

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0240.1
(1666 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
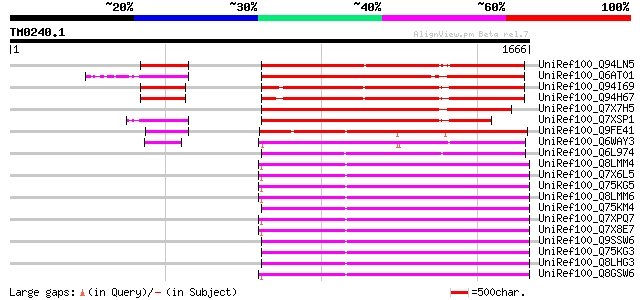
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sa... 918 0.0
UniRef100_Q6AT01 Putative polyprotein [Oryza sativa] 899 0.0
UniRef100_Q94I69 Putative retroelement [Oryza sativa] 877 0.0
UniRef100_Q94H67 Putative gag-pol protein [Oryza sativa] 858 0.0
UniRef100_Q7X7H5 OSJNBa0085C10.2 protein [Oryza sativa] 851 0.0
UniRef100_Q7XSP1 OSJNBa0070M12.15 protein [Oryza sativa] 789 0.0
UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic ... 706 0.0
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 644 0.0
UniRef100_Q6L974 GAG-POL [Vitis vinifera] 606 e-171
UniRef100_Q8LMM4 Putative gag-pol [Oryza sativa] 603 e-170
UniRef100_Q7X6L5 OSJNBb0093G06.3 protein [Oryza sativa] 601 e-170
UniRef100_Q75KG5 Putative polyprotein [Oryza sativa] 598 e-169
UniRef100_Q8LMM6 Putative gag-pol [Oryza sativa] 597 e-169
UniRef100_Q75KM4 Putative polyprotein [Oryza sativa] 596 e-168
UniRef100_Q7XPQ7 OSJNBa0053K19.16 protein [Oryza sativa] 595 e-168
UniRef100_Q7X8E7 OSJNBa0042F21.5 protein [Oryza sativa] 593 e-167
UniRef100_Q9SSW6 GAG-POL [Oryza sativa] 593 e-167
UniRef100_Q75KG3 Putative polyprotein [Oryza sativa] 592 e-167
UniRef100_Q8LHG3 GAG-POL [Oryza sativa] 591 e-167
UniRef100_Q8GSW6 Putative gag-pol polyprotein [Oryza sativa] 591 e-167
>UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1580
Score = 918 bits (2373), Expect = 0.0
Identities = 454/844 (53%), Positives = 593/844 (69%), Gaps = 19/844 (2%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
LP+K +P +QLPRR D+L +K EI+RL FIR RY +W++++VPVIKKNGK+
Sbjct: 576 LPLKPGVRPHQQLPRRCKADMLEPVKAEIKRLYDAGFIRPCRYAEWVSSIVPVIKKNGKV 635
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
RVCIDFR LN ATPKDEY M +A+ +VD+A+GH+ LS +DG +GYNQIF+AEED+ KT F
Sbjct: 636 RVCIDFRYLNKATPKDEYPMLVADQLVDAASGHKILSFMDGNAGYNQIFMAEEDIHKTTF 695
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
RCPGA+G +EWVV+ F LK+AGATYQR MN I+HD I ++VYIDD+VVKS +D +
Sbjct: 696 RCPGAIGLFEWVVITFVLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIA 755
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
LRK FER RKYGLKMNP KCAFGV AG FLGF+VH++GIE+ + AI PP K
Sbjct: 756 DLRKVFERTRKYGLKMNPTKCAFGVSAGQFLGFLVHERGIEVTQRSINAIKKIKPPEDKT 815
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+LQ ++GKINF+RRFI LS K + F+PLLRLK + F W AE QKA D +K YLSSPPV
Sbjct: 816 ELQEMIGKINFVRRFISILSGKLEPFTPLLRLKADQQFAWGAEQQKALDNIKEYLSSPPV 875
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSKERAIFYLSRVLNDAETRYTMIEKLCLC 1168
+ PP G P +LY+SA + +I S+L QE E KER +FYLSR L +AETRY+ +EKLCLC
Sbjct: 876 LIPPQKGIPFRLYLSAGDKSISSVLIQELER-KERVVFYLSRRLLNAETRYSPVEKLCLC 934
Query: 1169 LYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLKAVK 1228
LYFSC +L++Y+ + V D++K+MLS PIL RIGKW +LTE+ L Y KA+K
Sbjct: 935 LYFSCTRLRHYLLSNECTVICKADVVKYMLSAPILKGRIGKWIFSLTEFDLRYESPKAIK 994
Query: 1229 GQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKFKFRIK 1288
GQA+A+F+VD+ + + V V PW LFFDGS +G GIG+ I+SP G +F + IK
Sbjct: 995 GQAIANFIVDYR--DDSIGSVEVVPWTLFFDGSVCTHGCGIGLVIISPRGACFEFAYTIK 1052
Query: 1289 KKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKYYTKAN 1348
+NN+AEYEA++ L++L + A + + GDS LVI QL EY+C ++ L Y K
Sbjct: 1053 PYATNNQAEYEAVLKGLQLLKEVEADTIEIMGDSLLVISQLAGEYECKNDTLIVYNEKCQ 1112
Query: 1349 NLLAKFDDARLGHVSRIDNQEANELAQIASGYMVDKYRLKELIEVKEKLNPSDLNVLVID 1408
L+ +F L HVSR N EAN+LAQ ASGY K +K + + V VI
Sbjct: 1113 ELMREFRLVTLKHVSREQNIEANDLAQGASGY---KLMIKHV----------QIEVAVI- 1158
Query: 1409 NMAPKDWRKPIVDYLQNPVGTTDRKTKYRAMSYVIMGNELFKKNVDGTLLKCLSDDDALI 1468
DWR + YLQ+P + RK +Y+A+ Y+++ +EL+ + +DG LLKCLS D A +
Sbjct: 1159 --TADDWRYYVYQYLQDPSQSASRKLRYKALKYILLDDELYYRTIDGVLLKCLSTDQAKV 1216
Query: 1469 AISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASE 1528
AI VH+G+CG HQ+ KMKW+L G +WPT+++DC Y KGCQDCQ+ IQ PAS
Sbjct: 1217 AIGEVHEGICGTHQSAHKMKWLLRCAGYFWPTMLEDCFRYYKGCQDCQKFGAIQRAPASA 1276
Query: 1529 LHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFI 1588
++ IIKPWPFRGW +D+IG INP SS+ HK+I+VA DYFTKWVEAIPL+ V I F+
Sbjct: 1277 MNPIIKPWPFRGWGIDMIGMINPPSSKGHKFILVATDYFTKWVEAIPLKKVDSGDAIQFV 1336
Query: 1589 QNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLI 1648
Q HI+YR G+P+++TTDQG++FV + F +S GIKLL S+PYYAQANGQ EA+NK+LI
Sbjct: 1337 QEHIIYRVGIPQTITTDQGSIFVSDEFIQFADSMGIKLLNSSPYYAQANGQAEASNKSLI 1396
Query: 1649 SLIK 1652
LIK
Sbjct: 1397 KLIK 1400
Score = 124 bits (311), Expect = 2e-26
Identities = 60/152 (39%), Positives = 92/152 (60%), Gaps = 1/152 (0%)
Query: 421 ERAIFQRPTEQMKSHLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLI 480
E+ +F++P HLKPL++ V+ K ++K++VDGGA +NLMP +KLG+ DLI
Sbjct: 346 EQVVFEKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLI 405
Query: 481 PHDMVLSDYEGKTGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVP 540
+MVL D+ G ++G + + +TVG+ T F V+ K +Y+LLLGR+WIH +P
Sbjct: 406 KTNMVLKDFGGNPSETMGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIP 465
Query: 541 STLHQRISIWKADGFVENVQADQSYYLAETGY 572
ST+HQ + W+ D +E V AD + Y
Sbjct: 466 STIHQCLIQWQGDK-IEIVPADSQLKMENPSY 496
>UniRef100_Q6AT01 Putative polyprotein [Oryza sativa]
Length = 1739
Score = 899 bits (2324), Expect = 0.0
Identities = 449/844 (53%), Positives = 588/844 (69%), Gaps = 28/844 (3%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
LPIK +P +Q RR D+L +K EI+RL FIR RY W++++VPVIKKNGK+
Sbjct: 919 LPIKPGVRPRQQPLRRCKADMLEPVKAEIKRLYDAGFIRPWRYAKWVSSIVPVIKKNGKV 978
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
RVCIDFRDLN ATPKDEY MP+A+ +VD+A+G++ LS +DG +GYNQIF+AEED+ KTAF
Sbjct: 979 RVCIDFRDLNKATPKDEYPMPVADQLVDAASGNKILSFMDGNAGYNQIFMAEEDIHKTAF 1038
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
RCP A+G +EWVVM FGLK+AGATYQR MN I+HD I + VYIDD+VVKS +DQ+
Sbjct: 1039 RCPCAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVGVYIDDVVVKSKEIEDQIA 1098
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
LRK FER RKYGLKMNP KCAFGV AG FLGF+VH +GIE+ + AI PP +K
Sbjct: 1099 DLRKVFERTRKYGLKMNPTKCAFGVSAGQFLGFLVHDRGIEVTQRSVNAIKKIQPPENKT 1158
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+LQ ++GKI+F+RRFI NLS + + F+PLLRLK + F AE QKA D++K YLSSPPV
Sbjct: 1159 ELQEMIGKIHFVRRFISNLSGRLEPFTPLLRLKADQQFTCGAEQQKALDDIKEYLSSPPV 1218
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSKERAIFYLSRVLNDAETRYTMIEKLCLC 1168
+ PP G P +LY+SA +IGS+L QE E KER +FYLSR L +AETRY+ ++KLCLC
Sbjct: 1219 LIPPHKGIPFRLYLSAGEKSIGSVLIQELE-GKERVVFYLSRRLLNAETRYSPVKKLCLC 1277
Query: 1169 LYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLKAVK 1228
LYFSC +L++Y+ + V D++K+MLS PIL R+GKW +LTE+ Y KA+K
Sbjct: 1278 LYFSCTRLRHYLLSNECTVICKADVVKYMLSAPILKGRVGKWIFSLTEFDHRYESPKAIK 1337
Query: 1229 GQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKFKFRIK 1288
GQA+ADF+V+H + + V + PW LFFDGS +G GIG+ I+SP G +F + IK
Sbjct: 1338 GQAIADFIVEHR--DDSIGSVEIVPWTLFFDGSVCTHGCGIGLVIISPRGACFEFAYTIK 1395
Query: 1289 KKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKYYTKAN 1348
+NN+AEYEA++ L++L + A + + GDS LVI QL EY+C ++ L Y K
Sbjct: 1396 PYATNNQAEYEAVLKGLQLLKKVEADTIEIMGDSLLVISQLAGEYECKNDTLMVYNEKCQ 1455
Query: 1349 NLLAKFDDARLGHVSRIDNQEANELAQIASGYMVDKYRLKELIEVKEKLNPSDLNVLVID 1408
L+ +F R+ N EAN+LAQ ASGY K +I+ D+ V I
Sbjct: 1456 ELMKEF---------RLQNIEANDLAQGASGY-------KPMIK--------DVKV-EIA 1490
Query: 1409 NMAPKDWRKPIVDYLQNPVGTTDRKTKYRAMSYVIMGNELFKKNVDGTLLKCLSDDDALI 1468
M DWR + YL NP + RK +Y+A+ Y ++ +EL+ + +DG LLKCLS D A +
Sbjct: 1491 AMTADDWRYDVHRYLSNPSQSASRKLRYKALKYTLLDDELYYRTIDGVLLKCLSADQAKV 1550
Query: 1469 AISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASE 1528
AI VH+G+CG HQ+ KMKW+L R G +W T+++DC Y KGCQDCQ+ IQ PAS
Sbjct: 1551 AIGEVHEGICGTHQSAHKMKWLLRRAGYFWSTMLEDCFRYYKGCQDCQKFGAIQRAPASA 1610
Query: 1529 LHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFI 1588
++ IIKPWPFRGW +D+IG INP SS+ HK+I+VA DYFTKWVEAIPL+ V I F+
Sbjct: 1611 MNPIIKPWPFRGWGIDMIGMINPPSSKGHKFILVATDYFTKWVEAIPLKKVDSGDAIQFV 1670
Query: 1589 QNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLI 1648
Q +I+YRFG P+++TTDQG++FV + F +S IKLL S+PYYAQANGQ EA+NK+LI
Sbjct: 1671 QEYIIYRFGTPQTITTDQGSIFVSDEFVQFADSMSIKLLNSSPYYAQANGQAEASNKSLI 1730
Query: 1649 SLIK 1652
LIK
Sbjct: 1731 KLIK 1734
Score = 127 bits (319), Expect = 3e-27
Identities = 97/332 (29%), Positives = 153/332 (45%), Gaps = 45/332 (13%)
Query: 242 PKHHQKWSKIDLGQGSSYINNSRVSRSYSYGSGNYYAPEQMTRTQFRRFLRKRKAERERG 301
P H S +LG+ S + +S SRSY+ G+ R + R + +R
Sbjct: 540 PAGHTIPSTEELGKKSDIVESS--SRSYNRGN---------------RLRQTRVSVHQRL 582
Query: 302 GKFRSLWDIKPEDNPRNEPSVASIINQLDHAIKQGTVVPPNPAKEKGK-DVAEDEDEDML 360
G P + E + +++H +++ P + K + VA+D D
Sbjct: 583 G---------PVNQDHGEEDSVEVEQEINHRLRKAK--PRQEWRVKNQVPVADDVATDEA 631
Query: 361 DDFDDSDGEDLNIICIVSILPAEFDRILEVTENEEDYYDEEVPDDNPLCYYVMQKGSVED 420
I +V LP EF + + + D +EE K +
Sbjct: 632 KRLAKGKSVVTAPINMVFTLPVEFG----IDQADVDEVEEE-----------SAKLVLSP 676
Query: 421 ERAIFQRPTEQMKSHLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLI 480
