

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0238.4
(339 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
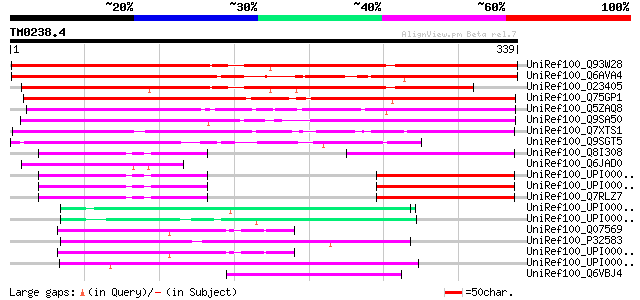
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q93W28 AT4g15540/dl3810w [Arabidopsis thaliana] 375 e-103
UniRef100_Q6AVA4 Hypothetical protein OJ1174_H11.9 [Oryza sativa] 318 2e-85
UniRef100_O23405 DNA chromosome 4, ESSA I CONTIG fragment NO. 4 ... 317 3e-85
UniRef100_Q75GP1 Expressed protein [Oryza sativa] 234 3e-60
UniRef100_Q5ZAQ8 Hypothetical protein B1143G03.16 [Oryza sativa] 221 2e-56
UniRef100_Q9SA50 F3O9.32 protein [Arabidopsis thaliana] 219 8e-56
UniRef100_Q7XTS1 OSJNBa0008M17.14 protein [Oryza sativa] 213 6e-54
UniRef100_Q9SGT5 T6H22.12 protein [Arabidopsis thaliana] 100 1e-19
UniRef100_Q8I308 Hypothetical protein PFI0705w [Plasmodium falci... 83 9e-15
UniRef100_Q6JAD0 Hypothetical protein [Zea mays] 80 8e-14
UniRef100_UPI00004675C5 UPI00004675C5 UniRef100 entry 80 1e-13
UniRef100_UPI000046BD7C UPI000046BD7C UniRef100 entry 79 1e-13
UniRef100_Q7RLZ7 Hypothetical protein [Plasmodium yoelii yoelii] 79 2e-13
UniRef100_UPI0000363EBD UPI0000363EBD UniRef100 entry 52 3e-05
UniRef100_UPI000042DF73 UPI000042DF73 UniRef100 entry 52 3e-05
UniRef100_Q07569 Myosin heavy chain [Entamoeba histolytica] 51 4e-05
UniRef100_P32583 Suppressor protein SRP40 [Saccharomyces cerevis... 51 5e-05
UniRef100_UPI00004983A5 UPI00004983A5 UniRef100 entry 50 7e-05
UniRef100_UPI0000362A11 UPI0000362A11 UniRef100 entry 50 9e-05
UniRef100_Q6VBJ4 Epa5p [Candida glabrata] 49 3e-04
>UniRef100_Q93W28 AT4g15540/dl3810w [Arabidopsis thaliana]
Length = 337
Score = 375 bits (963), Expect = e-103
Identities = 209/340 (61%), Positives = 254/340 (74%), Gaps = 18/340 (5%)
Query: 2 AAESGDLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKD 61
+A +G +FDLPD+L+QVLPSDPFEQLD+ARKITSIALS RV+ALESESS+LR +AEK+
Sbjct: 14 SAITGSRSFDLPDELLQVLPSDPFEQLDVARKITSIALSTRVSALESESSDLRELLAEKE 73
Query: 62 HLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRK 121
ELQS ++SL+A+LS L A+ +KE+LI ENASLSNTV++L RDVSKLE FRK
Sbjct: 74 KEFEELQSHVESLEASLSDAFHKLSLADGEKENLIRENASLSNTVKRLQRDVSKLEGFRK 133
Query: 122 TLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPS--ISDTGN 179
TLM SLQ++D N+ G ++AK D++ PS SS+ S S+
Sbjct: 134 TLMMSLQDDDQNA-GTTQIIAKPTP----------NDDDTPFQPSRHSSIQSQQASEAIE 182
Query: 180 SFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSIS 239
A D+E+DA +P + SL L SQ+TTPRLTPPGSPP LSAS +P TS+P+SPRRHS+S
Sbjct: 183 PAATDNENDAPKPSLSASLPLVSQTTTPRLTPPGSPPILSASGTPKTTSRPISPRRHSVS 242
Query: 240 FATSRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVK 299
FAT+RGM DD S S S S GSQT RTRVDGKEFFRQVR+RLSYEQFGAFL NVK
Sbjct: 243 FATTRGMFDDTRS-----SISISEPGSQTARTRVDGKEFFRQVRSRLSYEQFGAFLGNVK 297
Query: 300 ELNSHKQTREVTLQKADEIFGPENKDLYNIFEGLITRNVH 339
+LN+HKQTRE TL+KA+EIFG +N+DLY IFEGLITRN H
Sbjct: 298 DLNAHKQTREETLRKAEEIFGGDNRDLYVIFEGLITRNAH 337
>UniRef100_Q6AVA4 Hypothetical protein OJ1174_H11.9 [Oryza sativa]
Length = 317
Score = 318 bits (814), Expect = 2e-85
Identities = 184/340 (54%), Positives = 229/340 (67%), Gaps = 45/340 (13%)
Query: 2 AAESGDLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKD 61
AA F L +++ VLP DPFEQLD+ARKITSIAL++R+ LE+E + LRA++AE+D
Sbjct: 21 AAMVAAAEFGLTAEVMAVLPEDPFEQLDVARKITSIALASRLGRLEAEGARLRAQLAERD 80
Query: 62 HLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRK 121
+L +++ LDA L+ L RAE++KE+L +N+ LSNTVR+LNRDV+KLEVF+K
Sbjct: 81 AAAEDLSERVEQLDAALAVATGRLRRAEEEKEALQRDNSLLSNTVRRLNRDVAKLEVFKK 140
Query: 122 TLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSF 181
TLM+SLQE++D + P A+V S+ +S + G VP
Sbjct: 141 TLMQSLQEDEDPANTTPK--ARVSETSNFSSATSVG-------------VP--------- 176
Query: 182 AEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFA 241
RP P+ + L S ++TPR+TPP SPP AS+SP PRRHSIS
Sbjct: 177 ---------RPPRPH-VFLPSYNSTPRVTPPDSPPRSFASISP--------PRRHSISI- 217
Query: 242 TSRGMHDDRTSVFSSMSSSDS--GAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVK 299
TSR + DDR+S +S SS S AGS TGRTRVDGKEFFRQVRNRLSYEQF AFLANVK
Sbjct: 218 TSRNLFDDRSSAYSGHSSVTSPFDAGSHTGRTRVDGKEFFRQVRNRLSYEQFSAFLANVK 277
Query: 300 ELNSHKQTREVTLQKADEIFGPENKDLYNIFEGLITRNVH 339
ELNSHKQTRE TL+KADEIFGP+NKDLY IFEGLITRN+H
Sbjct: 278 ELNSHKQTREDTLRKADEIFGPDNKDLYTIFEGLITRNIH 317
>UniRef100_O23405 DNA chromosome 4, ESSA I CONTIG fragment NO. 4 [Arabidopsis
thaliana]
Length = 576
Score = 317 bits (812), Expect = 3e-85
Identities = 186/318 (58%), Positives = 225/318 (70%), Gaps = 32/318 (10%)
Query: 9 NFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQ 68
