

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
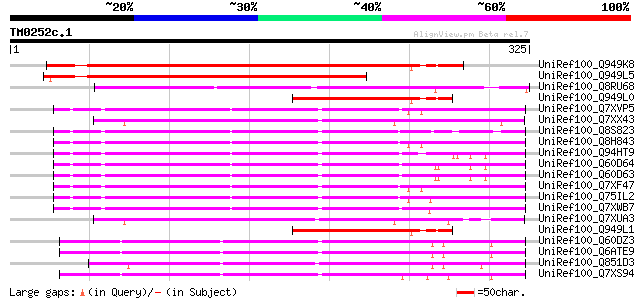
Query= TM0252c.1
(325 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q949K8 Gag polyprotein [Cicer arietinum] 264 3e-69
UniRef100_Q949L5 Gag polyprotein [Cicer arietinum] 191 2e-47
UniRef100_Q8RU68 Putative 22 kDa kafirin cluster; Ty3-Gypsy type... 123 8e-27
UniRef100_Q949L0 Putative gag polyprotein [Cicer arietinum] 119 1e-25
UniRef100_Q7XVP5 OSJNBa0023J03.1 protein [Oryza sativa] 119 1e-25
UniRef100_Q7XX43 OSJNBa0059D20.6 protein [Oryza sativa] 117 3e-25
UniRef100_Q8S823 Putative retroelement [Oryza sativa] 117 4e-25
UniRef100_Q8H843 Putative polyprotein [Oryza sativa] 115 1e-24
UniRef100_Q94HT9 Putative retroelement [Oryza sativa] 115 2e-24
UniRef100_Q60D64 Putative polyprotein [Oryza sativa] 114 3e-24
UniRef100_Q60D63 Putative polyprotein [Oryza sativa] 114 3e-24
UniRef100_Q7XF47 Putative polyprotein [Oryza sativa] 114 4e-24
UniRef100_Q75IL2 Putative polyprotein [Oryza sativa] 114 5e-24
UniRef100_Q7XWB7 OSJNBa0061A09.9 protein [Oryza sativa] 114 5e-24
UniRef100_Q7XUA3 OSJNBa0019D11.8 protein [Oryza sativa] 113 6e-24
UniRef100_Q949L1 Putative gag polyprotein [Cicer arietinum] 113 6e-24
UniRef100_Q60DZ3 Putative polyprotein [Oryza sativa] 113 8e-24
UniRef100_Q6ATE9 Putative polyprotein [Oryza sativa] 113 8e-24
UniRef100_Q851D3 Putative polyprotein [Oryza sativa] 112 1e-23
UniRef100_Q7XS94 OSJNBa0074B10.6 protein [Oryza sativa] 112 1e-23
>UniRef100_Q949K8 Gag polyprotein [Cicer arietinum]
Length = 277
Score = 264 bits (674), Expect = 3e-69
Identities = 138/264 (52%), Positives = 175/264 (66%), Gaps = 14/264 (5%)
Query: 24 NQLAEMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFT 83
+Q+AE + + ++ QT AA+ R+L R RE ++RGL +F R NPPKF
Sbjct: 16 DQMAEAMNNMAASVAAQT-------AAKTQRDLEKRGREIRAAESRGLVDFRRYNPPKFK 68
Query: 84 GGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVN 143
G ++AD WIQE+EKIF+++ G KV ATY+LLGDAEYWWR AR +M A H EVN
Sbjct: 69 GDEGSEKADQWIQEVEKIFDMINCQAGVKVSYATYMLLGDAEYWWRSARLLMGAAHEEVN 128
Query: 144 WNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPY 203
W SF+ FL+KYFP S R + FL L QGS+T+ EYAAK ESL++HFRFFR+++DEP+
Sbjct: 129 WESFKRKFLDKYFPMSTRTKLGDDFLKLHQGSLTVGEYAAKFESLSRHFRFFREEIDEPF 188
Query: 204 MCKRFVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAG---TGGPMRTSSR 260
MC RF GL+ +I+DSV PLGI RFQALVEK EVE MKN+R R G +GGP R
Sbjct: 189 MCHRFQDGLKYEIQDSVLPLGIQRFQALVEKCREVEDMKNKRASRVGNFNSGGPSRA--- 245
Query: 261 SYQGKGKLQRKKPYQRPTGEGFTP 284
+YQ +GK Q KPY RP P
Sbjct: 246 TYQNRGK-QVAKPYNRPQNNNGGP 268
>UniRef100_Q949L5 Gag polyprotein [Cicer arietinum]
Length = 209
Score = 191 bits (485), Expect = 2e-47
Identities = 100/205 (48%), Positives = 131/205 (63%), Gaps = 10/205 (4%)
Query: 22 NAN---QLAEMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQN 78
NAN Q+AE + + ++ QT AA+ R+L R RE ++RGL +F R N
Sbjct: 12 NANRNDQMAEAMNNMAASVAAQT-------AAKTQRDLEKRGREIRAAESRGLEDFRRYN 64
