Query: 624 SGSDKEQSLTAYYSAAMAFSDTPEEKKR---RENRSKRFDLGQGHRT------------E 668
++ + R R R KR DL R+ E
Sbjct: 583 YSGNECHPVGRRNPPPKGRGGRGAHMDRGRGRAQRGKRHDLAPTKRSRKKMAALECEDPE 642
Query: 669 NNHFRRKNAGA---GNLYSRRASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKRY 725
++K A G+ R LVL S + + D DW L + G C +I K Y
Sbjct: 643 RELKKQKRAARFQHGHSRRLRLEPLVLQMS---NLESSGADPDWQELQIVGTCPDITKHY 699
Query: 726 LRLTSAPDPATVRPEEVLEKALLMVQN---SQKNYLYKCDQLKSIRQDLTVQRIRNQLTV 782
LRLT APDP+TVRP VL+K+L MV++ +++Y + C+Q+KSIRQDLTVQ IR + TV
Sbjct: 700 LRLTCAPDPSTVRPVAVLKKSLCMVKSHWKEKQDYAFACEQMKSIRQDLTVQGIRTEFTV 759
Query: 783 KVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDLL 842
+VYETHAR A+E GD E+NQCQ+QL +LYA+ + G+ EF AY +L I + N+ D+
Sbjct: 760 EVYETHARIALEKGDHEEFNQCQTQLKSLYAENLAGNVGEFTAYRILYYIF-TKNSGDIT 818
Query: 843 SSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEKMRYKA 902
+ + L+ E K D V HALA+RAA GNY FF+LY AP ++ L+D + ++ R A
Sbjct: 819 TELAYLTREMKADPCVSHALALRAAWALGNYHRFFKLYCHAPCMSGYLVDKFADRERKAA 878
Query: 903 VSCMCRSY 910
+ M +++
Sbjct: 879 LKAMIKTH 886
>UniRef100_UPI00003C2180 UPI00003C2180 UniRef100 entry
Length = 799
Score = 238 bits (608), Expect = 5e-61
Identities = 166/444 (37%), Positives = 232/444 (51%), Gaps = 62/444 (13%)
Query: 576 NKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAHENGDASSGSDKEQSLTAY 635
N+++ ++LR F F T + LT AS + + S K + T+
Sbjct: 26 NRQAVSEELRALIFDSFQNGTIHTTDWANATLKSLTAASTSTKK----SLLKKRAAPTST 81
Query: 636 YSAAMAFSDTPEEKKRRENRSKRFDLGQGH-RTENNHFRRKNAGAGNLYSR----RASAL 690
+ +++ SD EE++R+E R+KRF+ Q R + + + +L SR RA A
Sbjct: 82 TTPSLSASDL-EEQQRKEKRAKRFEREQQEFRRQEDEMLETAIASTSLASRFGSIRAPAA 140
Query: 691 VLSKSFEDGVSKA--------------------------VED---IDWDALTVKGACQEI 721
V S+ S A V D IDWD TV G ++
Sbjct: 141 VGQPSWSAAASVAGTAPSHHASKYAASAPISSTQLTDTEVADPNVIDWDEHTVVGTSSKL 200
Query: 722 EKRYLRLTSAPDPATVRPEEVLEKALLMVQN---SQKNYLYKCDQLKSIRQDLTVQRIRN 778
EK YLRLTSAPDP TVRP L + L +++ ++ NY Y CDQ KS+RQDLTVQRI+N
Sbjct: 201 EKSYLRLTSAPDPKTVRPLSTLVQTLELLKKKWRTENNYSYICDQFKSMRQDLTVQRIKN 260
Query: 779 QLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNN 838
+ TVKVYE HAR A+E GDL EYNQCQSQL LYA GI G+ +EF AY +L ++H+ N
Sbjct: 261 EFTVKVYEIHARIALEMGDLGEYNQCQSQLRGLYAYGISGNAVEFLAYRIL-YLLHTKNR 319
Query: 839 R-------------------DLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRL 879
