

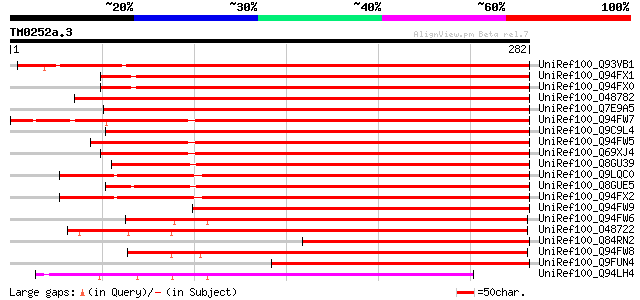
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0252a.3
(282 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q93VB1 Putative heme oxygenase 1 precursor [Pisum sati... 422 e-117
UniRef100_Q94FX1 Heme oxygenase 1 [Glycine max] 400 e-110
UniRef100_Q94FX0 Heme oxygenase 3 [Glycine max] 398 e-109
UniRef100_O48782 Heme oxygenase 1 precursor [Arabidopsis thaliana] 384 e-105
UniRef100_Q7E9A5 At2g26670/F18A8.4 [Arabidopsis thaliana] 383 e-105
UniRef100_Q94FW7 Heme oxygenase 1 [Lycopersicon esculentum] 382 e-105
UniRef100_Q9C9L4 Putative heme oxygenase; 43724-42483 [Arabidops... 364 2e-99
UniRef100_Q94FW5 Heme oxygenase 1 [Pinus taeda] 345 1e-93
UniRef100_Q69XJ4 Putative heme oxygenase 1 [Oryza sativa] 327 3e-88
UniRef100_Q8GU39 Heme oxygenase precursor [Physcomitrella patens] 316 4e-85
UniRef100_Q9LQC0 F19C14.8 protein [Arabidopsis thaliana] 301 1e-80
UniRef100_Q8GUE5 Heme oxygenase precursor [Ceratodon purpureus] 301 1e-80
UniRef100_Q94FX2 Heme oxygenase 4 [Arabidopsis thaliana] 296 4e-79
UniRef100_Q94FW9 Heme oxygenase 1 [Sorghum bicolor] 273 5e-72
UniRef100_Q94FW6 Heme oxygenase 2 [Lycopersicon esculentum] 236 6e-61
UniRef100_O48722 Heme oxygenase 2 precursor [Arabidopsis thaliana] 231 2e-59
UniRef100_Q84RN2 Putative heme oxygenase 1 [Momordica charantia] 221 1e-56
UniRef100_Q94FW8 Heme oxygenase 2 [Sorghum bicolor] 221 2e-56
UniRef100_Q9FUN4 Heme oxygenase 1-like protein [Arabidopsis thal... 210 4e-53
UniRef100_Q94LH4 Putative heme oxygenase 2 [Oryza sativa] 194 2e-48
>UniRef100_Q93VB1 Putative heme oxygenase 1 precursor [Pisum sativum]
Length = 283
Score = 422 bits (1086), Expect = e-117
Identities = 215/281 (76%), Positives = 240/281 (84%), Gaps = 7/281 (2%)
Query: 5 TVVSQIQSLYIIK---PRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAE 61
T + QI SL++ K P+LS + FRS +F +L + K VIVSAT +E
Sbjct: 7 TALYQIHSLFLYKTNHPQLSLSQT--QSFRSNFFFQRKLSFELPRMPRKETVIVSATTSE 64
Query: 62 TARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLV 121
+KR+PGE+KGFVEEMRFVAM+LHTKDQAKEGEKEV +PEE+AVTKWEPTVDGYL+FLV
Sbjct: 65 --KKRHPGEAKGFVEEMRFVAMRLHTKDQAKEGEKEVKKPEERAVTKWEPTVDGYLRFLV 122
Query: 122 DSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYA 181
DSK+VYDTLEKIVQDA +P YAEF+NTGLERSASL KDLEWFKEQGYTIP+PSSPGL YA
Sbjct: 123 DSKIVYDTLEKIVQDAAYPYYAEFKNTGLERSASLDKDLEWFKEQGYTIPEPSSPGLTYA 182
Query: 182 QYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQ 241
YL DLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIA +LLNN+ LEFYKWDGDLSQ+LQ
Sbjct: 183 VYLTDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIASQLLNNQALEFYKWDGDLSQMLQ 242
Query: 242 NVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
NVRDKLNKVAE+W+REEKNHCLEETEKSFK SGEILRLILS
Sbjct: 243 NVRDKLNKVAEEWSREEKNHCLEETEKSFKLSGEILRLILS 283