+FDLPD+L+QVLPSDPFEQLD+ARKITSIALS RV+ALESESS+LR +AEK+ ELQ
Sbjct: 257 SFDLPDELLQVLPSDPFEQLDVARKITSIALSTRVSALESESSDLRELLAEKEKEFEELQ 316
Query: 69 SQLDSLDATLSATADNLHRAEQDK-----ESLINENASLSNTVRKLNRDVSKLEVFRKTL 123
S ++SL+A+LS L A+ +K E+LI ENASLSNTV++L RDVSKLE FRKTL
Sbjct: 317 SHVESLEASLSDAFHKLSLADGEKVRPDPENLIRENASLSNTVKRLQRDVSKLEGFRKTL 376
Query: 124 MRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPS--ISDTGNSF 181
M SLQ++D N+ G ++AK D++ PS SS+ S S+
Sbjct: 377 MMSLQDDDQNA-GTTQIIAKPTP----------NDDDTPFQPSRHSSIQSQQASEAIEPA 425
Query: 182 AEDHESDAI---------RPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVS 232
A D+E+D I +P + SL L SQ+TTPRLTPPGSPP LSAS +P TS+P+S
Sbjct: 426 ATDNENDGIFVLCYIVAPKPSLSASLPLVSQTTTPRLTPPGSPPILSASGTPKTTSRPIS 485
Query: 233 PRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFG 292
PRRHS+SFAT+RGM DD S S S S GSQT RTRVDGKEFFRQVR+RLSYEQFG
Sbjct: 486 PRRHSVSFATTRGMFDDTRS-----SISISEPGSQTARTRVDGKEFFRQVRSRLSYEQFG 540
Query: 293 AFLANVKELNSHKQTREV 310
AFL NVK+LN+HKQTREV
Sbjct: 541 AFLGNVKDLNAHKQTREV 558
>UniRef100_Q75GP1 Expressed protein [Oryza sativa]
Length = 336
Score = 234 bits (596), Expect = 3e-60
Identities = 143/338 (42%), Positives = 205/338 (60%), Gaps = 19/338 (5%)
Query: 10 FDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQS 69
F LPD+++ V+P DP+EQLDLAR+IT++A++ RV LE E++ LR A+KD EL+
Sbjct: 8 FALPDEVLAVMPRDPYEQLDLARRITALAVAGRVTGLEREAARLRESAADKDRENGELRE 67
Query: 70 QLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQE 129
++ LD L T L A +D L E SL+ T +KL RD+ KLE F++ LM+SL++
Sbjct: 68 RVALLDRALQETNSRLRAALEDNIKLSKERDSLAQTSKKLARDLQKLESFKRHLMQSLRD 127
Query: 130 EDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDA 189
+ + D+ QS SS S+ GD S+ + ++ + + D G++ E +
Sbjct: 128 DSPSPQETVDITTCDQSISSKASSCGDGD---SITHTTTNLLSTSLDVGSTVQEVSKP-- 182
Query: 190 IRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDD 249
P Y+L S TPRLTP +P +S S SP R S +P+ S + + S+ +
Sbjct: 183 --PIQKYAL---SSHITPRLTPEATPKIMSTSASPRRMSTTATPKLMSGTTSPSKTRIEG 237
Query: 250 RTSVF--------SSMSSSDSGAGSQTGRT-RVDGKEFFRQVRNRLSYEQFGAFLANVKE 300
S+ SS ++S GRT R+DGKEFFRQ R+RLSYEQFGAFLAN+KE
Sbjct: 238 YMSMTPWYPSSKQSSAANSPPRGRPNPGRTPRIDGKEFFRQARSRLSYEQFGAFLANIKE 297
Query: 301 LNSHKQTREVTLQKADEIFGPENKDLYNIFEGLITRNV 338
LN+HKQ+RE TL+KA+EIFGP+NKDLY F+GL+ R++
Sbjct: 298 LNAHKQSREDTLKKAEEIFGPDNKDLYLSFQGLLNRSL 335
>UniRef100_Q5ZAQ8 Hypothetical protein B1143G03.16 [Oryza sativa]
Length = 345
Score = 221 bits (563), Expect = 2e-56
Identities = 144/335 (42%), Positives = 202/335 (59%), Gaps = 21/335 (6%)
Query: 12 LPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQL 71
LPD + LP DP+EQL++ARKIT++A++AR + LE E++ LR ++A+KD L AEL +
Sbjct: 23 LPDAIAAALPPDPYEQLEVARKITAVAVAARASRLELEAARLRQKLADKDRLAAELADRA 82
Query: 72 DSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEED 131
SL+ L + L A D L E SL++T +KL RD++KLE F++ LM+SL D
Sbjct: 83 ASLEQALRDSDARLRAALDDNAKLAKERDSLAHTSKKLARDLAKLETFKRHLMQSLG--D 140
Query: 132 DNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIR 191
DN P+ V + S+ S + D A V S++D G++ E + R
Sbjct: 141 DNP---PETVDIRTCEQSVAKASSWKDGVAH--SRHHHPVSSLAD-GSTEIESVNQEVAR 194
Query: 192 PRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDR- 250
P + L+ +PRLT + + +A+ SP R S VSP+ + ATS +
Sbjct: 195 P---FEQKLSVTHISPRLTSDPAAKTRTAATSPRRYSTAVSPKL--AASATSPRLEGHMA 249
Query: 251 ------TSVFSSMSSSDSGAGSQTGRT-RVDGKEFFRQVRNRLSYEQFGAFLANVKELNS 303
+S SS ++S A S +GRT RVDGKEFFRQ RNRLSYEQF AFLAN+KELN+
Sbjct: 250 MQPWLPSSKMSSAANSPPRAHSISGRTTRVDGKEFFRQARNRLSYEQFAAFLANIKELNA 309
Query: 304 HKQTREVTLQKADEIFGPENKDLYNIFEGLITRNV 338
H+Q+RE TLQKADEIFG ENKDL+ F+ L++R++
Sbjct: 310 HRQSREETLQKADEIFGSENKDLFMSFQSLLSRSL 344
>UniRef100_Q9SA50 F3O9.32 protein [Arabidopsis thaliana]
Length = 325
Score = 219 bits (558), Expect = 8e-56
Identities = 137/344 (39%), Positives = 198/344 (56%), Gaps = 38/344 (11%)
Query: 8 LNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAEL 67
L+F+LP++++ V+P DPFEQLDLARKITS+A+++RV+ L+SE ELR ++ K+ ++ EL
Sbjct: 6 LDFELPEEVLSVIPMDPFEQLDLARKITSMAIASRVSNLDSEVVELRQKLLGKESVVREL 65
Query: 68 QSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSL 127
+ + L+ L +D +L E SL+ TV KL RD++KLE F++ L++SL
Sbjct: 66 EEKASRLERDCREADSRLKVVLEDNMNLTKEKDSLAMTVTKLTRDLAKLETFKRQLIKSL 125
Query: 128 QEED------------DNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSIS 175
+E D G P ++ + S + S G + + P +S
Sbjct: 126 SDESGPQTEPVDIRTCDQPGSYPGKDGRINAHSIKQAYS--GSTDTNNPVVEASKY---- 179
Query: 176 DTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRR 235
TGN F+ + +PRLTP +P +S SVSP S SP+R
Sbjct: 180 -TGNKFS------------------MTSYISPRLTPTATPKIISTSVSPRGYSAAGSPKR 220
