

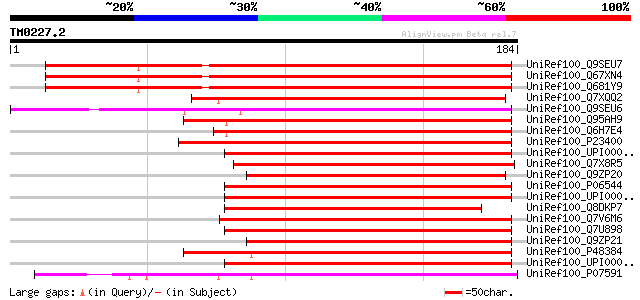
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0227.2
(184 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SEU7 Thioredoxin M-type 3, chloroplast precursor [Ar... 174 1e-42
UniRef100_Q67XN4 Putative thioredoxin M [Arabidopsis thaliana] 173 2e-42
UniRef100_Q681Y9 Putative thioredoxin M [Arabidopsis thaliana] 172 4e-42
UniRef100_Q7XQQ2 OSJNBa0084A10.3 protein [Oryza sativa] 132 4e-30
UniRef100_Q9SEU6 Thioredoxin M-type 4, chloroplast precursor [Ar... 121 8e-27
UniRef100_Q95AH9 Putative thioredoxin m2 [Pisum sativum] 121 1e-26
UniRef100_Q6H7E4 Putative Thioredoxin M-type, chloroplast [Oryza... 118 6e-26
UniRef100_P23400 Thioredoxin M-type, chloroplast precursor [Chla... 115 7e-25
UniRef100_UPI000033CED6 UPI000033CED6 UniRef100 entry 113 2e-24
UniRef100_Q7X8R5 OSJNBa0074L08.22 protein [Oryza sativa] 112 6e-24
UniRef100_Q9ZP20 Thioredoxin M-type, chloroplast precursor [Oryz... 110 2e-23
UniRef100_P06544 Thioredoxin 1 [Anabaena sp.] 109 3e-23
UniRef100_UPI000031FCAB UPI000031FCAB UniRef100 entry 109 4e-23
UniRef100_Q8DKP7 Thioredoxin [Synechococcus elongatus] 108 5e-23
UniRef100_Q7V6M6 Thioredoxin [Prochlorococcus marinus] 108 5e-23
UniRef100_Q7U898 Thioredoxin [Synechococcus sp.] 108 7e-23
UniRef100_Q9ZP21 Thioredoxin M-type, chloroplast precursor [Trit... 108 7e-23
UniRef100_P48384 Thioredoxin M-type, chloroplast precursor [Pisu... 108 7e-23
UniRef100_UPI00002987A9 UPI00002987A9 UniRef100 entry 108 9e-23
UniRef100_P07591 Thioredoxin M-type, chloroplast precursor [Spin... 108 9e-23
>UniRef100_Q9SEU7 Thioredoxin M-type 3, chloroplast precursor [Arabidopsis thaliana]
Length = 173
Score = 174 bits (440), Expect = 1e-42
Identities = 89/170 (52%), Positives = 122/170 (71%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+IA +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEIAGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YEIKAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNADNDLPVAEEYEIKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>UniRef100_Q67XN4 Putative thioredoxin M [Arabidopsis thaliana]
Length = 173
Score = 173 bits (439), Expect = 2e-42
Identities = 88/170 (51%), Positives = 122/170 (71%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+IA +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEIAGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YE+KAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNADNDLPVAEEYEVKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>UniRef100_Q681Y9 Putative thioredoxin M [Arabidopsis thaliana]
Length = 173
Score = 172 bits (436), Expect = 4e-42
Identities = 88/170 (51%), Positives = 121/170 (70%), Gaps = 3/170 (1%)
