

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0231b.3
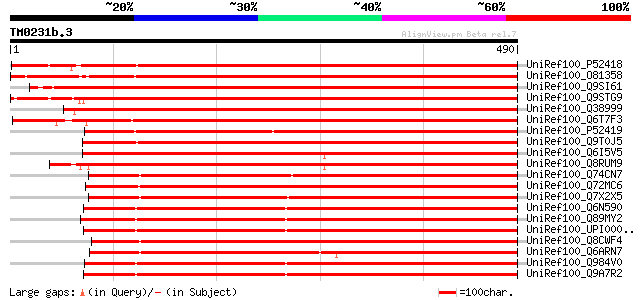
(490 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_P52418 Amidophosphoribosyltransferase, chloroplast pre... 773 0.0
UniRef100_O81358 Phosphoribosylpyrophosphate amidotransferase [V... 756 0.0
UniRef100_Q9SI61 Amidophosphoribosyltransferase [Arabidopsis tha... 744 0.0
UniRef100_Q9STG9 Amidophosphoribosyltransferase 2 [Arabidopsis t... 741 0.0
UniRef100_Q38999 Amidophosphoribosyltransferase [Arabidopsis tha... 732 0.0
UniRef100_Q6T7F3 5-phosphoribosyl-1-pyrophosphate amidotransfera... 730 0.0
UniRef100_P52419 Amidophosphoribosyltransferase, chloroplast pre... 716 0.0
UniRef100_Q9T0J5 Amidophosphoribosyltransferase-like protein [Ar... 639 0.0
UniRef100_Q6I5V5 Putative amidophosphoribosyltransferase [Oryza ... 561 e-158
UniRef100_Q8RUM9 Putative amidophosphoribosyltransferase [Oryza ... 559 e-158
UniRef100_Q74CN7 Amidophosphoribosyltransferase [Geobacter sulfu... 476 e-133
UniRef100_Q72MC6 Glutamine phosphoribosylpyrophosphate amidotran... 474 e-132
UniRef100_Q7X2X5 Putative amidophosphoribosyl transferase [uncul... 449 e-125
UniRef100_Q6N590 Amidophosphoribosyltransferase [Rhodopseudomona... 441 e-122
UniRef100_Q89MY2 Amidophosphoribosyltransferase [Bradyrhizobium ... 437 e-121
UniRef100_UPI00002E385F UPI00002E385F UniRef100 entry 432 e-119
UniRef100_Q8CWF4 Amidophosphoribosyltransferase [Bifidobacterium... 432 e-119
UniRef100_Q6ARN7 Probable amidophosphoribosyltransferase [Desulf... 432 e-119
UniRef100_Q984V0 Amidophosphoribosyltransferase [Rhizobium loti] 432 e-119
UniRef100_Q9A7R2 Amidophosphoribosyltransferase [Caulobacter cre... 430 e-119
>UniRef100_P52418 Amidophosphoribosyltransferase, chloroplast precursor [Glycine max]
Length = 569
Score = 773 bits (1997), Expect = 0.0
Identities = 405/493 (82%), Positives = 432/493 (87%), Gaps = 10/493 (2%)
Query: 2 AASNFSTLSSSS-SSFSKSKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPC-- 58
AASN STLSSSS S S S PF NTP LHN S PN +LSH P P C
Sbjct: 3 AASNLSTLSSSSLSKPSSSFPFRNTPTNNNASFLHNK-SLPNQNSLSHKLSSPLPLACNP 61
Query: 59 KNHNTTFPEPLCNDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVH 118
KN NT + DDEDQKPREECGVVGIYGDPEASRLC LALHALQHRGQEGAGIV VH
Sbjct: 62 KNQNTC----VFFDDEDQKPREECGVVGIYGDPEASRLCSLALHALQHRGQEGAGIVAVH 117
Query: 119 NNKIFQSVTGVGLVADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSV 178
+N + QSVTGVGLV+DVF ++KL +LPGT AIGHVRYSTAGQSMLKNVQPF+A YRF +V
Sbjct: 118 DNHL-QSVTGVGLVSDVFEQSKLSRLPGTSAIGHVRYSTAGQSMLKNVQPFLADYRFAAV 176
Query: 179 GVAHNGNLVNYRSLREKLEEN-GSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKG 237
VAHNGN VNYRSLR +LE N GSIFNT+SDTEVVLHLIATSK RPF+LRIVDACE L+G
Sbjct: 177 AVAHNGNFVNYRSLRARLEHNNGSIFNTTSDTEVVLHLIATSKHRPFLLRIVDACEHLQG 236
Query: 238 AYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVL 297
AYS VFVTEDKLVAVRDPFGFRPLVMG+R+NGAVV ASETCALDLIEA YEREVYPGEV+
Sbjct: 237 AYSLVFVTEDKLVAVRDPFGFRPLVMGRRTNGAVVLASETCALDLIEATYEREVYPGEVI 296
Query: 298 VVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECD 357
VVD G+QSLCLVSHPEPKQCIFEHIYF+LP+SVVFG+SVYESR+ FGEILA+ESPVECD
Sbjct: 297 VVDHTGIQSLCLVSHPEPKQCIFEHIYFALPNSVVFGRSVYESRKKFGEILASESPVECD 356
Query: 358 VVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAV 417
VVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRD GVKLKLSPV AV
Sbjct: 357 VVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDFGVKLKLSPVHAV 416
Query: 418 LEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELI 477
LEGKRVVVVDDSIVRGTTSSKIVRL+KEAGAKEVHMRIACPPI+ SCYYGVDTPSSEELI
Sbjct: 417 LEGKRVVVVDDSIVRGTTSSKIVRLLKEAGAKEVHMRIACPPIVASCYYGVDTPSSEELI 476
Query: 478 SNRMSVDEIREFI 490
SNRMSV+EIR+FI
Sbjct: 477 SNRMSVEEIRKFI 489
>UniRef100_O81358 Phosphoribosylpyrophosphate amidotransferase [Vigna unguiculata]
Length = 567
Score = 756 bits (1952), Expect = 0.0
Identities = 398/492 (80%), Positives = 423/492 (85%), Gaps = 8/492 (1%)
Query: 1 MAASNFSTLSSSSSSFSKSKPFTNTPCFPGN-KTLH-NPTSFPNPTTLSHAHKPPYPSPC 58
MAASN STL SSS S S PF NT F N TL N T+ T LSH+ + P PC
Sbjct: 1 MAASNLSTLCSSSHS-KPSFPFFNTQSFSVNTNTLFLNTTTNSFQTLLSHSTRYTLPLPC 59
Query: 59 KNHNTTFPEPLCNDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVH 118
TF NDDE KPREECGVVGIYGDPEASRLC LALHALQHRGQEGAGIV V
Sbjct: 60 TKTTNTFAS--FNDDE--KPREECGVVGIYGDPEASRLCSLALHALQHRGQEGAGIVAVD 115
Query: 119 NNKIFQSVTGVGLVADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSV 178
NN + +V GVGLV+DVF E KL QLPG+ AIGHVRYSTAG S L NVQPFVAGYRFGSV
Sbjct: 116 NN-VLHTVNGVGLVSDVFDETKLSQLPGSCAIGHVRYSTAGHSRLCNVQPFVAGYRFGSV 174
Query: 179 GVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGA 238
V+HNGN VNYRSLR KLE+NGSIFNT+SDT+VVLHLIATSK RPF+LRIVDACE LKGA