Query: 79 PPKFTGGTDPDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEAN 138
PPKF G ++AD WIQE+EKI ++++ G V ATY+LLGDAEYW R R +M A
Sbjct: 65 PPKFKGDESSEKADQWIQEVEKIIDMIKCQAGVTVSYATYMLLGDAEYWRRSGRLLMGAA 124
Query: 139 HVEVNWNSFRAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQ 198
H EVNW SF+ FL+KYFP S R + FL L QGSMT+ EYAAK SL++HFRF R++
Sbjct: 125 HEEVNWESFKRKFLDKYFPMSTRTKLGDDFLKLHQGSMTVGEYAAKFGSLSRHFRFVREE 184
Query: 199 VDEPYMCKRFVRGLRADIEDSVRPL 223
DEP+M F GL+ +I+DSV PL
Sbjct: 185 TDEPFMIHCFQDGLKYEIQDSVLPL 209
>UniRef100_Q8RU68 Putative 22 kDa kafirin cluster; Ty3-Gypsy type [Oryza sativa]
Length = 1230
Score = 123 bits (308), Expect = 8e-27
Identities = 86/284 (30%), Positives = 137/284 (47%), Gaps = 25/284 (8%)
Query: 54 RELHLRQREASLDQNR-GLNNFIRQNPPKFTGGTDPDEADLWIQEIEKIFEVLQTSEGAK 112
R++ + +R D NR GL F + PP F+G +P EA+ WI +EK FE + ++ K
Sbjct: 53 RQMEILERITKSDANRNGLGEFQKLKPPTFSGTANPLEAEEWIVAMEKSFEAMGCTDKEK 112
Query: 113 VGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAAFLEKYFPDSARDERESQFLTLR 172
+ ATY+L A WW A + + + W F+ AF +KYFP+S + +E +FL L+
Sbjct: 113 IIYATYMLQSSAFEWW-DAHKKSYSERIFITWELFKEAFYKKYFPESVKRMKEKEFLELK 171
Query: 173 QGSMTIPEYAAKLESLAKHFRFFRDQVD-EPYMCKRFVRGLRADIEDSVRPLGIMRFQAL 231
QG+ ++ EY + LA RF + V + +RF GLR ++ V + F+ +
Sbjct: 172 QGNKSVAEYEIEFSRLA---RFAPEFVQTDGSKARRFESGLRQPLKRRVEAFELTIFREV 228
Query: 232 VEKATEVEL-MKNRRMDRAGTGGPMRTSSRSYQGK------GKLQRKKPYQRPTGEGFTP 284
V KA +E +R++ +T++ QG+ G++QRK +
Sbjct: 229 VSKAQLLEKGYHEQRIEHGQPQKKFKTNNPQNQGRFRGNYSGQMQRKSSENQGRKCPICQ 288
Query: 285 GLYRPTIAAAGGAGSQAGSREITCFKCGEIGHYSTKCP---KGK 325
G + P+I CF+CGE GH +CP KGK
Sbjct: 289 GSHVPSICPNCWG---------RCFECGEAGHTRYQCPLLQKGK 323
>UniRef100_Q949L0 Putative gag polyprotein [Cicer arietinum]
Length = 115
Score = 119 bits (298), Expect = 1e-25
Identities = 62/103 (60%), Positives = 76/103 (73%), Gaps = 7/103 (6%)
Query: 178 IPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSVRPLGIMRFQALVEKATE 237
+ EYAAK ESL++HFRFFR+++DEP+MC RF GL+ +I+DSV PLGI RFQALVEK +
Sbjct: 1 VGEYAAKFESLSRHFRFFREEIDEPFMCHRFQDGLKYEIQDSVLPLGIQRFQALVEKCRK 60
Query: 238 VELMKNRRMDRAG---TGGPMRTSSRSYQGKGKLQRKKPYQRP 277
VE MKN+R R G +GGP R +YQ KGK Q KPY RP
Sbjct: 61 VEDMKNKRASRVGNFNSGGPSRV---NYQNKGK-QVAKPYNRP 99
>UniRef100_Q7XVP5 OSJNBa0023J03.1 protein [Oryza sativa]
Length = 1827
Score = 119 bits (297), Expect = 1e-25
Identities = 90/326 (27%), Positives = 141/326 (42%), Gaps = 37/326 (11%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+R PP F+ T+
Sbjct: 358 QMMAAMMQQMQ-QQHQQMHQRMMQHAEQQH--QQFGPPPPQSKLPEFLRVRPPTFSSTTN 414
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV AT+ L G A WW EV W F
Sbjct: 415 PMEANDWLHAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNHMATRPPG-TEVTWAEF 473
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+MT+ EY + LA++ D + ++
Sbjct: 474 CRSFRKAQVPDGVVAQKKREFRALHQGNMTVTEYLHEFNRLARYAP--EDVRTDAEKQEK 531
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDR------------------- 248