E+A+FQ+P HLKPL++ V+ K ++K++VDGGA +NLMP +KLG+ DLI
Sbjct: 677 EQAVFQKPEGTENRHLKPLYINGYVNGKLMSKMMVDGGAAVNLMPYATFRKLGRNAEDLI 736
Query: 481 PHDMVLSDYEGKTGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVP 540
+MVL D+ G + G + + +TVG+ T F+V+ K +Y+L LGR+WIH +P
Sbjct: 737 KTNMVLKDFGGNPSETKGVLNVGLTVGSKTIPTTFLVIDGKGSYSLFLGRDWIHANCCIP 796
Query: 541 STLHQRISIWKADGFVENVQADQSYYLAETGY 572
ST+HQ + W+ D VE V AD + Y
Sbjct: 797 STMHQCLIQWQGDK-VEIVPADSQLKMENPSY 827
>UniRef100_Q94I69 Putative retroelement [Oryza sativa]
Length = 2014
Score = 877 bits (2265), Expect = 0.0
Identities = 444/844 (52%), Positives = 578/844 (67%), Gaps = 29/844 (3%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
LPIK +P +Q RR D+L +K EI+RL FIR RY +W++++VPVIKKN
Sbjct: 1126 LPIKPGVRPHQQPLRRCKADMLEPVKVEIKRLYDACFIRPCRYAEWVSSIVPVIKKN--- 1182
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
DLN ATPKDEY MP+A+ +VD+A+G++ LS +DG GYNQIF+AEED+ KTAF
Sbjct: 1183 -------DLNKATPKDEYPMPVADQLVDAASGNKILSYMDGNVGYNQIFMAEEDIHKTAF 1235
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
RCPGA+G +EWVVM FGLK+AGATYQR MN I+HD I ++VYIDD+VVKS +D +
Sbjct: 1236 RCPGAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIA 1295
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
LRK FER RKYGLKMNP KCAFGV G FLGF+VH++GIE+ + AI PP +K
Sbjct: 1296 DLRKVFERTRKYGLKMNPTKCAFGVSVGQFLGFLVHERGIEVTQRSVNAIKKIQPPENKT 1355
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+LQ + GKINF+RRFI LS + + F+PLLRLK + F W AE QKA D++K YLSSPPV
Sbjct: 1356 ELQEMNGKINFVRRFISYLSGRLEPFTPLLRLKADQQFTWGAEQQKALDDIKEYLSSPPV 1415
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSKERAIFYLSRVLNDAETRYTMIEKLCLC 1168
+ PP G +LY+SA +IGS+L QE + KER +FYLSR L D ETRY+ +EKLCLC
Sbjct: 1416 LIPPQKGISFRLYLSAGEKSIGSVLIQE-LDGKERVVFYLSRRLLDVETRYSPMEKLCLC 1474
Query: 1169 LYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLKAVK 1228
LYFSC +L +Y+ + V D+IK+MLS PIL R+GKW +LTE+ L Y KA+K
Sbjct: 1475 LYFSCTRLSHYLLSNECTVICKADVIKYMLSAPILKGRVGKWIFSLTEFDLRYESPKAIK 1534
Query: 1229 GQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKFKFRIK 1288
GQA+ADF+V+H + + V V PW FFDGS + GIG+ I+SP G KF + IK
Sbjct: 1535 GQAIADFIVEHC--DDSIGSVEVVPWTSFFDGSVCTHDCGIGLVIISPRGACFKFAYTIK 1592
Query: 1289 KKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKYYTKAN 1348
+NN+AEYEA++ L++L + A + + GDS LVI QL EY+C S+ L Y K
Sbjct: 1593 PYATNNQAEYEAVLKGLQLLKEVEADAIEIMGDSLLVISQLAGEYECKSDTLMVYNEKCQ 1652
Query: 1349 NLLAKFDDARLGHVSRIDNQEANELAQIASGYMVDKYRLKELIEVKEKLNPSDLNVLVID 1408
L+ +F L HVSR N EAN+LAQ ASGY K +K +VK + I
Sbjct: 1653 ELMQEFRLVTLKHVSREQNIEANDLAQGASGY---KPMIK---DVKAE----------IA 1696
Query: 1409 NMAPKDWRKPIVDYLQNPVGTTDRKTKYRAMSYVIMGNELFKKNVDGTLLKCLSDDDALI 1468
+ DWR + YL NP + RK +Y+A+ Y ++ +EL+ + +DG LLKCLS D A +
Sbjct: 1697 AITAGDWRYDVHQYLHNPSQSASRKLRYKALKYTLLDDELYYRTIDGVLLKCLSADQAKV 1756
Query: 1469 AISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASE 1528
AI VH+G+CG HQ+ KMKW+L R +WPT+++DC +Y KGCQDCQ+ IQ PAS
Sbjct: 1757 AIGEVHEGICGTHQSAHKMKWLLRRARYFWPTMLEDCFKYYKGCQDCQKFGAIQRAPASA 1816
Query: 1529 LHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFI 1588
++ IIKPWPFRGW +D+IG I+ SS+ HK+I+VA DYFTK VEAIPL+ V I F+
Sbjct: 1817 MNPIIKPWPFRGWGIDMIGMISRPSSKGHKFILVATDYFTKLVEAIPLKKVDSGDAIQFV 1876
Query: 1589 QNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLI 1648
Q HI+YRFG+P+++TTDQG++FV + F +S GIKLL S+PYYAQANGQ EA+NK+LI
Sbjct: 1877 QEHIIYRFGIPQTITTDQGSIFVSDEFVQFVDSMGIKLLNSSPYYAQANGQAEASNKSLI 1936
Query: 1649 SLIK 1652
LIK
Sbjct: 1937 KLIK 1940
Score = 117 bits (292), Expect = 4e-24
Identities = 60/142 (42%), Positives = 87/142 (61%), Gaps = 1/142 (0%)
Query: 421 ERAIFQRPTEQMKSHLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLI 480
E+AIF++P HLKPL++ V+ K ++K++VDGGA +NLMP +KLG+ DLI
Sbjct: 899 EQAIFEKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAEVNLMPYATFRKLGRNVEDLI 958
Query: 481 PHDMVLSDYEGKTGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVP 540
+MVL D+ G + G + +TVG T F V+ +Y+LLLGR+WIH +P
Sbjct: 959 KTNMVLIDFGGNLSETKGVSNVELTVGNKTIPTTFFVIDGNGSYSLLLGRDWIHANCCIP 1018
Query: 541 STLHQRISIWKADGFVENVQAD 562
ST+HQ + W+ D +E V AD
Sbjct: 1019 STMHQCLIQWQ-DDKIEIVPAD 1039
>UniRef100_Q94H67 Putative gag-pol protein [Oryza sativa]
Length = 2273
Score = 858 bits (2217), Expect = 0.0
Identities = 437/844 (51%), Positives = 569/844 (66%), Gaps = 38/844 (4%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
LPIK +P +Q RR D+L +K EI+RL FIR RY +W++N
Sbjct: 1394 LPIKPGVRPHQQPLRRCKADMLEPVKVEIKRLYDACFIRPCRYAEWVSN----------- 1442
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
LN ATPKDEY MP+A+ +VD+A+G++ LS +DG GYNQIF+AEED+ KTAF
Sbjct: 1443 --------LNKATPKDEYPMPVADQLVDAASGNKILSYMDGNVGYNQIFMAEEDIHKTAF 1494
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
RCPGA+G +EWVVM FGLK+AGATYQR MN I+HD I ++VYIDD+VVKS +D +
Sbjct: 1495 RCPGAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSKEIEDHIA 1554
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
LRK FER RKYGLKMNP KCAFGV G FLGF+VH++GIE+ + AI PP +K
Sbjct: 1555 DLRKVFERTRKYGLKMNPTKCAFGVSVGQFLGFLVHERGIEVTQRSVNAIKKIQPPENKT 1614
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+LQ + GKINF+RRFI LS + + F+PLLRLK + F W AE QKA D++K YLSSPPV
Sbjct: 1615 ELQEMNGKINFVRRFISYLSGRLEPFTPLLRLKADQQFTWGAEQQKALDDIKEYLSSPPV 1674
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSKERAIFYLSRVLNDAETRYTMIEKLCLC 1168
+ PP G +LY+SA +IGS+L QE + KER +FYLSR L D ETRY+ +EKLCLC
Sbjct: 1675 LIPPQKGISFRLYLSAGEKSIGSVLIQE-LDGKERVVFYLSRRLLDVETRYSPMEKLCLC 1733
Query: 1169 LYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLKAVK 1228
LYFSC +L +Y+ + V D+IK+MLS PIL R+GKW +LTE+ L Y KA+K
Sbjct: 1734 LYFSCTRLSHYLLSNECTVICKADVIKYMLSAPILKGRVGKWIFSLTEFDLRYESPKAIK 1793
Query: 1229 GQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKFKFRIK 1288
GQA+ADF+V+H + + V V PW FFDGS + GIG+ I+SP G KF + IK
Sbjct: 1794 GQAIADFIVEHC--DDSIGSVEVVPWTSFFDGSVCTHDCGIGLVIISPRGACFKFAYTIK 1851
Query: 1289 KKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKYYTKAN 1348
+NN+AEYEA++ L++L + A + + GDS LVI QL EY+C S+ L Y K
Sbjct: 1852 PYATNNQAEYEAVLKGLQLLKEVEADAIEIMGDSLLVISQLAGEYECKSDTLMVYNEKCQ 1911
Query: 1349 NLLAKFDDARLGHVSRIDNQEANELAQIASGYMVDKYRLKELIEVKEKLNPSDLNVLVID 1408
L+ +F L HVSR N EAN+LAQ ASGY K +K +VK + I
Sbjct: 1912 ELMQEFRLVTLKHVSREQNIEANDLAQGASGY---KPMIK---DVKAE----------IA 1955
Query: 1409 NMAPKDWRKPIVDYLQNPVGTTDRKTKYRAMSYVIMGNELFKKNVDGTLLKCLSDDDALI 1468
+ DWR + YL NP + RK +Y+A+ Y ++ +EL+ + +DG LLKCLS D A +
Sbjct: 1956 AITAGDWRYDVHQYLHNPSQSASRKLRYKALKYTLLDDELYYRTIDGVLLKCLSADQAKV 2015
Query: 1469 AISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASE 1528
AI VH+G+CG HQ+ KMKW+L R +WPT+++DC +Y KGCQDCQ+ IQ PAS
Sbjct: 2016 AIGEVHEGICGTHQSAHKMKWLLRRARYFWPTMLEDCFKYYKGCQDCQKFGAIQRAPASA 2075
Query: 1529 LHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFI 1588
++ IIKPWPFRGW +D+IG I+ SS+ HK+I+VA DYFTK VEAIPL+ V I F+
Sbjct: 2076 MNPIIKPWPFRGWGIDMIGMISRPSSKGHKFILVATDYFTKLVEAIPLKKVDSGDAIQFV 2135
Query: 1589 QNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLI 1648
Q HI+YRFG+P+++TTDQG++FV + F +S GIKLL S+PYYAQANGQ EA+NK+LI
Sbjct: 2136 QEHIIYRFGIPQTITTDQGSIFVSDEFVQFVDSMGIKLLNSSPYYAQANGQAEASNKSLI 2195
Query: 1649 SLIK 1652
LIK
Sbjct: 2196 KLIK 2199
Score = 117 bits (292), Expect = 4e-24
Identities = 60/142 (42%), Positives = 87/142 (61%), Gaps = 1/142 (0%)
Query: 421 ERAIFQRPTEQMKSHLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLI 480
E+AIF++P HLKPL++ V+ K ++K++VDGGA +NLMP +KLG+ DLI
Sbjct: 1152 EQAIFEKPEGTENRHLKPLYINGYVNGKPMSKMMVDGGAEVNLMPYATFRKLGRNVEDLI 1211
Query: 481 PHDMVLSDYEGKTGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVP 540
+MVL D+ G + G + +TVG T F V+ +Y+LLLGR+WIH +P
Sbjct: 1212 KTNMVLIDFGGNLSETKGVSNVELTVGNKTIPTTFFVIDGNGSYSLLLGRDWIHANCCIP 1271
Query: 541 STLHQRISIWKADGFVENVQAD 562
ST+HQ + W+ D +E V AD
Sbjct: 1272 STMHQCLIQWQ-DDKIEIVPAD 1292
>UniRef100_Q7X7H5 OSJNBa0085C10.2 protein [Oryza sativa]
Length = 861
Score = 851 bits (2198), Expect = 0.0
Identities = 422/803 (52%), Positives = 557/803 (68%), Gaps = 19/803 (2%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
LPIK +P +Q PRR D+L +K EI+RL FI RY +W++++VPVIKKNGK+
Sbjct: 76 LPIKPGVRPRQQPPRRCKADMLEPVKAEIKRLYDAGFIHPYRYAEWVSSIVPVIKKNGKV 135
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
RVCIDFRDLN ATPKDEY MP+A+ + D+A+G++ LS +D +GYNQIF+AEED+ KTAF
Sbjct: 136 RVCIDFRDLNKATPKDEYPMPVADQLFDAASGNKILSFMDRNTGYNQIFMAEEDIHKTAF 195
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
RCP A+G +EWVVM F LK+AGATYQR MN I+HD I + ++V IDD+VVKS DD +
Sbjct: 196 RCPSAIGLFEWVVMTFDLKSAGATYQRAMNYIYHDLIGSLVEVDIDDVVVKSKEVDDHIA 255
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
LRK FER RKYGLKMNP KCAFGV A FLGF+VH++GIE+ + AI PP +K
Sbjct: 256 DLRKVFERTRKYGLKMNPTKCAFGVSARQFLGFLVHERGIEVTQRSVNAIQKIKPPENKT 315
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+LQ ++GKINF+RRFI NLS + + F+PLLRLK + F W AE QKA D +K YLSSPPV