Query: 14 SSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSF-PFHLRITHKPLHLQNPPHLPRSHKIRS 72
SSSSSS + + H SS S L P SF P LR + + L + R +
Sbjct: 4 SSSSSSICFNPTRFHTARHISSPSRLFPVTSFSPRSLRFSDRRSLLSSSASRLRLSPL-- 61
Query: 73 LRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
+++RA +T+ WE+S+LKS+ PVLVEFY SWCGPCRMVHR+ID+I +YAG+LNC+L
Sbjct: 62 CVRDSRAAEVTQRSWEDSVLKSETPVLVEFYTSWCGPCRMVHRIIDEITGDYAGKLNCYL 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+N D D+ +A++YEIKAVPVV+LFK+G+K ++++GTMPKEFY++AIERVL
Sbjct: 122 LNADNDLPVAEEYEIKAVPVVLLFKNGEKRESIMGTMPKEFYISAIERVL 171
>UniRef100_Q7XQQ2 OSJNBa0084A10.3 protein [Oryza sativa]
Length = 175
Score = 132 bits (332), Expect = 4e-30
Identities = 59/117 (50%), Positives = 84/117 (71%), Gaps = 3/117 (2%)
Query: 67 SHKIRSLR---QETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKE 123
+H+IR++ A IT W ++ SDIPVL+EF+ASWCGPCRMVHR++D+IA+E
Sbjct: 52 AHRIRTVSCAYSPRGAKTITACSWNEYVICSDIPVLIEFWASWCGPCRMVHRIVDEIAQE 111
Query: 124 YAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
YAGR+ C+ ++TD Q+A Y I+ +P V+LFKDG+K ++ GT+PK YV AIE+
Sbjct: 112 YAGRIKCYKLDTDDYPQVATSYSIERIPTVLLFKDGEKTHSITGTLPKAVYVRAIEK 168
>UniRef100_Q9SEU6 Thioredoxin M-type 4, chloroplast precursor [Arabidopsis thaliana]
Length = 193
Score = 121 bits (304), Expect = 8e-27
Identities = 68/194 (35%), Positives = 109/194 (56%), Gaps = 15/194 (7%)
Query: 1 MASISTSISLTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPSLSFPFHLRITHKPLHLQN 60
MAS+ S+++T S ++S SSS S SR ++P+ F + + L Q+
Sbjct: 1 MASLLDSVTVTRVFSLPIAASVSSSSAAP---SVSRRRISPARFLEFRGLKSSRSLVTQS 57
Query: 61 PP-------HLPRSHKIRSLRQETRATPI-----TKDLWENSILKSDIPVLVEFYASWCG 108
+ R +I Q+T A + + W+ +L+SD+PVLVEF+A WCG
Sbjct: 58 ASLGANRRTRIARGGRIACEAQDTTAAAVEVPNLSDSEWQTKVLESDVPVLVEFWAPWCG 117
Query: 109 PCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGT 168
PCRM+H ++D +AK++AG+ + +NTD A+ Y I++VP V++FK G+K D++IG
Sbjct: 118 PCRMIHPIVDQLAKDFAGKFKFYKINTDESPNTANRYGIRSVPTVIIFKGGEKKDSIIGA 177
Query: 169 MPKEFYVNAIERVL 182
+P+E IER L
Sbjct: 178 VPRETLEKTIERFL 191
>UniRef100_Q95AH9 Putative thioredoxin m2 [Pisum sativum]
Length = 180
Score = 121 bits (303), Expect = 1e-26
Identities = 53/121 (43%), Positives = 81/121 (66%), Gaps = 2/121 (1%)
Query: 64 LPRSHKIRSLRQET--RATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIA 121
+PR ++ ++T IT W++ +++SD PVLVEF+A WCGPCRM+H +ID++A
Sbjct: 60 VPRGGRVLCEARDTAVEVASITDGNWQSLVIESDTPVLVEFWAPWCGPCRMMHPIIDELA 119
Query: 122 KEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERV 181
KEY G+ C+ +NTD A Y I+++P V+ FKDG+K DA+IG++PK + IE+
Sbjct: 120 KEYVGKFKCYKLNTDESPSTATRYGIRSIPTVIFFKDGEKKDAIIGSVPKASLITTIEKF 179
Query: 182 L 182
L
Sbjct: 180 L 180
>UniRef100_Q6H7E4 Putative Thioredoxin M-type, chloroplast [Oryza sativa]