Query: 236 HSISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGRT-RVDGKEFFRQVRNRLSYEQFGAF 294
S + + ++ +S SS ++S + RT R+DGKEFFRQ R+RLSYEQF +F
Sbjct: 221 TSGAVSPTKATLWYPSSQQSSAANSPPRNRTLPARTPRMDGKEFFRQARSRLSYEQFSSF 280
Query: 295 LANVKELNSHKQTREVTLQKADEIFGPENKDLYNIFEGLITRNV 338
LAN+KELN+ KQTRE TL+KADEIFG ENKDLY F+GL+ RN+
Sbjct: 281 LANIKELNAQKQTREETLRKADEIFGEENKDLYLSFQGLLNRNM 324
>UniRef100_Q7XTS1 OSJNBa0008M17.14 protein [Oryza sativa]
Length = 312
Score = 213 bits (542), Expect = 6e-54
Identities = 133/335 (39%), Positives = 198/335 (58%), Gaps = 30/335 (8%)
Query: 3 AESGDLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDH 62
A +G ++F LPD+++ V+P+DP+EQLD+ARKITS+A+++RV+ LE++++ LR +A++D
Sbjct: 6 AAAGVVDFHLPDEILAVIPTDPYEQLDVARKITSMAIASRVSRLEADAARLRRDLADRDR 65
Query: 63 LIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKT 122
A+L+++L DA L A D + L E SL++T +K+ R+++KLE F+K
Sbjct: 66 AEADLRARLADSDARLLAALD-------ENAKLAKERDSLASTAKKMARNLAKLEAFKKQ 118
Query: 123 LMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFA 182
LM+SL E++ + ++LT+ +E S S +S+ S T A
Sbjct: 119 LMKSLSEDNLLQLSEIGDDRDFDANNNLTARVPSWKDEVS--SSRTSADSSSRSTMTESA 176
Query: 183 EDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFAT 242
++H+ PY T P+LTP +P LS SPT++ V HS T
Sbjct: 177 QEHQFSV----TPY--------TAPKLTPGSTPKFLSGPTSPTKSLSEV----HS----T 216
Query: 243 SRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELN 302
H + +S+ +S S GR R+DGKEFFRQ R RLSYEQFGAFLAN+KE N
Sbjct: 217 FSSWHGSSSHQYSAPTSPPQHR-SFAGRPRIDGKEFFRQARTRLSYEQFGAFLANIKEFN 275
Query: 303 SHKQTREVTLQKADEIFGPENKDLYNIFEGLITRN 337
+ KQ+RE TL KA+EIFG E+KDLY F+ ++ RN
Sbjct: 276 AQKQSREDTLSKAEEIFGTEHKDLYISFQNMLNRN 310
>UniRef100_Q9SGT5 T6H22.12 protein [Arabidopsis thaliana]
Length = 309
Score = 99.8 bits (247), Expect = 1e-19
Identities = 86/280 (30%), Positives = 132/280 (46%), Gaps = 46/280 (16%)
Query: 1 MAAESGDLNFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEK 60
M+ GD F+L D+++ V+P+DP++QLDLARKITS+A+++RV+ LES+ S LR ++ EK
Sbjct: 1 MSQSGGD--FNLSDEILAVIPTDPYDQLDLARKITSMAIASRVSNLESQVSGLRQKLLEK 58
Query: 61 DHLIAELQSQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFR 120
D L+ EL+ ++ S + +L + L E SL+ T +KL RD +K
Sbjct: 59 DRLVHELEDRVSSFERLYHEADSSLKNVVDENMKLTQERDSLAITAKKLGRDYAK----- 113
Query: 121 KTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGN- 179
Q E + P KV S+ + + V SV + + N
Sbjct: 114 -------QTETADVRMVPR--GKVSSRVACS--------------DVIYSVCIVDENSNG 150
Query: 180 SFAEDHESDAIRPRVPYSLLLASQSTTPR----LTPPGSPPSLSASVSPTRTSKPVSPRR 235
S++ + R R QS TP+ TP G+P LS + SP S SP+
Sbjct: 151 SYSNNEGLSEARQR---------QSMTPQFSPAFTPSGTPKILSTAASPRSYSAASSPKL 201
Query: 236 HSISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVDG 275
S + + + +D R ++SS S S A S V G
Sbjct: 202 FSGAASPTSSHYDIR--MWSSTSQQSSVANSPPRSHSVSG 239
>UniRef100_Q8I308 Hypothetical protein PFI0705w [Plasmodium falciparum]
Length = 249
Score = 83.2 bits (204), Expect = 9e-15
Identities = 43/112 (38%), Positives = 65/112 (57%)
Query: 226 RTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNR 285
RT ++ S ++ ++ D ++ + +S G DG+ FFR R+R
Sbjct: 137 RTMLEMNGNEDSCQSIINKIVNSDTHNIINGSFNSPYIGNVPLGEKNTDGRAFFRNARSR 196
Query: 286 LSYEQFGAFLANVKELNSHKQTREVTLQKADEIFGPENKDLYNIFEGLITRN 337
LSYEQF FL+N+K+LN+H+Q RE TL+KA IFG N DLY F+ LI+++
Sbjct: 197 LSYEQFNQFLSNIKKLNNHQQKREETLKKAQAIFGEANLDLYEEFKVLISKH 248
Score = 71.6 bits (174), Expect = 3e-11
Identities = 41/113 (36%), Positives = 64/113 (56%), Gaps = 7/113 (6%)
Query: 20 LPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLS 79
LPSD EQL L KI + A RV + E+E L+ ++ EK ++ +Q + +L+ L
Sbjct: 12 LPSDADEQLALGFKIVTNAYKTRVTSQEAEIRSLKGQLTEKQEQLSSIQKKYSNLEVQL- 70
Query: 80 ATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDD 132
++ R Q L +EN L T++KLNRD+ +LE +K ++ S+QEE D
Sbjct: 71 --IESTQRGNQ----LADENKQLITTIKKLNRDIDRLENLKKAVLNSIQEEHD 117
>UniRef100_Q6JAD0 Hypothetical protein [Zea mays]
Length = 161
Score = 80.1 bits (196), Expect = 8e-14
Identities = 52/136 (38%), Positives = 73/136 (53%), Gaps = 28/136 (20%)
Query: 9 NFDLPDDLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQ 68
+F LPD+L+ LP DP+EQLDLAR+IT++A++ RV+ LE E+ LRA A KD AEL+
Sbjct: 10 DFALPDELLAALPRDPYEQLDLARRITALAVAGRVSGLEREAGRLRAEAAGKDRESAELR 69
Query: 69 SQLDSLDATLSAT--------ADNLHRAEQD--------------------KESLINENA 100
++ LDA L T DN+ RA + + L E
Sbjct: 70 ERVALLDAALQETNARLRAALEDNVGRARPEFNLSAFASVWVSETGACVLVQIKLSKERD 129
Query: 101 SLSNTVRKLNRDVSKL 116
SL+ T +KL RD+ K+
Sbjct: 130 SLAQTSKKLARDLHKV 145
>UniRef100_UPI00004675C5 UPI00004675C5 UniRef100 entry