Sbjct: 175 AVSHNGNFVNYRSLRAKLEDNGSIFNTTSDTDVVLHLIATSKHRPFLLRIVDACENLKGA 234
Query: 239 YSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLV 298
YS VF+TEDKLVAV+DPFGFRPLVMG+RSNGAVVFASETCALDLIEA YEREV PGEV+V
Sbjct: 235 YSLVFLTEDKLVAVKDPFGFRPLVMGRRSNGAVVFASETCALDLIEATYEREVNPGEVVV 294
Query: 299 VDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDV 358
VD G+QSLCLV+HPEPKQCIFEHIYF+LP+SVVFG+SVYESRR FGEILATESPVECDV
Sbjct: 295 VDHTGIQSLCLVTHPEPKQCIFEHIYFALPNSVVFGRSVYESRRKFGEILATESPVECDV 354
Query: 359 VIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVL 418
VIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRD GVKLKL PV+ VL
Sbjct: 355 VIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDFGVKLKLFPVRGVL 414
Query: 419 EGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELIS 478
EGKRVVVVD+SIVRGTT SKIVRLIKEAGAKEVHMRIACPPI+GSCYYGVDTPS EELIS
Sbjct: 415 EGKRVVVVDESIVRGTTLSKIVRLIKEAGAKEVHMRIACPPIVGSCYYGVDTPSKEELIS 474
Query: 479 NRMSVDEIREFI 490
NRMSV+EIREFI
Sbjct: 475 NRMSVEEIREFI 486
>UniRef100_Q9SI61 Amidophosphoribosyltransferase [Arabidopsis thaliana]
Length = 566
Score = 744 bits (1921), Expect = 0.0
Identities = 378/472 (80%), Positives = 415/472 (87%), Gaps = 5/472 (1%)
Query: 20 KPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCKNH-NTTFPEPLCNDDEDQKP 78
KPF P L +P S P P +L H + SP + N+ P ND++D+KP
Sbjct: 32 KPFLKPPHL---SLLPSPLSSP-PPSLIHGVSSYFSSPSPSEDNSHTPFDYHNDEDDEKP 87
Query: 79 REECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVFTE 138
REECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTV K+ Q++TGVGLV++VF E
Sbjct: 88 REECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVSPEKVLQTITGVGLVSEVFNE 147
Query: 139 AKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLEE 198
+KLDQLPG AIGHVRYSTAG SMLKNVQPFVAGYRFGS+GVAHNGNLVNY++LR LEE
Sbjct: 148 SKLDQLPGEFAIGHVRYSTAGASMLKNVQPFVAGYRFGSIGVAHNGNLVNYKTLRAMLEE 207
Query: 199 NGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGF 258
NGSIFNTSSDTEVVLHLIA SK RPF +RI+DACEKL+GAYS VFVTEDKLVAVRDP+GF
Sbjct: 208 NGSIFNTSSDTEVVLHLIAISKARPFFMRIIDACEKLQGAYSMVFVTEDKLVAVRDPYGF 267
Query: 259 RPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHPEPKQC 318
RPLVMG+RSNGAVVFASETCALDLIEA YEREVYPGEVLVVDK+G++S CL+ EPKQC
Sbjct: 268 RPLVMGRRSNGAVVFASETCALDLIEATYEREVYPGEVLVVDKDGVKSQCLMPKFEPKQC 327
Query: 319 IFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGYAAKAG 378
IFEHIYFSLP+S+VFG+SVYESR +FGEILATESPVECDVVIAVPDSGVVAALGYAAK+G
Sbjct: 328 IFEHIYFSLPNSIVFGRSVYESRHVFGEILATESPVECDVVIAVPDSGVVAALGYAAKSG 387
Query: 379 VPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTSSK 438
VPFQQGLIRSHYVGRTFIEPSQKIRD GVKLKLSPV+ VLEGKRVVVVDDSIVRGTTSSK
Sbjct: 388 VPFQQGLIRSHYVGRTFIEPSQKIRDFGVKLKLSPVRGVLEGKRVVVVDDSIVRGTTSSK 447
Query: 439 IVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
IVRL++EAGAKEVHMRIA PPI+ SCYYGVDTPSSEELISNR+SV+EI EFI
Sbjct: 448 IVRLLREAGAKEVHMRIASPPIVASCYYGVDTPSSEELISNRLSVEEINEFI 499
>UniRef100_Q9STG9 Amidophosphoribosyltransferase 2 [Arabidopsis thaliana]
Length = 561
Score = 741 bits (1913), Expect = 0.0
Identities = 390/500 (78%), Positives = 430/500 (86%), Gaps = 15/500 (3%)
Query: 1 MAASNFSTLSSSSSSFSK-SKPFTNTPCFPGNKTLHNPTSFPNPTTLSHAHKPPYPSPCK 59
MAA+ S++SSS S +K +K N ++ L NP F NP++ S + P S
Sbjct: 1 MAAT--SSISSSLSLNAKPNKLSNNNNNNKPHRFLRNP--FLNPSSSSFSPLPASISSSS 56
Query: 60 NHNTTFP----EPLC-----NDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQE 110
+ + +FP PL NDD D+KPREECGVVGIYGD EASRLCYLALHALQHRGQE
Sbjct: 57 S-SPSFPLRVSNPLTLLAADNDDYDEKPREECGVVGIYGDSEASRLCYLALHALQHRGQE 115
Query: 111 GAGIVTVHNNKIFQSVTGVGLVADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFV 170
GAGIVTV +K+ Q++TGVGLV++VF+E+KLDQLPG +AIGHVRYSTAG SMLKNVQPFV
Sbjct: 116 GAGIVTVSKDKVLQTITGVGLVSEVFSESKLDQLPGDIAIGHVRYSTAGSSMLKNVQPFV 175
Query: 171 AGYRFGSVGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVD 230
AGYRFGSVGVAHNGNLVNY LR LEENGSIFNTSSDTEVVLHLIA SK RPF +RIVD
Sbjct: 176 AGYRFGSVGVAHNGNLVNYTKLRADLEENGSIFNTSSDTEVVLHLIAISKARPFFMRIVD 235
Query: 231 ACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYERE 290
ACEKL+GAYS VFVTEDKLVAVRDP GFRPLVMG+RSNGAVVFASETCALDLIEA YERE
Sbjct: 236 ACEKLQGAYSMVFVTEDKLVAVRDPHGFRPLVMGRRSNGAVVFASETCALDLIEATYERE 295
Query: 291 VYPGEVLVVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILAT 350
VYPGEVLVVDK+G++ CL+ HPEPKQCIFEHIYFSLP+S+VFG+SVYESR +FGEILAT
Sbjct: 296 VYPGEVLVVDKDGVKCQCLMPHPEPKQCIFEHIYFSLPNSIVFGRSVYESRHVFGEILAT 355
Query: 351 ESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLK 410
ESPV+CDVVIAVPDSGVVAALGYAAKAGV FQQGLIRSHYVGRTFIEPSQKIRD GVKLK
Sbjct: 356 ESPVDCDVVIAVPDSGVVAALGYAAKAGVAFQQGLIRSHYVGRTFIEPSQKIRDFGVKLK 415
Query: 411 LSPVKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDT 470