Sbjct: 316 ELQEMIGKINFVRRFISNLSGRLEPFTPLLRLKADQQFTWGAEQQKALDNIKEYLSSPPV 375
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSKERAIFYLSRVLNDAETRYTMIEKLCLC 1168
+ PP G P +LY+SA + GS+L QE E KER +FY+SR L DAETRY+ +EKLCLC
Sbjct: 376 LIPPQKGIPFRLYLSAGEKSNGSVLIQELE-GKERVVFYISRRLLDAETRYSPMEKLCLC 434
Query: 1169 LYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLKAVK 1228
LYF C +L++Y+ + V D++++MLS PIL +IG W +LTE+ L Y KA+K
Sbjct: 435 LYFLCTRLRHYLLSNECTVICKADVVRYMLSAPILKGQIGNWIFSLTEFDLRYESPKAIK 494
Query: 1229 GQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKFKFRIK 1288
GQA+ADF+V+H + + V + PW LFFDGS +G GIG+ I+SP G +F + IK
Sbjct: 495 GQAIADFIVEHR--DDSIGSVEIVPWTLFFDGSVCTHGCGIGLVIISPRGACFEFAYTIK 552
Query: 1289 KKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKYYTKAN 1348
+NN+AEYEA++ L++L + A + + GDS LVI QL EY+C S+ Y K
Sbjct: 553 SYATNNQAEYEAVLKGLQLLKEVEANIIEIMGDSLLVISQLAGEYECKSDTFIVYNEKCL 612
Query: 1349 NLLAKFDDARLGHVSRIDNQEANELAQIASGYMVDKYRLKELIEVKEKLNPSDLNVLVID 1408
L+ +F L HVSR N EAN+LAQ ASGY K +I+ D+ V I
Sbjct: 613 ELMKEFRLVTLKHVSREQNLEANDLAQGASGY-------KPMIK--------DVKV-EIA 656
Query: 1409 NMAPKDWRKPIVDYLQNPVGTTDRKTKYRAMSYVIMGNELFKKNVDGTLLKCLSDDDALI 1468
M+ DWR + YL NP + RK +Y+A+ Y ++G+EL+ + +DG LLKCLS D A +
Sbjct: 657 AMSADDWRYDVHQYLSNPSQSASRKLRYKALKYTLLGDELYYRVIDGVLLKCLSADQAKV 716
Query: 1469 AISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASE 1528
AI VH+G+CG HQ+ KMKW+L R G +WPT+++ Y KGCQDCQ+ IQ PAS
Sbjct: 717 AIGEVHEGICGTHQSAHKMKWLLRRTGYFWPTMLEVYFRYYKGCQDCQKFGAIQRAPASA 776
Query: 1529 LHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFI 1588
++ IIKPWPFRGW +D+IG INP SS+ HK+I+VA DYFTKWVEAIPL+ V I F+
Sbjct: 777 MNPIIKPWPFRGWEIDMIGMINPPSSKGHKFILVATDYFTKWVEAIPLKKVDSGDAIQFV 836
Query: 1589 QNHIVYRFGLPESLTTDQGTVFV 1611
Q HI+Y+FGLP+++T DQG++F+
Sbjct: 837 QEHIIYQFGLPQTITMDQGSIFM 859
>UniRef100_Q7XSP1 OSJNBa0070M12.15 protein [Oryza sativa]
Length = 1685
Score = 789 bits (2037), Expect = 0.0
Identities = 390/739 (52%), Positives = 512/739 (68%), Gaps = 19/739 (2%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
LP+K +P +Q PRR D+L +K EI+RL FI RY +W++++VPVIKKNGK+
Sbjct: 964 LPLKPGIRPHQQPPRRCKADMLEPVKAEIKRLYDAGFIHPCRYAEWVSSIVPVIKKNGKV 1023
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
RVCIDFRDLN ATPKDEY MP+A+ +VD+A+GH+ LS +DG +GYNQIF+ EED+ KT F
Sbjct: 1024 RVCIDFRDLNKATPKDEYPMPVADQLVDAASGHKILSFMDGNAGYNQIFMTEEDIHKTVF 1083
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
RCPGA+G +EWVVM FGLK+AGATYQR MN I+HD I ++VYIDD+VVKS +D +
Sbjct: 1084 RCPGAIGLFEWVVMTFGLKSAGATYQRAMNYIYHDLIGWLVEVYIDDVVVKSREIEDHIA 1143
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
LRK FER RKYGLKMNP KCAFGV AG FLGF+VH++GIEI + AI PP +K
Sbjct: 1144 DLRKVFERTRKYGLKMNPTKCAFGVSAGQFLGFLVHERGIEIPQRSINAIKKIKPPGNKT 1203
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+LQ ++GKINF+RRFI NLS K + F+PLLRL+ + F W AE QKA D +K YLSSPPV
Sbjct: 1204 ELQEMIGKINFVRRFISNLSGKLEPFTPLLRLRADQQFTWGAEQQKALDNIKEYLSSPPV 1263
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSKERAIFYLSRVLNDAETRYTMIEKLCLC 1168
+ PP G P LY+SA + +IGS+L QE E KER +FYLSR L DAETRY+ +EKLCLC
Sbjct: 1264 LIPPQKGIPFWLYLSAGDKSIGSVLIQELE-GKERVVFYLSRRLLDAETRYSPVEKLCLC 1322
Query: 1169 LYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLKAVK 1228
LYFSC KL++Y+ + V D+IK+MLS PIL R+GKW +LTE+ L + KA+K
Sbjct: 1323 LYFSCTKLRHYLLSNECTVVCKADVIKYMLSAPILKVRVGKWIFSLTEFDLRHESPKAIK 1382
Query: 1229 GQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKFKFRIK 1288
GQAVADF+V H + V + PW LFFDGS +G GIG+ I++P G +F + I
Sbjct: 1383 GQAVADFIVGHR--DGSIGLVDIMPWVLFFDGSVCSHGYGIGLVIIAPWGASFEFAYAIT 1440
Query: 1289 KKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKYYTKAN 1348
+NN+AEYEA++ L++L + A + + GDS LVI QL EY+C + L YY K
Sbjct: 1441 HHVTNNQAEYEAVLKGLQLLQEVAADAIEIMGDSLLVISQLAGEYECKDDTLMVYYEKCR 1500
Query: 1349 NLLAKFDDARLGHVSRIDNQEANELAQIASGYMVDKYRLKELIEVKEKLNPSDLNVLVID 1408
L+++F L H+SR N EAN+L+Q ASGY K K+ IE++ I
Sbjct: 1501 TLISEFRLVTLRHISREQNIEANDLSQGASGY---KPMTKD-IEIE------------IA 1544
Query: 1409 NMAPKDWRKPIVDYLQNPVGTTDRKTKYRAMSYVIMGNELFKKNVDGTLLKCLSDDDALI 1468
+ DWR + YLQNP + RK +Y+A+ Y ++ ++L+ + +DG LLKCLS D A +
Sbjct: 1545 TITADDWRYDVFQYLQNPSQSALRKLRYKALKYTLLDDDLYYRTIDGVLLKCLSADQAKV 1604
Query: 1469 AISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASE 1528
AI +H+G+CG HQ+ KMKW+L R G +WPT+++DC Y KGCQDCQ+ IQ PAS
Sbjct: 1605 AIGEIHEGICGTHQSAHKMKWLLRRAGYFWPTMLEDCFRYYKGCQDCQKFGAIQRAPASA 1664
Query: 1529 LHSIIKPWPFRGWALDLIG 1547
++ II+PWPFRGW +D+IG
Sbjct: 1665 MNPIIQPWPFRGWGIDMIG 1683
Score = 128 bits (322), Expect = 1e-27
Identities = 74/199 (37%), Positives = 110/199 (55%), Gaps = 16/199 (8%)
Query: 374 ICIVSILPAEFDRILEVTENEEDYYDEEVPDDNPLCYYVMQKGSVEDERAIFQRPTEQMK 433
I +V LPAEF + + + D +EE K + ERA+F++P
Sbjct: 690 INMVFTLPAEFG----IDQADVDELEEE-----------SAKLVLSPERAVFEKPEGTEN 734
Query: 434 SHLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLIPHDMVLSDYEGKT 493
HLKPL++ V+ K ++K++VDGGA +NLMP +KLG+T DL+ +MVL D+ G
Sbjct: 735 RHLKPLYINGYVNGKPMSKMMVDGGAAVNLMPYTTFRKLGRTTEDLMKTNMVLKDFGGNP 794
Query: 494 GSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRISIWKAD 553
+ G + + +TVG+ T F V+ K +Y+LLLGR+WIH +PST+HQ + W+ D
Sbjct: 795 SETKGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD 854
Query: 554 GFVENVQADQSYYLAETGY 572
VE V AD + Y
Sbjct: 855 K-VEIVPADSRLKMKNPSY 872
>UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic DNA, chromosome 1, PAC
clone:P0433F09 [Oryza sativa]
Length = 2876
Score = 706 bits (1821), Expect = 0.0
Identities = 377/894 (42%), Positives = 545/894 (60%), Gaps = 45/894 (5%)
Query: 802 SRIGGIELPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPV 861
SR+ +L I +PVKQ PRR P+ ++ E++RL+ FI+ +Y WLAN+VPV
Sbjct: 1829 SRVATHKLAIDPQFRPVKQPPRRLRPEFQDQVIAEVDRLINVGFIKEIQYPRWLANIVPV 1888
Query: 862 IKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEE 921
KKNG++RVC+DFRDLN A PKD++ +PI EM+VDS G+ LS GYNQI +
Sbjct: 1889 EKKNGQVRVCVDFRDLNRACPKDDFPLPITEMVVDSTTGYGALS------GYNQIKMDLL 1942
Query: 922 DVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSP 981
D TAFR P G + + VMPFGLKNAGATYQR M + D I ++ Y+DD+VVK+
Sbjct: 1943 DAFDTAFRTPK--GNFYYTVMPFGLKNAGATYQRAMQFVLDDLIHHSVECYVDDMVVKTK 2000
Query: 982 SRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDT 1041
+ LR FER+R++ LKMNPLKCAF V +G FLGFV+ +GIEI K KAIL+
Sbjct: 2001 DHEHHQEDLRIVFERLRRHQLKMNPLKCAFAVQSGVFLGFVIRHRGIEIEPKKIKAILNM 2060
Query: 1042 NPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKA 1101
PP K L+ L GK+ ++RRFI NLS + + FS L+ KK F W+ E Q FD +K
Sbjct: 2061 PPPQELKDLRKLQGKLAYIRRFISNLSGRIQPFSKLM--KKGTPFVWDEECQNGFDSIKR 2118
Query: 1102 YLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENSKERAIFYLSRVLNDAETRYTM 1161
YL +PPV+A P+ G+P+ LYI+ +IG++LAQ ++ KE A +YLSR + AE Y+
Sbjct: 2119 YLLNPPVLAAPVKGRPLILYIATQPASIGALLAQHNDEGKEVACYYLSRTMVGAEQNYSP 2178
Query: 1162 IEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTY 1221
IEKLCL L F+ KL++Y+ + + + D I+++LS+P+L R+GKWAL + EY +T+
Sbjct: 2179 IEKLCLALIFALKKLRHYMLAHQIQLIARADPIRYVLSQPVLTGRLGKWALLMMEYDITF 2238
Query: 1222 APLKAVKGQAVADFLVDHTLP-----------KEIVTYVGVQPWKLFFDGSSHKN----G 1266
P KA+KGQA+A+FL H +P +EI T + W+L+FDG+S K+ G
Sbjct: 2239 VPQKAIKGQALAEFLATHPMPDDSPLIANLPDEEIFTAELQEQWELYFDGASRKDINPDG 2298
Query: 1267 T-----GIGMFIVSPGGTPTKFKFRI-KKKCSNNEAEYEALISSLEILIALGARNVVVKG 1320
T G G+ +P G F + K++CSNNEAEYEALI L + +++ R++ G
Sbjct: 2299 TPRRRAGAGLVFKTPQGGVIYHSFSLLKEECSNNEAEYEALIFGLLLALSMEVRSLRAHG 2358
Query: 1321 DSELVIKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANELAQIASGY 1380
DS L+I+Q+ Y+ L YYT A L+ KF+ + HV R N A+ LA++A+
Sbjct: 2359 DSRLIIRQINNIYEVRKPELVPYYTVARRLMDKFEHIEVIHVPRSKNAPADALAKLAAAL 2418
Query: 1381 MVDKYRLKELIEVKEK--------LNPSDLNVLVIDNMAPKDWRKPIVDYLQN---PVGT 1429
+ +++ V+E+ L P ++N+++ ++ +DWR+P +DY ++ P
Sbjct: 2419 VFQGDNPAQIV-VEERWLLPAVLELIPEEVNIIITNSAEEEDWRQPFLDYFKHGSLPEDP 2477
Query: 1430 TDRKTKYRAM-SYVIMGNELFKKNV-DGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKM 1487
+R+ R + SY+ L+K++ LL+C+ +A + VH G+CG HQ+G KM
Sbjct: 2478 VERRQLQRRLPSYIYKAGVLYKRSYGQEVLLRCVDRSEANRVLQEVHHGVCGGHQSGPKM 2537
Query: 1488 KWILFRQGMYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIKPWPFRGWALDLIG 1547
+ G YWP IM DC++ AK C CQ H +H P + LH + WPF W +D+IG
Sbjct: 2538 YHSIRLVGYYWPGIMADCLKTAKTCHGCQIHDNFKHQPPAPLHPTVPSWPFDAWGIDVIG 2597
Query: 1548 EINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQG 1607
INP SSR H++I+ A DYF+KW EA+PL+ V VI+F++ HI+YRFG+P +T+D
Sbjct: 2598 LINPPSSRGHRFILTATDYFSKWAEAVPLREVKSSDVINFLERHIIYRFGVPHRITSDNA 2657
Query: 1608 TVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLKD 1661
F QK+ F E + IK ST YY QANG EA NKTL ++K + ++ +D
Sbjct: 2658 KAFKSQKIYRFMEKYKIKWNYSTGYYPQANGMAEAFNKTLGKILKKTVDKHRRD 2711
Score = 90.9 bits (224), Expect = 3e-16
Identities = 43/138 (31%), Positives = 76/138 (54%)
Query: 435 HLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLIPHDMVLSDYEGKTG 494
H +PL++ + + ++L+D G+ +N++P L + G T DL P D+V+ ++ +
Sbjct: 1378 HNRPLYIEGNIGSAHLRRILIDLGSAVNILPVRSLTRAGFTTKDLEPIDVVICGFDNQGK 1437