Length = 173
Score = 118 bits (296), Expect = 6e-26
Identities = 50/110 (45%), Positives = 83/110 (75%), Gaps = 2/110 (1%)
Query: 75 QET--RATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFL 132
QET + +TK W++ +++S++PVLVEF+ASWCGPC+M+ VI ++KEY G+LNC+
Sbjct: 62 QETSFQVPDVTKSTWQSLVVESELPVLVEFWASWCGPCKMIDPVIGKLSKEYEGKLNCYK 121
Query: 133 VNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+NTD + IA + I+++P +M+FK+G+K DAVIG +P+ V++I++ +
Sbjct: 122 LNTDENPDIATQFGIRSIPTMMIFKNGEKKDAVIGAVPESTLVSSIDKYI 171
>UniRef100_P23400 Thioredoxin M-type, chloroplast precursor [Chlamydomonas
reinhardtii]
Length = 140
Score = 115 bits (287), Expect = 7e-25
Identities = 49/121 (40%), Positives = 78/121 (63%)
Query: 62 PHLPRSHKIRSLRQETRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIA 121
P R+ RS+ A + D ++N +L+S +PVLV+F+A WCGPCR++ V+D+IA
Sbjct: 19 PAFARAAPRRSVVVRAEAGAVNDDTFKNVVLESSVPVLVDFWAPWCGPCRIIAPVVDEIA 78
Query: 122 KEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERV 181
EY +L C +NTD +A +Y I+++P +M+FK G+KC+ +IG +PK V +E+
Sbjct: 79 GEYKDKLKCVKLNTDESPNVASEYGIRSIPTIMVFKGGKKCETIIGAVPKATIVQTVEKY 138
Query: 182 L 182
L
Sbjct: 139 L 139
>UniRef100_UPI000033CED6 UPI000033CED6 UniRef100 entry
Length = 107
Score = 113 bits (283), Expect = 2e-24
Identities = 48/104 (46%), Positives = 72/104 (69%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A P+T +E +L+SD+PVLV+F+A WCGPCRMV ++D+IAKE+ G++ F +NTD +
Sbjct: 4 AAPVTDASFEQDVLQSDVPVLVDFWAPWCGPCRMVAPIVDEIAKEFEGQIKVFKLNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+A Y I+++P +M+FK GQK D V+G +PK I + L
Sbjct: 64 PNVASQYGIRSIPTLMVFKGGQKVDTVVGAVPKATLSGTISKYL 107
>UniRef100_Q7X8R5 OSJNBa0074L08.22 protein [Oryza sativa]
Length = 180
Score = 112 bits (279), Expect = 6e-24
Identities = 42/102 (41%), Positives = 77/102 (75%)
Query: 82 ITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQI 141
+TK W++ +++S++PVLV ++A+WCGPC+M+ V+ ++KEY G+L C+ +NTD + I
Sbjct: 78 VTKSTWQSLVMESELPVLVGYWATWCGPCKMIDPVVGKLSKEYEGKLKCYKLNTDENPDI 137
Query: 142 ADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVLK 183
A Y ++++P +M+FK+G+K DAVIG +P+ + +IE+ ++
Sbjct: 138 ASQYGVRSIPTMMIFKNGEKKDAVIGAVPESTLIASIEKFVE 179
>UniRef100_Q9ZP20 Thioredoxin M-type, chloroplast precursor [Oryza sativa]
Length = 172
Score = 110 bits (274), Expect = 2e-23
Identities = 46/94 (48%), Positives = 69/94 (72%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
W++ +L S+ PVLVEF+A WCGPCRM+ VID++AKEY G++ C VNTD IA +Y
Sbjct: 75 WDSMVLGSEAPVLVEFWAPWCGPCRMIAPVIDELAKEYVGKIKCCKVNTDDSPNIATNYG 134
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
I+++P V++FK+G+K ++VIG +PK I++
Sbjct: 135 IRSIPTVLMFKNGEKKESVIGAVPKTTLATIIDK 168
>UniRef100_P06544 Thioredoxin 1 [Anabaena sp.]