>UniRef100_Q94FX1 Heme oxygenase 1 [Glycine max]
Length = 250
Score = 400 bits (1027), Expect = e-110
Identities = 194/233 (83%), Positives = 216/233 (92%), Gaps = 2/233 (0%)
Query: 50 KSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKW 109
++ VIVSA AET +K+ GESKGFVEEMRFVAM+LHT+DQA+EGEKEV +PEEKAVTKW
Sbjct: 20 RAAVIVSAATAETPKKK--GESKGFVEEMRFVAMRLHTRDQAREGEKEVKQPEEKAVTKW 77
Query: 110 EPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYT 169
+P+V+GYLKFLVDSK+VYDTLEKIVQ+A HPSYAEFRNTGLERSASLA+DLEWFKEQGYT
Sbjct: 78 DPSVEGYLKFLVDSKLVYDTLEKIVQEAPHPSYAEFRNTGLERSASLAEDLEWFKEQGYT 137
Query: 170 IPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEF 229
IP+PSSPGL YAQYL++LS DPQAFICHFYNIYFAHSAGGRMIGKK+A +LLNNK LEF
Sbjct: 138 IPEPSSPGLTYAQYLKELSVKDPQAFICHFYNIYFAHSAGGRMIGKKVAEKLLNNKALEF 197
Query: 230 YKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
YKWD DL +LLQNVRDKLNKVAE W+REEK+HCLEETEKSFK SGEILRLILS
Sbjct: 198 YKWDDDLPRLLQNVRDKLNKVAEPWSREEKDHCLEETEKSFKLSGEILRLILS 250
>UniRef100_Q94FX0 Heme oxygenase 3 [Glycine max]
Length = 249
Score = 398 bits (1022), Expect = e-109
Identities = 193/233 (82%), Positives = 213/233 (90%), Gaps = 2/233 (0%)
Query: 50 KSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKW 109
++ VIVSA AET +K+ GESKGFVEEMRFVAM+LHT+DQA+EGEKEV +PEEKAVTKW
Sbjct: 19 RTAVIVSAATAETPKKK--GESKGFVEEMRFVAMRLHTRDQAREGEKEVKQPEEKAVTKW 76
Query: 110 EPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYT 169
P+V+GYLKFLVDSK+VYDTLEKIV +A HP YAEFRNTGLERSASL +DL+WFKEQGYT
Sbjct: 77 NPSVEGYLKFLVDSKLVYDTLEKIVHEAPHPFYAEFRNTGLERSASLVEDLDWFKEQGYT 136
Query: 170 IPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEF 229
IP+PSSPGL YAQYL++LS DPQAFICHFYNIYFAHSAGGRMIGKK+A +LLNNK LEF
Sbjct: 137 IPEPSSPGLTYAQYLKELSVKDPQAFICHFYNIYFAHSAGGRMIGKKVAEKLLNNKALEF 196
Query: 230 YKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
YKWDGDL QLLQNVRDKLNKVAE WTREEK+HCLEETEKSFK SGEILRLILS
Sbjct: 197 YKWDGDLPQLLQNVRDKLNKVAEPWTREEKDHCLEETEKSFKLSGEILRLILS 249
>UniRef100_O48782 Heme oxygenase 1 precursor [Arabidopsis thaliana]
Length = 282
Score = 384 bits (987), Expect = e-105
Identities = 180/247 (72%), Positives = 213/247 (85%)
Query: 36 PTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGE 95
P ++L + VV+ + TAAE +KRYPGESKGFVEEMRFVAM+LHTKDQAKEGE
Sbjct: 36 PRIQILSMTMNKSPSLVVVAATTAAEKQKKRYPGESKGFVEEMRFVAMRLHTKDQAKEGE 95
Query: 96 KEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSAS 155
KE EE+ V KWEPTV+GYL+FLVDSK+VYDTLE I+QD+ P+YAEF+NTGLER+
Sbjct: 96 KETKSIEERPVAKWEPTVEGYLRFLVDSKLVYDTLELIIQDSNFPTYAEFKNTGLERAEK 155
Query: 156 LAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGK 215
L+ DLEWFKEQGY IP+P++PG Y+QYL++L++ DPQAFICHFYNIYFAHSAGGRMIG+
Sbjct: 156 LSTDLEWFKEQGYEIPEPTAPGKTYSQYLKELAEKDPQAFICHFYNIYFAHSAGGRMIGR 215