Query: 495 SSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRISIWKADG 554
+LGAI + I + T + F V+ + +Y+ LLGR WIH VPSTLHQ + +G
Sbjct: 1438 PTLGAITIKIQMSTFSFKVRFFVIEANTSYSALLGRPWIHKYRVVPSTLHQCLKFLDGNG 1497
Query: 555 FVENVQADQSYYLAETGY 572
+ + ++ S Y + Y
Sbjct: 1498 VQQRITSNFSPYTIQESY 1515
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 644 bits (1662), Expect = 0.0
Identities = 342/902 (37%), Positives = 529/902 (57%), Gaps = 50/902 (5%)
Query: 798 AWPQSRIGGIE-------LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTAR 850
AW + G++ LP++E VKQ RR PD+ KIKEE+++ F+
Sbjct: 1201 AWSYQDMPGLDTDIVVHRLPLREGCPSVKQKLRRTSPDMATKIKEEVQKQWDAGFLAVTS 1260
Query: 851 YVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGY 910
Y W+AN+VPV KK+GK+R+C+D+RDLN A+PKD++ +P +++VD+ A S +DG+
Sbjct: 1261 YPPWMANIVPVPKKDGKVRMCVDYRDLNRASPKDDFPLPHIDVLVDNTAQSSVFSFMDGF 1320
Query: 911 SGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQ 970
SGYNQI +A ED+ KT F P GT+ + VMPFGLKNAGATYQR M T+FHD + ++
Sbjct: 1321 SGYNQIKMAPEDMEKTTFITPW--GTFCYKVMPFGLKNAGATYQRAMTTLFHDMMHKEIE 1378
Query: 971 VYIDDIVVKSPSRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEI 1030
VY+DD++ KS + ++ L++L+K F+R+RK+ L++NP KC FGV +G LGF+V +KGIE+
Sbjct: 1379 VYVDDMIAKSQTEEEHLVNLQKLFDRLRKFKLRLNPNKCTFGVRSGKLLGFIVSEKGIEV 1438
Query: 1031 NKNKAKAILDTNPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEA 1090
+ K KAI + P ++KQ++ LG++N++ RFI +L+ + LLR K +W
Sbjct: 1439 DPAKVKAIQEMPEPKTEKQVRGFLGRLNYIARFISHLTATCEPIFKLLR--KNQAIKWND 1496
Query: 1091 EHQKAFDELKAYLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENS-KERAIFYLS 1149
+ QKAFD++K YL PP++ PP+ G+P+ +Y+S T ++G +L + DE+ KE AI+YLS
Sbjct: 1497 DCQKAFDKIKEYLQKPPILIPPVPGRPLIMYLSVTENSMGCVLGRHDESGRKEHAIYYLS 1556
Query: 1150 RVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGK 1209
+ D ETRY+++EK C L ++ +L+ Y+ ++ S D +K++ KP L R+ +
Sbjct: 1557 KKFTDCETRYSLLEKTCCALAWAARRLRQYMLNHTTLLISKMDPVKYIFEKPALTGRVAR 1616
Query: 1210 WALALTEYSLTYAPLKAVKGQAVADFLVDHTL----------PKEIVTYV---------- 1249
W + LTEY + Y KA+KG ++D+L + + P E + Y+
Sbjct: 1617 WQMILTEYDIQYTSQKAIKGSILSDYLAEQPIEDYQPMMFEFPDEDIMYLKMKDCKEPLV 1676
Query: 1250 --GVQP---WKLFFDGSSHKNGTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISS 1304
G P W L FDG+ + NG G+G +++P G F R+ +NNEAEYEA I
Sbjct: 1677 EEGPDPDDKWTLMFDGAVNMNGNGVGAVLINPKGAHMPFSARLTFDVTNNEAEYEACIMG 1736
Query: 1305 LEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSR 1364
+E I L + + + GDS LV+ Q+ ++ +L Y +L F +L HV R
Sbjct: 1737 IEEAIDLRIKTLDIFGDSALVVNQVNGDWNTNQPHLIPYRDYTRRILTFFKKVKLYHVPR 1796
Query: 1365 IDNQEANELAQIASGYMVDKYRLKELIEVKEKLNPSDL---NVLVIDNMAPKDWRKPIVD 1421
+NQ A+ LA ++S V+ + + V P+ + +VID K W I +
Sbjct: 1797 DENQMADALATLSSMIKVNWWNHVPHVAVNRLERPAYVFAAESVVIDE---KPWYYDIKN 1853
Query: 1422 YLQN---PVGTT--DRKTKYRAMS--YVIMGNELFKKNVDGTLLKCLSDDDALIAISAVH 1474
+L+ P G + D+KT R Y+ + L+K+N D LL+C+ +A + + VH
Sbjct: 1854 FLKTQEYPEGASKNDKKTLRRLAGSFYLNQDDVLYKRNFDMVLLRCMDRPEADMLMQEVH 1913
Query: 1475 DGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIK 1534
+G G H G M L R G YW T+ DC +YA+ C CQ +A HVP S L+ +
Sbjct: 1914 EGSFGTHAGGHAMAKKLLRAGYYWMTMESDCFKYARKCHKCQIYADRVHVPPSPLNVMNS 1973
Query: 1535 PWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVY 1594
PWPF W +D+IG+I P +S H++I+VAIDYFTKWVEA N+T+ V FI+ I+
Sbjct: 1974 PWPFAMWGIDMIGKIEPTASNGHRFILVAIDYFTKWVEAASYANITKQVVTRFIKKEIIC 2033
Query: 1595 RFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNI 1654
R+G+PE + TD G+ + + + + I+ S+PY + NG VEAANK + +++ +
Sbjct: 2034 RYGVPERIITDNGSNLNNKMMKELCKDFKIEHHNSSPYRPKMNGAVEAANKNIKKIVRKM 2093
Query: 1655 LV 1656
+V
Sbjct: 2094 VV 2095
Score = 78.2 bits (191), Expect = 2e-12
Identities = 40/119 (33%), Positives = 68/119 (56%), Gaps = 1/119 (0%)
Query: 433 KSHLKPLFVWAKVDEKGVNKVLVDGGATINLMPKFMLKKLGKTEADLIPHDMVLSDYEGK 492
++H K L + + ++ VLVD G+++N++PK +LKK+ L P D+++ ++
Sbjct: 823 RNHNKALHITMECKGAVLSHVLVDTGSSLNVLPKQILKKIDVEGFVLTPSDLIVRAFDRS 882
Query: 493 TGSSLGAIMLNITVGTVARSTLFIVVPSKANYNLLLGREWIHGVGAVPSTLHQRIS-IW 550
S G + L + +G +F V+ + Y+ LLGR WIH GAV STLHQ++ +W
Sbjct: 883 KRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAVSSTLHQKLKYVW 941
>UniRef100_Q6L974 GAG-POL [Vitis vinifera]
Length = 1027
Score = 606 bits (1563), Expect = e-171
Identities = 324/857 (37%), Positives = 507/857 (58%), Gaps = 19/857 (2%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
L + +PV+Q RRFHPD I+ EI++LL FIR Y DWLANVV V KK GK
Sbjct: 13 LNVVSTARPVRQRIRRFHPDRQRVIRNEIDKLLEAGFIREVSYPDWLANVVVVPKKEGKW 72
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
RVC+D+ +LN A PKD + +P + +VDS +G LS LD +SGY+QI ++ +D K AF
Sbjct: 73 RVCVDYTNLNNACPKDSFPLPRIDQIVDSTSGQGMLSFLDAFSGYHQIPMSPDDEEKIAF 132
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
P L Y+ VMPFGLKNAGATYQR+M IF I ++VYIDDIVVKS +R+ +L
Sbjct: 133 ITPHDLYCYK--VMPFGLKNAGATYQRLMTKIFKPLIGHSVEVYIDDIVVKSKTREQHIL 190
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
HL++ F +R+YG+K+NP KCAFGV A FLGF+V ++GIE++ ++ KA+++T PP +KK
Sbjct: 191 HLQEVFYLLRRYGMKLNPSKCAFGVSARKFLGFMVSQRGIEVSPDQVKAVMETPPPRNKK 250
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+LQ L GK+ L RFI ++ + F L ++K T W Q A + +K YL PP+
Sbjct: 251 ELQRLTGKLVALGRFIARFIDELRPF--FLAIRKAGTHGWTDNCQNALERIKHYLMQPPI 308
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSKERAIFYLSRVLNDAETRYTMIEKLCLC 1168
++ PI + + +Y++ + I ++L + +++ I+Y+SR L D ETRY+ +E + L
Sbjct: 309 LSSPIPKEKLYMYLAVSEWAISAVLFRCPSPKEQKPIYYVSRALADVETRYSKMELISLA 368
Query: 1169 LYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAPLKAVK 1228
L + KL+ Y + V+V + ++++L KP L R+ +WA+ L+E+ + + P ++K
Sbjct: 369 LRSAAQKLRPYFQAHPVIVLTDQP-LRNILHKPDLTGRMLQWAIELSEFGIEFQPRLSMK 427
Query: 1229 GQAVADFLVDHT-LPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKFKFRI 1287
GQ +ADF+++++ P + + W L DG+S +G+G+G+ + SP G + R+
Sbjct: 428 GQVMADFVLEYSRKPGQHEGSRKKEWWTLRVDGASRSSGSGVGLLLQSPTGEHLEQAIRL 487
Query: 1288 KKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKYYTKA 1347
SNNEAEYEA++S L++ +AL + + DS+LV+K + +EY+ +A+Y K
Sbjct: 488 GFSASNNEAEYEAILSGLDLALALSVSKLRIFSDSQLVVKHVQEEYEAKDARMARYLAKV 547
Query: 1348 NNLLAKFDDARLGHVSRIDNQEANELAQIASGYMVDKYRLKELIEVKEKLNPSDLNVLVI 1407
N L +F + + + R DN+ A+ LA IA+ + + L+ + + NPS + +
Sbjct: 548 RNTLQQFTEWTIEKIKRADNRRADALAGIAASLSIKE---AILLPIHVQTNPSVSEISIC 604
Query: 1408 DNM-AP----KDWRKPIVDYLQNPVGTTD----RKTKYRAMSYVIMGNELFKKNVDGTLL 1458
AP ++W I +Y++ D K + +A + ++G L+K++ G L
Sbjct: 605 STTEAPQADDQEWMNDITEYIRTGTLPGDPKQAHKVRVQAARFTLIGGHLYKRSFTGPYL 664
Query: 1459 KCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQDCQRH 1518
+CL +A ++ +H+G+ G H G + QG YWPT+ K+ Y K C CQR+
Sbjct: 665 RCLGHSEAQYVLAELHEGIYGNHSGGRSLAHRAHSQGYYWPTMKKEAAAYVKRCDKCQRY 724
Query: 1519 AGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEAIPLQN 1578
A I H+P++ L SI PWPF W +D++ + P + Q K+++VA DYF+KWVEA +
Sbjct: 725 APIPHMPSTTLKSISGPWPFAQWGMDIVRPL-PTAPAQKKFLLVATDYFSKWVEAEAYAS 783
Query: 1579 VTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYYAQANG 1638
V F+ +I+ RFG+P+++ D G F +F I+ STP Y Q+NG
Sbjct: 784 TKDKDVTKFVWKNIICRFGIPQTIIADNGPQFDSIAFRNFCSELNIRNSYSTPRYPQSNG 843
Query: 1639 QVEAANKTLISLIKNIL 1655
Q EA NKTLI+ +K L
Sbjct: 844 QAEATNKTLITALKKRL 860
>UniRef100_Q8LMM4 Putative gag-pol [Oryza sativa]
Length = 2017
Score = 603 bits (1555), Expect = e-170
Identities = 338/891 (37%), Positives = 507/891 (55%), Gaps = 32/891 (3%)
Query: 798 AWPQSRIGGI-------ELPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTAR 850
AW S + GI L +KED KP+KQ RRF D IKEE+ +LL FI+
Sbjct: 980 AWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVL 1039
Query: 851 YVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGY 910
+ DWLAN V V KK G+ R+C+D+ DLN + PKD + +P + +VDS AG E LS LD Y
Sbjct: 1040 HPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCY 1099
Query: 911 SGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQ 970
SGY+QI + E D KT+F P G Y +V MPFGLKNAGATYQR++ F I ++
Sbjct: 1100 SGYHQIRLKESDCLKTSFITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1157
Query: 971 VYIDDIVVKSPSRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEI 1030
Y+DD+VVK+ +DD +L L ++F +R + +K+NP KC FGV +G LGF+V +GI+
Sbjct: 1158 AYVDDVVVKTKQKDDLILDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1217
Query: 1031 NKNKAKAILDTNPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEA 1090
N K AIL+ PP+++K +Q L G + L RF+ L E+ F LL KK D F+W
Sbjct: 1218 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDDFQWGP 1275
Query: 1091 EHQKAFDELKAYLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAI 1145
E QKAF++ K L+ PPV+A P +P+ LY+SAT+ + ++L E E +R I
Sbjct: 1276 EAQKAFEDFKKLLTEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPI 1335
Query: 1146 FYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHS 1205
+++S VL D++TRY ++KL + + KL +Y + V V + + + +L +
Sbjct: 1336 YFVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDILHNREANG 1394
Query: 1206 RIGKWALALTEYSLTYAPLKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKN 1265