Length = 106
Score = 109 bits (273), Expect = 3e-23
Identities = 47/104 (45%), Positives = 71/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T ++ +L SD+PVLV+F+A WCGPCRMV V+D+IA++Y G++ VNTD +
Sbjct: 3 AAQVTDSTFKQEVLDSDVPVLVDFWAPWCGPCRMVAPVVDEIAQQYEGKIKVVKVNTDEN 62
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
Q+A Y I+++P +M+FK GQK D V+G +PK +E+ L
Sbjct: 63 PQVASQYGIRSIPTLMIFKGGQKVDMVVGAVPKTTLSQTLEKHL 106
>UniRef100_UPI000031FCAB UPI000031FCAB UniRef100 entry
Length = 107
Score = 109 bits (272), Expect = 4e-23
Identities = 46/104 (44%), Positives = 70/104 (67%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T ++ +L+SD+PVLV+F+A WCGPCRMV ++D+IAKE+ GR+ +NTD +
Sbjct: 4 AAAVTDASFDQDVLQSDVPVLVDFWAPWCGPCRMVAPIVDEIAKEFEGRIKVVKLNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+A Y I+++P +M+FK GQK D V+G +PK I + L
Sbjct: 64 PNVASQYGIRSIPTLMVFKSGQKVDTVVGAVPKTTLAGTISKYL 107
>UniRef100_Q8DKP7 Thioredoxin [Synechococcus elongatus]
Length = 107
Score = 108 bits (271), Expect = 5e-23
Identities = 47/93 (50%), Positives = 67/93 (71%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T +E +L SDIPVLV+F+A WCGPCRMV V+D+IA EY GR+ VNTD +
Sbjct: 4 ALSVTDATFEEEVLNSDIPVLVDFWAPWCGPCRMVAPVVDEIANEYQGRVKVVKVNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPK 171
++A D+ I+++P +M+FK GQK D ++G +PK
Sbjct: 64 SKVATDFGIRSIPTLMIFKGGQKVDILVGAVPK 96
>UniRef100_Q7V6M6 Thioredoxin [Prochlorococcus marinus]
Length = 107
Score = 108 bits (271), Expect = 5e-23
Identities = 46/106 (43%), Positives = 71/106 (66%)
Query: 77 TRATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTD 136
+ A +T +E +L+SD+PVLV+F+A WCGPCRMV ++D+IAKE+ ++ F +NTD
Sbjct: 2 SNAAAVTDASFEQDVLQSDVPVLVDFWAPWCGPCRMVAPIVDEIAKEFESKIKVFKLNTD 61
Query: 137 TDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+ +A Y I+++P +M+FK GQK D V+G +PK I + L
Sbjct: 62 ENPNVASQYGIRSIPTLMVFKGGQKVDTVVGAVPKATLSGTISKYL 107
>UniRef100_Q7U898 Thioredoxin [Synechococcus sp.]
Length = 107
Score = 108 bits (270), Expect = 7e-23
Identities = 46/104 (44%), Positives = 71/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T +E +L+SD+PVLV+F+A WCGPCRMV ++++IAKE+ G++ F +NTD +
Sbjct: 4 AAAVTDASFEQDVLQSDVPVLVDFWAPWCGPCRMVAPIVEEIAKEFDGQIKVFKLNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+A Y I+++P +M+FK GQK D V+G +PK I + L
Sbjct: 64 PNVASQYGIRSIPTLMVFKGGQKVDTVVGAVPKATLSGTISKYL 107
>UniRef100_Q9ZP21 Thioredoxin M-type, chloroplast precursor [Triticum aestivum]
Length = 175
Score = 108 bits (270), Expect = 7e-23
Identities = 45/96 (46%), Positives = 68/96 (69%)
Query: 87 WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTDMQIADDYE 146
W+N ++ + PVLVEF+A WCGPCRM+ VID++AK+Y G++ C VNTD IA Y
Sbjct: 78 WDNMVIACESPVLVEFWAPWCGPCRMIAPVIDELAKDYVGKIKCCKVNTDDCPNIASTYG 137
Query: 147 IKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
I+++P V++FKDG+K ++VIG +PK I++ +