R D+ + M L++E K + AV+HAL VRAA+ +GNY +FF+L
Sbjct: 320 RDRDDRLSDLAPLRITFAFPDVNALMAELTEEHKAEPAVEHALQVRAALVTGNYHSFFQL 379
Query: 880 YKEAPNLNTCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVANGGSDEKD 939
Y +APN+N +MD +VE+ R A+ M + YRP++ +S+I+ L F A A+ K
Sbjct: 380 YTDAPNMNAYIMDHFVERERVNALHIMSKCYRPSIALSFIAEELAFQDVAAADEFLTAKG 439
Query: 940 TEAFEECLEWLKAHGASIMTDNNK 963
+ E A AS T N K
Sbjct: 440 AAVYLEPTPAELAALASSQTTNGK 463
>UniRef100_Q96PV6 KIAA1932 protein [Homo sapiens]
Length = 795
Score = 236 bits (602), Expect = 3e-60
Identities = 208/738 (28%), Positives = 328/738 (44%), Gaps = 79/738 (10%)
Query: 221 SGAATTSVQYQQQQQNPWADYYNQTEVSCAP-GTENLSVTSSSTFGGPIPAATS-GYATP 278
+GA + +NP + Q S + G S SSS GP+ +A A
Sbjct: 37 AGAGRENGMETPMHENPEWEKARQALASISKSGAAGGSAKSSSN--GPVASAQYVSQAEA 94
Query: 279 NSQPQQSYPPYWRQDSSASAVPSFQPAAINSGDHDGYWNHGAQTSQIHQTNPIQP----- 333
++ QQ Y +++Q + A + P G D ++ A Q+ P QP
Sbjct: 95 SALQQQQYYQWYQQYNYAYPYSYYYPMPPVPG-MDESMSYQAPPQQLPSAQPPQPSNPPH 153
Query: 334 -----NYQSPLDLKSSYDNFQDQQKTVSSQGTNLYLPPPPPLPPPPQQVVSTPIQSAPSL 388
N + + Q T + G + Y P P Q + ++ AP
Sbjct: 154 GAHTLNSGPQPGTAPATQHSQAGPATGQAYGPHTYTEPAKP---KKGQQLWNRMKPAPGT 210
Query: 389 DAKRVSKMQIPTNPRIASNLTYGQPKIEKDSSTTNTALKPAYLAVSLPKSTEKISSNDAA 448
K I P + ++G ++ ++ P P+ + S + A
Sbjct: 211 GGL---KFNIQKRPFAVTTQSFGS-----NAEGQHSGFGPQ----PNPEKVQNHSGSSAR 258
Query: 449 NSIL-KPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEP 507
++ KP +P+ ++ YVER T C+ ++ + ++KE++ DG+ T +W EP
Sbjct: 259 GNLSGKPDDWPQDMKEYVERCFTACESEEDKDRTEKLLKEVLQARLQDGSAYTIDWSREP 318
Query: 508 L-----FPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSA 562
L P+ +S + SS H S +R P ++ P + + ++
Sbjct: 319 LPGLTREPVAESPKKKRWEAASSLHPPRGAGSATRGGGAPSQR--GTPGAGGAGRARGNS 376
Query: 563 WVPNVKDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAHENGDA 622
+ + +N S D R+ R+SS++P R F++ S +H + D+
Sbjct: 377 FTKFGNRNVFMKDNSSSSSTDSRS--------RSSSRSPTRHFRR------SDSHSDSDS 422
Query: 623 SSGSDKEQSLTAYYSAAMAFSDTPEEKKR---RENRSKRFDLGQGHRT------------ 667
S ++ + R R R KR DL R+
Sbjct: 423 SYSGNECHPVGRRNPPPKGRGGRGAHMDRGRGRAQRGKRHDLAPTKRSRKKMAALECEDP 482
Query: 668 ENNHFRRKNAGA---GNLYSRRASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKR 724
E ++K A G+ R LVL S + + D DW L + G C +I K
Sbjct: 483 ERELKKQKRAARFQHGHSRRLRLEPLVLQMS---SLESSGADPDWQELQIVGTCPDITKH 539