F+ GL ++ + + F+ LV+KA E +N+ MDR
Sbjct: 532 FMAGLDDELTNQLISGDYADFERLVDKAIRQEDQRNK-MDRKRKAAQFRSNQGSQHRPRF 590
Query: 249 --AGTGGPMR---------TSSRSYQGKGKLQRKKPYQRPTGEGFTPGLYRPTIAAAGGA 297
GGP SS +QG Q + Q G P + A A
Sbjct: 591 TPGQQGGPTTMIVRQHRPFNSSNFHQGTSGSQNHQGGQPNRGAAPRPPMAPAQSAQPAQA 650
Query: 298 GSQAGSREITCFKCGEIGHYSTKCPK 323
+ G++ +CF CGE+GH++ KCPK
Sbjct: 651 KKETGAKPGSCFNCGELGHFADKCPK 676
>UniRef100_Q7XX43 OSJNBa0059D20.6 protein [Oryza sativa]
Length = 1470
Score = 117 bits (294), Expect = 3e-25
Identities = 79/281 (28%), Positives = 129/281 (45%), Gaps = 13/281 (4%)
Query: 53 ARELHLRQREASLDQNRG---LNNFIRQNPPKFTGGTDPDEADLWIQEIEKIFEVLQTSE 109
A + L Q AS NRG F+R PP F +P +A+ W++ IEK +++ E
Sbjct: 27 ATQTQLLQAIASNQGNRGGSSFGEFMRTKPPTFATAEEPMDAEDWLRIIEKKLTLVRVRE 86
Query: 110 GAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAAFLEKYFPDSARDERESQFL 169
KV A L G A WW + E + E NW F AF + + P + ++++F
Sbjct: 87 ADKVIFAANQLEGPAGDWWDTYKEAREEDAGEPNWEEFTTAFRDNFVPAAVMRMKKNEFR 146
Query: 170 TLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSVRPLGIMRFQ 229
LRQG+ T+ EY + LA++ RD DE +F+ GL ++ + FQ
Sbjct: 147 KLRQGNTTVQEYLNRFTQLARYAT--RDLADEEEKIDKFIEGLNDELRGPMIGQDHDSFQ 204
Query: 230 ALVEKATEVE----LMKNRRMDRAGTGGPMRTSSRSYQGKGKLQRKKPYQRPTGEGFTPG 285
+L+ K +E ++++ R R P +T+ + +G + + +
Sbjct: 205 SLINKVVRLENDRKVVEHNRKRRLAMNRPPQTAPQRPKGTTPSAWRPTVVTTSRPAASSN 264
Query: 286 LYRP-TIAAAGGAGSQAGSREI---TCFKCGEIGHYSTKCP 322
+RP TI +QA + + +CF CGE GH++ KCP
Sbjct: 265 FHRPVTIQNRAPTPNQAATGSVRPGSCFNCGEYGHFANKCP 305
>UniRef100_Q8S823 Putative retroelement [Oryza sativa]
Length = 1425
Score = 117 bits (293), Expect = 4e-25
Identities = 85/296 (28%), Positives = 138/296 (45%), Gaps = 16/296 (5%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+ PP F+ T+
Sbjct: 52 QMMAAMMQQMQ-QQHQQMHQRMLQHAEQQH--QQFGPPPPQSKLPEFLLVRPPTFSSTTN 108
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV AT+ L G A WW EV W F
Sbjct: 109 PMEANDWLHAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNHMATRPPG-TEVTWAEF 167
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+ T+ EY + LA++ D + ++
Sbjct: 168 CRSFKKAQVPDGVVAQKKREFRALHQGNRTVTEYLHEFNRLARYAP--EDVRTDAEKQEK 225
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQGKGK 267
F+ GL ++ + + F+ LV+KA E +N +MDR + S+++ G G+
Sbjct: 226 FLAGLDDELTNQLISGDYADFERLVDKAIRQEDQRN-KMDRKSNFPQGASGSQNHHG-GQ 283
Query: 268 LQRKKPYQRPTGEGFTPGLYRPTIAAAGGAGSQAGSREITCFKCGEIGHYSTKCPK 323
R + P P P+ A G++ GS CF CGE+GH++ KCPK
Sbjct: 284 PNRGAAPRPP----MAPAQSGPSAQAKKETGAKPGS----CFNCGELGHFADKCPK 331
>UniRef100_Q8H843 Putative polyprotein [Oryza sativa]
Length = 1796
Score = 115 bits (289), Expect = 1e-24
Identities = 88/326 (26%), Positives = 139/326 (41%), Gaps = 37/326 (11%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+R PP F+ T+
Sbjct: 327 QMMAAMMQQMQ-QQHQQMHQRMMQHAEQQH--QQFGPPPPQSKLPEFLRVRPPTFSSTTN 383
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV AT+ L G A WW EV W F