Sbjct: 138 IRSIPTVLMFKDGEKKESVIGAVPKTTLCTIIDKYI 173
>UniRef100_P48384 Thioredoxin M-type, chloroplast precursor [Pisum sativum]
Length = 172
Score = 108 bits (270), Expect = 7e-23
Identities = 46/120 (38%), Positives = 76/120 (63%), Gaps = 1/120 (0%)
Query: 64 LPRSHKIRSLRQETRATPITKDL-WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAK 122
L +H + R+ + D W+ ++ S+ PVLV+F+A WCGPCRM+ +ID++AK
Sbjct: 53 LIENHVLLHAREAVNEVQVVNDSSWDELVIGSETPVLVDFWAPWCGPCRMIAPIIDELAK 112
Query: 123 EYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
EYAG++ C+ +NTD A Y I+++P V+ FK+G++ D+VIG +PK +E+ +
Sbjct: 113 EYAGKIKCYKLNTDESPNTATKYGIRSIPTVLFFKNGERKDSVIGAVPKATLSEKVEKYI 172
>UniRef100_UPI00002987A9 UPI00002987A9 UniRef100 entry
Length = 107
Score = 108 bits (269), Expect = 9e-23
Identities = 45/104 (43%), Positives = 71/104 (68%)
Query: 79 ATPITKDLWENSILKSDIPVLVEFYASWCGPCRMVHRVIDDIAKEYAGRLNCFLVNTDTD 138
A +T ++ +L+SD+PVLV+F+A WCGPCRMV ++D+I+KE+ GR+ +NTD +
Sbjct: 4 AAAVTDASFDQDVLQSDVPVLVDFWAPWCGPCRMVAPIVDEISKEFEGRIKVVKLNTDEN 63
Query: 139 MQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIERVL 182
+A Y I+++P +M+FK GQK D V+G +PK + I + L
Sbjct: 64 PNVASQYGIRSIPTLMVFKGGQKVDTVVGAVPKTTLASTISKYL 107
>UniRef100_P07591 Thioredoxin M-type, chloroplast precursor [Spinacia oleracea]
Length = 181
Score = 108 bits (269), Expect = 9e-23
Identities = 60/184 (32%), Positives = 107/184 (57%), Gaps = 18/184 (9%)
Query: 10 LTSSSSSSSSSSSSSSHLHDPFLSSSRSHLNPS--LSFPFH--LRITHKPLHLQNPPHLP 65
L S+S+S + + SH+H HL PS ++ P L+ + L P
Sbjct: 7 LQLSTSASVGTVAVKSHVH---------HLQPSSKVNVPTFRGLKRSFPALSSSVSSSSP 57
Query: 66 RSHKIRSLR-QETRATPITKDL----WENSILKSDIPVLVEFYASWCGPCRMVHRVIDDI 120
R + S+ + + A +D+ W+ +L+S++PV+V+F+A WCGPC+++ VID++
Sbjct: 58 RQFRYSSVVCKASEAVKEVQDVNDSSWKEFVLESEVPVMVDFWAPWCGPCKLIAPVIDEL 117
Query: 121 AKEYAGRLNCFLVNTDTDMQIADDYEIKAVPVVMLFKDGQKCDAVIGTMPKEFYVNAIER 180
AKEY+G++ + +NTD IA Y I+++P V+ FK+G++ +++IG +PK ++IE+
Sbjct: 118 AKEYSGKIAVYKLNTDEAPGIATQYNIRSIPTVLFFKNGERKESIIGAVPKSTLTDSIEK 177
Query: 181 VLKP 184
L P
Sbjct: 178 YLSP 181
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.131 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 297,678,602
Number of Sequences: 2790947
Number of extensions: 12032614
Number of successful extensions: 166650
Number of sequences better than 10.0: 2940
Number of HSP's better than 10.0 without gapping: 2339
Number of HSP's successfully gapped in prelim test: 619
Number of HSP's that attempted gapping in prelim test: 129836
Number of HSP's gapped (non-prelim): 15895
length of query: 184
length of database: 848,049,833
effective HSP length: 120
effective length of query: 64
effective length of database: 513,136,193
effective search space: 32840716352
effective search space used: 32840716352
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0227.2