LSPV+ VLEGKRVVVVDDSIVRGTTSSKIVRL++EAGAKEVHMRIA PPII SCYYGVDT
Sbjct: 416 LSPVRGVLEGKRVVVVDDSIVRGTTSSKIVRLLREAGAKEVHMRIASPPIIASCYYGVDT 475
Query: 471 PSSEELISNRMSVDEIREFI 490
PSS ELISNRMSVDEIR++I
Sbjct: 476 PSSNELISNRMSVDEIRDYI 495
>UniRef100_Q38999 Amidophosphoribosyltransferase [Arabidopsis thaliana]
Length = 511
Score = 732 bits (1889), Expect = 0.0
Identities = 367/441 (83%), Positives = 401/441 (90%), Gaps = 3/441 (0%)
Query: 53 PYPSPCKNH---NTTFPEPLCNDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQ 109
P SP +H N+ P ND++D+KPREECGVVGIYGDPEASRL YLALHALQHRGQ
Sbjct: 4 PPTSPLLHHPKNNSHAPFDYHNDEDDEKPREECGVVGIYGDPEASRLFYLALHALQHRGQ 63
Query: 110 EGAGIVTVHNNKIFQSVTGVGLVADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPF 169
EGAGIVTV K+ Q++TGVGLV++VF E+KLDQLPG AI HVRYSTAG SMLKNVQPF
Sbjct: 64 EGAGIVTVSPEKVLQTITGVGLVSEVFNESKLDQLPGEFAIAHVRYSTAGASMLKNVQPF 123
Query: 170 VAGYRFGSVGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIV 229
VAGYRFGS+GVAHNGNLVNY++LR LEENGSIFNTSSDTEVVLHLIA SK RPF +RI+
Sbjct: 124 VAGYRFGSIGVAHNGNLVNYKTLRAMLEENGSIFNTSSDTEVVLHLIAISKARPFFMRII 183
Query: 230 DACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYER 289
DACEKL+GAYS VFVTEDKLVAVRDP+GFRPLVMG+RSNGAVVFASETCALDLIEA YER
Sbjct: 184 DACEKLQGAYSMVFVTEDKLVAVRDPYGFRPLVMGRRSNGAVVFASETCALDLIEATYER 243
Query: 290 EVYPGEVLVVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILA 349
EVYPGEVLVVDK+G++S CL+ EPKQCIFEHIYFSLP+S+VFG+SVYESR +FGEILA
Sbjct: 244 EVYPGEVLVVDKDGVKSQCLMPKFEPKQCIFEHIYFSLPNSIVFGRSVYESRHVFGEILA 303
Query: 350 TESPVECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKL 409
TESPVECDVVIAVPDSGVVAALGYAAK+GVPFQQGLIRSHYVGRTFIEPSQKIRD GVKL
Sbjct: 304 TESPVECDVVIAVPDSGVVAALGYAAKSGVPFQQGLIRSHYVGRTFIEPSQKIRDFGVKL 363
Query: 410 KLSPVKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVD 469
KLSPV+ VLEGKRVVVVDDSIVRGTTSSKIVRL++EAGAKEVHMRIA PPI+ SCYYGVD
Sbjct: 364 KLSPVRGVLEGKRVVVVDDSIVRGTTSSKIVRLLREAGAKEVHMRIASPPIVASCYYGVD 423
Query: 470 TPSSEELISNRMSVDEIREFI 490
TPSSEELISNR+SV+EI EFI
Sbjct: 424 TPSSEELISNRLSVEEINEFI 444
>UniRef100_Q6T7F3 5-phosphoribosyl-1-pyrophosphate amidotransferase [Nicotiana
tabacum]
Length = 573
Score = 730 bits (1885), Expect = 0.0
Identities = 382/497 (76%), Positives = 426/497 (84%), Gaps = 15/497 (3%)
Query: 3 ASNFSTLSSSSSSFSK-SKPFTNTPCFPGNKTLH-NPTSFPNP--TTLSHAHKPPYPSPC 58
A+ ST S+++++ S S+P C K L +P + P P T ++ + K P
Sbjct: 2 AATVSTASAAATNKSPLSQPLDKPFCSLSQKLLSLSPKTHPKPYRTLITASSKNPL---- 57
Query: 59 KNHNTTFPEPLCND-----DEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAG 113
N +F + N D+D KPREECGVVGIYGD EASRLCYLALHALQHRGQEGAG
Sbjct: 58 -NDVISFKKSADNTLDSYFDDDDKPREECGVVGIYGDSEASRLCYLALHALQHRGQEGAG 116
Query: 114 IVTVHNNKIFQSVTGVGLVADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGY 173
IV V N+ + +S+TGVGLV+DVF E+KLDQLPG +AIGHVRYSTAG SMLKNVQPFVA Y
Sbjct: 117 IVAV-NDDVLKSITGVGLVSDVFNESKLDQLPGDMAIGHVRYSTAGSSMLKNVQPFVASY 175
Query: 174 RFGSVGVAHNGNLVNYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACE 233
+FGSVGVAHNGNLVNY+ LR +LEENGSIFNTSSDTEVVLHLIA SK RPF+LRIV+ACE
Sbjct: 176 KFGSVGVAHNGNLVNYKLLRSELEENGSIFNTSSDTEVVLHLIAISKARPFLLRIVEACE 235
Query: 234 KLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYP 293
K++GAYS VFVTEDKLVAVRDP GFRPLVMG+RSNGAVVFASETCALDLIEA YEREV P
Sbjct: 236 KIEGAYSMVFVTEDKLVAVRDPHGFRPLVMGRRSNGAVVFASETCALDLIEATYEREVNP 295
Query: 294 GEVLVVDKNGLQSLCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESP 353
GEV+VVDK+G+QS+CL+ HPE K CIFEHIYF+LP+SVVFG+SVYESRR FGEILATE+P
Sbjct: 296 GEVVVVDKDGVQSICLMPHPERKSCIFEHIYFALPNSVVFGRSVYESRRAFGEILATEAP 355
Query: 354 VECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSP 413
VECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRD GVKLKLSP
Sbjct: 356 VECDVVIAVPDSGVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDFGVKLKLSP 415
Query: 414 VKAVLEGKRVVVVDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSS 473
V+AVLEGKRVVVVDDSIVRGTTSSKIVRL+KEAGAKEVHMRIA PPII SCYYGVDTPSS
Sbjct: 416 VRAVLEGKRVVVVDDSIVRGTTSSKIVRLLKEAGAKEVHMRIASPPIIASCYYGVDTPSS 475
Query: 474 EELISNRMSVDEIREFI 490
+ELISNRMSV+EI+EFI
Sbjct: 476 DELISNRMSVEEIKEFI 492
>UniRef100_P52419 Amidophosphoribosyltransferase, chloroplast precursor [Vigna
aconitifolia]
Length = 485
Score = 716 bits (1847), Expect = 0.0
Identities = 360/418 (86%), Positives = 386/418 (92%), Gaps = 2/418 (0%)
Query: 73 DEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLV 132
++D+KPREECGVVGIYGDPEASRLC LALHALQHRGQEGAGIV VH+N +F V GVGLV
Sbjct: 10 NDDEKPREECGVVGIYGDPEASRLCSLALHALQHRGQEGAGIVAVHDN-LFHQVNGVGLV 68
Query: 133 ADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSL 192
+DVF EAKL +LPG+ AIGHVRYSTAG S L NVQPFVAGYRFGSV VAHNGN VNYRSL
Sbjct: 69 SDVFNEAKLSELPGSCAIGHVRYSTAGHSKLVNVQPFVAGYRFGSVAVAHNGNFVNYRSL 128