Query: 216 KIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGE 275
K+A +L+NK LEFYKWDG+LSQLLQNVR+KLNKVAE+WTREEKNHCLEETEKSFK+SGE
Sbjct: 216 KVAERILDNKELEFYKWDGELSQLLQNVREKLNKVAEEWTREEKNHCLEETEKSFKYSGE 275
Query: 276 ILRLILS 282
ILRLILS
Sbjct: 276 ILRLILS 282
>UniRef100_Q7E9A5 At2g26670/F18A8.4 [Arabidopsis thaliana]
Length = 240
Score = 383 bits (983), Expect = e-105
Identities = 178/231 (77%), Positives = 208/231 (89%)
Query: 52 VVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEP 111
VV+ + TAAE +KRYPGESKGFVEEMRFVAM+LHTKDQAKEGEKE EE+ V KWEP
Sbjct: 10 VVVAATTAAEKQKKRYPGESKGFVEEMRFVAMRLHTKDQAKEGEKETKSIEERPVAKWEP 69
Query: 112 TVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIP 171
TV+GYL+FLVDSK+VYDTLE I+QD+ P+YAEF+NTGLER+ L+ DLEWFKEQGY IP
Sbjct: 70 TVEGYLRFLVDSKLVYDTLELIIQDSNFPTYAEFKNTGLERAEKLSTDLEWFKEQGYEIP 129
Query: 172 QPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYK 231
+P++PG Y+QYL++L++ DPQAFICHFYNIYFAHSAGGRMIG+K+A +L+NK LEFYK
Sbjct: 130 EPTAPGKTYSQYLKELAEKDPQAFICHFYNIYFAHSAGGRMIGRKVAERILDNKELEFYK 189
Query: 232 WDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
WDG+LSQLLQNVR+KLNKVAE+WTREEKNHCLEETEKSFK+SGEILRLILS
Sbjct: 190 WDGELSQLLQNVREKLNKVAEEWTREEKNHCLEETEKSFKYSGEILRLILS 240
>UniRef100_Q94FW7 Heme oxygenase 1 [Lycopersicon esculentum]
Length = 278
Score = 382 bits (980), Expect = e-105
Identities = 196/284 (69%), Positives = 227/284 (79%), Gaps = 8/284 (2%)
Query: 1 MASATVVSQIQSLYIIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFRQMKS--VVIVSAT 58
MAS + +SQ Q L + KP+ + QF SI P +R Q KS VV+ + T
Sbjct: 1 MASISPLSQSQPL-LEKPQFTVLNKSQNQFFSI--PFSRFTQSCNLSLKKSRMVVVSATT 57
Query: 59 AAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLK 118
AAE + KRYPGE+KGFVEEMRFVAMKLHTKDQAKEGEKE P ++ + KWEP+V+GYLK
Sbjct: 58 AAEKSNKRYPGEAKGFVEEMRFVAMKLHTKDQAKEGEKE---PVDQPLAKWEPSVEGYLK 114
Query: 119 FLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGL 178
FLVDSK+VYDTLE+IV+ A P YAEFRNTGLERS LAKDLEWF++QG+ IP+PS+PG+
Sbjct: 115 FLVDSKLVYDTLERIVEKAPFPEYAEFRNTGLERSEVLAKDLEWFRQQGHAIPEPSTPGV 174
Query: 179 NYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQ 238
YA YL +LS+ DPQAFICHFYN YFAHSAGGRMIGKK+A ++L+ K LEFYKWDGDLSQ
Sbjct: 175 TYASYLEELSEKDPQAFICHFYNTYFAHSAGGRMIGKKVAEKVLDKKELEFYKWDGDLSQ 234
Query: 239 LLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
LLQNVRDKLNKVAE WTREEKNHCLEETEKSFK+SG ILRLI S
Sbjct: 235 LLQNVRDKLNKVAENWTREEKNHCLEETEKSFKFSGAILRLIFS 278
>UniRef100_Q9C9L4 Putative heme oxygenase; 43724-42483 [Arabidopsis thaliana]
Length = 285
Score = 364 bits (934), Expect = 2e-99
Identities = 171/230 (74%), Positives = 199/230 (86%)
Query: 53 VIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPT 112
V+ +A E +K+YPGESKGFVEEMRFVAM+LHTKDQA+EGEKE PEE V KWEPT
