

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
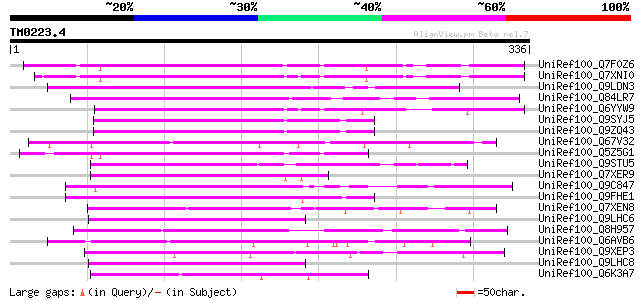
Query= TM0223.4
(336 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7F0Z6 Myb family protein-like [Oryza sativa] 129 1e-28
UniRef100_Q7XNI0 OSJNBb0032D24.13 protein [Oryza sativa] 127 4e-28
UniRef100_Q9LDN3 Gb|AAF34841.1 [Arabidopsis thaliana] 125 2e-27
UniRef100_Q84LR7 Hypothetical protein [Arabidopsis thaliana] 119 2e-25
UniRef100_Q6YYW9 Ribosomal protein-like [Oryza sativa] 118 2e-25
UniRef100_Q9SYJ5 Hypothetical protein T7B11.25 [Arabidopsis thal... 117 3e-25
UniRef100_Q9ZQ43 Hypothetical protein At2g22710 [Arabidopsis tha... 117 4e-25
UniRef100_Q67V32 NAM-like protein [Oryza sativa] 108 2e-22
UniRef100_Q5Z5G1 Myb protein-like [Oryza sativa] 106 8e-22
UniRef100_Q9STU5 Hypothetical protein T23J7.10 [Arabidopsis thal... 104 3e-21
UniRef100_Q7XER9 Hypothetical protein [Oryza sativa] 102 2e-20
UniRef100_Q9C847 Hypothetical protein F8D11.7 [Arabidopsis thali... 100 1e-19
UniRef100_Q9FHE1 Glutathione transferase-like [Arabidopsis thali... 100 1e-19
UniRef100_Q7XEN8 Putative NAM-like protein [Oryza sativa] 100 1e-19
UniRef100_Q9LHC6 Gb|AAF34841.1 [Arabidopsis thaliana] 94 4e-18
UniRef100_Q8H957 A protein [Oryza sativa] 92 3e-17
UniRef100_Q6AVB6 Hypothetical protein OJ1212_C10.14 [Oryza sativa] 92 3e-17
UniRef100_Q9XEP3 Hypothetical protein [Sorghum bicolor] 92 3e-17
UniRef100_Q9LHC8 Gb|AAF34841.1 [Arabidopsis thaliana] 91 3e-17
UniRef100_Q6K3A7 Cytokinin inducibl protein-like [Oryza sativa] 90 8e-17
>UniRef100_Q7F0Z6 Myb family protein-like [Oryza sativa]
Length = 417
Score = 129 bits (323), Expect = 1e-28
Identities = 94/329 (28%), Positives = 158/329 (47%), Gaps = 25/329 (7%)
Query: 10 SQAPPTGSTGSKVSDTQCEATPDDTQHEGLDDIDLEDEDQSSGKKRTR--WRVKDDLLLV 67
+Q + S+ S +QC A ++ +++ D Q +G++ TR W +++L L+
Sbjct: 106 NQVVGSASSHGSESASQCPARQEENNLVNIEESS--DNSQETGRRGTRVNWTEEENLRLL 163
Query: 68 QSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKMLKCRFGKLSKDIQFFT 127
SWLN S D G D+ +W + +F+ + RT K +G + KDI F
Sbjct: 164 SSWLNNSLDSINGNDKKGEYYWRDVAAEFNGNASSNNRKRTVVQCKTHWGGVKKDIAKFC 223
Query: 128 GCYNKVTTPWKSGHSEKDIMAEAHALFQVDHK-KDFTHENVWRMVKDEPKWKGQSMKTNS 186
G Y++ W SG S+ IM +AHAL++ ++ K FT E +WR +KD+PKW Q +
Sbjct: 224 GAYSRARRTWSSGFSDDMIMEKAHALYKSENNDKTFTLEYMWRELKDQPKW--QRILEED 281
Query: 187 RGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKR--KAKTTDTASSTLSFV 244
K++ G T S+S E E + RP+G+K K K K A S L
Sbjct: 282 SKNKRTKISESGAYT--SSSNQETEEETSRKEKRPEGQKKAKAKLKGKGKKPAPSPLGDQ 339
Query: 245 PHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLKQSELQLRQEELKFKKAENFRAYM 304
P D + +A +++ A E L+ A+ K +E + Q + E ++ Y+
Sbjct: 340 PSQDFVLFNEA-VKLRA--------EAVLKSAEATTKSAEAKKEQTRM-----EKYQTYL 385