Query: 193 REKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAV 252
R KLE+NGSIFNT+SDTEVVLHLIATSK RPF+LR+VDACE LKGAYS VF+TEDKLVAV
Sbjct: 129 RAKLEDNGSIFNTTSDTEVVLHLIATSKHRPFLLRVVDACENLKGAYSLVFLTEDKLVAV 188
Query: 253 RDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSH 312
RD FGFRPLVMG+R NGAVVFASETCALDLI+A YEREV PGEV+VVD G+QSLCLV+H
Sbjct: 189 RD-FGFRPLVMGRRKNGAVVFASETCALDLIDATYEREVNPGEVVVVDHTGIQSLCLVTH 247
Query: 313 PEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALG 372
EPKQCIFEHIYF+LP+SVVFG+SVYESRR FGEILATESPVECDVVIAVPDSGVVAALG
Sbjct: 248 QEPKQCIFEHIYFALPNSVVFGRSVYESRRKFGEILATESPVECDVVIAVPDSGVVAALG 307
Query: 373 YAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVR 432
YAAKAGVPFQQGLIRSH+VGRTFIEPSQKIRD GVKLKL PV+ VLEGKRVVVVDDSIVR
Sbjct: 308 YAAKAGVPFQQGLIRSHHVGRTFIEPSQKIRDFGVKLKLFPVRGVLEGKRVVVVDDSIVR 367
Query: 433 GTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
GTTSSKIVRLIKEAGAKEVHMRIACPPI+ SCYYGVDTPS EELISNRM V+EIR+FI
Sbjct: 368 GTTSSKIVRLIKEAGAKEVHMRIACPPIVASCYYGVDTPSKEELISNRMDVEEIRKFI 425
>UniRef100_Q9T0J5 Amidophosphoribosyltransferase-like protein [Arabidopsis thaliana]
Length = 532
Score = 639 bits (1649), Expect = 0.0
Identities = 317/421 (75%), Positives = 372/421 (88%), Gaps = 2/421 (0%)
Query: 71 NDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVG 130
N DED K EECGVVGI+GDPEASRL YLALHALQHRGQEGAGIV + N + +S+TGVG
Sbjct: 66 NVDEDDKLHEECGVVGIHGDPEASRLSYLALHALQHRGQEGAGIVAANQNGL-ESITGVG 124
Query: 131 LVADVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYR 190
LV+DVFTE+KL+ LPG +AIGHVRYST+G SMLKNVQPF+A + GS+ VAHNGN VNY+
Sbjct: 125 LVSDVFTESKLNNLPGDIAIGHVRYSTSGASMLKNVQPFIASCKLGSLAVAHNGNFVNYK 184
Query: 191 SLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLV 250
L+ KLEE GSIF TSSDTE+VLHLIA SK + F+LR++DACEKL+GAYS VFV EDKL+
Sbjct: 185 QLKTKLEEMGSIFITSSDTELVLHLIAKSKAKTFLLRVIDACEKLRGAYSMVFVFEDKLI 244
Query: 251 AVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKN-GLQSLCL 309
AVRDPFGFRPLVMG+RSNGAVVFASETCALDLI+A YEREV PGE++VVD+N G S+ +
Sbjct: 245 AVRDPFGFRPLVMGRRSNGAVVFASETCALDLIDATYEREVCPGEIVVVDRNHGDSSMFM 304
Query: 310 VSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVA 369
+SHPE KQC+FEH YFS P+S+VFG+SVYE+RR++GEILAT +PV+CDVVIAVPDSG VA
Sbjct: 305 ISHPEQKQCVFEHGYFSQPNSIVFGRSVYETRRMYGEILATVAPVDCDVVIAVPDSGTVA 364
Query: 370 ALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDS 429
ALGYAAKAGVPFQ GL+RSHY RTFIEP+Q+IRD VK+KLSPV+AVLEGKRVVVVDDS
Sbjct: 365 ALGYAAKAGVPFQIGLLRSHYAKRTFIEPTQEIRDFAVKVKLSPVRAVLEGKRVVVVDDS 424
Query: 430 IVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREF 489
IVRGTTS KIVR++++AGAKEVHMRIA PP+I SCYYGVDTP S+ELIS++MSV+ I++
Sbjct: 425 IVRGTTSLKIVRMLRDAGAKEVHMRIALPPMIASCYYGVDTPRSQELISSKMSVEAIQKH 484
Query: 490 I 490
I
Sbjct: 485 I 485
>UniRef100_Q6I5V5 Putative amidophosphoribosyltransferase [Oryza sativa]
Length = 541
Score = 561 bits (1446), Expect = e-158
Identities = 285/425 (67%), Positives = 337/425 (79%), Gaps = 5/425 (1%)
Query: 71 NDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVG 130
+D+ D PREECG+VG+ GDP+AS LCYL L LQHRG+EGAGIV V + +SVTG+G
Sbjct: 69 DDESDDHPREECGLVGVVGDPDASSLCYLGLQKLQHRGEEGAGIVAVGGDGKLKSVTGLG 128
Query: 131 LVADVFTE-AKLDQLPGTLAIGHVRYSTAGQSM-LKNVQPFVAGYRFGSVGVAHNGNLVN 188
LVADVF + A+L LPG AIGHVRYSTAG + L+NVQPF+AGYRFG V VAHNGNLVN
Sbjct: 129 LVADVFGDPARLASLPGPAAIGHVRYSTAGAAASLRNVQPFLAGYRFGQVAVAHNGNLVN 188
Query: 189 YRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDK 248
Y++LR KLE GSIFNTSSDTEV+LHLIATS RP + RI DACE+L GAYS +F+T DK
Sbjct: 189 YQALRNKLEARGSIFNTSSDTEVILHLIATSLSRPLLSRICDACERLAGAYSLLFLTADK 248
Query: 249 LVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKN--GLQS 306
+ AVRDP GFRPLV+G+R NG V FASETCALDLIEA YEREV PGEV+VVD+ + S
Sbjct: 249 MFAVRDPHGFRPLVLGRRRNGTVAFASETCALDLIEATYEREVEPGEVVVVDRRDMSVSS 308
Query: 307 LCLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVE-CDVVIAVPDS 365
CLV H + C+FEHIYF+LP+SVVF +V+E R FG LA ESP DVVI VPDS
Sbjct: 309 ACLVPHRPRRSCVFEHIYFALPNSVVFSHAVHERRTAFGRALAEESPAAGADVVIPVPDS 368
Query: 366 GVVAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVV 425
G AALG+A +G+ FQQGLIR HY GR+FI+P+Q IRDL VKLKL+PV V+ GK VVV
Sbjct: 369 GFYAALGFARASGLEFQQGLIRWHYSGRSFIQPTQAIRDLAVKLKLAPVHGVIRGKSVVV 428
Query: 426 VDDSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDE 485
VDDS+VRGTTSSKIVRL+++AGA+EVHMRIA PP++GSC YG+DTPS ELISNRM ++
Sbjct: 429 VDDSLVRGTTSSKIVRLLRDAGAREVHMRIASPPVVGSCLYGIDTPSEGELISNRMDLEG 488
Query: 486 IREFI 490
+R I
Sbjct: 489 VRREI 493
>UniRef100_Q8RUM9 Putative amidophosphoribosyltransferase [Oryza sativa]
Length = 551
Score = 559 bits (1440), Expect = e-158
Identities = 292/468 (62%), Positives = 352/468 (74%), Gaps = 20/468 (4%)