Sbjct: 384 PMEANDWLHAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNHMATRPPG-TEVTWAEF 442
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+ T+ EY + LA++ D + ++
Sbjct: 443 CRSFRKAQVPDGVVAQKKREFRALHQGNRTVTEYLQEFNRLARYAP--EDVRTDAEKQEK 500
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDR------------------- 248
F+ GL ++ + + F+ LV+KA E +N +MDR
Sbjct: 501 FMAGLDDELTNQLISGDYADFERLVDKAIRQEDQRN-KMDRKRKAAQFRAPQGSHQRPRF 559
Query: 249 --AGTGGPMR---------TSSRSYQGKGKLQRKKPYQRPTGEGFTPGLYRPTIAAAGGA 297
GGP SS +QG Q Q G P + + A
Sbjct: 560 TPGQQGGPTTMIVRQHRPFNSSNFHQGSSGSQNHHGGQPNRGAAPRPPMAPAQSGPSAQA 619
Query: 298 GSQAGSREITCFKCGEIGHYSTKCPK 323
+ G++ +CF CGE+GH++ KCPK
Sbjct: 620 KKETGAKPGSCFNCGELGHFADKCPK 645
>UniRef100_Q94HT9 Putative retroelement [Oryza sativa]
Length = 1714
Score = 115 bits (288), Expect = 2e-24
Identities = 91/330 (27%), Positives = 146/330 (43%), Gaps = 45/330 (13%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+R PP F+ T+
Sbjct: 245 QMIAAMMQQMQ-QQHQQMHQRMMQHAEQQH--QQFGPPPPQSKLPEFLRVRPPTFSSTTN 301
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV AT+ L G A WW EV W F
Sbjct: 302 PMEANDWLHAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNHMATRPPG-TEVTWAEF 360
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+ T+ EY + LA++ D + ++
Sbjct: 361 CRSFRKAQVPDGVVAQKKREFRALHQGNRTVTEYLHEFNRLARYAP--EDVRTDAEKQEK 418
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQGKGK 267
F+ GL ++ + + F+ LV+KA E +N+ MDR R ++QG +
Sbjct: 419 FMAGLDDELTNQLISGDYADFERLVDKAIRQEDQRNK-MDRKRKAAQFR----AHQGSHQ 473
Query: 268 LQRKKPYQR--PTG------EGFTPGLY---------------------RPTIAAAGG-- 296
R P Q+ PT F P + RP++A A
Sbjct: 474 RPRFTPGQQGGPTTMIVRQHRPFNPSNFHQGTSGSQNHHGSQPNRGAAPRPSVAPAQSGQ 533
Query: 297 ---AGSQAGSREITCFKCGEIGHYSTKCPK 323
A + G++ +CF CGE+GH++ KCPK
Sbjct: 534 PAQAKKETGAKPGSCFNCGELGHFADKCPK 563
>UniRef100_Q60D64 Putative polyprotein [Oryza sativa]
Length = 1777
Score = 114 bits (286), Expect = 3e-24
Identities = 88/326 (26%), Positives = 145/326 (43%), Gaps = 37/326 (11%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+R PP F+ T+
Sbjct: 308 QMMAAMMQQMQ-QQHQQMHQRMMQHAEQQH--QQFGPPPPQSKLPEFLRVRPPTFSSTTN 364
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV AT+ L G A WW EV W F
Sbjct: 365 PMEANDWLHAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNHMATRPPG-TEVTWAEF 423
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+ T+ EY + LA++ D + ++
Sbjct: 424 CRSFRKAQVPDGVVAQKKREFRALHQGNRTVTEYLHEFNGLARYAP--EDVRTDAEKQEK 481
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQGK-- 265
F+ GL ++ + + F+ LV+KA E +N+ MDR R S+Q
Sbjct: 482 FMAGLDDELTNQLISGDYADFERLVDKAIRQEDQRNK-MDRKRKAAQFRAPQGSHQRPRF 540
Query: 266 --GK--------LQRKKPYQRPTGEGFTPGLY-------------RPTIAAAGG-----A 297
G+ +++ +P+ T G RP++A A A
Sbjct: 541 TPGQQGGPTTMIVRQHRPFNPSNFHQGTSGSQNHHGGQPNRGAAPRPSVAPAQSGQPAQA 600
Query: 298 GSQAGSREITCFKCGEIGHYSTKCPK 323
+ G++ +CF CGE+GH++ KCPK
Sbjct: 601 KKETGAKPGSCFNCGELGHFADKCPK 626
>UniRef100_Q60D63 Putative polyprotein [Oryza sativa]