Query: 725 YLRLTSAPDPATVRPEEVLEKALLMVQ---NSQKNYLYKCDQLKSIRQDLTVQRIRNQLT 781
YLRLT APDP+TVRP VL+K+L MV+ +++Y + C+Q+KSIRQDLTVQ IR + T
Sbjct: 540 YLRLTCAPDPSTVRPVAVLKKSLCMVKCHWKEKQDYAFACEQMKSIRQDLTVQGIRTEFT 599
Query: 782 VKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDL 841
V+VYETHAR A+E GD E+NQCQ+QL +LYA+ + G+ EF AY +L I + N+ D+
Sbjct: 600 VEVYETHARIALEKGDHEEFNQCQTQLKSLYAENLPGNVGEFTAYRILYYIF-TKNSGDI 658
Query: 842 LSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEKMRYK 901
+ + L+ E K D V HALA+R A GNY FFRLY AP ++ L+D + ++ R
Sbjct: 659 TTELAYLTRELKADPCVAHALALRTAWALGNYHRFFRLYCHAPCMSGYLVDKFADRERKV 718
Query: 902 AVSCMCRSYRPTVPVSYI 919
A+ M ++Y VP S +
Sbjct: 719 ALKAMIKTY--VVPSSLL 734
>UniRef100_UPI000036C09F UPI000036C09F UniRef100 entry
Length = 539
Score = 234 bits (597), Expect = 1e-59
Identities = 162/493 (32%), Positives = 246/493 (49%), Gaps = 48/493 (9%)
Query: 453 KPGMFPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPL---- 508
KP +P+ ++ YVER T C+ ++ + ++KE++ DG+ T +W EPL
Sbjct: 12 KPDDWPQDMKEYVERCFTACESEEDKDRTEKLLKEVLQARLQDGSAYTIDWSREPLPGLT 71
Query: 509 -FPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDNPVSISNDTVKYSAWVPNV 567
P+ +S + SS H S +R P ++ P + + +++
Sbjct: 72 REPVAESPKKKRWEAASSLHPPRGAGSATRGGGAPSQR--GTPGAGGAGRARGNSFTKFG 129
Query: 568 KDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTDASIAHENGDASSGSD 627
+ +N S D R+ R+SS++P R F++ S +H + D+S +
Sbjct: 130 NRNVFMKDNSSSSSTDSRS--------RSSSRSPTRHFRR------SDSHSDSDSSYSGN 175
Query: 628 KEQSLTAYYSAAMAFSDTPEEKKR---RENRSKRFDLGQGHRT------------ENNHF 672
+ + R R R KR DL R+ E
Sbjct: 176 ECHPVGRRNPPPKGRGGRGAHMDRGRGRAQRGKRHDLAPTKRSRKKMAALECEDPERELK 235
Query: 673 RRKNAGA---GNLYSRRASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKRYLRLT 729
++K A G+ R LVL S + + D DW L + G C +I K YLRLT
Sbjct: 236 KQKRAARFQHGHSRRLRLEPLVLQMS---SLESSGADPDWQELQIVGTCPDITKHYLRLT 292
Query: 730 SAPDPATVRPEEVLEKALLMVQ---NSQKNYLYKCDQLKSIRQDLTVQRIRNQLTVKVYE 786
APDP+TVRP VL+K+L MV+ +++Y + C+Q+KSIRQDLTVQ IR + TV+VYE
Sbjct: 293 CAPDPSTVRPVAVLKKSLCMVKCHWKEKQDYAFACEQMKSIRQDLTVQGIRTEFTVEVYE 352
Query: 787 THARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDLLSSMT 846
THAR A+E GD E+NQCQ+QL +LYA+ + G+ EF AY +L I + N+ D+ + +
Sbjct: 353 THARIALEKGDHEEFNQCQTQLKSLYAENLPGNVGEFTAYRILYYIF-TKNSGDITTELA 411