Sbjct: 56 VVTAAAITEKQQKKYPGESKGFVEEMRFVAMRLHTKDQAREGEKESRSPEEGPVAKWEPT 115
Query: 113 VDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQ 172
V+GYL FLVDSK+VYDTLE I+ + P+YA F+NTGLER+ SL KDLEWFKEQGY IP+
Sbjct: 116 VEGYLHFLVDSKLVYDTLEGIIDGSNFPTYAGFKNTGLERAESLRKDLEWFKEQGYEIPE 175
Query: 173 PSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKW 232
P +PG Y++YL+DL++NDPQAFICHFYNIYFAHSAGG+MIG K++ ++L+NK LEFYKW
Sbjct: 176 PMAPGKTYSEYLKDLAENDPQAFICHFYNIYFAHSAGGQMIGTKVSKKILDNKELEFYKW 235
Query: 233 DGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
DG LSQLLQNVR KLNKVAE WTREEK+HCLEETEKSFK+SGEILRLILS
Sbjct: 236 DGQLSQLLQNVRQKLNKVAEWWTREEKSHCLEETEKSFKFSGEILRLILS 285
>UniRef100_Q94FW5 Heme oxygenase 1 [Pinus taeda]
Length = 316
Score = 345 bits (884), Expect = 1e-93
Identities = 159/238 (66%), Positives = 203/238 (84%), Gaps = 3/238 (1%)
Query: 45 RFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEK 104
R + +V+ +A AE RK YPGE+KGFVEEMRFVAMKLHT+D AKEG+KE P+ +
Sbjct: 82 RGNSLGGLVVKAAATAEKPRKNYPGEAKGFVEEMRFVAMKLHTRDVAKEGKKE---PDAQ 138
Query: 105 AVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFK 164
+ KWEPT++GYLKFLVDSKVVYDTLE +VQ AT+P+YAEFRNTGLERS LAKDLEWF+
Sbjct: 139 PIGKWEPTIEGYLKFLVDSKVVYDTLEDVVQKATYPAYAEFRNTGLERSEKLAKDLEWFR 198
Query: 165 EQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNN 224
+QG+ IP+PSSPGL+Y+++L++LS+ DP AF+CHFYN+YFAH+AGGRMIG+K+A ++L+
Sbjct: 199 QQGHIIPEPSSPGLSYSKHLQELSEKDPPAFLCHFYNVYFAHTAGGRMIGRKVAEKILDG 258
Query: 225 KGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
+ LEFYKW+ DL+ LL VR+K+N VAE W+REEKNHCLEETEKSF +SG++LRLI+S
Sbjct: 259 RELEFYKWEEDLTTLLNRVREKINLVAEGWSREEKNHCLEETEKSFDYSGKLLRLIIS 316
>UniRef100_Q69XJ4 Putative heme oxygenase 1 [Oryza sativa]
Length = 289
Score = 327 bits (837), Expect = 3e-88
Identities = 157/233 (67%), Positives = 190/233 (81%), Gaps = 3/233 (1%)
Query: 50 KSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKW 109
+ +V+ +ATAAE A E K FVEEMR VAM+LHTKDQAKEGEKE P+ V +W
Sbjct: 60 RRMVVAAATAAEMAPAASGEEGKPFVEEMRAVAMRLHTKDQAKEGEKE---PQAPPVARW 116
Query: 110 EPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYT 169
EP+VDGYL+FLVDSK+V++TLE IV A P YAEFRNTGLERS L KDLEWFKEQG+T
Sbjct: 117 EPSVDGYLRFLVDSKLVFETLETIVDRAAVPWYAEFRNTGLERSEQLKKDLEWFKEQGHT 176
Query: 170 IPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEF 229
IP+PS+PG YA YL +L++ D QAFICHFYN+YFAH+AGGRMIGKK++ +LN K LEF
Sbjct: 177 IPEPSAPGTTYASYLEELAEKDSQAFICHFYNVYFAHTAGGRMIGKKVSENILNKKELEF 236
Query: 230 YKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
YKW+G+LSQLLQNVR+KLN+VA WTREEK+HCL+ETEKSF +SG++LR I +
Sbjct: 237 YKWEGNLSQLLQNVRNKLNEVASSWTREEKDHCLDETEKSFSYSGDLLRHIFT 289
>UniRef100_Q8GU39 Heme oxygenase precursor [Physcomitrella patens]
Length = 310
Score = 316 bits (810), Expect = 4e-85