Length = 1765
Score = 114 bits (286), Expect = 3e-24
Identities = 88/326 (26%), Positives = 145/326 (43%), Gaps = 37/326 (11%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+R PP F+ T+
Sbjct: 308 QMMAAMMQQMQ-QQHQQMHQRMMQHAEQQH--QQFGPPPPQSKLPEFLRVRPPTFSSTTN 364
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV AT+ L G A WW EV W F
Sbjct: 365 PMEANDWLHAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNHMATRPPG-TEVTWAEF 423
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+ T+ EY + LA++ D + ++
Sbjct: 424 CRSFRKAQVPDGVVAQKKREFRALHQGNRTVTEYLHEFNGLARYAP--EDVRTDAEKQEK 481
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQGK-- 265
F+ GL ++ + + F+ LV+KA E +N+ MDR R S+Q
Sbjct: 482 FMAGLDDELTNQLISGDYADFERLVDKAIRQEDQRNK-MDRKRKAAQFRAPQGSHQRPRF 540
Query: 266 --GK--------LQRKKPYQRPTGEGFTPGLY-------------RPTIAAAGG-----A 297
G+ +++ +P+ T G RP++A A A
Sbjct: 541 TPGQQGGPTTMIVRQHRPFNPSNFHQGTSGSQNHHGGQPNRGAAPRPSVAPAQSGQPAQA 600
Query: 298 GSQAGSREITCFKCGEIGHYSTKCPK 323
+ G++ +CF CGE+GH++ KCPK
Sbjct: 601 KKETGAKPGSCFNCGELGHFADKCPK 626
>UniRef100_Q7XF47 Putative polyprotein [Oryza sativa]
Length = 2162
Score = 114 bits (285), Expect = 4e-24
Identities = 88/326 (26%), Positives = 138/326 (41%), Gaps = 37/326 (11%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+R PP F+ T+
Sbjct: 646 QMMAAMMQQMQ-QQHQQMHQRMMQHAEQQH--QQFGPPPPQSKLPEFLRVRPPTFSSTTN 702
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV AT+ L G A WW EV W F
Sbjct: 703 PMEANDWLHAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNHMATRPPG-TEVTWVEF 761
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+ T+ EY + LA++ D + ++
Sbjct: 762 CRSFRKAQVPDGVMAQKKREFRALHQGNRTVTEYLHEFNRLARYAP--EDVRTDAEKQEK 819
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDR------------------- 248
F+ GL ++ + + F+ LV+KA E +N +MDR
Sbjct: 820 FMAGLDDELTNQLISGDYADFEWLVDKAIRQEDQRN-KMDRKRKAAQFRAPQGSHQRPRF 878
Query: 249 --AGTGGPMR---------TSSRSYQGKGKLQRKKPYQRPTGEGFTPGLYRPTIAAAGGA 297
GGP SS +QG Q Q G P + A
Sbjct: 879 TPGQQGGPTTMIVRQHRPFNSSNFHQGSSGSQNHHGGQPNRGAAPRPPMAPAQSGPPAQA 938
Query: 298 GSQAGSREITCFKCGEIGHYSTKCPK 323
+ G++ +CF CGE+GH++ KCPK
Sbjct: 939 KKETGAKPGSCFNCGELGHFADKCPK 964
>UniRef100_Q75IL2 Putative polyprotein [Oryza sativa]
Length = 1730
Score = 114 bits (284), Expect = 5e-24
Identities = 88/326 (26%), Positives = 137/326 (41%), Gaps = 37/326 (11%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+R PP F+ T+
Sbjct: 316 QMMAAMMQQMQ-QQHQQMYQRMLQHAEQQH--QQFGPPPPQSKLPEFLRVRPPTFSSTTN 372
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV T+ L G A WW EV W F
Sbjct: 373 PMEANDWLHAIEKKLNLLQCNDQEKVAFTTHQLQGPASAWWDNHMATRPPG-TEVTWAEF 431
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+ T+ EY + LA++ D + +R
Sbjct: 432 CRSFRKAQVPDGVVAQKKREFRALHQGNRTVTEYLHEFNRLARYAP--EDVRTDAEKQER 489
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDR------------------- 248
F+ GL ++ + + F+ LV+KA E +N +MDR