Length = 249
Score = 79.7 bits (195), Expect = 1e-13
Identities = 40/92 (43%), Positives = 58/92 (62%)
Query: 246 MHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHK 305
++ D S+ + +S + DGK FFR R+RL+YEQF FL+N+K+LN+H+
Sbjct: 157 VNSDAHSIINGNFNSPYLGNTPVIEKNTDGKAFFRNARSRLTYEQFNQFLSNIKKLNNHQ 216
Query: 306 QTREVTLQKADEIFGPENKDLYNIFEGLITRN 337
Q RE TL+KA IFG N DLY F+ LI+++
Sbjct: 217 QKREETLKKAQIIFGETNADLYEEFKILISKH 248
Score = 68.2 bits (165), Expect = 3e-10
Identities = 41/113 (36%), Positives = 63/113 (55%), Gaps = 7/113 (6%)
Query: 20 LPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLS 79
LPSD EQL L KI + A RV + E+E L+ ++ EK ++ +Q + +L+ L
Sbjct: 12 LPSDADEQLALGFKIVTNAYKTRVTSQEAEIRSLKGQLTEKLEQLSSIQKKYSNLEVQL- 70
Query: 80 ATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDD 132
++ R Q L EN L T++KLNRD+ +LE +K ++ S+QEE D
Sbjct: 71 --IESTQRGNQ----LSEENKQLIFTIKKLNRDIDRLENLKKAVLNSIQEEHD 117
>UniRef100_UPI000046BD7C UPI000046BD7C UniRef100 entry
Length = 249
Score = 79.3 bits (194), Expect = 1e-13
Identities = 39/92 (42%), Positives = 58/92 (62%)
Query: 246 MHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHK 305
++ D S+ + +S + DGK FFR R+RL+YEQF FL+N+K+LN+H+
Sbjct: 157 VNSDAHSIINGNFNSPYLGNTSVIEKNTDGKAFFRNARSRLTYEQFNQFLSNIKKLNNHQ 216
Query: 306 QTREVTLQKADEIFGPENKDLYNIFEGLITRN 337
Q R+ TL+KA IFG N DLY F+ LI+++
Sbjct: 217 QKRDETLKKAQIIFGETNADLYEEFKVLISKH 248
Score = 68.2 bits (165), Expect = 3e-10
Identities = 41/113 (36%), Positives = 63/113 (55%), Gaps = 7/113 (6%)
Query: 20 LPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLS 79
LPSD EQL L KI + A RV + E+E L+ ++ EK ++ +Q + +L+ L
Sbjct: 12 LPSDADEQLALGFKIVTNAYKTRVTSQEAEIRSLKGQLTEKLEQLSSIQKKYSNLEVQL- 70
Query: 80 ATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDD 132
++ R Q L EN L T++KLNRD+ +LE +K ++ S+QEE D
Sbjct: 71 --IESTQRGNQ----LSEENKQLIFTIKKLNRDIDRLENLKKAVLNSIQEEHD 117
>UniRef100_Q7RLZ7 Hypothetical protein [Plasmodium yoelii yoelii]
Length = 249
Score = 79.0 bits (193), Expect = 2e-13
Identities = 39/92 (42%), Positives = 58/92 (62%)
Query: 246 MHDDRTSVFSSMSSSDSGAGSQTGRTRVDGKEFFRQVRNRLSYEQFGAFLANVKELNSHK 305
++ D S+ + +S + DGK FFR R+RL+YEQF FL+N+K+LN+H+
Sbjct: 157 VNSDAHSIINGNFNSPYLGNTPVIEKNTDGKAFFRNARSRLTYEQFNQFLSNIKKLNNHQ 216
Query: 306 QTREVTLQKADEIFGPENKDLYNIFEGLITRN 337
Q R+ TL+KA IFG N DLY F+ LI+++
Sbjct: 217 QKRDETLKKAQIIFGETNADLYEEFKVLISKH 248
Score = 68.2 bits (165), Expect = 3e-10
Identities = 41/113 (36%), Positives = 63/113 (55%), Gaps = 7/113 (6%)
Query: 20 LPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLS 79
LPSD EQL L KI + A RV + E+E L+ ++ EK ++ +Q + +L+ L
Sbjct: 12 LPSDADEQLALGFKIVTNAYKTRVTSQEAEIRSLKGQLTEKLEQLSSIQKKYSNLEVQL- 70
Query: 80 ATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDD 132
++ R Q L EN L T++KLNRD+ +LE +K ++ S+QEE D
Sbjct: 71 --IESTQRGNQ----LSEENKQLIFTIKKLNRDIDRLENLKKAVLNSIQEEHD 117
>UniRef100_UPI0000363EBD UPI0000363EBD UniRef100 entry
Length = 287
Score = 51.6 bits (122), Expect = 3e-05
Identities = 57/243 (23%), Positives = 95/243 (38%), Gaps = 7/243 (2%)
Query: 35 TSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKES 94
+S+A S+ V++ + SS + A + S ++ S+++ + S
Sbjct: 13 SSLASSSSVSSSSASSSSASSSSASSSSTSSSSASSSSPSSSSASSSSTSSSSTSSSSAS 72
Query: 95 LINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQS------QS 148
++S S++ + D S S D+S A A S S
Sbjct: 73 SSASSSSASSSSTSSSSDSSSSASSSSASSSSTSSSSDSSSSASSSSASSSSASSSSASS 132
Query: 149 SLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPR 208
S TS+S + AS + SSS S SD+ +S + S + S AS S+
Sbjct: 133 SSTSSSSDSSSSASSSSASSSSTSSSSDSSSSASSSSASSSSASSSSASSSSASSSSASS 192
Query: 209 LTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQT 268
+ SP + S+S S + TS + + S A+S TS S SSS S + + +
Sbjct: 193 -SSASSPSASSSSASSSSTSSSSTSSSSASSSASSSSASSSSTSSSSDSSSSASSSSASS 251
Query: 269 GRT 271
T
Sbjct: 252 SST 254
Score = 47.0 bits (110), Expect = 7e-04
Identities = 51/234 (21%), Positives = 92/234 (38%), Gaps = 5/234 (2%)
Query: 35 TSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKES 94
+S++ S+ ++L S SS ++ + S S +T S++A + + S
Sbjct: 4 SSVSSSSTSSSLASSSS-----VSSSSASSSSASSSSASSSSTSSSSASSSSPSSSSASS 58
Query: 95 LINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTS 154
++S S++ + S + +S + + SS S+S
Sbjct: 59 SSTSSSSTSSSSASSSASSSSASSSSTSSSSDSSSSASSSSASSSSTSSSSDSSSSASSS 118
Query: 155 QFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGS 214
+ AS + SSS S SD+ +S + S + S AS S+ + S
Sbjct: 119 SASSSSASSSSASSSSTSSSSDSSSSASSSSASSSSTSSSSDSSSSASSSSASSSSASSS 178
Query: 215 PPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQT 268