Query: 847 RLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEKMRYKAVSCM 906
L+ E K D V HALA+R A GNY FFRLY AP ++ L+D + ++ R A+ M
Sbjct: 412 YLTRELKADPCVAHALALRTAWALGNYHRFFRLYCHAPCMSGYLVDKFADRERKVALKAM 471
Query: 907 CRSYRPTVPVSYI 919
++Y VP S +
Sbjct: 472 IKTY--VVPSSLL 482
>UniRef100_Q7S9S1 Hypothetical protein [Neurospora crassa]
Length = 517
Score = 218 bits (554), Expect = 1e-54
Identities = 111/221 (50%), Positives = 151/221 (68%), Gaps = 4/221 (1%)
Query: 714 VKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQNSQK---NYLYKCDQLKSIRQD 770
V G C+ +EKRYLRLT+ P P+ VRPE VL + L +++ K NY Y CDQ KS+RQD
Sbjct: 250 VVGTCETLEKRYLRLTAPPVPSVVRPERVLRQTLELLKRKWKKEQNYSYICDQFKSMRQD 309
Query: 771 LTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLC 830
LTVQRI+N+ TV+VYE HAR A+E GDL EYNQCQ+QL ALY GI+G +EF AY +L
Sbjct: 310 LTVQRIKNEFTVEVYEIHARIALEKGDLGEYNQCQTQLKALYKMGIKGKSIEFKAYRILY 369
Query: 831 VIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCL 890
I H+ N L + L+ K++EA+KHAL VR+A+ GNY FFRLY + PN+ L
Sbjct: 370 FI-HTANRTALNDVLADLTAAEKEEEAIKHALDVRSALALGNYHRFFRLYNDTPNMGAYL 428
Query: 891 MDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVA 931
MD++V + R A+ +C++Y+P VP+ +++ L F + A A
Sbjct: 429 MDMFVGRERLAALCNICKAYKPDVPLRFVTEELYFESDADA 469
Score = 40.8 bits (94), Expect = 0.21
Identities = 36/146 (24%), Positives = 55/146 (37%), Gaps = 16/146 (10%)
Query: 365 PPPPPLPPPPQQVVSTPIQSAPSLDAKRVSKMQIPTNPRIASNLTYGQPKIEKDSSTTNT 424
PPPP PP S ++ P P YG I +T
Sbjct: 8 PPPPGTQPPAAYPYSVQVRQTFGQPYSAAPPASYPPPP------AYGPLPINPPPATPLH 61
Query: 425 ALKPAYLAVSLPKSTEKISSNDAANSILKPGMFPKSLRGYVERALTRCKDDKQMKACQ-- 482
A L + P +T++ + K +P+S+R YV+R+ D + +
Sbjct: 62 TNPAAPLTAATPTTTQEPAK--------KKIDWPESVRNYVQRSFQTSNLDPSVTRAEME 113
Query: 483 AVMKEMITKATADGTLCTRNWDMEPL 508
A +KE I+ A G + T NW+ PL
Sbjct: 114 AKLKETISYANDQGHMYTVNWEKMPL 139
>UniRef100_UPI000042C24F UPI000042C24F UniRef100 entry
Length = 480
Score = 215 bits (547), Expect = 6e-54
Identities = 145/445 (32%), Positives = 225/445 (49%), Gaps = 64/445 (14%)
Query: 562 AWVPNVKDRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRPFKKQRLTD--------- 612
AWV N R NK++ ++L+ F Q T + + Q L
Sbjct: 12 AWVQNCLARATA-SNKDAVNNELKKVLFEAHAQGTINTTDWSKVELQALKSQAQRTTYTP 70
Query: 613 ------ASIAHENGDASSGSDKEQSL-----------TAYYSAAMAFSDTPEEKKRRENR 655
+S A ++S K+ +L T+ + + FS + EE++ + R