RI KWAL L L++ P ++K QA+ADF+ + T +E T ++ W + FDGS +
Sbjct: 1395 RIAKWALELMSLDLSFKPRISIKSQALADFVAEWTECQEDTTVKKMEHWTMHFDGSKRLS 1454
Query: 1266 GTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELV 1325
GTG G+ ++SP G + I S+N AEYEAL+ L I I+LG + ++V+GDS+LV
Sbjct: 1455 GTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLV 1514
Query: 1326 IKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASG 1379
+ Q+ KE+ C+ +N+ Y + L KFD L HV R DN+ A+ LA ++A
Sbjct: 1515 VNQVMKEWSCLDDNMKAYRQEVRKLEDKFDGLELSHVLRHDNEAADRLANFGSKREVAPS 1574
Query: 1380 YMVDKYRLKELIEVKEKLNPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTK 1435
+ ++ + K+ + ++ + M DWR+P++ +L + D+ +
Sbjct: 1575 DVFVEHLYTPTVPHKDTTQAAGIHDVA---MVETDWREPLIRFLTSQELPQDKDEAERIS 1631
Query: 1436 YRAMSYVIMGNELFKKNVDGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQG 1495
R+ YV+ EL+KK+ G L +C+S ++ + +H G+CG H A + +RQG
Sbjct: 1632 RRSKLYVMHEAELYKKSPSGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQG 1691
Query: 1496 MYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSR 1555
+WPT + D + + C+ CQ A H+PA EL +I WPF W LD++G
Sbjct: 1692 FFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG- 1750
Query: 1556 QHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKV 1615
+ ++ VAID F+KW+EA P+ +T D DF N IV+RFG+P + TD GT F G
Sbjct: 1751 GYTHLFVAIDKFSKWIEAKPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVF 1809
Query: 1616 ASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
F E +GIK+ ++ + +NGQVE AN ++ IK + + LK W
Sbjct: 1810 KDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDRLKPYAGKW 1860
>UniRef100_Q7X6L5 OSJNBb0093G06.3 protein [Oryza sativa]
Length = 1986
Score = 601 bits (1549), Expect = e-170
Identities = 337/891 (37%), Positives = 505/891 (55%), Gaps = 32/891 (3%)
Query: 798 AWPQSRIGGI-------ELPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTAR 850
AW S + GI L +KED KP+KQ RRF D IKEE+ +LL FI+
Sbjct: 949 AWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVL 1008
Query: 851 YVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGY 910
+ DWLAN V V KK G+ R+C+D+ DLN + PKD + +P + +VDS AG E LS LD Y
Sbjct: 1009 HPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCY 1068
Query: 911 SGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQ 970
SGY+QI + E D KT+F P G Y +V MPFGLKNAGATYQR++ F I ++
Sbjct: 1069 SGYHQIRLKESDCLKTSFITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1126
Query: 971 VYIDDIVVKSPSRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEI 1030
Y+DD+VVK+ +DD + L ++F +R + +K+NP KC FGV +G LGF+V +GI+
Sbjct: 1127 AYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1186
Query: 1031 NKNKAKAILDTNPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEA 1090
N K AIL+ PP+++K +Q L G + L RF+ L E+ F LL KK D F+W
Sbjct: 1187 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDNFQWGP 1244
Query: 1091 EHQKAFDELKAYLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAI 1145
E QKAF++ K L+ PPV+A P +P+ LY+SAT+ + ++L E E +R I
Sbjct: 1245 EAQKAFEDFKQLLTKPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPI 1304
Query: 1146 FYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHS 1205
+++S VL D++TRY ++KL + + KL +Y + V V + + + +L +
Sbjct: 1305 YFVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDVLHNREANG 1363
Query: 1206 RIGKWALALTEYSLTYAPLKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKN 1265
RI KWAL L +++ P ++K QA+ADF+ + T +E ++ W + FDGS +
Sbjct: 1364 RIAKWALELMSLDISFKPRTSIKSQALADFVAEWTECQEDTPVEKIEHWTMHFDGSKRLS 1423
Query: 1266 GTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELV 1325
GTG G+ ++SP G + I S+N AEYEAL+ L I I+LG + ++V+GDS+LV
Sbjct: 1424 GTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLV 1483
Query: 1326 IKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASG 1379
+ Q+ KE+ CI +N+ Y + L KFD L HV R +N+ A+ LA + A
Sbjct: 1484 VNQVMKEWSCIDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREAAPS 1543
Query: 1380 YMVDKYRLKELIEVKEKLNPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTK 1435
+ ++ + K+ +D + +V M DWR+P + +L + D+ +
Sbjct: 1544 DVFVEHLYTPTVPHKDTTQDADTHDVV---MVEADWREPFIRFLSSQELPQDKDEAERIS 1600
Query: 1436 YRAMSYVIMGNELFKKNVDGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQG 1495
R+ YV+ +EL+KK+ G L +C+S ++ + +H G+CG H A + +RQG
Sbjct: 1601 RRSKLYVMHESELYKKSPSGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQG 1660
Query: 1496 MYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSR 1555
+WPT + D + + C+ CQ A H+PA EL +I WPF W LD++G
Sbjct: 1661 FFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG- 1719
Query: 1556 QHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKV 1615
+ ++ VAID F+KW+EA P+ +T D DF N IV+RFG+P + TD GT F G
Sbjct: 1720 GYTHLFVAIDKFSKWIEAKPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVF 1778
Query: 1616 ASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
F E +GIK+ ++ + +NGQVE AN ++ IK + + LK W
Sbjct: 1779 KDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDRLKPYAGKW 1829
>UniRef100_Q75KG5 Putative polyprotein [Oryza sativa]
Length = 1991
Score = 598 bits (1542), Expect = e-169
Identities = 335/891 (37%), Positives = 503/891 (55%), Gaps = 32/891 (3%)
Query: 798 AWPQSRIGGI-------ELPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTAR 850
AW S + GI L +KED KP+KQ RRF D IKEE+ +LL FI+
Sbjct: 954 AWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVL 1013
Query: 851 YVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGY 910
+ DWLAN V V KK G+ R+C+D+ DLN + PKD + +P + +VDS AG E LS LD Y
Sbjct: 1014 HPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCY 1073
Query: 911 SGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQ 970
SGY+QI + E D KT+F P G Y +V MPFGLKNAGATYQR++ F I ++
Sbjct: 1074 SGYHQIRLKESDCLKTSFITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1131
Query: 971 VYIDDIVVKSPSRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEI 1030
Y+DD+VVK+ +DD +L L ++F +R + +K+NP KC FGV +G LGF+V +GI+
Sbjct: 1132 AYVDDVVVKTKQKDDLILDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1191
Query: 1031 NKNKAKAILDTNPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEA 1090
N K AIL+ PP+++K +Q L G + L RF+ L E+ F LL KK D F+W
Sbjct: 1192 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDNFQWGP 1249
Query: 1091 EHQKAFDELKAYLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAI 1145
E QKAF++ K L+ PPV+A P +P+ LY+SAT+ + ++L E E +R I
Sbjct: 1250 EAQKAFEDFKQLLTKPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPI 1309
Query: 1146 FYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHS 1205
+++S VL D++TRY ++KL + + KL +Y + V V + + + +L +
Sbjct: 1310 YFVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDVLHNREANG 1368
Query: 1206 RIGKWALALTEYSLTYAPLKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKN 1265
RI KWAL L +++ P ++K QA+ADF+ + T +E ++ W + FDGS +
Sbjct: 1369 RIAKWALELMSLDISFKPRTSIKSQALADFVAEWTECQEDTPVEKIEHWTMHFDGSKRLS 1428
Query: 1266 GTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELV 1325
GTG G+ ++SP G + I S+N AEYEAL+ L I I+LG + ++V+GDS+LV
Sbjct: 1429 GTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLV 1488
Query: 1326 IKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASG 1379
+ Q+ KE+ CI +N+ Y + L KFD L HV R +N+ A+ LA + A
Sbjct: 1489 VNQVMKEWSCIDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREAAPS 1548
Query: 1380 YMVDKYRLKELIEVKEKLNPSDLNVLVIDNMAPKDWRKPIVDYLQN----PVGTTDRKTK 1435
+ ++ + K+ +D + + M DWR+P++ +L + +
Sbjct: 1549 DVFVEHLYSPTVPHKDATQVADTHDIA---MVEADWREPLIRFLTSQELPQYKDEAERIS 1605
Query: 1436 YRAMSYVIMGNELFKKNVDGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQG 1495
R+ YV+ EL+KK+ G L +C+S ++ + +H G+CG H A + +RQG
Sbjct: 1606 RRSKLYVMHEAELYKKSPSGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQG 1665
Query: 1496 MYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSR 1555
+WPT + D + + C+ CQ A H+PA EL +I WPF W LD++G
Sbjct: 1666 FFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG- 1724
Query: 1556 QHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKV 1615
+ ++ +AID F+KW+EA P+ +T D DF N IV+RFG+P + TD GT F G
Sbjct: 1725 GYTHLFMAIDKFSKWIEAKPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVF 1783
Query: 1616 ASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
F E +GIK+ ++ + +NGQVE AN ++ IK + + LK W
Sbjct: 1784 KDFCEDFGIKICYASVAHPMSNGQVERANGMILQRIKARVFDRLKPYAGKW 1834
>UniRef100_Q8LMM6 Putative gag-pol [Oryza sativa]
Length = 1986
Score = 597 bits (1540), Expect = e-169
Identities = 335/891 (37%), Positives = 502/891 (55%), Gaps = 32/891 (3%)
Query: 798 AWPQSRIGGI-------ELPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTAR 850
AW S + GI L +KED KP+KQ RRF D IKEE+ +LL FI+
Sbjct: 949 AWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVL 1008
Query: 851 YVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGY 910
+ DWLAN V V KK G+ R+C+D+ DLN + PKD + +P + +VDS AG E LS LD Y
Sbjct: 1009 HPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCY 1068