S S++ S + +S S S S A+S TS S+ SS+ S + S +
Sbjct: 179 SASSSSASSSSASSSSASSPSASSSSASSSSTSSSSTSSSSASSSASSSSASSS 232
Score = 41.6 bits (96), Expect = 0.031
Identities = 54/226 (23%), Positives = 90/226 (38%), Gaps = 11/226 (4%)
Query: 35 TSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKES 94
+S + S+ ++ S SS+ + + + S DS + S++A + + S
Sbjct: 73 SSASSSSASSSSTSSSSDSSSSASSSSASSSSTSSSSDSSSSASSSSASSSSASSSSASS 132
Query: 95 LINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTS 154
++S S++ + S T S +S A A S SS +++S
Sbjct: 133 SSTSSSSDSSSSASSSSASSS-----STSSSSDSSSSASSSSASSSSASSSSASSSSASS 187
Query: 155 QFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGS 214
+ ++ PS SSS S S T +S + + S AS S+T + S
Sbjct: 188 SSASSSSASSPSASSSSASSSSTSSSSTSSSSASS-----SASSSSASSSSTSSSSDSSS 242
Query: 215 PPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSS 260
S S++ S + +S S S S A+S TS F S SSS
Sbjct: 243 SASSSSASSSSTSSSSDSSSSASSSSASSSSASSSSTS-FPSASSS 287
Score = 39.3 bits (90), Expect = 0.16
Identities = 44/182 (24%), Positives = 74/182 (40%), Gaps = 4/182 (2%)
Query: 88 AEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQ 147
A S + ++SL+++ + S + S S +P + S
Sbjct: 1 ASSSSVSSSSTSSSLASSSSVSSSSASSSSASSSSASSSSTSSSSASSSSPSSSSASSSS 60
Query: 148 SSLTST-SQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTT 206
+S +ST S + AS + SSS S SD+ +S + S + S AS S+
Sbjct: 61 TSSSSTSSSSASSSASSSSASSSSTSSSSDSSSSASSSSASSSSTSSSSDSSSSASSSSA 120
Query: 207 PRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGS 266
+ S S S++ S + +S S S + ++S D +S SS S+S S A S
Sbjct: 121 SSSSASSSSASSSSTSSSSDSSSSAS---SSSASSSSTSSSSDSSSSASSSSASSSSASS 177
Query: 267 QT 268
+
Sbjct: 178 SS 179
Score = 35.8 bits (81), Expect = 1.7
Identities = 34/109 (31%), Positives = 46/109 (42%), Gaps = 2/109 (1%)
Query: 168 SSSVPSISDTGNSFAEDHE--SDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPT 225
SSS S S T +S A S + S +S ST+ SP S SAS S T
Sbjct: 2 SSSSVSSSSTSSSLASSSSVSSSSASSSSASSSSASSSSTSSSSASSSSPSSSSASSSST 61
Query: 226 RTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGRTRVD 274
+S S S + ++S +S SS S+S S A S + + D
Sbjct: 62 SSSSTSSSSASSSASSSSASSSSTSSSSDSSSSASSSSASSSSTSSSSD 110
>UniRef100_UPI000042DF73 UPI000042DF73 UniRef100 entry
Length = 547
Score = 51.6 bits (122), Expect = 3e-05
Identities = 58/241 (24%), Positives = 95/241 (39%), Gaps = 27/241 (11%)
Query: 35 TSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKES 94
TS+++SA + S SS + S + D T S++A + + S
Sbjct: 164 TSLSVSATTTSSSSTSS-------------SSSSSTPSTSDVTTSSSASSSPSSTTSSSS 210
Query: 95 LINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTS 154
++S + T S T S+ N+ + + SS T++S
Sbjct: 211 STAFSSSTTETSSSATSSSS-------TTSSSISSTQSNTSSS----SNTSFSSSTTASS 259
Query: 155 QFGDNEASL---PPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTP 211
F + +S PS ++S SIS T +SF ++ A P S + S +T+ +
Sbjct: 260 SFSSSTSSSFSPSPSSTTSSSSISSTSSSFTTSSDTSASSSVSPSSSVSPSSTTSSSSSF 319
Query: 212 PGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGRT 271
S SL+ + S T +S P S S S + S D TS SS + S + + T
Sbjct: 320 SSSSSSLTITSSSTTSSIPSSSEVSSTSTSASSSSSDSSTSTSSSSETDQSSSSTTQSST 379
Query: 272 R 272
R
Sbjct: 380 R 380
Score = 37.0 bits (84), Expect = 0.77
Identities = 51/245 (20%), Positives = 91/245 (36%), Gaps = 7/245 (2%)
Query: 28 LDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHR 87
L+ +KI + + L S S+ +R + S ++L +T N
Sbjct: 6 LNQPKKILQLKIRENSTDLLSTSTSFSSRTTQGSTTAPSSSSIQQQQTSSLMSTNQNSFS 65
Query: 88 AEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQ 147
+ N + + + T + S T S+ + ++ + SQ
Sbjct: 66 TTTTSQFTSNSSPNSAQTFSSSPENSSSR---MSTPTTSISTQPASTTSIQQSQSLQTSQ 122
Query: 148 SSLTSTSQFGDNEASLPPSVSSSVPS--ISDTGNSFAEDHESD-AIRPRVPYSLLLASQS 204
S S Q ++ S P+ +S PS +SD SF ++ ++ S +S S
Sbjct: 123 QSQPSQQQSQQSQQSQSPTSTSQAPSSTMSDNNTSFVSPSDTSLSVSATTTSSSSTSSSS 182
Query: 205 TTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFA-TSRGMHDDRTSVFSSMSSSDSG 263
++ + S SAS SP+ T+ S S S TS ++ SS+SS+ S
Sbjct: 183 SSSTPSTSDVTTSSSASSSPSSTTSSSSSTAFSSSTTETSSSATSSSSTTSSSISSTQSN 242
Query: 264 AGSQT 268
S +
Sbjct: 243 TSSSS 247
>UniRef100_Q07569 Myosin heavy chain [Entamoeba histolytica]
Length = 2139
Score = 51.2 bits (121), Expect = 4e-05
Identities = 38/162 (23%), Positives = 80/162 (48%), Gaps = 10/162 (6%)
Query: 33 KITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDK 92
K+ L + L S+ + L A + +KD IAE+Q +D + ++ ++ E++K
Sbjct: 958 KLKGEDLEVEITELNSQINTLNATVDKKDKTIAEMQESIDEKEDEITKLKGDIKLLEEEK 1017
Query: 93 ESLINENASLSNT----VRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQS 148