Query: 39 SFPNPTTLSHAHKPPYPSPCKNHNTTFPE-----PLCNDDE------DQKPREECGVVGI 87
S P+ L+ H+ P+P + FPE P D E D PREECGV G+
Sbjct: 36 SAAKPSPLALRHRAGRPAPLQ----AFPEYDRVTPFDYDGEVDGGDGDDHPREECGVFGV 91
Query: 88 YGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVFTE-AKLDQLPG 146
GDP+A+ LCYL L LQHRG+EGAGI ++ + G+GLV DVF + A+L +LPG
Sbjct: 92 VGDPDATSLCYLGLQKLQHRGEEGAGIAAAGDDGTIKLERGLGLVGDVFGDPARLGKLPG 151
Query: 147 TLAIGHVRYSTAGQSM-LKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLEENGSIFNT 205
AIGHVRYSTAG + L+NVQPF+AGYRFG + VAHNGNLVNY++LR KLE GSIF+T
Sbjct: 152 QAAIGHVRYSTAGAAASLRNVQPFLAGYRFGQLAVAHNGNLVNYQALRNKLEAQGSIFST 211
Query: 206 SSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGFRPLVMGK 265
SSDTEV+LHLIATS RP + RI DACE+L GAYS +F+T DKL+AVRDPFGFRPLVMG+
Sbjct: 212 SSDTEVILHLIATSLSRPLLARICDACERLAGAYSLLFLTADKLLAVRDPFGFRPLVMGR 271
Query: 266 RSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKN--GLQSLCLVSHPEPKQCIFEHI 323
R+NGA+VFASETCALDLI+A YEREV PGEV+VVD+ + S CLV H K C+FEHI
Sbjct: 272 RANGAIVFASETCALDLIDATYEREVEPGEVVVVDRRDMSVSSACLVPHRPRKSCVFEHI 331
Query: 324 YFSLPSSVVFGKSVYESRRLFGEILATESPV-ECDVVIAVPDSGVVAALGYAAKAGVPFQ 382
YF+LP+SVVFG +V+E R +G LA ESP DVVI VPDSG AALG++ +G+ FQ
Sbjct: 332 YFALPNSVVFGHAVHERRNAYGRALAEESPAPTADVVIPVPDSGFYAALGFSQTSGLEFQ 391
Query: 383 QGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTSSKIVRL 442
QGLIR HY GR+FI+PSQ IRDL VKLKL+PV V+ GK VVVVDDS+VRGTTSSKIVRL
Sbjct: 392 QGLIRWHYSGRSFIQPSQAIRDLAVKLKLAPVHGVIRGKSVVVVDDSLVRGTTSSKIVRL 451
Query: 443 IKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
+++AGA+EVHMRIA PP+IGSC YG+DTPS ELISNRM ++ +R I
Sbjct: 452 LRDAGAREVHMRIASPPVIGSCLYGIDTPSEGELISNRMDLEGVRRAI 499
>UniRef100_Q74CN7 Amidophosphoribosyltransferase [Geobacter sulfurreducens]
Length = 467
Score = 476 bits (1224), Expect = e-133
Identities = 239/415 (57%), Positives = 309/415 (73%), Gaps = 3/415 (0%)
Query: 77 KPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVF 136
+P EECG+ GIYG PEA+ L YL L+ALQHRGQE GIV+ ++ +GLVADVF
Sbjct: 6 RPTEECGIFGIYGHPEAANLTYLGLYALQHRGQEACGIVSSDGRSLYTH-RSMGLVADVF 64
Query: 137 TEAKL-DQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREK 195
++ LPG AIGHVRYST G S+LKNVQP Y GS+ +AHNGNLVN + ++++
Sbjct: 65 GNQEIFGNLPGESAIGHVRYSTTGDSVLKNVQPIKVDYSRGSIAIAHNGNLVNAQYIKDE 124
Query: 196 LEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDP 255
LE GSIF T+ DTEV+LHL+ATSK RI+DA ++KGAY +F+TE ++VA RDP
Sbjct: 125 LEAWGSIFQTTMDTEVILHLLATSKHSSLEDRIIDALGRIKGAYCLLFLTETRMVAARDP 184
Query: 256 FGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHPEP 315
GFRPL +GK + VV ASE+CALDLIEAE+ RE+ PGE++V+ K+GL S EP
Sbjct: 185 HGFRPLCLGKLGDAWVV-ASESCALDLIEAEFVREIEPGEIVVITKDGLTSHFPHKKIEP 243
Query: 316 KQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGYAA 375
CIFE +YF+ P S +FGK+VY+ R+ FG L E ++ D+VI VPDSGV AALGYA
Sbjct: 244 APCIFEFVYFARPDSYIFGKNVYQVRKDFGRQLCREHRIDADIVIPVPDSGVPAALGYAQ 303
Query: 376 KAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTT 435
+AG+PF+ GLIR+HYVGRTFIEP Q IR GVK+KL+PV+ VL+GKRVVV+DDSIVRGTT
Sbjct: 304 EAGIPFELGLIRNHYVGRTFIEPQQSIRHFGVKIKLNPVREVLKGKRVVVIDDSIVRGTT 363
Query: 436 SSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
S KIV++++ AGA EVH+RI+ PP CYYG+DTP+ +ELIS+ +++EIR++I
Sbjct: 364 SRKIVKMVRNAGASEVHVRISSPPTSYPCYYGIDTPTRKELISSSHTIEEIRKYI 418
>UniRef100_Q72MC6 Glutamine phosphoribosylpyrophosphate amidotransferase [Leptospira
interrogans]
Length = 490
Score = 474 bits (1219), Expect = e-132
Identities = 226/417 (54%), Positives = 307/417 (73%), Gaps = 1/417 (0%)
Query: 74 EDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVA 133
+D KP+EEC + GI+ EAS YL L+++QHRGQE +GIV+ +++ G+GLVA
Sbjct: 15 QDDKPKEECAIFGIFNSSEASNFTYLGLYSMQHRGQESSGIVSSDGEHLYR-YAGMGLVA 73
Query: 134 DVFTEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLR 193
+FTE KL +L G AIGH RYST G S L+N QP G+V +AHNGNLVN LR
Sbjct: 74 HIFTETKLKELQGNAAIGHNRYSTTGASFLRNAQPLRVESHLGAVSLAHNGNLVNSWELR 133
Query: 194 EKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVR 253
+LE+ GSIF T+ D+EV++HL+A S + F+ + A +K++GAYS V +T+ +L+AVR
Sbjct: 134 SQLEKEGSIFQTTIDSEVIVHLMARSGETDFLSALSSALKKVRGAYSLVILTKTQLIAVR 193
Query: 254 DPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHP 313
DP GFRPLVMG+R +G++VFASETCA D+ + +YER+V PGE++VVDKNG+ S
Sbjct: 194 DPNGFRPLVMGRREDGSIVFASETCAFDITDTKYERDVEPGEMIVVDKNGMNSYYPFPKA 253
Query: 314 EPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGY 373
P CIFE+IYF+ P S +FG+SVY+ R+ G LA E PV DVVI VPDS +AALGY
Sbjct: 254 SPSLCIFEYIYFARPDSSIFGESVYKVRKNLGRFLARELPVPADVVIPVPDSANIAALGY 313
Query: 374 AAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRG 433
A ++G+ +Q GLIRSHY+GRTFIEP QKIRD G K+K + V+ V+EGKRV+VVDDSI+RG