Sbjct: 490 FMAGLDDELTNQLISGDYADFERLVDKAIRQEDQRN-KMDRKRKAAQFRAPQGSHQRPRF 548
Query: 249 --AGTGGPMRTSSRSY---------QGKGKLQRKKPYQRPTGEGFTPGLYRPTIAAAGGA 297
GGP R Y QG Q Q G P + + A
Sbjct: 549 APGQQGGPTTMIVRQYRPFHPSNFPQGASGSQNHHGGQPNRGAAPRPPMAPAQSGPSAQA 608
Query: 298 GSQAGSREITCFKCGEIGHYSTKCPK 323
+ G++ +CF CGE+GH++ KCPK
Sbjct: 609 RKETGAKPGSCFNCGELGHFADKCPK 634
>UniRef100_Q7XWB7 OSJNBa0061A09.9 protein [Oryza sativa]
Length = 1653
Score = 114 bits (284), Expect = 5e-24
Identities = 86/326 (26%), Positives = 139/326 (42%), Gaps = 37/326 (11%)
Query: 28 EMVATLVQAMTVQTNDNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTD 87
+M+A ++Q M Q + +R + A + H Q+ L F+R PP F+ T+
Sbjct: 250 QMMAAMMQQMQ-QQHQQMHQRMMQYAEQQH--QQFGPPPPQSKLPEFLRVRPPTFSSTTN 306
Query: 88 PDEADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSF 147
P EA+ W+ IEK +LQ ++ KV AT+ L G A WW EV W F
Sbjct: 307 PMEANDWLHAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNHMATRPPG-TEVTWAEF 365
Query: 148 RAAFLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKR 207
+F + PD +++ +F L QG+ T+ EY + LA++ D + ++
Sbjct: 366 CRSFRKAQVPDGVVAQKKREFRALHQGNRTVTEYLHEFNRLARYAP--EDVRTDAEKQEK 423
Query: 208 FVRGLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRS------ 261
F+ GL ++ + + F+ LV+KA E +N+ MDR R++ S
Sbjct: 424 FMAGLDDELTNQLISGDYADFERLVDKAIRQEDQRNK-MDRKRKAAQFRSNQGSQHRPRF 482
Query: 262 ------------------------YQGKGKLQRKKPYQRPTGEGFTPGLYRPTIAAAGGA 297
+QG Q + Q G P + A A
Sbjct: 483 TPGQQGGPTTMIVRQHRPFHSGNFHQGASGSQNHQGGQPNRGAAPRPPMAPAQSAQPAQA 542
Query: 298 GSQAGSREITCFKCGEIGHYSTKCPK 323
+ G++ +CF CGE+GH++ KCPK
Sbjct: 543 KKETGAKPGSCFNCGELGHFADKCPK 568
>UniRef100_Q7XUA3 OSJNBa0019D11.8 protein [Oryza sativa]
Length = 1470
Score = 113 bits (283), Expect = 6e-24
Identities = 84/290 (28%), Positives = 124/290 (41%), Gaps = 31/290 (10%)
Query: 53 ARELHLRQREASLDQNRG---LNNFIRQNPPKFTGGTDPDEADLWIQEIEKIFEVLQTSE 109
A + L Q AS NRG F+R PP F +P EA+ W++ IEK +++ E
Sbjct: 27 ATQTQLLQAIASNQGNRGGSSFGEFMRTKPPTFATADEPMEAEDWLRIIEKKLTLVRVRE 86
Query: 110 GAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAAFLEKYFPDSARDERESQFL 169
KV A L G A WW + E E W F AF E + P + ++++F
Sbjct: 87 ADKVIFAVNQLEGPAGDWWDTYKEAREDGAGEPTWEEFTVAFRENFVPAAVMRMKKNEFR 146
Query: 170 TLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSVRPLGIMRFQ 229
LRQG+ T+ EY K LA++ D DE +F+ GL ++ + FQ
Sbjct: 147 RLRQGNTTVQEYLNKFTQLARY--AIGDLADEEEKIDKFIEGLNDELRGPMIGQDHESFQ 204
Query: 230 ALVEKATEVE----LMKNRRMDRAGTGGPMRTSSRSYQGKGKLQRKKP------------ 273
+L+ K +E +++ R R +T + +G K P
Sbjct: 205 SLINKVVRLENDQRTVEHNRKRRLAMSRLPQTVPQRLKGATPSGWKPPVMVTNRPAALSN 264
Query: 274 YQRPTG-EGFTPGLYRPTIAAAGGAGSQAGSREITCFKCGEIGHYSTKCP 322
+ RP + TP PT+AA G + + CF CGE GHY+ CP
Sbjct: 265 FNRPVALQNRTP---TPTLAAPG------AKKNVDCFNCGEYGHYANNCP 305
>UniRef100_Q949L1 Putative gag polyprotein [Cicer arietinum]
Length = 116
Score = 113 bits (283), Expect = 6e-24
Identities = 61/103 (59%), Positives = 74/103 (71%), Gaps = 7/103 (6%)