+ L + A +S T +KLN+ + E + + + QE +D D+ ++Q Q+
Sbjct: 1018 DDLEQDRADVSATKDDIAKKLNKITIECEDAKDEIAKLEQELEDEENKNKDLTNELQ-QT 1076
Query: 149 SLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAI 190
L + G+ E SL V+++ SD ++ +++ E++ +
Sbjct: 1077 QL----KLGETEKSLAAQVAAT-KKASDERDTLSQNLENEKL 1113
>UniRef100_P32583 Suppressor protein SRP40 [Saccharomyces cerevisiae]
Length = 406
Score = 50.8 bits (120), Expect = 5e-05
Identities = 59/245 (24%), Positives = 102/245 (41%), Gaps = 17/245 (6%)
Query: 35 TSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKES 94
+S + S+ ++ S SS + D +E S S ++ S+++D+ +E D S
Sbjct: 43 SSSSSSSGESSSSSSSSSSSSSSDSSDSSDSESSSSSSSSSSSSSSSSDSESSSESDSSS 102
Query: 95 LINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTS 154
+ ++S S++ + S+ E ++ E DN AK + +SS +S S
Sbjct: 103 SGSSSSSSSSSDESSSESESEDETKKRA------RESDNEDAKETKKAKTEPESSSSSES 156
Query: 155 QFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPG- 213
+ +S S S S + +S + D ESD+ S +S S++ +
Sbjct: 157 SSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSSSSSSSSDSSSDSDSSSSD 216
Query: 214 ----------SPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSG 263
S S S S S + +S S S ++S D+ TS SS S SDS
Sbjct: 217 SSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDSSSDESTSSDSSDSDSDSD 276
Query: 264 AGSQT 268
+GS +
Sbjct: 277 SGSSS 281
Score = 40.0 bits (92), Expect = 0.091
Identities = 48/183 (26%), Positives = 75/183 (40%), Gaps = 15/183 (8%)
Query: 89 EQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQS 148
E+ S + ++S S++ + S E + S D+S + D + S S
Sbjct: 23 EKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSSSSSSSSSSSSDSSDSS-DSESSSSSSS 81
Query: 149 SLTSTSQFGDNEASLPPSVSSSVPSISDTGN---SFAEDHESDAIRPRVPYSLLLASQST 205
S +S+S D+E+S SSS S S + + S +E D + R S ++ T
Sbjct: 82 SSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSESESEDETKKRARESDNEDAKET 141
Query: 206 TPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAG 265
T P S S +S S + S S + G D S SS SSSDS +
Sbjct: 142 KKAKTEPESSSSSESSSSGS-----------SSSSESESGSESDSDSSSSSSSSSDSESD 190
Query: 266 SQT 268
S++
Sbjct: 191 SES 193
Score = 38.9 bits (89), Expect = 0.20
Identities = 47/173 (27%), Positives = 70/173 (40%), Gaps = 17/173 (9%)
Query: 112 DVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSV 171
+V KL V K + EE +S + + S SS +S+S G++ +S S SSS
Sbjct: 10 EVPKLSVKEKEI-----EEKSSSSSSSSSSSSSSSSSSSSSSSSSGESSSS---SSSSSS 61
Query: 172 PSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPV 231
S SD+ +S D ES + S +S S + + S S S+S S + S
Sbjct: 62 SSSSDSSDS--SDSESSSSSSSSSSSSSSSSDSESSSESDSSSSGSSSSSSSSSDESSSE 119
Query: 232 SPRRHSISFATSRGMHDD-------RTSVFSSMSSSDSGAGSQTGRTRVDGKE 277
S ++D +T SS SS S +GS + G E
Sbjct: 120 SESEDETKKRARESDNEDAKETKKAKTEPESSSSSESSSSGSSSSSESESGSE 172
>UniRef100_UPI00004983A5 UPI00004983A5 UniRef100 entry
Length = 2151
Score = 50.4 bits (119), Expect = 7e-05
Identities = 38/162 (23%), Positives = 80/162 (48%), Gaps = 10/162 (6%)
Query: 33 KITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDK 92
K+ L + L S+ + L A + +KD IAE+Q +D + ++ ++ E++K
Sbjct: 970 KLKGEDLEDEITELNSQINTLNATVDKKDKTIAEMQESIDEKEDEITKLKGDIKLLEEEK 1029
Query: 93 ESLINENASLSNT----VRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQS 148
+ L + A +S T +KLN+ + E + + + QE +D D+ ++Q Q+
Sbjct: 1030 DDLEQDRADVSATKDDIAKKLNKITIECEDAKDEIAKLEQELEDEENKNKDLTNELQ-QT 1088
Query: 149 SLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAI 190
L + G+ E SL V+++ SD ++ +++ E++ +
Sbjct: 1089 QL----KLGETEKSLAAQVAAT-KKASDERDTLSQNLENEKL 1125
>UniRef100_UPI0000362A11 UPI0000362A11 UniRef100 entry
Length = 302
Score = 50.1 bits (118), Expect = 9e-05
Identities = 53/243 (21%), Positives = 104/243 (41%), Gaps = 3/243 (1%)
Query: 34 ITSIALSARVNALESESSELRARIAEKDHLIAE--LQSQLDSLDATLSATADNLHRAEQD 91
I+S + S+ ++ S SS + + I+ I+ + S S+ ++ S+++ + +
Sbjct: 44 ISSSSSSSSSSSSISSSSSISSSISSSSSSISSSRISSSSSSISSSSSSSSSSSSXSSSS 103
Query: 92 KESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLT 151
S + ++S S++ + S + S+ +S + + S SS +
Sbjct: 104 SSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSSSSISSSSSSSSSSISSSSSSSSSSSSS 163
Query: 152 STSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTP 211
S+S + S S SSS S S + +S + + A V S+ A+Q +
Sbjct: 164 SSSSSSSSSISSSSSSSSSSSSSSSSSSSSSSSSSTAAAAAAVAASVAAAAQQQQQQQQH 223
Query: 212 PG-SPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQTGR 270
S S S+S+S + +S +S S S ++S +S SS SSS S + S +
Sbjct: 224 SSISSSSSSSSISSSSSSSSISSSSSSSSSSSSISSSSSSSSSSSSSSSSISSSSSSSSS 283
Query: 271 TRV 273
+ +
Sbjct: 284 SSI 286
Score = 44.3 bits (103), Expect = 0.005