Sbjct: 314 AEESGISYQSGLIRSHYIGRTFIEPDQKIRDFGAKIKYNVVRNVVEGKRVIVVDDSIMRG 373
Query: 434 TTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
TTS KI+++I+ AGAKE+H+R++ PP I CYYG+D P+ ELI+ +++EIR+++
Sbjct: 374 TTSRKIIKMIRNAGAKEIHLRVSAPPTISPCYYGIDIPTHNELIAATHTIEEIRKYL 430
>UniRef100_Q7X2X5 Putative amidophosphoribosyl transferase [uncultured Acidobacteria
bacterium]
Length = 489
Score = 449 bits (1156), Expect = e-125
Identities = 234/414 (56%), Positives = 296/414 (70%), Gaps = 2/414 (0%)
Query: 77 KPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVF 136
K REECGV GI+G EAS L YL L+ALQHRGQE AGI + I S VG V DVF
Sbjct: 4 KFREECGVFGIFGHAEASNLTYLGLYALQHRGQESAGIAASDGSLIRVS-KAVGYVNDVF 62
Query: 137 TEAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKL 196
L +LPGTLA+GHVRYSTAG S + N QP V G + +AHNGNLVN +R+ L
Sbjct: 63 NGDTLAKLPGTLAVGHVRYSTAGDSGVANAQPIVVDSFHGQLALAHNGNLVNAGEVRDAL 122
Query: 197 EENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPF 256
G+IF TS+D+EV++HL A SK I+DA +++GA+S V +T+D+LV VRDP
Sbjct: 123 IREGAIFQTSADSEVLVHLFARSKGVTAEAAIIDAISQVRGAFSLVMMTKDRLVGVRDPH 182
Query: 257 GFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHPEPK 316
GFRPLV+GK + A V +SETCALDLI A Y R+V PGE++VV GL+S +
Sbjct: 183 GFRPLVLGKLRD-AWVLSSETCALDLIGASYVRDVEPGEIVVVSDQGLKSSKPFAPARRS 241
Query: 317 QCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGYAAK 376
QC+FEH+YF+ P S VFG+SV E R FG LA ES V DVV+ +PDSGV AA+G+A
Sbjct: 242 QCVFEHVYFARPDSYVFGESVNEVRTEFGRRLARESGVAADVVVPIPDSGVCAAVGFAEA 301
Query: 377 AGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTS 436
+G+P + GLIR+HYVGRTFIEP Q IR GV++KL+PV+++LEG+RVV+VDDSIVRGTTS
Sbjct: 302 SGIPMRFGLIRNHYVGRTFIEPQQSIRHFGVRVKLNPVRSILEGRRVVLVDDSIVRGTTS 361
Query: 437 SKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
KIVR+++ AGA EVHMRI+CPP I C+YGVDTP ELI+ +++EIR ++
Sbjct: 362 RKIVRMVRSAGATEVHMRISCPPTISPCFYGVDTPRRSELIAATHTIEEIRRYL 415
>UniRef100_Q6N590 Amidophosphoribosyltransferase [Rhodopseudomonas palustris]
Length = 509
Score = 441 bits (1134), Expect = e-122
Identities = 224/420 (53%), Positives = 303/420 (71%), Gaps = 3/420 (0%)
Query: 72 DDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGL 131
D + REECGV GI+G P+A+ + L LHALQHRGQE AGIV+ N + F S +GL
Sbjct: 30 DTDGDTLREECGVFGIFGHPDAAAITALGLHALQHRGQEAAGIVSFDNGR-FHSERRLGL 88
Query: 132 VADVFTEAK-LDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYR 190
V D F+ A + +LPG AIGHVRYST G+++L+NVQP A G V HNGNL N
Sbjct: 89 VGDTFSRADVIARLPGNSAIGHVRYSTTGETILRNVQPLFAELNAGGFAVGHNGNLTNGL 148
Query: 191 SLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLV 250
+LR +L +G+I +++DTEV+LHL+A SK+ FI R +++ L+GAYS V +T KLV
Sbjct: 149 TLRRELIRSGAIMQSTTDTEVILHLVAHSKKTRFIERFIESLRALEGAYSLVSLTNKKLV 208
Query: 251 AVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLV 310
RDP G RPLV+G+ +G + ASETCALD+I A Y R+V PGEV+V D++G+ S
Sbjct: 209 GARDPLGIRPLVLGEL-DGCPILASETCALDIIGAHYVRDVEPGEVIVFDRHGVTSHKPF 267
Query: 311 SHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAA 370
P+ CIFE+IYF+ P SVV G+SVY+ R+ FG LA ES VE DVV+ VPDSGV AA
Sbjct: 268 PPQPPRPCIFEYIYFARPDSVVGGRSVYDVRKAFGAQLALESHVESDVVVPVPDSGVPAA 327
Query: 371 LGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSI 430
+GY+ ++GVPF+ G+IR+HYVGRTFI+P+Q +R+LGV++K S +A +EGKR+V++DDS+
Sbjct: 328 IGYSRQSGVPFELGIIRNHYVGRTFIQPTQSVRELGVRMKHSANRAAIEGKRIVLIDDSL 387
Query: 431 VRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
VRGTTS KIV+++++AGA+EVH RIA PPI YYG+DTP L++ S++E+R+ I
Sbjct: 388 VRGTTSKKIVKMMRDAGAREVHFRIASPPITHPDYYGIDTPDRAGLLAATHSLEEMRDLI 447
>UniRef100_Q89MY2 Amidophosphoribosyltransferase [Bradyrhizobium japonicum]
Length = 507
Score = 437 bits (1125), Expect = e-121
Identities = 222/423 (52%), Positives = 303/423 (71%), Gaps = 3/423 (0%)
Query: 69 LCNDDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTG 128
L +D E REECGV GIYG P+A+ + L LHALQHRGQE AGIV+ ++ F S
Sbjct: 25 LQDDLEGDTLREECGVFGIYGHPDAAAITALGLHALQHRGQEAAGIVSYDGSR-FHSERR 83
Query: 129 VGLVADVFTEAK-LDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLV 187
+GLV D F+ + +D+LPG +A+GHVRYST G ++L+NVQP A G + VAHNGNL
Sbjct: 84 LGLVGDTFSRREVIDRLPGNMAVGHVRYSTTGATILRNVQPLFAELNAGGLAVAHNGNLT 143
Query: 188 NYRSLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTED 247
N +LR +L NG++ +++DTEV+LHL+A S++ FI R +DA +++GAY+ V +T
Sbjct: 144 NGLTLRRELVRNGAMMQSTTDTEVILHLVARSRRSRFIERYIDALREIEGAYALVSLTNK 203
Query: 248 KLVAVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSL 307
KLV RDP G RPLV+G+ +G + SETCALD+I A + R++ PGEV+V D+NG
Sbjct: 204 KLVGARDPRGIRPLVLGEL-DGCPILTSETCALDIIGARFIRDIEPGEVIVFDENGQDIH 262
Query: 308 CLVSHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGV 367
P+ CIFE+IYFS P S+V G+SVYE R+ FG LA ES V DVV+ VPDSGV