Query: 178 IPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSVRPLGIMRFQALVEKATE 237
+ EYAAK ESL++HFRFFR++ EP+MC RF GL+ +I+DSV PLGI RFQALVEK E
Sbjct: 1 VGEYAAKFESLSRHFRFFREKDYEPFMCHRFQDGLKYEIQDSVLPLGIQRFQALVEKCRE 60
Query: 238 VELMKNRRMDRAG---TGGPMRTSSRSYQGKGKLQRKKPYQRP 277
VE MKN+R R G +GGP R +YQ +GK Q KPY RP
Sbjct: 61 VEDMKNKRASRVGNFNSGGPSRA---NYQNRGK-QVAKPYNRP 99
>UniRef100_Q60DZ3 Putative polyprotein [Oryza sativa]
Length = 1374
Score = 113 bits (282), Expect = 8e-24
Identities = 87/316 (27%), Positives = 136/316 (42%), Gaps = 28/316 (8%)
Query: 32 TLVQAMTVQTN-DNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTDPDE 90
TL Q + QT N + A++ + + Q QN+ L +F+ PP F+ T+P E
Sbjct: 28 TLAQILAQQTQLINLLVQQAQNQQGNNQNQNPPPPPQNK-LADFLHVRPPTFSSTTNPVE 86
Query: 91 ADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAA 150
A W+ +EK +++Q +E KV A++ L G A WW R A + W F AA
Sbjct: 87 AGDWLHAVEKKLDLIQCTEQEKVSFASHQLHGPAAEWWDHFR-QGRAEGEPITWQEFTAA 145
Query: 151 FLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVR 210
F + + P ++ +F L QGS ++ EY LA++ D + ++F+
Sbjct: 146 FKKTHIPSGVVALKKREFRALNQGSRSVTEYLHDFNRLARYAP--EDVRTDEERQEKFLE 203
Query: 211 GLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQ------- 263
GL ++ ++ FQ LV+KA E NR + + S Q
Sbjct: 204 GLNDELSYALMSTDFRDFQQLVDKAIRQEDKYNRMEQKKRRAAQFKAQQGSNQRPRLVTG 263
Query: 264 ---------GKGKLQR--KKPYQRPTGEGFTPGLYRPTIAAAGGAGSQ-----AGSREIT 307
G + R ++ Y TG AA A +Q GS+ +T
Sbjct: 264 PQAPSYPQGGSSSVVRPQRQFYNNNTGNRGNDNRNMVVHPAATSAQNQPVRKEQGSKTVT 323
Query: 308 CFKCGEIGHYSTKCPK 323
CF CG++GHY+ KCPK
Sbjct: 324 CFNCGDLGHYADKCPK 339
>UniRef100_Q6ATE9 Putative polyprotein [Oryza sativa]
Length = 1472
Score = 113 bits (282), Expect = 8e-24
Identities = 87/316 (27%), Positives = 136/316 (42%), Gaps = 28/316 (8%)
Query: 32 TLVQAMTVQTN-DNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTDPDE 90
TL Q + QT N + A++ + + Q QN+ L +F+ PP F+ T+P E
Sbjct: 28 TLAQILAQQTQLINLLVQQAQNQQGNNQNQNPPPPPQNK-LADFLHVRPPTFSSTTNPVE 86
Query: 91 ADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAA 150
A W+ +EK +++Q +E KV A++ L G A WW R A + W F AA
Sbjct: 87 AGDWLHAVEKKLDLIQCTEQEKVSFASHQLHGPAAEWWDHFR-QGRAEGEPITWQEFTAA 145
Query: 151 FLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVR 210
F + + P ++ +F L QGS ++ EY LA++ D + ++F+
Sbjct: 146 FKKTHIPSGVVALKKREFRALNQGSRSVTEYLHDFNRLARYAP--EDVRTDEERQEKFLE 203
Query: 211 GLRADIEDSVRPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQ------- 263
GL ++ ++ FQ LV+KA E NR + + S Q
Sbjct: 204 GLNDELSYALMSTDFRDFQQLVDKAIRQEDKYNRMEQKKRRAAQFKAQQGSNQRPRLVTG 263
Query: 264 ---------GKGKLQR--KKPYQRPTGEGFTPGLYRPTIAAAGGAGSQ-----AGSREIT 307
G + R ++ Y TG AA A +Q GS+ +T
Sbjct: 264 PQAPSYPQGGSSSVVRPQRQFYNNNTGNRGNDNRNMVVHPAATSAQNQPVRKEQGSKTVT 323
Query: 308 CFKCGEIGHYSTKCPK 323
CF CG++GHY+ KCPK
Sbjct: 324 CFNCGDLGHYADKCPK 339
>UniRef100_Q851D3 Putative polyprotein [Oryza sativa]
Length = 1770
Score = 112 bits (281), Expect = 1e-23
Identities = 81/301 (26%), Positives = 133/301 (43%), Gaps = 30/301 (9%)