Query: 305 DILNKNTSGMNDEELRTHNALRAFALSEL 333
+L+K+T+ +D +L+ H A+ +EL
Sbjct: 386 KLLDKDTANFSDAKLKRHEAVLEKLATEL 414
>UniRef100_Q7XNI0 OSJNBb0032D24.13 protein [Oryza sativa]
Length = 399
Score = 127 bits (319), Expect = 4e-28
Identities = 95/322 (29%), Positives = 161/322 (49%), Gaps = 26/322 (8%)
Query: 17 STGSKVSDTQCEATPDDTQHEGLDDIDLEDEDQSSGKKRTR--WRVKDDLLLVQSWLNIS 74
S GSK S +QC A ++ +++ D Q +G++ T W +++L L+ SWLN S
Sbjct: 96 SHGSK-SASQCPARQEENNLVNIEESS--DNSQETGRRGTHVNWTEEENLRLLSSWLNNS 152
Query: 75 KDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKMLKCRFGKLSKDIQFFTGCYNKVT 134
D G D+ +W + +F+ + RT K +G + KDI F G Y++
Sbjct: 153 LDSINGNDKKGEYYWRDVAAEFNGNASSNNRKRTVVQCKTHWGGVKKDIAKFCGAYSRAR 212
Query: 135 TPWKSGHSEKDIMAEAHALFQVDHK-KDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSG 193
W SG S+ IM +AHAL++ ++ K FT E +WR +KD+PKW+ + ++ +S+ K++
Sbjct: 213 RTWSSGFSDDMIMEKAHALYKSENNDKTFTLEYMWRELKDQPKWR-RILEEDSK-NKRTK 270
Query: 194 AGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKR--KAKTTDTASSTLSFVPHPDVLA 251
G T S+S E E + RP+G+K K K K A S L P D +
Sbjct: 271 ISESGAYT--SSSNQETEEETSRKEKRPEGQKKAKAKLKGKGKKPAPSPLGDQPSQDFVL 328
Query: 252 MGKAKMEMMANFREIRNRELDLQQADQQLKQSELQLRQEELKFKKAENFRAYMDILNKNT 311
+A +++ A E L+ A+ K +E + Q + E ++ Y+ +L+K+T
Sbjct: 329 FNEA-VKLRA--------EAVLKSAEATTKSAEAKKEQTRM-----EKYQTYLKLLDKDT 374
Query: 312 SGMNDEELRTHNALRAFALSEL 333
+ +D +L+ H A+ +EL
Sbjct: 375 ANFSDAKLKRHEAVLEKLATEL 396
>UniRef100_Q9LDN3 Gb|AAF34841.1 [Arabidopsis thaliana]
Length = 287
Score = 125 bits (313), Expect = 2e-27
Identities = 82/270 (30%), Positives = 128/270 (47%), Gaps = 13/270 (4%)
Query: 25 TQCEATPDDTQHEGLDDIDLEDEDQSSGK-KRTRWRVKDDLLLVQSWLNISKDPTVGTDQ 83
T+ A EG D EDE + R +W K+D++LV +WLN SKDP +G DQ
Sbjct: 9 TEVPAFSSQLSEEGSDLEGSEDEVKPKQSISRKKWTAKEDIVLVSAWLNTSKDPVIGNDQ 68
Query: 84 TAAKFWDRIRDQFDEYRDFD-TPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHS 142
FW RI D P R K R+GK++K + F GCY TT SG S
Sbjct: 69 QGQSFWKRIAAYVAASPSLDGLPKREHAKCKHRWGKVNKSVTKFVGCYKTTTTHKTSGQS 128
Query: 143 EKDIMAEAHALFQVDHKKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTD 202
E D+M A+ ++ D KK+FT ++ WR ++ + KW + + + K+ G DG ++
Sbjct: 129 EDDVMKLAYEIYFNDTKKNFTLDHAWRELRYDQKWCEATSRKGDKNAKRKKCG-DGNASS 187
Query: 203 PSASIDCDEYEATPPTTRPKGKKAEKRKAKTTDTASSTLSFVPHPDVLAMGKAK-MEMMA 261
++ D + PP G KA K KA+ S+T+ P + + +E
Sbjct: 188 QPIHVEDDSVMSRPP-----GVKAAKAKAR----KSATIKEGKKPATVKDDSGQSVEHFQ 238
Query: 262 NFREIRNRELDLQQADQQLKQSELQLRQEE 291
N E++ ++ D ++ + +Q E L + E
Sbjct: 239 NLWELKEKDWDRKEKQSKHQQLERLLSKTE 268
>UniRef100_Q84LR7 Hypothetical protein [Arabidopsis thaliana]
Length = 392
Score = 119 bits (297), Expect = 2e-25