Identities = 152/227 (66%), Positives = 183/227 (79%), Gaps = 3/227 (1%)
Query: 56 SATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPTVDG 115
S+ AAE A+ RYPGE KGFVEEMRFVAMKLHTKDQ+KEGEKE + + + +W+PT++G
Sbjct: 85 SSAAAEKAKARYPGEKKGFVEEMRFVAMKLHTKDQSKEGEKEA---DVQPIGQWQPTIEG 141
Query: 116 YLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSS 175
YLKFLVDSK VYDT+E IVQ A HPSY FRNTGLERS LAKDLEWF QG+ IP+
Sbjct: 142 YLKFLVDSKEVYDTMETIVQKAAHPSYELFRNTGLERSERLAKDLEWFASQGHAIPKAGV 201
Query: 176 PGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGD 235
G YA+YL +LS+ D AFICHFYN+YFAHSAGG IGKK+A +L+ + LEFYKWDG+
Sbjct: 202 DGTTYAKYLTELSETDVPAFICHFYNVYFAHSAGGYFIGKKVAKMILDGRELEFYKWDGE 261
Query: 236 LSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
L QLL V++ LNKVAE+W+R+EKN+CL+ETE SFK+SG+ILRLI S
Sbjct: 262 LFQLLGAVKENLNKVAEEWSRDEKNNCLKETENSFKYSGKILRLIAS 308
>UniRef100_Q9LQC0 F19C14.8 protein [Arabidopsis thaliana]
Length = 283
Score = 301 bits (772), Expect = 1e-80
Identities = 148/255 (58%), Positives = 192/255 (75%), Gaps = 5/255 (1%)
Query: 28 RQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHT 87
R R+I P L+++ Q V +V+A A E +RYP E GFVEEMRFV MK+H
Sbjct: 34 RYVRTIAAPRRHLVRRANEDQTLVVNVVAA-AGEKPERRYPREPNGFVEEMRFVVMKIHP 92
Query: 88 KDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRN 147
+DQ KEG+ + + V+ W T++GYLKFLVDSK+V++TLE+I+ ++ +YA +N
Sbjct: 93 RDQVKEGKSDSND----LVSTWNFTIEGYLKFLVDSKLVFETLERIINESAIQAYAGLKN 148
Query: 148 TGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHS 207
TGLER+ +L++DLEWFKEQGY IP+ PG Y+QYL+++++ DP AFICHFYNI FAHS
Sbjct: 149 TGLERAENLSRDLEWFKEQGYEIPESMVPGKAYSQYLKNIAEKDPPAFICHFYNINFAHS 208
Query: 208 AGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETE 267
AGGRMIG K+A ++L+NK LEFYKWDG LS+LLQNV ++LNKVAE WTREEKNHCLEETE
Sbjct: 209 AGGRMIGTKVAEKILDNKELEFYKWDGQLSELLQNVSEELNKVAELWTREEKNHCLEETE 268
Query: 268 KSFKWSGEILRLILS 282
KSFK+ EI R +LS
Sbjct: 269 KSFKFYWEIFRYLLS 283
>UniRef100_Q8GUE5 Heme oxygenase precursor [Ceratodon purpureus]
Length = 306
Score = 301 bits (771), Expect = 1e-80
Identities = 148/231 (64%), Positives = 186/231 (80%), Gaps = 5/231 (2%)
Query: 53 VIVSATAAETARKRYPGESKGFVEEMRFVAMKLHTKDQAKEGEKEVTEPEEKAVTKWEPT 112
V S+ A E ++ R PGE KGFVEEMRFVAMKLHT+DQ+KEGEKE + + V +W+P+
Sbjct: 79 VRASSVAPEKSKSR-PGEKKGFVEEMRFVAMKLHTRDQSKEGEKEA---DVQPVGQWKPS 134
Query: 113 VDGYLKFLVDSKVVYDTLEKIVQDATHPSYAE-FRNTGLERSASLAKDLEWFKEQGYTIP 171
+ GY+KFLVDSK VYDT+E IV+ A+HPS E FRNTGLERSA LAKDLEWF QG+ IP
Sbjct: 135 IPGYIKFLVDSKKVYDTMETIVEKASHPSSDEYFRNTGLERSARLAKDLEWFASQGHEIP 194
Query: 172 QPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYK 231
+ G+ YA L +LS++D AFICHFYN+YFAHSAGGR IGK++A ++L+ + LEFYK
Sbjct: 195 EAGPDGITYANLLTELSESDVPAFICHFYNVYFAHSAGGRFIGKQVAQQILDGRELEFYK 254
Query: 232 WDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLILS 282