Query: 50 AEDARELHLRQREASLDQNRGLNN---------FIRQNPPKFTGGTDPDEADLWIQEIEK 100
A+ A+ +++ ++ QN+G N+ F+R PP F+ T+P EA W+ IEK
Sbjct: 311 AQQAQLMNMMMQQLQNQQNQGNNHAPPQNKLAEFLRVRPPIFSSTTNPVEAGDWLHAIEK 370
Query: 101 IFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAAFLEKYFPDSA 160
++LQ ++ KV A++ L G A WW R + + W F AAF + + P
Sbjct: 371 KLDLLQCTDQEKVSFASHQLHGPASEWWDHFR-LNRTTAEPITWLEFTAAFRKTHIPSGV 429
Query: 161 RDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVRGLRADIEDSV 220
++ +F +L QGS ++ EY + LA++ D + +RF+ GL ++ +
Sbjct: 430 VSLKKKEFRSLNQGSRSVTEYLHEFNRLARY--ALEDVRTDEERQERFLEGLNDELSYPL 487
Query: 221 RPLGIMRFQALVEKATEVELMKNRRMDRAGTGGPMRTSSRSYQ--------------GKG 266
FQ LV+KA E NR + +T + Q G
Sbjct: 488 MTGDYPDFQKLVDKAIRQEDKYNRMEQKKRRIAQFKTQQGNNQRLRLTLGPQSMPQGGSS 547
Query: 267 KLQR--KKPYQRPTGEGFTPGLYRPTIAAA--GGAGSQAGSREITCFKCGEIGHYSTKCP 322
+ R ++ + G RP A+ A + GS+ + CF CG+ GHY+ KCP
Sbjct: 548 SVVRPQRQFFNNHAGNNIRNQAPRPVAASTQQQPAKREQGSKPVVCFNCGDPGHYADKCP 607
Query: 323 K 323
K
Sbjct: 608 K 608
>UniRef100_Q7XS94 OSJNBa0074B10.6 protein [Oryza sativa]
Length = 1447
Score = 112 bits (281), Expect = 1e-23
Identities = 91/316 (28%), Positives = 139/316 (43%), Gaps = 28/316 (8%)
Query: 32 TLVQAMTVQTN-DNAQRRAAEDARELHLRQREASLDQNRGLNNFIRQNPPKFTGGTDPDE 90
TL Q + QT N + A++ + + Q QN+ L +F+R PP F+ T+P E
Sbjct: 20 TLAQILAQQTQLINLLVQQAQNQQANNQNQNPPPPPQNK-LADFLRVRPPTFSSTTNPVE 78
Query: 91 ADLWIQEIEKIFEVLQTSEGAKVGLATYLLLGDAEYWWRGARGMMEANHVEVNWNSFRAA 150
A W+ +EK +++Q +E KV A++ L G A WW R A + W F AA
Sbjct: 79 AGDWLHAVEKKLDLIQCTEQEKVSFASHQLHGPAAEWWDHFR-QGRAEGEPITWQEFTAA 137
Query: 151 FLEKYFPDSARDERESQFLTLRQGSMTIPEYAAKLESLAKHFRFFRDQVDEPYMCKRFVR 210
F + + P ++ +F L QGS ++ EY LA++ D + ++F+
Sbjct: 138 FKKTHIPAGVVALKKREFRALNQGSRSVTEYLHDFNRLARYAP--EDVRTDEERQEKFLE 195
Query: 211 GLRADIEDSVRPLGIMRFQALVEKATEVELMKNR---RMDRAGTGGPMRTSSR------- 260
GL ++ ++ FQ LV+KA E NR +M RA + S++
Sbjct: 196 GLNDELSYALMSTDFWDFQQLVDKAIRQEDKYNRMEQKMRRAAQFKAQQGSNQRPRLVTG 255
Query: 261 ----SYQGKGKLQRKKP----YQRPTGEGFTPGLYRPTIAAAGGAGSQ-----AGSREIT 307
SY G +P Y TG AA A +Q GS+ +
Sbjct: 256 PQAPSYSQGGSSSVIRPQRQFYNNNTGNRGNDNRNMVARPAATSAQNQPVRREQGSKPVV 315
Query: 308 CFKCGEIGHYSTKCPK 323
CF CG+ GHY+ KCPK
Sbjct: 316 CFNCGDPGHYADKCPK 331
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 534,066,528
Number of Sequences: 2790947
Number of extensions: 22451792
Number of successful extensions: 61780
Number of sequences better than 10.0: 880
Number of HSP's better than 10.0 without gapping: 563
Number of HSP's successfully gapped in prelim test: 317
Number of HSP's that attempted gapping in prelim test: 60021
Number of HSP's gapped (non-prelim): 1357
length of query: 325
length of database: 848,049,833
effective HSP length: 127
effective length of query: 198
effective length of database: 493,599,564
effective search space: 97732713672
effective search space used: 97732713672
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0252c.1