Identities = 81/292 (27%), Positives = 137/292 (46%), Gaps = 25/292 (8%)
Query: 40 DDIDLEDEDQSSGKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEY 99
D LE+ ++R +W K+D++LV SWLN SKD +G +Q A FW RI +D
Sbjct: 86 DSPSLEEHVPKVKRERMKWSAKEDMVLVSSWLNTSKDAVIGNEQKANTFWSRIAAYYDAS 145
Query: 100 RDFD-TPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDH 158
+ R +K R+ K++ + F G Y + SG ++ D+++ AH +F D+
Sbjct: 146 PQLNGLRKRMQGNIKQRWAKINDGVCKFVGSYEAASREKSSGQNDNDVISLAHEIFNNDY 205
Query: 159 KKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPT 218
F E+ WR+++ + KW Q+ S+ +K A TS PS ++
Sbjct: 206 GYKFPLEHAWRVLRHDQKWCSQA-SVMSKRRKCDKAAQPSTSQPPSHGVE-------EAM 257
Query: 219 TRPKGKKAEKRKAKTTDTASSTLSFVPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQ 278
+RP G KA K KAK T T ++T V G A +E+ + + +++Q D
Sbjct: 258 SRPIGVKAAKAKAKKTVTKTTT--------VEDKGNAMLEIQSIW--------EIKQKDW 301
Query: 279 QLKQSELQLRQEELKFKKAENFRAYMDILNKNTSGMNDEELRTHNALRAFAL 330
+L+Q + + +E+ + K + ++ L + D E+ N L F L
Sbjct: 302 ELRQKDREQEKEDFEKKDRLSKTTLLESLIAKKEPLTDNEVTLKNKLIDFLL 353
>UniRef100_Q6YYW9 Ribosomal protein-like [Oryza sativa]
Length = 303
Score = 118 bits (296), Expect = 2e-25
Identities = 83/283 (29%), Positives = 139/283 (48%), Gaps = 25/283 (8%)
Query: 56 TRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKMLKCR 115
+ W +++L L+ SWLN S D G D+ +W + +F+ + RT K
Sbjct: 38 SNWTEEENLRLLSSWLNNSLDSINGNDKKGEYYWRDVAAEFNGNTSSNNRKRTVVQCKTH 97
Query: 116 FGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHK-KDFTHENVWRMVKDE 174
+G + KDI F G Y++ W SG S+ IM +AHAL++ ++ K FT E +WR +KD+
Sbjct: 98 WGGVKKDIAKFCGAYSRARRTWSSGFSDDMIMEKAHALYKSENNDKTFTLEYMWRELKDQ 157
Query: 175 PKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAE--KRKAK 232
PKW+ + ++ +S+ K++ G T S+S E E + RP+G+K K K K
Sbjct: 158 PKWR-RILEEDSK-NKRTKISESGAYT--SSSNQETEEETSRKEKRPEGQKKAKVKLKGK 213
Query: 233 TTDTASSTLSFVPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLKQSELQLRQEEL 292
A S L P D + + +A +A+ LK +E + E
Sbjct: 214 GKKHAPSPLGDQPSQDFVLLNEAVK----------------LRAEAVLKSTEATTKSAEA 257
Query: 293 KFK--KAENFRAYMDILNKNTSGMNDEELRTHNALRAFALSEL 333
K + + E ++ Y+ +L+K+T+ ND +L+ H A+ +EL
Sbjct: 258 KKEQTRMEKYQTYLKLLDKDTANFNDVKLKRHEAILEKLATEL 300
>UniRef100_Q9SYJ5 Hypothetical protein T7B11.25 [Arabidopsis thaliana]
Length = 302
Score = 117 bits (294), Expect = 3e-25
Identities = 67/183 (36%), Positives = 94/183 (50%), Gaps = 5/183 (2%)
Query: 55 RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFD-TPPRTGKMLK 113
R +W K+D++L+ +WLN SKDP VG +Q A FW RI D D P R K
Sbjct: 62 RKKWAAKEDIVLISAWLNTSKDPVVGNEQKAPAFWKRIASYVAANPDLDGVPKRASAQCK 121
Query: 114 CRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKKDFTHENVWRMVKD 173
R+ K+++ + F GCY T SG +E D+M A+ LF+ D KK F+ ++ WR+++