Sbjct: 263 KPFPPMAPRPCIFEYIYFSRPDSIVHGRSVYEVRKAFGAQLARESHVPIDVVVPVPDSGV 322
Query: 368 VAALGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVD 427
AA+GY+ +GVPF+ G+IR+HYVGRTFI+P+Q IR+ GV++K S +A +EGKR++++D
Sbjct: 323 PAAVGYSQHSGVPFELGIIRNHYVGRTFIQPTQAIRESGVRMKHSANRAAIEGKRIILID 382
Query: 428 DSIVRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIR 487
DS+VRGTTS KIVR++++AGAKEVH R+A PPI+ YYG+D P L++ S++E+R
Sbjct: 383 DSLVRGTTSKKIVRMMRDAGAKEVHFRLASPPILYPDYYGIDLPDRGGLLAATHSLEEMR 442
Query: 488 EFI 490
E I
Sbjct: 443 EII 445
>UniRef100_UPI00002E385F UPI00002E385F UniRef100 entry
Length = 483
Score = 432 bits (1111), Expect = e-119
Identities = 218/420 (51%), Positives = 298/420 (70%), Gaps = 3/420 (0%)
Query: 72 DDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGL 131
D + K EECGV GI+G P+AS L L LHALQHRGQEGAGIVT K F V GL
Sbjct: 5 DTSEDKLHEECGVFGIFGSPDASTLTALGLHALQHRGQEGAGIVTFDGEK-FHGVRRPGL 63
Query: 132 VADVFTEAK-LDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYR 190
V D F + + +D+L G+ AIGHVRYST G S+LKN+QP A G AHNGNL N
Sbjct: 64 VGDHFNKTEVIDRLLGSNAIGHVRYSTTGNSILKNIQPLYADLSSGGFACAHNGNLTNAS 123
Query: 191 SLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLV 250
LR+ L ++G+IF ++SDTE +L L+A S ++ + +++DA K++GAYS V +T KL+
Sbjct: 124 ILRDSLVKDGAIFQSTSDTETILQLVAKSSRQQTVDKLIDALFKIQGAYSLVILTNKKLI 183
Query: 251 AVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLV 310
VRDP+G RPLV+G NGA V SETCALD+I A+Y R+V GE++++ ++G++S+
Sbjct: 184 GVRDPYGIRPLVLGDL-NGAPVLTSETCALDIIGAKYVRDVEHGEIIIISEDGMESIKPF 242
Query: 311 SHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAA 370
+ + CIFE IYFS P S+V GK+VY R+ G+ LA E+ ++ DV+I VPDSGV AA
Sbjct: 243 PKVQERPCIFERIYFSRPDSIVGGKTVYTYRKNLGQELAKENHIDADVIIPVPDSGVPAA 302
Query: 371 LGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSI 430
LGY+ ++ +PF+ G+IR+HYVGRTFIEPSQ IR GVKLK + + ++ K+V+++DDSI
Sbjct: 303 LGYSQESKIPFELGIIRNHYVGRTFIEPSQSIRQFGVKLKHNANSSYIKNKKVILIDDSI 362
Query: 431 VRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
VRGTT+ KI++++ +AGAKEVH+ IACPPI +YG+DTP ELI+ + S++EI I
Sbjct: 363 VRGTTAKKIIKMVYDAGAKEVHLGIACPPIKHPDFYGIDTPDYNELIAAKSSLEEINNII 422
>UniRef100_Q8CWF4 Amidophosphoribosyltransferase [Bifidobacterium longum]
Length = 503
Score = 432 bits (1111), Expect = e-119
Identities = 211/412 (51%), Positives = 302/412 (73%), Gaps = 2/412 (0%)
Query: 80 EECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVF-TE 138
EECG+ G++G P+ASRL Y LHALQHRGQEGAGIV+ N + G+GL+ VF E
Sbjct: 10 EECGIFGVWGHPDASRLTYFGLHALQHRGQEGAGIVSNDNGHLIGH-RGLGLLTQVFGDE 68
Query: 139 AKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLEE 198
++++L G AIGHVRY+TAG N+QPF+ + G V + HNGNL N SLR KLE+
Sbjct: 69 REIERLKGNCAIGHVRYATAGSGTTDNIQPFIFRFHDGDVALCHNGNLTNCPSLRRKLED 128
Query: 199 NGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFGF 258
G+IF+++SDTEV++HLI S QR F+ ++ +A + G ++++ +TED ++ DP GF
Sbjct: 129 EGAIFHSNSDTEVLMHLIRRSMQRTFMDKLKEALNTVHGGFAYLLMTEDAMIGALDPNGF 188
Query: 259 RPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHPEPKQC 318
RPL +GK NGA V ASETCALD++ AE R + PGE++VV+ +G + + ++ + C
Sbjct: 189 RPLSLGKMKNGAYVLASETCALDVVGAELVRNIRPGEIVVVNDHGYKIVQYTNNTQLAIC 248
Query: 319 IFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGYAAKAG 378
E+IYF+ P S ++G +V+ +R+ G LA ESPVE D+VI VP+S + AA GYA AG
Sbjct: 249 SMEYIYFARPDSDIYGVNVHSARKRMGARLAAESPVEADMVIGVPNSSLSAASGYAEAAG 308
Query: 379 VPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTTSSK 438
+P + GLI++ YV RTFI+P+Q++R+ GV++KLS V++V++GKRV+V+DDSIVRGTTS +
Sbjct: 309 LPNEMGLIKNQYVARTFIQPTQELREQGVRMKLSAVRSVVKGKRVIVIDDSIVRGTTSKR 368
Query: 439 IVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
IV+L+KEAGA EVHMRI+ PP+ C+YG+D +++ELI+ +MSV+EIRE+I
Sbjct: 369 IVQLLKEAGAAEVHMRISSPPLKYPCFYGIDISTTKELIAAKMSVEEIREYI 420
>UniRef100_Q6ARN7 Probable amidophosphoribosyltransferase [Desulfotalea psychrophila]
Length = 479
Score = 432 bits (1111), Expect = e-119
Identities = 214/415 (51%), Positives = 296/415 (70%), Gaps = 4/415 (0%)
Query: 78 PREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLVADVFT 137
P ECGV GI+ +A++L Y L+ALQHRGQE AGIVT ++F +GLV ++FT
Sbjct: 14 PTHECGVCGIFDHEDAAKLTYFGLYALQHRGQESAGIVTSTGKEVFLH-KDMGLVPEIFT 72
Query: 138 EAKLDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRSLREKLE 197
E L L G ++IGHVRYST G S N QP + ++ ++ VAHNGNLVN LR +LE
Sbjct: 73 EDILQGLKGDMSIGHVRYSTTGASNFTNTQPLMVHHKNQTLSVAHNGNLVNSTELRNRLE 132
Query: 198 ENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVAVRDPFG 257
E+GSIF T+ D+E VLHL+ + + + LKGAYS +++TED LVAVRDP G
Sbjct: 133 ESGSIFQTTMDSEAVLHLMVRNSGPDLDKTLSETFRALKGAYSLLYMTEDTLVAVRDPDG 192
Query: 258 FRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVSHPE--P 315
FRPL +G+ + G V ASETCALDLIEA+Y R++ PGEVL++ K G + + P+ P