Identities = 38/141 (26%), Positives = 65/141 (45%), Gaps = 4/141 (2%)
Query: 128 QEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHES 187
Q++ +S + + + S SS +S+S + +S+ S+SSS SIS + S S
Sbjct: 28 QQQHSSSSSSSSSSSSISSSSSSSSSSSSISSSSSISSSISSSSSSISSSRIS----SSS 83
Query: 188 DAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMH 247
+I S +S S++ + S S S+S S + +S +S S S ++
Sbjct: 84 SSISSSSSSSSSSSSXSSSSSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSSSSISSSS 143
Query: 248 DDRTSVFSSMSSSDSGAGSQT 268
+S SS SSS S + S +
Sbjct: 144 SSSSSSISSSSSSSSSSSSSS 164
Score = 43.5 bits (101), Expect = 0.008
Identities = 46/203 (22%), Positives = 87/203 (42%), Gaps = 10/203 (4%)
Query: 69 SQLDSLDATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQ 128
S S ++ S+++ +Q + S + ++S S+++ + S + S
Sbjct: 7 SSSSSSSSSSSSSSSTAAAQQQQQHSSSSSSSSSSSSISSSSSSSSSSSSISSSSSISSS 66
Query: 129 EEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESD 188
+S + ++ S SS++S+S + +S S SSS S S + +S + S
Sbjct: 67 ISSSSSSISSSRISS--SSSSISSSSSSSSSSSSXSSSSSSSSSSSSSSSSSSSSSSSSS 124
Query: 189 AIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHD 248
S + +S S++ + S S S+S+S + +S S S S ++S
Sbjct: 125 --------SSISSSSSSSSSSSISSSSSSSSSSISSSSSSSSSSSSSSSSSSSSSSISSS 176
Query: 249 DRTSVFSSMSSSDSGAGSQTGRT 271
+S SS SSS S + S + T
Sbjct: 177 SSSSSSSSSSSSSSSSSSSSSST 199
Score = 42.0 bits (97), Expect = 0.024
Identities = 48/239 (20%), Positives = 96/239 (40%), Gaps = 9/239 (3%)
Query: 35 TSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKES 94
T+ A + ++ S SS + I+ + S S+ ++ S ++ + S
Sbjct: 22 TAAAQQQQQHSSSSSSSSSSSSISSS----SSSSSSSSSISSSSSISSSISSSSSSISSS 77
Query: 95 LINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTS 154
I+ ++S ++ + S + S +S + + + S SS +S+S
Sbjct: 78 RISSSSSSISSSSSSSSSSSSXSSSSSSSSSSSSSSSSSSSSSSSSSSSISSSSSSSSSS 137
Query: 155 QFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGS 214
+ +S S+SSS S S + +S + S +I S +S S++ + S
Sbjct: 138 SISSSSSSSSSSISSSSSSSSSSSSSSSSSSSSSSISSSSSSSSSSSSSSSSSSSSSSSS 197
Query: 215 PPS-----LSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQT 268
+ ++ASV+ + + SIS ++S +S S SSS S + S +
Sbjct: 198 STAAAAAAVAASVAAAAQQQQQQQQHSSISSSSSSSSISSSSSSSSISSSSSSSSSSSS 256
Score = 40.8 bits (94), Expect = 0.053
Identities = 48/221 (21%), Positives = 91/221 (40%), Gaps = 28/221 (12%)
Query: 48 SESSELRARIAEKDHLIAELQSQLDSLDATLSATADNLHRAEQDKESL--INENASLSNT 105
S SS + + A Q Q S ++ S+++ ++ + S I+ ++S+S++
Sbjct: 7 SSSSSSSSSSSSSSSTAAAQQQQQHSSSSSSSSSSSSISSSSSSSSSSSSISSSSSISSS 66
Query: 106 VRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPP 165
+ + +S + S + + + S SS +S S + +S
Sbjct: 67 ISSSSSSISSSRI---------------SSSSSSISSSSSSSSSSSSXSSSSSSSSSSSS 111
Query: 166 SVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPT 225
S SSS S S + +S + S + S+ +S S++ ++ S S S+S S +
Sbjct: 112 SSSSSSSSSSSSSSSISSSSSSSSSS-----SISSSSSSSSSSISSSSSSSSSSSSSSSS 166
Query: 226 RTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGS 266
+S SIS ++S +S SS SSS S +
Sbjct: 167 SSSSS------SISSSSSSSSSSSSSSSSSSSSSSSSSTAA 201
Score = 34.7 bits (78), Expect = 3.8
Identities = 31/113 (27%), Positives = 51/113 (44%), Gaps = 3/113 (2%)
Query: 159 NEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSL 218
+ +S S SSS S S T + + S + S + +S S++ + S S+
Sbjct: 4 SSSSSSSSSSSSSSSSSSTAAAQQQQQHSSSSSSSSSSSSISSSSSSSSSSSSISSSSSI 63
Query: 219 SASVSPTRTS---KPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQT 268
S+S+S + +S +S SIS ++S S SS SSS S + S +
Sbjct: 64 SSSISSSSSSISSSRISSSSSSISSSSSSSSSSSSXSSSSSSSSSSSSSSSSS 116
Score = 33.9 bits (76), Expect = 6.5
Identities = 29/104 (27%), Positives = 50/104 (47%), Gaps = 1/104 (0%)
Query: 166 SVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPT 225
S SSS S S + +S + + A + + +S +S S++ ++ S S S+S+S +
Sbjct: 1 SSSSSSSSSSSSSSSSSSSSSTAAAQQQQQHSSSSSSSSSSSSISSSSSSSSSSSSISSS 60
Query: 226 RT-SKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQT 268
+ S +S SIS + +S SS SSS S + S +
Sbjct: 61 SSISSSISSSSSSISSSRISSSSSSISSSSSSSSSSSSXSSSSS 104
>UniRef100_Q6VBJ4 Epa5p [Candida glabrata]
Length = 1218
Score = 48.5 bits (114), Expect = 3e-04
Identities = 38/117 (32%), Positives = 56/117 (47%)
Query: 146 SQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQST 205
S SS +S+S + +S S SSS PS S + +S + S + P S +S S+
Sbjct: 349 SSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSS 408
Query: 206 TPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDS 262
+ + S S S+S S + +S SP R S S ++S +S SS SSS S
Sbjct: 409 SSSSSSSSSSSSSSSSSSSSSSSSSPSPSRSSSSSSSSSSSSSSSSSSSSSSSSSSS 465
Score = 45.8 bits (107), Expect = 0.002