Sbjct: 71 QPTYAVSSPALSASSSASTLTKDTTLKKKKKKNNGTSTSSFPSPYLFSGSAEEEEAKARR 130
Query: 656 SKRFDLGQGHRTENNHFRRK------------NAGAGNLYSRRASALVLSKS---FEDGV 700
+ RF + N+ + G G + + ++ K + V
Sbjct: 131 AARFQKPAATPSSNHAAASGGIGTWFVDDAIGSGGLGMVPGQVGKRKIMGKGNLGYGGAV 190
Query: 701 SKAVED--IDWDALTVKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQNSQKN-- 756
+ V+ IDWDA T++G ++EK YLRLTS P PA +RP VLE+ L ++++ KN
Sbjct: 191 VQEVDPNVIDWDAYTIRGTSTKLEKSYLRLTSEPSPADIRPLPVLEQTLELLKSRWKNEH 250
Query: 757 -YLYKCDQLKSIRQDLTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADG 815
Y Y DQ KS+RQDLTVQRI+N TVKVYE HAR A+EA DL EYNQCQS L LY G
Sbjct: 251 NYAYALDQFKSMRQDLTVQRIKNDFTVKVYEIHARIALEAKDLGEYNQCQSMLRQLYELG 310
Query: 816 IEGSYMEFAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVA 875
+ G EF +Y ++ ++H+ N D+ + + +L++ K+ AVKH+L V AA+++ NY
Sbjct: 311 LHGHPEEFLSYRIM-YLLHTRNRSDMATLLAQLTEAEKQHPAVKHSLDVHAALSTSNYHR 369
Query: 876 FFRLYKEAPNLNTCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVANGGS 935
FFRL+ APN++ +MD +VE+ R A++ M ++Y T+P+ YI H L F +
Sbjct: 370 FFRLFITAPNMSGYIMDHFVERERMSALAVMSKAYM-TLPLDYIFHTLAFDSE------- 421
Query: 936 DEKDTEAFEECLEWLKAHGASIMTD 960
+E ++L H A++ T+
Sbjct: 422 --------DETHQFLTEHNAAVYTN 438
>UniRef100_UPI0000248E94 UPI0000248E94 UniRef100 entry
Length = 383
Score = 214 bits (545), Expect = 1e-53
Identities = 127/309 (41%), Positives = 179/309 (57%), Gaps = 24/309 (7%)
Query: 670 NHFRRKNAGAGNLYSR-RASALVLSKSFEDGVSKAVEDIDWDALTVKGACQEIEKRYLRL 728
N +K + A +++ R LVL+ + D + E + WD + G CQ+I K YLRL
Sbjct: 94 NREAKKQSRAARFHTKLRTEPLVLNINVFDLPNGTQEGLSWDDCPIVGTCQDITKNYLRL 153
Query: 729 TSAPDPATVRPEEVLEKALLMVQ---NSQKNYLYKCDQLKSIRQDLTVQRIRNQLTVKVY 785
T APDP+TVRP VL K+L+ V+ S ++Y+Y C+Q+KSIRQDLTVQ +R TV+VY
Sbjct: 154 TCAPDPSTVRPVPVLRKSLIAVKAHWKSNQDYVYACEQMKSIRQDLTVQGVRTDFTVEVY 213
Query: 786 ETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLCVIMHSNNNRDLLSSM 845
ETHAR A+E GD E+NQCQ+QL ALY D + EF AY L+ I + N+ DL + +
Sbjct: 214 ETHARIALEKGDHEEFNQCQTQLKALYKDCPSDNVGEFTAYRLIYYIF-TKNSGDLTTEL 272
Query: 846 TRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCLMDLYVEKMRYKAVSC 905
L+ E + D V HAL +R A GN+ FFRLY++AP + L+D +VE+ R A+
Sbjct: 273 VYLTTELRADPCVAHALELRTAWALGNFHRFFRLYQKAPRMAAYLIDKFVERERNIALRA 332
Query: 906 MCRSYRPTVPVSYISHVLGFSTSAVANGGSDEKDTEAFEECLEWLKAHGASIM-TDNNKD 964