WDG+LS+LL V+ LNKVAE+WTRE+K+HCL+ETE SFK+SG+ILRLI+S
Sbjct: 255 WDGELSELLGAVKVNLNKVAEEWTREQKDHCLKETENSFKYSGKILRLIVS 305
>UniRef100_Q94FX2 Heme oxygenase 4 [Arabidopsis thaliana]
Length = 283
Score = 296 bits (758), Expect = 4e-79
Identities = 147/255 (57%), Positives = 190/255 (73%), Gaps = 5/255 (1%)
Query: 28 RQFRSIYFPTTRLLQQHRFRQMKSVVIVSATAAETARKRYPGESKGFVEEMRFVAMKLHT 87
R R+I P L+++ Q V +V+A A E +RYP E GFVEEMRFV MK+H
Sbjct: 34 RYVRTIAAPRRHLVRRANEDQTLVVNVVAA-AGEKPERRYPREPNGFVEEMRFVVMKIHQ 92
Query: 88 KDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRN 147
+ Q KEG+ + V+ W T++GYLKFLVDSK+V++TLE+I+ ++ +YA +N
Sbjct: 93 EIQVKEGKSVSND----LVSTWNFTIEGYLKFLVDSKLVFETLERIINESAIQAYAGLKN 148
Query: 148 TGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHS 207
TGLER+ +L++DLEWFKEQGY IP+ PG Y+QYL+++++ DP AFICHFYNI FAHS
Sbjct: 149 TGLERAENLSRDLEWFKEQGYEIPESMVPGKAYSQYLKNIAEKDPPAFICHFYNINFAHS 208
Query: 208 AGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETE 267
AGGRMIG K+A ++L+NK LEFYKWDG LS+LLQNV ++LNKVAE WTREEKNHCLEETE
Sbjct: 209 AGGRMIGTKVAEKILDNKELEFYKWDGQLSELLQNVSEELNKVAELWTREEKNHCLEETE 268
Query: 268 KSFKWSGEILRLILS 282
KSFK+ EI R +LS
Sbjct: 269 KSFKFYWEIFRYLLS 283
>UniRef100_Q94FW9 Heme oxygenase 1 [Sorghum bicolor]
Length = 184
Score = 273 bits (697), Expect = 5e-72
Identities = 126/183 (68%), Positives = 154/183 (83%)
Query: 100 EPEEKAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKD 159
E E V KWEP+V+GYL+FLVDSK+V+ TLE IV+ A P YAEFRNTGLERS +L KD
Sbjct: 2 EAEAPPVAKWEPSVEGYLRFLVDSKLVFQTLEDIVERAAVPWYAEFRNTGLERSEALKKD 61
Query: 160 LEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAG 219
LEWF++QG+TIP+PS PG YA L +LS+ DPQAFICHFYN+YFAH+AGGRMIGKK++
Sbjct: 62 LEWFRQQGHTIPEPSPPGTTYASLLEELSEKDPQAFICHFYNVYFAHTAGGRMIGKKVSE 121
Query: 220 ELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRL 279
++LN K LEFYKW+G+LSQLLQNVR+KLN+VA W+REEK+HCLEETEKSF +SG +LR
Sbjct: 122 KILNKKELEFYKWEGNLSQLLQNVRNKLNEVASSWSREEKDHCLEETEKSFAYSGGLLRH 181
Query: 280 ILS 282
I +
Sbjct: 182 IFT 184
>UniRef100_Q94FW6 Heme oxygenase 2 [Lycopersicon esculentum]
Length = 368
Score = 236 bits (601), Expect = 6e-61
Identities = 112/236 (47%), Positives = 160/236 (67%), Gaps = 18/236 (7%)
Query: 64 RKRYPGESKGFVEEMRFVAMKLHTK---------DQAKEGEKEVTEPEEKAV-------- 106
RK+YPGE KG EEMRFVAMK D+ K+ E +E V
Sbjct: 132 RKQYPGEKKGITEEMRFVAMKFRNSKGKKKSESDDEMKDDGYESVSSDEDDVGGGGSGGG 191
Query: 107 -TKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKE 165
W+P+++G+LK+LVDSK+V+ T+E+IV +++ SYA FR TGLER+ ++KDL+WF +
Sbjct: 192 EATWQPSIEGFLKYLVDSKLVFSTIERIVDESSDVSYAYFRRTGLERTECISKDLKWFGQ 251
Query: 166 QGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNK 225
QG+ IP+PS PG+ YA YL +L++ P+ F+ HFYNIYF+H AGG++I KK LL K