Identities = 39/143 (27%), Positives = 62/143 (43%)
Query: 126 SLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDH 185
S +S + + S SS +S+S + +S PS SSS S S + +S +
Sbjct: 338 SPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSPS 397
Query: 186 ESDAIRPRVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRG 245
S + S +S S++ + S S S+S SP+R+S S S S ++S
Sbjct: 398 PSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSRSSSSSSSSSSSSSSSSSSS 457
Query: 246 MHDDRTSVFSSMSSSDSGAGSQT 268
+S S SSS S + S +
Sbjct: 458 SSSSSSSSSPSPSSSSSSSSSSS 480
Score = 40.8 bits (94), Expect = 0.053
Identities = 36/119 (30%), Positives = 55/119 (45%), Gaps = 7/119 (5%)
Query: 150 LTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRL 209
L + SQ ++ +S PS SSS S S + +S + S + S +S S++
Sbjct: 322 LPTPSQDINSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSS-------SSSSSSSSSSSSS 374
Query: 210 TPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQT 268
P S S S+S S + +S SP S S ++S +S SS SSS S + S +
Sbjct: 375 PSPSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 433
Score = 40.0 bits (92), Expect = 0.091
Identities = 37/130 (28%), Positives = 55/130 (41%)
Query: 133 NSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRP 192
+S + + S SS S S + +S S SSS S S + +S + S
Sbjct: 378 SSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPS 437
Query: 193 RVPYSLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTS 252
R S +S S++ + S S S+S SP+ +S S S S ++S +S
Sbjct: 438 RSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSPSPSSSS 497
Query: 253 VFSSMSSSDS 262
SS SSS S
Sbjct: 498 SSSSSSSSSS 507
Score = 39.7 bits (91), Expect = 0.12
Identities = 40/145 (27%), Positives = 61/145 (41%), Gaps = 4/145 (2%)
Query: 126 SLQEEDDNSGGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDH 185
S +S +P + S SS +S+S +S S SSS S S + +S +
Sbjct: 365 SSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSS 424
Query: 186 ESDAIRPRVPY----SLLLASQSTTPRLTPPGSPPSLSASVSPTRTSKPVSPRRHSISFA 241
S + P S +S S++ + S S S+S SP+ +S S S S +
Sbjct: 425 SSSSSSSSSPSPSRSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSS 484
Query: 242 TSRGMHDDRTSVFSSMSSSDSGAGS 266
+S +S SS SSS S + S
Sbjct: 485 SSSSSSPSPSSSSSSSSSSSSSSPS 509
Score = 38.5 bits (88), Expect = 0.27
Identities = 42/179 (23%), Positives = 68/179 (37%)
Query: 90 QDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNSGGAPDMVAKVQSQSS 149
QD S + + S S++ + S + S +S +P + S SS
Sbjct: 327 QDINSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSS 386
Query: 150 LTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRVPYSLLLASQSTTPRL 209
+S+S S S SSS S S + +S + S + S S++
Sbjct: 387 SSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSRSSSSSSSS 446
Query: 210 TPPGSPPSLSASVSPTRTSKPVSPRRHSISFATSRGMHDDRTSVFSSMSSSDSGAGSQT 268
+ S S S+S S + +S P S S ++S +S S SSS S + S +
Sbjct: 447 SSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSS 505
Score = 37.4 bits (85), Expect = 0.59
Identities = 46/221 (20%), Positives = 91/221 (40%), Gaps = 20/221 (9%)
Query: 15 DLVQVLPSDPFEQLDLARKITSIALSARVNALESESSELRARIAEKDHLIAELQSQLDSL 74
D+ + LP+ P + ++ + + S+ ++ S SS + + + S S
Sbjct: 317 DVCEPLPT-PSQDINSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSP 375
Query: 75 DATLSATADNLHRAEQDKESLINENASLSNTVRKLNRDVSKLEVFRKTLMRSLQEEDDNS 134
+ S+++ + + S ++S S++ + S + S +S
Sbjct: 376 SPSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSS------SSSSSSSSSSSSS 429
Query: 135 GGAPDMVAKVQSQSSLTSTSQFGDNEASLPPSVSSSVPSISDTGNSFAEDHESDAIRPRV 194
+ ++ S SS +S+S + +S S SSS PS S + +S + S +
Sbjct: 430 SSSSPSPSRSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSS----- 484
Query: 195 PYSLLLASQSTTPRLTPPGSPPSLSASVSPT--RTSKPVSP 233
+S S++P + S S S+S SP+ +SKPV P
Sbjct: 485 ------SSSSSSPSPSSSSSSSSSSSSSSPSIQPSSKPVDP 519
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.125 0.332
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 500,111,538
Number of Sequences: 2790947
Number of extensions: 19357145
Number of successful extensions: 114424
Number of sequences better than 10.0: 2428
Number of HSP's better than 10.0 without gapping: 481
Number of HSP's successfully gapped in prelim test: 1989
Number of HSP's that attempted gapping in prelim test: 107329
Number of HSP's gapped (non-prelim): 7337
length of query: 339
length of database: 848,049,833
effective HSP length: 128
effective length of query: 211
effective length of database: 490,808,617
effective search space: 103560618187
effective search space used: 103560618187
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0238.4