Sbjct: 193 FRPLCLGRLTTGGWVIASETCALDLIEADYVRDIEPGEVLIM-KRGEEMRSIFPWPKQTP 251
Query: 316 KQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAALGYAA 375
CIFE +YF+ P S VFG +VY+SR+ G+ILA E+ ++ D V+ PDSG AA+GYA
Sbjct: 252 HHCIFEQVYFARPDSDVFGINVYQSRKQMGKILAREAKIDADFVMPFPDSGNYAAIGYAQ 311
Query: 376 KAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIVRGTT 435
++G+P + G+IR+HYVGRTFIEPSQ +RD VK+KL+PV+++L+GKRV++V+DSI+RGTT
Sbjct: 312 ESGIPMEMGVIRNHYVGRTFIEPSQSMRDFNVKVKLNPVRSLLKGKRVIIVEDSIIRGTT 371
Query: 436 SSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
VR ++EAGAKE+HM ++CPP + +CYYG+D P+S +LI+ +V+EI E++
Sbjct: 372 GKSRVRALREAGAKEIHMVVSCPPTMHACYYGIDFPTSSQLIACNKNVEEIAEYL 426
>UniRef100_Q984V0 Amidophosphoribosyltransferase [Rhizobium loti]
Length = 483
Score = 432 bits (1110), Expect = e-119
Identities = 214/419 (51%), Positives = 301/419 (71%), Gaps = 3/419 (0%)
Query: 73 DEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGLV 132
+ D +ECGV GI+G +A+ + L LHALQHRGQE AGIV+ ++ F VGL+
Sbjct: 5 EADDHFHDECGVFGIFGRQDAAAIVTLGLHALQHRGQEAAGIVSYDGSQ-FHVERHVGLI 63
Query: 133 ADVFTEAK-LDQLPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYRS 191
D FT+ + +D L G AIGH RY+T G + ++N+QPF A G VAHNGNL N +
Sbjct: 64 GDTFTKQRVIDSLQGNRAIGHTRYATTGGAGMRNIQPFFAELADGGFAVAHNGNLTNAMT 123
Query: 192 LREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLVA 251
++ L++ G+IF+++SDTE +LHL+ATSK+R R +DA +++GA+S V +T K++
Sbjct: 124 VQRALQKQGAIFSSTSDTETLLHLVATSKERDLNSRFIDAVRQVEGAFSLVAMTAKKMIG 183
Query: 252 VRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLVS 311
RDP G RPLV+G +GA + ASETCALD+I A + R++ PGE++VV G++SL
Sbjct: 184 CRDPLGIRPLVLGDL-DGAWILASETCALDIIGARFVRDLKPGEMVVVTSKGIESLFPFE 242
Query: 312 HPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAAL 371
+ + CIFE++YF+ P S V G++VYE R+ G LA E+PVE D+V+ VPDSG AA+
Sbjct: 243 PQKTRFCIFEYVYFARPDSSVEGRNVYEVRKRIGAELAQENPVEADIVVPVPDSGTPAAI 302
Query: 372 GYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSIV 431
G++ AG+PF+ G+IR+HYVGRTFI+P IR +GVKLK + + ++EGKRVV+VDDSIV
Sbjct: 303 GFSQAAGIPFELGIIRNHYVGRTFIQPGDSIRHMGVKLKHNANRRMIEGKRVVLVDDSIV 362
Query: 432 RGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
RGTTS KIV+++++AGA+EVHMRIA PP SC+YGVDTP +L+++RMSV+E+ EFI
Sbjct: 363 RGTTSQKIVQMVRDAGAREVHMRIASPPTSASCFYGVDTPEKSKLLASRMSVEEMAEFI 421
>UniRef100_Q9A7R2 Amidophosphoribosyltransferase [Caulobacter crescentus]
Length = 500
Score = 430 bits (1106), Expect = e-119
Identities = 217/420 (51%), Positives = 297/420 (70%), Gaps = 3/420 (0%)
Query: 72 DDEDQKPREECGVVGIYGDPEASRLCYLALHALQHRGQEGAGIVTVHNNKIFQSVTGVGL 131
D ED R ECGV G++G +A+ + L LHALQHRGQE GI N+ F + +G
Sbjct: 18 DPEDDSLRLECGVFGVFGVRDAAAIAALGLHALQHRGQEACGIAAFDGNR-FHTERHMGH 76
Query: 132 VADVFTEAKLDQ-LPGTLAIGHVRYSTAGQSMLKNVQPFVAGYRFGSVGVAHNGNLVNYR 190
V D FT A L Q LPG +AIGH RYSTAG S ++NVQP A G V +AHNGNL N+
Sbjct: 77 VGDAFTGADLVQRLPGNMAIGHTRYSTAGGSFIRNVQPMFADLETGGVAIAHNGNLTNFL 136
Query: 191 SLREKLEENGSIFNTSSDTEVVLHLIATSKQRPFILRIVDACEKLKGAYSFVFVTEDKLV 250
+LRE+L + G+IF ++SD+E +LHLIA S++ + R VDA +++G Y+ V +T K++
Sbjct: 137 TLRERLVQEGAIFQSTSDSEAILHLIARSRKAKIVDRFVDAISQIEGGYALVAITNKKMI 196
Query: 251 AVRDPFGFRPLVMGKRSNGAVVFASETCALDLIEAEYEREVYPGEVLVVDKNGLQSLCLV 310
VRDP G RPLV+G +G V ASETCALD+I A + R++ GE++V+++ G+ S
Sbjct: 197 GVRDPLGIRPLVLGDL-DGKPVLASETCALDMIGARFVRDIEHGEMVVIEETGITSTKPF 255
Query: 311 SHPEPKQCIFEHIYFSLPSSVVFGKSVYESRRLFGEILATESPVECDVVIAVPDSGVVAA 370
+ C+FE++YF+ P SVV G+SVY R+ G LA E+ VE DVV+ VPDSGV AA
Sbjct: 256 QTAPARPCVFEYVYFARPDSVVNGRSVYGVRKRMGMNLAKETGVEADVVVPVPDSGVPAA 315
Query: 371 LGYAAKAGVPFQQGLIRSHYVGRTFIEPSQKIRDLGVKLKLSPVKAVLEGKRVVVVDDSI 430
LGYA ++G+PF+ G+IR+HYVGRTFI+P+Q +R+LGV++K SP +AVL GKRVV++DDSI
Sbjct: 316 LGYAQQSGLPFEMGIIRNHYVGRTFIQPTQGVRELGVRMKHSPNRAVLAGKRVVLIDDSI 375
Query: 431 VRGTTSSKIVRLIKEAGAKEVHMRIACPPIIGSCYYGVDTPSSEELISNRMSVDEIREFI 490
VRGTTS KIVR+++EAGAKEVH+R A PPI +YG+D P E+L++ S++E+ F+
Sbjct: 376 VRGTTSLKIVRMVREAGAKEVHLRSASPPIKWPDFYGIDMPEREQLLAANKSLEEMARFL 435
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 837,103,264
Number of Sequences: 2790947
Number of extensions: 37114468
Number of successful extensions: 109758
Number of sequences better than 10.0: 2552
Number of HSP's better than 10.0 without gapping: 1885
Number of HSP's successfully gapped in prelim test: 668
Number of HSP's that attempted gapping in prelim test: 104177
Number of HSP's gapped (non-prelim): 2761
length of query: 490
length of database: 848,049,833
effective HSP length: 131
effective length of query: 359
effective length of database: 482,435,776
effective search space: 173194443584
effective search space used: 173194443584
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0231b.3