Sbjct: 122 QRWAKMNELVMKFVGCYATTTNQKASGQTENDVMLFANELFENDMKKKFSLDHAWRLLRH 181
Query: 174 EPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAKT 233
+ KW S +G K GT S ID ++ + RP G KA K KAK
Sbjct: 182 DQKWL-ISNAPKGKGIAKRRKVRVGTQAASSQPIDLEDDDV---MRRPPGVKAAKAKAKK 237
Query: 234 TDT 236
T T
Sbjct: 238 TPT 240
>UniRef100_Q9ZQ43 Hypothetical protein At2g22710 [Arabidopsis thaliana]
Length = 300
Score = 117 bits (293), Expect = 4e-25
Identities = 67/183 (36%), Positives = 94/183 (50%), Gaps = 5/183 (2%)
Query: 55 RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFD-TPPRTGKMLK 113
R +W K+D++L+ +WLN SKDP VG +Q A FW RI D D P R K
Sbjct: 60 RKKWAAKEDIVLISAWLNTSKDPVVGNEQKAPAFWKRIASYVAASPDLDGVPKRASAQCK 119
Query: 114 CRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKKDFTHENVWRMVKD 173
R+ K+++ + F GCY T SG +E D+M A+ LF+ D KK F+ ++ WR+++
Sbjct: 120 QRWAKMNELVMKFVGCYATTTNQKASGQTENDVMLFANELFKNDMKKKFSLDHAWRLLRH 179
Query: 174 EPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAKT 233
+ KW S +G K GT S ID ++ + RP G KA K KAK
Sbjct: 180 DQKWL-ISNAPKGKGIAKRRKVRVGTQAASSQPIDLEDDDV---MRRPPGVKAAKAKAKM 235
Query: 234 TDT 236
T T
Sbjct: 236 TPT 238
>UniRef100_Q67V32 NAM-like protein [Oryza sativa]
Length = 376
Score = 108 bits (271), Expect = 2e-22
Identities = 96/345 (27%), Positives = 155/345 (44%), Gaps = 50/345 (14%)
Query: 13 PPTGSTGSKVSD-------TQCEATPDDTQHEGLDDIDLEDEDQSS-------------- 51
PP G G K T E D TQ D+ + D D+S
Sbjct: 40 PPIGVDGWKAQGLFIGEQVTDEEVCTDGTQSVYFTDLLVNDVDESQFAAPTSDPISNGAP 99
Query: 52 -----GKKRTR-WRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTP 105
+ RT+ + ++D+LLV +WLN+ DP G DQ+ +W RI + F ++F++
Sbjct: 100 TAAKISQGRTKNFTTQEDILLVSAWLNVGMDPIQGVDQSQGTYWARIHEYFHANKEFES- 158
Query: 106 PRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKK--DFT 163
R+ L R+ + D+ F GC +++ +SG D +A A LF+ + KK F
Sbjct: 159 TRSESSLLNRWSAIQHDVNIFCGCMSRIEARNQSGSRVDDKIANACELFKEEDKKHRKFN 218
Query: 164 HENVWRMVKDEPKWKGQSMKTN-----SRGQKKSGAGADGTSTDPSASIDCDEY-EATPP 217
+ W ++KD+PKW K S ++K+ A + TS +P+ D D Y P
Sbjct: 219 LMHCWNILKDKPKWMDNRKKVGCAKKPSNKKQKTVANSSPTSVEPA---DLDVYCSDAQP 275
Query: 218 TTRPKGKKAEK---RKAKTTDTASSTLSFVPHPDVL-AMGKAKM--EMMANFREIRNREL 271
+ RP GKKA K R+ +T + + DV+ + K +M + A E R
Sbjct: 276 SVRPDGKKAAKQKLRQGRTIEAVDYLMEKKKEADVVRELKKEEMCKKAFALQEERCKRAF 335
Query: 272 DLQQADQQLKQSELQLRQEEL-KFKKAENFRAYMDILNKNTSGMN 315
LQ+ +L++ + + +++E K +K E R IL + S MN
Sbjct: 336 ALQEERNKLEREKFEFQKKEAEKAEKVEEER----ILGLDLSTMN 376
>UniRef100_Q5Z5G1 Myb protein-like [Oryza sativa]
Length = 409
Score = 106 bits (265), Expect = 8e-22
Identities = 69/231 (29%), Positives = 119/231 (50%), Gaps = 14/231 (6%)
Query: 7 QFQSQAPPTGSTGSKVSDTQCEATPDDTQHEGLDDIDLEDEDQSS--GKKRTR--WRVKD 62