+ +S+RP+V V Y+ L F + CL +L G S +D +K
Sbjct: 333 ILKSFRPSVSVEYVQSSLAFPD---------------LDTCLAFLTGLGISFTPSDPSK- 376
Query: 965 ILVDTKVSS 973
+D KVSS
Sbjct: 377 --IDCKVSS 383
Score = 40.8 bits (94), Expect = 0.21
Identities = 14/52 (26%), Positives = 32/52 (60%)
Query: 457 FPKSLRGYVERALTRCKDDKQMKACQAVMKEMITKATADGTLCTRNWDMEPL 508
+P++++ YV+R T C+ ++ + ++K+++ DG+ T +W+ EPL
Sbjct: 1 WPQAMKEYVQRCFTACESEEDKDRTEKMLKDLLQSRLQDGSAYTIDWNREPL 52
>UniRef100_UPI000021B5A6 UPI000021B5A6 UniRef100 entry
Length = 508
Score = 208 bits (530), Expect = 6e-52
Identities = 137/414 (33%), Positives = 218/414 (52%), Gaps = 34/414 (8%)
Query: 539 EPLPEEKPVDNPVSISNDTVKYSAWVPNVKDRKVVMENKESKEDDLRNTKFSPFIQRTSS 598
+P + K +D P+S+ + +++P+ D V E+K + + + T+
Sbjct: 60 QPTEQSKKIDWPLSVRQYVAR--SFLPDNMDHTVDRTEMEAKLKETISNAHQTGVMYTTD 117
Query: 599 ----KAPQRPFKKQRL-----TDASIAHENGDASSGSDKEQSLTAYYSAAMAFSDTPEEK 649
PQ K++RL T ++ + + ++ + K++ + S + + P +
Sbjct: 118 WDKYPLPQELIKQERLASLMSTTVALPNTTWNVATPTQKKRKSSDMSSEGLPGATAPWQS 177
Query: 650 KRRENRSKRFDLGQGHRTENNHFRRKNAGAGNLYSRRASALVLSKSFE------DGVSKA 703
+ R ++T + +R GA S +A+ K E DG K+
Sbjct: 178 AKSNPLESRVS----YQTTD---KRPAPGAS---SNKAAMSKFQKQLEKRQRRFDGGYKS 227
Query: 704 VEDIDWDALT---VKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQNS---QKNY 757
+ + G CQ +EKRYLRLT P + VRPE +L + L +++ + NY
Sbjct: 228 TYRSPSPPPSDGPIVGTCQTLEKRYLRLTGPPVISQVRPEPILHQTLDLLKKKWRKESNY 287
Query: 758 LYKCDQLKSIRQDLTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIE 817
Y CDQ KS+RQDLTVQ I+N+ TV VYE HAR A+E GDL EYNQCQ+QL ALY G++
Sbjct: 288 SYICDQFKSLRQDLTVQHIKNEFTVSVYEIHARIALEKGDLGEYNQCQTQLRALYQLGLK 347
Query: 818 GSYMEFAAYNLLCVIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFF 877
G+ +EF AY +L I H+ N L +M L+ K++ +KHAL VR+A+ GNY FF
Sbjct: 348 GNPLEFKAYRILYFI-HTANRSALNDAMADLTPAEKEERPLKHALNVRSALALGNYHRFF 406
Query: 878 RLYKEAPNLNTCLMDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVA 931
+LY + PN+ LMD++V + R A+ +CR+Y+P V + +++ LGF + A A
Sbjct: 407 QLYLDTPNMGAYLMDMFVGRERLAALCNICRAYKPDVNLRFVTEELGFESDAEA 460
>UniRef100_UPI000023CC0B UPI000023CC0B UniRef100 entry
Length = 509
Score = 207 bits (526), Expect = 2e-51
Identities = 102/221 (46%), Positives = 150/221 (67%), Gaps = 4/221 (1%)
Query: 714 VKGACQEIEKRYLRLTSAPDPATVRPEEVLEKALLMVQNSQK---NYLYKCDQLKSIRQD 770