Query: 911 SGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQ 970
SGY+QI + E D KT+F P G Y +V MPFGLKNAGATYQR++ F I ++
Sbjct: 1069 SGYHQIRLKESDCLKTSFITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1126
Query: 971 VYIDDIVVKSPSRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEI 1030
Y+DD+VVK+ +DD + L ++F +R + +K+NP KC FGV +G LGF+V +GI+
Sbjct: 1127 AYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1186
Query: 1031 NKNKAKAILDTNPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEA 1090
N K AIL+ PP+++K +Q L G + L RF+ L E+ F LL KK D F+W
Sbjct: 1187 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDNFQWGP 1244
Query: 1091 EHQKAFDELKAYLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAI 1145
E QKAF++ K L+ PPV+A P +P+ LY+SAT+ + ++L E E +R I
Sbjct: 1245 EAQKAFEDFKQLLTKPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPI 1304
Query: 1146 FYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHS 1205
+++S VL D++ RY ++KL + + KL +Y + V V + + + +L +
Sbjct: 1305 YFVSEVLADSKARYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDVLHNREANG 1363
Query: 1206 RIGKWALALTEYSLTYAPLKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKN 1265
RI KWAL L +++ P ++K QA+ADF+ + T +E ++ W + FDGS +
Sbjct: 1364 RIAKWALELMSLDISFKPRTSIKSQALADFVAEWTECQEDTPVEKIEHWTMHFDGSKRLS 1423
Query: 1266 GTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELV 1325
GTG G+ ++SP G + I S+N AEYEAL+ L I I+LG + ++V+GDS+LV
Sbjct: 1424 GTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLV 1483
Query: 1326 IKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASG 1379
+ Q+ KE+ CI +N+ Y + L KFD L HV R +N+ A+ LA + A
Sbjct: 1484 VNQVMKEWSCIDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKRETAPS 1543
Query: 1380 YMVDKYRLKELIEVKEKLNPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTK 1435
+ ++ + K+ +D + M DWR+P + +L + D+ +
Sbjct: 1544 DVFVEHLYTPTVPHKDTTQDADTRNIA---MVEADWREPFIRFLTSQELPQDKDEAERIS 1600
Query: 1436 YRAMSYVIMGNELFKKNVDGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQG 1495
R+ YV+ +EL+KK+ G L +C+S ++ + +H G+CG H A + +RQG
Sbjct: 1601 RRSKLYVLHESELYKKSPSGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQG 1660
Query: 1496 MYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSR 1555
+WPT + D + + C+ CQ A H+PA EL +I WPF W LD++G
Sbjct: 1661 FFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG- 1719
Query: 1556 QHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKV 1615
+ ++ VAID F+KW+EA P+ +T D DF N IV+RFG+P + TD GT F G
Sbjct: 1720 GYTHLFVAIDKFSKWIEAKPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVF 1778
Query: 1616 ASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
F E +GIK+ ++ + +NGQVE AN ++ IK + + LK W
Sbjct: 1779 KDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDRLKPYAGKW 1829
>UniRef100_Q75KM4 Putative polyprotein [Oryza sativa]
Length = 2004
Score = 596 bits (1536), Expect = e-168
Identities = 330/873 (37%), Positives = 497/873 (56%), Gaps = 25/873 (2%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
L +KED KP+KQ RRF D IKEE+ +LL FI+ + DWLAN V V KK G+
Sbjct: 985 LHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVLHPDWLANPVLVRKKTGQW 1044
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
R+C+D+ DLN + PKD + +P + +VDS AG E LS LD YSGY+QI + E D KT+F
Sbjct: 1045 RMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCYSGYHQIRLKESDCLKTSF 1104
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
P G Y +V MPFGLKNAGATYQR++ F I ++ Y+DD+VVK+ +DD +L
Sbjct: 1105 ITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVEAYVDDVVVKTKQKDDLIL 1162
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
L ++F +R + +K+NP KC FGV +G LGF+V +GI+ N K AIL+ PP+++K
Sbjct: 1163 DLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQANPEKVTAILNMKPPSTQK 1222
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+Q L G + L RF+ L E+ F LL KK D F+W E QKAF++ K L+ PPV
Sbjct: 1223 DVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDNFQWGPEAQKAFEDFKQLLTKPPV 1280
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAIFYLSRVLNDAETRYTMIE 1163
+A P +P+ LY+SAT+ + ++L E E +R I+++S VL D++TRY ++
Sbjct: 1281 LASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYFVSEVLADSKTRYPQVQ 1340
Query: 1164 KLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAP 1223
KL + + KL +Y + V V + + + +L + RI KWAL L +++ P
Sbjct: 1341 KLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDVLHNREANGRIAKWALELMSLDISFKP 1399
Query: 1224 LKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKF 1283
++K QA+ADF+ + T +E ++ W + FDGS +GTG G+ ++SP G +
Sbjct: 1400 RTSIKSQALADFVAEWTECQEDTPVEKIEHWTMHFDGSKRLSGTGAGVVLISPTGERLSY 1459
Query: 1284 KFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKY 1343
I S+N AEYEAL+ L I I+LG + ++V+GDS+LV+ Q+ KE+ CI +N+ Y
Sbjct: 1460 VLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCIDDNMMAY 1519
Query: 1344 YTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASGYMVDKYRLKELIEVKEKL 1397
+ L KFD L HV R +N+ A+ LA + A + ++ + K+
Sbjct: 1520 RQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREAAPSDVFVEHLYSPTVPHKDAT 1579
Query: 1398 NPSDLNVLVIDNMAPKDWRKPIVDYLQN----PVGTTDRKTKYRAMSYVIMGNELFKKNV 1453
+D + + M DWR+P++ +L + + R+ YV+ EL+KK+
Sbjct: 1580 QVADTHDIA---MVEADWREPLIRFLTSQELPQYKDEAERISRRSKLYVMHEAELYKKSP 1636
Query: 1454 DGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQ 1513
G L +C+S ++ + +H G+CG H A + +RQG +WPT + D + + C+
Sbjct: 1637 SGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCE 1696
Query: 1514 DCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEA 1573
CQ A H+PA EL +I WPF W LD++G + ++ +AID F+KW+EA
Sbjct: 1697 GCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG-GYTHLFMAIDKFSKWIEA 1755
Query: 1574 IPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYY 1633
P+ +T D DF N IV+RFG+P + TD GT F G F E +GIK+ ++ +
Sbjct: 1756 KPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAH 1814
Query: 1634 AQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
+NGQVE AN ++ IK + + LK W
Sbjct: 1815 PMSNGQVERANGMILQRIKARVFDRLKPYAGKW 1847
>UniRef100_Q7XPQ7 OSJNBa0053K19.16 protein [Oryza sativa]
Length = 2010
Score = 595 bits (1534), Expect = e-168
Identities = 333/891 (37%), Positives = 504/891 (56%), Gaps = 32/891 (3%)
Query: 798 AWPQSRIGGI-------ELPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTAR 850
AW S + GI L +KED KP+KQ RRF D IKEE+ +LL FI+
Sbjct: 973 AWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVL 1032
Query: 851 YVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGY 910
+ DWLAN V V KK G+ R+C+D+ DLN + PKD + +P + +VDS AG E LS LD Y
Sbjct: 1033 HPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGRELLSFLDCY 1092
Query: 911 SGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQ 970
SGY+QI + E D KT+F P G Y +V MPFGLKNAGATYQR++ F I ++
Sbjct: 1093 SGYHQIRLKESDCLKTSFITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1150
Query: 971 VYIDDIVVKSPSRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEI 1030
Y+DD+VVK+ +DD + L ++F +R + +K+NP KC FGV +G +GF+V +GI+
Sbjct: 1151 AYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLVGFMVSHRGIQA 1210
Query: 1031 NKNKAKAILDTNPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEA 1090
N K AIL+ PP+++K +Q L G + L RF+ L E+ F LL KK D F+W
Sbjct: 1211 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDNFQWGP 1268
Query: 1091 EHQKAFDELKAYLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAI 1145
E QKAF++ K L+ PPV+A P +P+ LY+SAT+ + ++L E E +R I
Sbjct: 1269 EAQKAFEDFKKLLTEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPI 1328
Query: 1146 FYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHS 1205
+++S VL D++TRY ++KL + + KL +Y + V V + + + +L +
Sbjct: 1329 YFVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDILHNREANG 1387
Query: 1206 RIGKWALALTEYSLTYAPLKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKN 1265
RI KWAL L +++ P ++K QA+ADF+ + T +E ++ W + FDGS +
Sbjct: 1388 RIAKWALELMSLDISFKPRISIKSQALADFVAEWTECQEDTPAENMEHWTMHFDGSKRLS 1447
Query: 1266 GTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELV 1325
GTG G+ ++SP G + I S+N AEYEAL+ L I I+LG + ++V+GDS+LV
Sbjct: 1448 GTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLV 1507
Query: 1326 IKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASG 1379
+ Q+ KE+ C+ +N+ Y + L KFD L HV R +N+ A+ LA ++A
Sbjct: 1508 VNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREVAPS 1567
Query: 1380 YMVDKYRLKELIEVKEKLNPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTK 1435
+ ++ + K+ + + + M DWR+P++ +L + D+ +
Sbjct: 1568 DVFVEHLYTPTVPHKDTTQVAGTHDVA---MVETDWREPLIRFLTSQELPQDKDEAERIS 1624
Query: 1436 YRAMSYVIMGNELFKKNVDGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQG 1495
R+ YV+ EL+KK+ G L +C+S ++ + +H G+CG H A + +RQG
Sbjct: 1625 RRSKLYVMHEAELYKKSPSGILQRCVSLEEGRQLLQDIHSGICGNHAAARTIVGKAYRQG 1684
Query: 1496 MYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSR 1555
+WPT + D + + C+ CQ A H+PA EL +I WPF W LD++G
Sbjct: 1685 FFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG- 1743