Sbjct: 252 QGHEIPEPSIPGVTYANYLEELAEKTPRLFLSHFYNIYFSHIAGGQVIAKKAFERLLEEK 311
Query: 226 GLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRLIL 281
LEFYKW+GD +LL++VRD N +A+ W+R++KN L E K+F++ G+I+RLI+
Sbjct: 312 ELEFYKWEGDEEKLLRDVRDSFNMLAKHWSRDDKNKSLREVTKAFRFMGQIVRLII 367
>UniRef100_O48722 Heme oxygenase 2 precursor [Arabidopsis thaliana]
Length = 299
Score = 231 bits (588), Expect = 2e-59
Identities = 118/274 (43%), Positives = 176/274 (64%), Gaps = 24/274 (8%)
Query: 32 SIYFP----TTRLLQQHRFRQMKSVVIVSATAAETA-----RKRYPGESKGFVEEMRFVA 82
SI FP T R Q+H +S S A RK+YPGE+ G EEMRFVA
Sbjct: 25 SISFPFQISTQRKPQKHLLNLCRSTPTPSQQKASQRKRTRYRKQYPGENIGITEEMRFVA 84
Query: 83 MKLH---------------TKDQAKEGEKEVTEPEEKAVTKWEPTVDGYLKFLVDSKVVY 127
M+L T+ + +E E++ + +E W+P+ +G+LK+LVDSK+V+
Sbjct: 85 MRLRNVNGKKLDLSEDKTDTEKEEEEEEEDDDDDDEVKEETWKPSKEGFLKYLVDSKLVF 144
Query: 128 DTLEKIVQDATHPSYAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDL 187
DT+E+IV ++ + SYA FR TGLER S+ KDL+W +EQ IP+PS+ G++YA+YL +
Sbjct: 145 DTIERIVDESENVSYAYFRRTGLERCESIEKDLQWLREQDLVIPEPSNVGVSYAKYLEEQ 204
Query: 188 SQNDPQAFICHFYNIYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKL 247
+ F+ HFY+IYF+H AGG+++ ++++ +LL K LEF +W+GD LL+ VR+KL
Sbjct: 205 AGESAPLFLSHFYSIYFSHIAGGQVLVRQVSEKLLEGKELEFNRWEGDAQDLLKGVREKL 264
Query: 248 NKVAEQWTREEKNHCLEETEKSFKWSGEILRLIL 281
N + E W+R+EKN CL+ET K+FK+ G+I+RLI+
Sbjct: 265 NVLGEHWSRDEKNKCLKETAKAFKYMGQIVRLII 298
>UniRef100_Q84RN2 Putative heme oxygenase 1 [Momordica charantia]
Length = 129
Score = 221 bits (564), Expect = 1e-56
Identities = 101/123 (82%), Positives = 116/123 (94%)
Query: 160 LEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSAGGRMIGKKIAG 219
LE+ KEQGY IP+PSSPG+ YA+YL +LS+NDPQAFICHFYNIYFAH+AGGRMIG+K+A
Sbjct: 7 LEFLKEQGYAIPEPSSPGITYARYLEELSENDPQAFICHFYNIYFAHTAGGRMIGRKVAE 66
Query: 220 ELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEKSFKWSGEILRL 279
++LNNK LEFYKWDGDLSQLLQNVR+KLNKVAE W+REEKNHCLEETEKSFK+SG+ILRL
Sbjct: 67 KILNNKELEFYKWDGDLSQLLQNVREKLNKVAESWSREEKNHCLEETEKSFKYSGDILRL 126
Query: 280 ILS 282
ILS
Sbjct: 127 ILS 129
>UniRef100_Q94FW8 Heme oxygenase 2 [Sorghum bicolor]
Length = 328
Score = 221 bits (562), Expect = 2e-56
Identities = 108/253 (42%), Positives = 165/253 (64%), Gaps = 36/253 (14%)
Query: 65 KRYPGESKGFVEEMRFVAMKLH--------------------TKDQAKEGEKEVTEPE-- 102
K+YPGES G EEMRFVAM+L ++D+ +E E+E E E
Sbjct: 75 KQYPGESVGVAEEMRFVAMRLRNPKRTTIKDKAGTENADAGASEDEDEEEEEEEQEEEDG 134
Query: 103 --------------EKAVTKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPSYAEFRNT 148
E +W P+++G++++LVDSK+V+ T+E++V ++T +Y FR +
Sbjct: 135 GGGVKEEHEKEEEGELEAGEWMPSMEGFVRYLVDSKLVFGTIERVVAESTDVAYVYFRKS 194
Query: 149 GLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNIYFAHSA 208
GLERSAS++KDLEWF++QG IP+PS+ G YA YL +L++++ AF+ H+YNIYFAH