+ G C+ +EK+YLRLT+ P P+ VRPE +L + L +++ K NY Y CDQ KS+RQD
Sbjct: 242 IVGTCEVLEKKYLRLTAPPVPSKVRPESILRQTLDLLKKKWKRESNYSYICDQFKSMRQD 301
Query: 771 LTVQRIRNQLTVKVYETHARFAMEAGDLSEYNQCQSQLTALYADGIEGSYMEFAAYNLLC 830
LTVQ I+N TV VYE HAR A+E GD+ EYNQCQ+QL +LY G++G+ +EF AY +L
Sbjct: 302 LTVQHIKNDFTVSVYEIHARIALEKGDIGEYNQCQTQLRSLYGMGLKGNPIEFKAYRILY 361
Query: 831 VIMHSNNNRDLLSSMTRLSDEAKKDEAVKHALAVRAAVTSGNYVAFFRLYKEAPNLNTCL 890
I H+ N L ++ L+ K+++A+KHAL VR+++ GNY FF+LY + PN+ L
Sbjct: 362 FI-HTANRTGLNDTLADLTTAEKEEKAIKHALNVRSSLALGNYHRFFQLYLDTPNMGAYL 420
Query: 891 MDLYVEKMRYKAVSCMCRSYRPTVPVSYISHVLGFSTSAVA 931
MD++V + R A+ +C++Y+P V + +I+ LGF + A A
Sbjct: 421 MDMFVVRERLAALCNICKAYKPDVKLRFITEELGFESDADA 461
Score = 39.7 bits (91), Expect = 0.46
Identities = 53/236 (22%), Positives = 91/236 (38%), Gaps = 12/236 (5%)
Query: 375 QQVVSTPIQSAPSLDAKRVSKMQIPTNPRIASNLTYGQPKIEKDSSTTNTALKPAYLAVS 434
QQ S Q+ P+ A + IP R N YG P + ++P + S
Sbjct: 8 QQAPSVTQQTVPASYAYPTNPAFIPVAVRQGFN-QYGVPPPYAGYAPPPPPVQPQMSSAS 66
Query: 435 LPKSTEKISSNDAANSILKPGMFPKSLRGYVERALTRCKDDKQMKACQ--AVMKEMITKA 492
P + + A + + +P+S+R YV+R+ D+ + + A +K+ I A
Sbjct: 67 -PVNGVTATPEQAKSKV----DWPESVRSYVQRSFLPQYDEPTVPRAEIEAKLKDTIGTA 121
Query: 493 TADGTLCTRNWDMEPL---FPMPDSDVVNKDSVLSSTHKRSPRRSKSRWEPLPEEKPVDN 549
A+GTL T +WD PL + D K S + + + + + N
Sbjct: 122 KANGTLYTLDWDNMPLPQALVKAERDASTKQSFQVPLNSKKRKSTDFACNDNSQPPWRTN 181
Query: 550 PVSISNDTVKYSAWVPNVK-DRKVVMENKESKEDDLRNTKFSPFIQRTSSKAPQRP 604
S D + +S+ D + +K KE + R +F + S +P P
Sbjct: 182 SRSSLEDRISFSSPEKRASMDEPLPKSSKFQKEANKRKRRFENEYKSLRSPSPPPP 237
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.125 0.365
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,795,995,220
Number of Sequences: 2790947
Number of extensions: 81183221
Number of successful extensions: 337253
Number of sequences better than 10.0: 3072
Number of HSP's better than 10.0 without gapping: 173
Number of HSP's successfully gapped in prelim test: 3067
Number of HSP's that attempted gapping in prelim test: 317218
Number of HSP's gapped (non-prelim): 11695
length of query: 1003
length of database: 848,049,833
effective HSP length: 137
effective length of query: 866
effective length of database: 465,690,094
effective search space: 403287621404
effective search space used: 403287621404
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0260.13