Query: 1556 QHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKV 1615
+ ++ VAID F+KW+EA P+ +T D DF N IV+RFG+P + TD GT F G
Sbjct: 1744 GYTHLFVAIDKFSKWIEAKPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVF 1802
Query: 1616 ASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
F E +GIK+ ++ + +NGQVE AN ++ IK + + LK W
Sbjct: 1803 KDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDRLKPYAGKW 1853
>UniRef100_Q7X8E7 OSJNBa0042F21.5 protein [Oryza sativa]
Length = 1950
Score = 593 bits (1530), Expect = e-167
Identities = 332/891 (37%), Positives = 503/891 (56%), Gaps = 32/891 (3%)
Query: 798 AWPQSRIGGI-------ELPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTAR 850
AW S + GI L +KED KP+KQ RRF D IKEE+ +LL FI+
Sbjct: 913 AWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVL 972
Query: 851 YVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGY 910
+ DWLAN V V KK G+ R+C+D+ DLN + PKD + +P + +VDS AG E LS LD Y
Sbjct: 973 HPDWLANPVLVRKKTGQWRMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCY 1032
Query: 911 SGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQ 970
SGY+QI + E D KT+F P G Y +V MPFGLKNAGATYQR++ F I ++
Sbjct: 1033 SGYHQIRLKESDCLKTSFITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1090
Query: 971 VYIDDIVVKSPSRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEI 1030
Y+DD+VVK+ +DD + L ++F +R + +K+NP KC FGV +G LGF+V +GI+
Sbjct: 1091 AYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1150
Query: 1031 NKNKAKAILDTNPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEA 1090
N K AIL+ PP+++K +Q L G + L RF+ L E+ F LL KK D F+W
Sbjct: 1151 NPEKVTAILNMKPPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDNFQWGP 1208
Query: 1091 EHQKAFDELKAYLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAI 1145
E QKAF++ K L+ PP++A P +P+ LY+SAT+ + ++L E E +R I
Sbjct: 1209 EAQKAFEDFKKLLTEPPILASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPI 1268
Query: 1146 FYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHS 1205
+++S VL D++TRY ++KL + + KL +Y + V V + + + +L +
Sbjct: 1269 YFVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDILHNREANG 1327
Query: 1206 RIGKWALALTEYSLTYAPLKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKN 1265
RI KWAL L +++ P ++K QA+ADF+ + T +E ++ W + FDGS +
Sbjct: 1328 RIAKWALELMSLDISFKPRISIKSQALADFVAEWTECQEDTPAENMEHWTMHFDGSKRLS 1387
Query: 1266 GTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELV 1325
GTG G+ ++SP G + I S+N AEYEAL+ L I I+LG + ++V+GDS+LV
Sbjct: 1388 GTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLV 1447
Query: 1326 IKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASG 1379
+ Q+ KE+ C+ +N+ Y + L KFD L HV R +N+ A+ LA ++A
Sbjct: 1448 VNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREMAPS 1507
Query: 1380 YMVDKYRLKELIEVKEKLNPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTK 1435
+ ++ + K+ +D + + + DWR+P + +L + D+ +
Sbjct: 1508 DVFVEHLYTPTVPHKDTTQDADTHDVA---LVEADWREPFIRFLTSQELPQDKDEAERIS 1564
Query: 1436 YRAMSYVIMGNELFKKNVDGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQG 1495
R+ Y + EL+KK+ G L +C+S ++ + +H G+CG H A + +RQG
Sbjct: 1565 RRSKLYAMHEAELYKKSPSGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQG 1624
Query: 1496 MYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSR 1555
+WPT + D + + C+ CQ A H+PA EL +I WPF W LD++G
Sbjct: 1625 FFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG- 1683
Query: 1556 QHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKV 1615
+ ++ VAID F+KW+EA P+ +T D DF N IV+RFG+P + TD GT F G
Sbjct: 1684 GYTHLFVAIDKFSKWIEAKPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVF 1742
Query: 1616 ASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
F E +GIK+ ++ + +NGQVE AN ++ IK + + LK W
Sbjct: 1743 KDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDRLKPYAGKW 1793
>UniRef100_Q9SSW6 GAG-POL [Oryza sativa]
Length = 1032
Score = 593 bits (1529), Expect = e-167
Identities = 330/873 (37%), Positives = 498/873 (56%), Gaps = 25/873 (2%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
L +KED KP+KQ RRF D IKEE+ +LL FI+ + DWLAN V V KK G+
Sbjct: 13 LHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVLHPDWLANPVLVRKKTGQW 72
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
R+C+D+ DLN + PKD + +P + +VDS AG E LS LD YSGY+QI + E D KT+F
Sbjct: 73 RMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCYSGYHQIRLKESDCLKTSF 132
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
P G Y +V MPFGLKNAGATYQR++ F I ++ Y+DD+VVK+ +DD +
Sbjct: 133 ITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVEAYVDDVVVKTKQKDDLIS 190
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
L ++F +R + +K+NP KC FGV +G LGF+V +GI+ N K AIL+ PP+++K
Sbjct: 191 DLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQANPEKVTAILNMKPPSTQK 250
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+Q L G + L RF+ L E+ F LL KK D F+W E QKAF++ K L+ PPV
Sbjct: 251 DVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDNFQWGPEAQKAFEDFKKLLTEPPV 308
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAIFYLSRVLNDAETRYTMIE 1163
+A P +P+ LY+SAT+ + ++L E E +R I+++S VL D++TRY ++
Sbjct: 309 LASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYFVSEVLADSKTRYPQVQ 368
Query: 1164 KLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAP 1223
KL + + KL +Y + V V + + + +L + RI KWAL L +++ P
Sbjct: 369 KLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDILHNREANGRIAKWALELMSLDISFKP 427
Query: 1224 LKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKF 1283
++K QA+ADF+ + T +E ++ W + FDGS +GTG G+ ++SP G +
Sbjct: 428 RISIKSQALADFVAEWTECQEDTPAEKMEHWTMHFDGSKRLSGTGAGVVLISPTGERLSY 487
Query: 1284 KFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKY 1343
I S+N AEYEAL+ L I I+LG + ++V+GDS+LV+ Q+ KE+ C+ +N+ Y
Sbjct: 488 VLWIHFSTSHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAY 547
Query: 1344 YTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASGYMVDKYRLKELIEVKEKL 1397
+ L KFD L HV R +N+ A+ LA + A + ++ + K+
Sbjct: 548 RQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREAAPSDVFVEHLYTPTVPHKDTT 607
Query: 1398 NPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTKYRAMSYVIMGNELFKKNV 1453
+D + + M DWR+P++ +L + D+ + R+ YV+ EL+KK+
Sbjct: 608 QVADTHDIA---MVEADWREPLIRFLTSQELPRDKDEAERISRRSKLYVMHEAELYKKSP 664
Query: 1454 DGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQ 1513
G L +C+S ++ + +H G+CG H A + +RQG +WPT + D + + C+
Sbjct: 665 SGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCE 724
Query: 1514 DCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEA 1573
CQ A H+PA EL +I WPF W LD++G + ++ VAID F+KW+EA
Sbjct: 725 GCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG-GYTHLFVAIDKFSKWIEA 783
Query: 1574 IPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYY 1633
P+ +T D DF N IV+RFG+P + TD GT F G F E +GIK+ ++ +
Sbjct: 784 KPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAH 842
Query: 1634 AQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
+NGQVE AN ++ IK + + LK W
Sbjct: 843 PMSNGQVERANGMILQGIKARVFDRLKPYAGKW 875
>UniRef100_Q75KG3 Putative polyprotein [Oryza sativa]
Length = 2000
Score = 592 bits (1527), Expect = e-167
Identities = 329/873 (37%), Positives = 498/873 (56%), Gaps = 25/873 (2%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
L +KED KP+KQ RRF D IKEE+ +LL FI+ + DWLAN V V KK G+
Sbjct: 981 LHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVLHPDWLANPVLVRKKTGQW 1040
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
R+C+D+ DLN + PKD + +P + +VDS AG E LS LD YSGY+QI + E D KT+F
Sbjct: 1041 RMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCYSGYHQIRLKESDCLKTSF 1100
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
P G Y +V MPFGLKNAGATYQR++ F I ++ Y+DD+VVK+ +DD +
Sbjct: 1101 ITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVEAYVDDVVVKTKQKDDLIS 1158
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
L ++F +R + +K+NP KC FGV +G LGF+V +GI+ N K AIL+ PP+++K
Sbjct: 1159 DLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQANPEKVTAILNMKPPSTQK 1218
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+Q L G + L RF+ L E+ F LL KK D F+W E QKAF++ K L+ PPV
Sbjct: 1219 DVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDDFQWGPEAQKAFEDFKKLLTEPPV 1276
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAIFYLSRVLNDAETRYTMIE 1163
+A P +P+ LY+SAT+ + ++L E E +R I+++S VL D++TRY ++
Sbjct: 1277 LASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYFVSEVLADSKTRYPQVQ 1336
Query: 1164 KLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAP 1223
KL + + KL +Y + V V + + + +L + RI KWAL L +++ P
Sbjct: 1337 KLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDILHNRKANGRIAKWALELMSLDISFKP 1395
Query: 1224 LKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKF 1283
++K QA+ADF+ + T +E ++ W + FDGS +GTG G+ ++SP G +
Sbjct: 1396 RISIKSQALADFVAEWTECQEDTPAENMEHWTMHFDGSKRLSGTGAGVVLISPTGERLSY 1455
Query: 1284 KFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKY 1343