Sbjct: 195 GLERSASISKDLEWFRKQGIAIPEPSTSGSTYAAYLTELAESNAPAFLSHYYNIYFAHIT 254
Query: 209 GGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHCLEETEK 268
GG IG KI ++L + LEFYKWD D LL++ R+KLN++++ WTR+++N CL+E K
Sbjct: 255 GGVAIGNKICKKILEGRELEFYKWDTDAELLLKDAREKLNELSKHWTRKDRNLCLKEAAK 314
Query: 269 SFKWSGEILRLIL 281
F++ G+++RLI+
Sbjct: 315 CFQYLGKMVRLII 327
>UniRef100_Q9FUN4 Heme oxygenase 1-like protein [Arabidopsis thaliana]
Length = 140
Score = 210 bits (534), Expect = 4e-53
Identities = 97/140 (69%), Positives = 118/140 (84%)
Query: 143 AEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYNI 202
A +NTGLER+ +L++DLEWFKEQGY IP+ PG Y+QYL+++++ DP AFICHFYNI
Sbjct: 1 AGLKNTGLERAENLSRDLEWFKEQGYEIPESMVPGKAYSQYLKNIAEKDPPAFICHFYNI 60
Query: 203 YFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAEQWTREEKNHC 262
FAHSAGGRMIG K+A ++L+NK LEFYKWDG LS+LLQNV ++LNKVAE WTREEKNHC
Sbjct: 61 NFAHSAGGRMIGTKVAEKILDNKELEFYKWDGQLSELLQNVSEELNKVAELWTREEKNHC 120
Query: 263 LEETEKSFKWSGEILRLILS 282
LEETEKSFK+ EI R +LS
Sbjct: 121 LEETEKSFKFYWEIFRYLLS 140
>UniRef100_Q94LH4 Putative heme oxygenase 2 [Oryza sativa]
Length = 354
Score = 194 bits (493), Expect = 2e-48
Identities = 107/291 (36%), Positives = 167/291 (56%), Gaps = 55/291 (18%)
Query: 15 IIKPRLSPPPPPHRQFRSIYFPTTRLLQQHRFR-----------------QMKSVVIVSA 57
++ PR PPPPP R+ R + T +L + + ++V + A
Sbjct: 12 VVPPR--PPPPPPRRARPLRSFTGLILTRDLAALTVARCAPSPPAPAAEAEAEAVAVDEA 69
Query: 58 TAAETARKRYP----GESKGFVEEMRFVAMKLHT-------------------------- 87
A+ +RYP GE+ G EEMRFVAM+L
Sbjct: 70 PPAKPRPRRYPRQYPGEAVGVAEEMRFVAMRLRNPKRTTLKMDDTGAEEEVGDGVSEDAS 129
Query: 88 ---KDQAKEGEKEVTEPEEKAV---TKWEPTVDGYLKFLVDSKVVYDTLEKIVQDATHPS 141
+++ +E + +V E EE+ +W P+++G++K+LVDSK+V+DT+E+IV ++T +
Sbjct: 130 ASEEEEEEEDDDDVVEEEEEGAGLEGEWMPSMEGFVKYLVDSKLVFDTVERIVAESTDVA 189
Query: 142 YAEFRNTGLERSASLAKDLEWFKEQGYTIPQPSSPGLNYAQYLRDLSQNDPQAFICHFYN 201
Y FR +GLERSA + KDLEWF QG +P+PS+ G YA YL +L++++ AF+ H+YN
Sbjct: 190 YVYFRKSGLERSARITKDLEWFGGQGIAVPEPSTAGSTYATYLTELAESNAPAFLSHYYN 249
Query: 202 IYFAHSAGGRMIGKKIAGELLNNKGLEFYKWDGDLSQLLQNVRDKLNKVAE 252
IYFAH+ GG IG KI+ ++L + LEFYKWD D+ LL++ R+KLN++++
Sbjct: 250 IYFAHTTGGVAIGNKISKKILEGRELEFYKWDSDVELLLKDTREKLNELSK 300
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 490,010,616
Number of Sequences: 2790947
Number of extensions: 20566213
Number of successful extensions: 76538
Number of sequences better than 10.0: 123
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 76466
Number of HSP's gapped (non-prelim): 131
length of query: 282
length of database: 848,049,833
effective HSP length: 126
effective length of query: 156
effective length of database: 496,390,511
effective search space: 77436919716
effective search space used: 77436919716
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0252a.3