I S+N AEYEAL+ L I I+LG + ++V+GDS+LV+ Q+ KE+ C+ +N+ Y
Sbjct: 1456 VLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAY 1515
Query: 1344 YTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASGYMVDKYRLKELIEVKEKL 1397
+ L KFD L HV R +N+ A+ LA + A + ++ + K+
Sbjct: 1516 RQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREAAPSDVFVEHLYTPTVPHKDTT 1575
Query: 1398 NPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTKYRAMSYVIMGNELFKKNV 1453
+D + + M DWR+P++ +L + D+ + R+ YV+ EL+KK+
Sbjct: 1576 QDADTHDVA---MVEADWREPLIRFLTSQELPQDKDEAERISRRSKLYVLHEAELYKKSP 1632
Query: 1454 DGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQ 1513
G L +C+S ++ + +H G+CG H A + +RQG +WPT + D + + C+
Sbjct: 1633 SGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCE 1692
Query: 1514 DCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEA 1573
CQ A H+PA EL +I WPF W LD++G + ++ VAID F+KW+EA
Sbjct: 1693 GCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG-GYTHLFVAIDKFSKWIEA 1751
Query: 1574 IPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYY 1633
P+ +T D DF + IV+RFG+P + TD GT F G F E +GIK+ ++ +
Sbjct: 1752 KPVVTITADNARDFFIS-IVHRFGVPNRIITDNGTQFTGGVFKDFCEDFGIKICYASVAH 1810
Query: 1634 AQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
+NGQVE AN ++ IK + + LK W
Sbjct: 1811 PMSNGQVERANGMILQGIKARVFDRLKPYAGKW 1843
>UniRef100_Q8LHG3 GAG-POL [Oryza sativa]
Length = 1032
Score = 591 bits (1524), Expect = e-167
Identities = 328/873 (37%), Positives = 498/873 (56%), Gaps = 25/873 (2%)
Query: 809 LPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTARYVDWLANVVPVIKKNGKM 868
L +KED KP+KQ RRF D IKEE+ +LL FI+ + DWLAN V V KK G+
Sbjct: 13 LHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVHHPDWLANPVLVRKKTGQW 72
Query: 869 RVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGYSGYNQIFIAEEDVSKTAF 928
R+C+D+ DLN + PKD + +P + +VDS AG+E LS LD YSGY+QI + E D KT+F
Sbjct: 73 RMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGYELLSFLDCYSGYHQIRLKESDCLKTSF 132
Query: 929 RCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQVYIDDIVVKSPSRDDQLL 988
P G Y +V MPFGLKNAGATYQR++ F I ++ Y+DD+VVK+ +DD +
Sbjct: 133 ITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVEAYVDDVVVKTKQKDDLIS 190
Query: 989 HLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEINKNKAKAILDTNPPTSKK 1048
L ++F +R + +K+NP KC FGV +G LGF+V +GI+ N K AIL+ PP+++K
Sbjct: 191 DLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQANPEKVTAILNMKPPSTQK 250
Query: 1049 QLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEAEHQKAFDELKAYLSSPPV 1108
+Q L G + L RF+ L E+ F LL KK D F+W E QKAF++ K L+ PPV
Sbjct: 251 DVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDDFQWGPEAQKAFEDFKKLLTEPPV 308
Query: 1109 MAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAIFYLSRVLNDAETRYTMIE 1163
+A P +P+ LY+SAT+ + ++L E E +R I+++S VL D++TRY ++
Sbjct: 309 LASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPIYFVSEVLADSKTRYPQVQ 368
Query: 1164 KLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHSRIGKWALALTEYSLTYAP 1223
KL + + KL +Y + V V + + + +L + RI KWAL L +++ P
Sbjct: 369 KLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDILHNREANGRIAKWALELMSLDISFKP 427
Query: 1224 LKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKNGTGIGMFIVSPGGTPTKF 1283
++K QA+ADF+ + T +E ++ W + FDGS +GTG G+ ++SP G +
Sbjct: 428 RISIKSQALADFVAEWTECQEDTPAEKMEHWTMHFDGSKRLSGTGAGVVLISPTGERLSY 487
Query: 1284 KFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELVIKQLTKEYKCISENLAKY 1343
I S+N AEYEAL+ L I I+LG + ++V+GDS+LV+ Q+ KE+ C+ +N+ Y
Sbjct: 488 VLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLVVNQVMKEWSCLDDNMMAY 547
Query: 1344 YTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASGYMVDKYRLKELIEVKEKL 1397
+ L KFD L HV R +N+ A+ LA ++A + ++ + K+
Sbjct: 548 RQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREVAPSDVFVEHLYTPTVPHKDTT 607
Query: 1398 NPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTKYRAMSYVIMGNELFKKNV 1453
+ + + M DWR+P++ +L + D+ + R+ YV+ EL+KK+
Sbjct: 608 QVAGTHDVA---MVAADWREPLIRFLTSQELPQDKDEAERISRRSKLYVMHEAELYKKSP 664
Query: 1454 DGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQGMYWPTIMKDCMEYAKGCQ 1513
G L +C+S ++ + +H G+CG H A + +RQG +WPT + D + + C+
Sbjct: 665 SGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQGFFWPTAVSDADKIVRTCE 724
Query: 1514 DCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSRQHKYIIVAIDYFTKWVEA 1573
CQ A H+PA EL +I WPF W LD++G + ++ VAID F+KW+EA
Sbjct: 725 GCQFFARQIHLPAQELQTIPLSWPFVVWGLDMVGPFKKAVG-GYTHLFVAIDKFSKWIEA 783
Query: 1574 IPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKVASFTESWGIKLLTSTPYY 1633
P+ +T D DF N IV+RFG+P + TD G F G F E +GIK+ ++ +
Sbjct: 784 KPVVTITADNARDFFIN-IVHRFGVPNRIITDNGRQFTGGVFKDFCEDFGIKICYASVAH 842
Query: 1634 AQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
+NGQVE AN ++ IK + + LK W
Sbjct: 843 PMSNGQVERANGMILQGIKARVFDRLKPYAGKW 875
>UniRef100_Q8GSW6 Putative gag-pol polyprotein [Oryza sativa]
Length = 2012
Score = 591 bits (1523), Expect = e-167
Identities = 333/891 (37%), Positives = 502/891 (55%), Gaps = 32/891 (3%)
Query: 798 AWPQSRIGGI-------ELPIKEDKKPVKQLPRRFHPDVLVKIKEEIERLLRCKFIRTAR 850
AW S + GI L +KED KP+KQ RRF D IKEE+ +LL FI+
Sbjct: 975 AWKPSDMPGIPREVIEHSLHVKEDAKPIKQRLRRFAQDRKDAIKEELTKLLAAGFIKEVL 1034
Query: 851 YVDWLANVVPVIKKNGKMRVCIDFRDLNAATPKDEYHMPIAEMMVDSAAGHEYLSLLDGY 910
+ DWLAN V V KK G+ +C+D+ DLN + PKD + +P + +VDS AG E LS LD Y
Sbjct: 1035 HPDWLANPVLVRKKTGQWLMCVDYTDLNKSCPKDPFGLPRIDQVVDSTAGCELLSFLDCY 1094
Query: 911 SGYNQIFIAEEDVSKTAFRCPGALGTYEWVVMPFGLKNAGATYQRVMNTIFHDFIETFMQ 970
SGY+QI + E D KT+F P G Y +V MPFGLKNAGATYQR++ F I ++
Sbjct: 1095 SGYHQIRLKESDCLKTSFITP--FGAYCYVTMPFGLKNAGATYQRMIQRCFSTQIGRNVE 1152
Query: 971 VYIDDIVVKSPSRDDQLLHLRKSFERMRKYGLKMNPLKCAFGVIAGDFLGFVVHKKGIEI 1030
Y+DD+VVK+ +DD + L ++F +R + +K+NP KC FGV +G LGF+V +GI+
Sbjct: 1153 AYVDDVVVKTKQKDDLISDLEETFASIRAFRMKLNPEKCTFGVPSGKLLGFMVSHRGIQA 1212
Query: 1031 NKNKAKAILDTNPPTSKKQLQSLLGKINFLRRFIVNLSEKTKSFSPLLRLKKEDTFRWEA 1090
N K AIL+ P+++K +Q L G + L RF+ L E+ F LL KK D+FRW
Sbjct: 1213 NPEKVTAILNMKSPSTQKDVQKLTGCMAALSRFVSRLGERGMPFFKLL--KKTDSFRWGP 1270
Query: 1091 EHQKAFDELKAYLSSPPVMAPPIGGKPMKLYISATNGTIGSMLAQEDENSK-----ERAI 1145
E QKAF++ K L+ PPV+A P +P+ LY+SAT+ + ++L E E +R I
Sbjct: 1271 EAQKAFEDFKKLLTEPPVLASPHPQEPLLLYVSATSQVVSTVLVVEREEEGHVQKVQRPI 1330
Query: 1146 FYLSRVLNDAETRYTMIEKLCLCLYFSCVKLKYYIKPIDVMVFSHYDIIKHMLSKPILHS 1205
+++S VL D++TRY ++KL + + KL +Y + V V + + + +L +
Sbjct: 1331 YFVSEVLADSKTRYPQVQKLLYGILITTRKLSHYFQGHSVTVVTSFPL-GDILHNREANG 1389
Query: 1206 RIGKWALALTEYSLTYAPLKAVKGQAVADFLVDHTLPKEIVTYVGVQPWKLFFDGSSHKN 1265
RI KWAL L +++ P ++K QA+ADF+ + T +E ++ W + FDGS +
Sbjct: 1390 RIAKWALELMSLDISFKPRISIKSQALADFVAEWTECQEDTPAENMEHWTMHFDGSKRLS 1449
Query: 1266 GTGIGMFIVSPGGTPTKFKFRIKKKCSNNEAEYEALISSLEILIALGARNVVVKGDSELV 1325
GTG G+ ++SP G + I S+N AEYEAL+ L I I+LG + ++V+GDS+LV
Sbjct: 1450 GTGAGVVLISPTGERLSYVLWIHFSASHNVAEYEALLHGLRIAISLGIKRLIVRGDSQLV 1509
Query: 1326 IKQLTKEYKCISENLAKYYTKANNLLAKFDDARLGHVSRIDNQEANELA------QIASG 1379
+ Q+ KE+ C+ +N+ Y + L KFD L HV R +N+ A+ LA + A
Sbjct: 1510 VNQVMKEWSCLDDNMMAYRQEVRKLEDKFDGLELSHVLRHNNEAADRLANFGSKREAAPS 1569
Query: 1380 YMVDKYRLKELIEVKEKLNPSDLNVLVIDNMAPKDWRKPIVDYLQNPVGTTDR----KTK 1435
+ ++ + K+ + + + M DWR+P++ +L + D+ +
Sbjct: 1570 DVFVEHLYSPTVPHKDATQAAGAHDVA---MVEADWREPLIRFLTSQELPQDKDEAERIS 1626
Query: 1436 YRAMSYVIMGNELFKKNVDGTLLKCLSDDDALIAISAVHDGLCGAHQAGIKMKWILFRQG 1495
R+ YV+ EL+KK+ G L +C+S ++ + +H G+CG H A + +RQG
Sbjct: 1627 RRSKLYVLHEAELYKKSPSGILQRCVSLEEGRQLLKDIHSGICGNHAAARTIVGKAYRQG 1686
Query: 1496 MYWPTIMKDCMEYAKGCQDCQRHAGIQHVPASELHSIIKPWPFRGWALDLIGEINPCSSR 1555
+WPT + D + + C+ CQ A H+PA EL +I WPF W LD++G
Sbjct: 1687 FFWPTAVSDADKIVRTCEGCQFFARQIHLPAQELQTIPLSWPFAVWGLDMVGPFKKAVG- 1745
Query: 1556 QHKYIIVAIDYFTKWVEAIPLQNVTQDTVIDFIQNHIVYRFGLPESLTTDQGTVFVGQKV 1615
+ ++ VAID F+KW+EA P+ +T D DF N IV+RFG+P + TD GT F G
Sbjct: 1746 GYTHLFVAIDKFSKWIEAKPVVTITADNARDFFIN-IVHRFGVPNRIITDNGTQFTGGVF 1804
Query: 1616 ASFTESWGIKLLTSTPYYAQANGQVEAANKTLISLIKNILVENLKDGIKHW 1666
F E +GIK+ ++ + +NGQVE AN ++ IK + + LK W
Sbjct: 1805 KDFCEDFGIKICYASVAHPMSNGQVERANGMILQGIKARVFDRLKPYAGKW 1855
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.325 0.140 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,864,466,048
Number of Sequences: 2790947
Number of extensions: 127407078
Number of successful extensions: 489766
Number of sequences better than 10.0: 28833
Number of HSP's better than 10.0 without gapping: 2966
Number of HSP's successfully gapped in prelim test: 25873
Number of HSP's that attempted gapping in prelim test: 447024
Number of HSP's gapped (non-prelim): 37660
length of query: 1666
length of database: 848,049,833
effective HSP length: 141
effective length of query: 1525
effective length of database: 454,526,306
effective search space: 693152616650
effective search space used: 693152616650
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 82 (36.2 bits)
Lotus: description of TM0240.1