Q+ + T +T S S A+P T+H + +++E+ SS G++ TR W D
Sbjct: 113 QYSTSIRATANTSSHGS-----ASPCHTRHNEKEVVEVEEASDSSEEGRRGTRINWTEDD 167
Query: 63 DLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKMLKCRFGKLSKD 122
++ L+ SWLN S DP G D+ + ++W + +F+ + R K + + + +D
Sbjct: 168 NIRLMSSWLNNSVDPIKGNDKKSEQYWKAVAREFNSNMPSNGNKRNPKQCRTHWDNVKRD 227
Query: 123 IQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQV-DHKKDFTHENVWRMVKDEPKWKGQS 181
+ F G Y+K T + SG+S+ IM +A ++ +++K FT E +W+ +KD+PKW+
Sbjct: 228 VTKFCGFYSKARTTFTSGYSDDMIMEKAREWYKKHNNQKPFTLEYMWKDLKDQPKWR--R 285
Query: 182 MKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKAK 232
+ S K+S G T S+S E E RP+G+KA K++ K
Sbjct: 286 VLEESSHNKRSKISESGAYT--SSSNQDTEEETERKEKRPEGQKAAKQRQK 334
>UniRef100_Q9STU5 Hypothetical protein T23J7.10 [Arabidopsis thaliana]
Length = 302
Score = 104 bits (260), Expect = 3e-21
Identities = 69/245 (28%), Positives = 123/245 (50%), Gaps = 19/245 (7%)
Query: 53 KKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFD-TPPRTGKM 111
++R +W +DL+LV +WLN SKD +G +Q FW RI + + R
Sbjct: 45 RERRKWSAGEDLVLVSAWLNTSKDAVIGNEQKGYAFWSRIAAYYGASPKLNGVEKRETGH 104
Query: 112 LKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKKDFTHENVWRMV 171
+K R+ K+++ + F G Y T SG ++ D++A AH ++ +H K FT E+ WR++
Sbjct: 105 IKQRWTKINEGVGKFVGSYEAATKQKSSGQNDDDVVALAHEIYNSEHGK-FTLEHAWRVL 163
Query: 172 KDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTTRPKGKKAEKRKA 231
+ E KW S K+ + +D +++ +EA +RP G KA K KA
Sbjct: 164 RFEQKWL-------SAPSTKATVMSKRRKSDKASTSQPQTHEAEEAMSRPIGVKAAKAKA 216
Query: 232 KTTDTASSTLSFVPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQLKQSELQLRQEE 291
K + ++T V G +E+ + + EI+ ++ +L+Q D++ ++ + + +QE
Sbjct: 217 KKAVSKTTT--------VEDKGNVMLEIQSIW-EIKQKDWELRQKDREQEKEDFE-KQER 266
Query: 292 LKFKK 296
L K
Sbjct: 267 LSRTK 271
>UniRef100_Q7XER9 Hypothetical protein [Oryza sativa]
Length = 300
Score = 102 bits (253), Expect = 2e-20
Identities = 57/170 (33%), Positives = 88/170 (51%), Gaps = 16/170 (9%)
Query: 53 KKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKML 112
+ R W +++L L+ W++ S DP G D+ + +W + D F+ + R+ K L
Sbjct: 62 RTRINWTEEENLHLLSYWIHYSTDPVKGIDRKSEFYWKAVADVFNTSAPTNGHKRSVKQL 121
Query: 113 KCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHK-KDFTHENVWRMV 171
K +G + +DI F G Y ++ WKSG S+ +M AHA+F+ ++K K FT E +WR V
Sbjct: 122 KTHWGDVKRDITKFCGVYGRLKATWKSGQSDDMVMNSAHAIFEKENKDKPFTLEYMWREV 181
Query: 172 KDEPKW-------------KGQSMKTNSR--GQKKSGAGADGTSTDPSAS 206
KD PKW K +++ R GQKK+ AG G D + S
Sbjct: 182 KDLPKWCRIVQEDSGNKRTKEETISKEKRPDGQKKAKAGLKGKGKDAAPS 231
>UniRef100_Q9C847 Hypothetical protein F8D11.7 [Arabidopsis thaliana]
Length = 579
Score = 99.8 bits (247), Expect = 1e-19
Identities = 81/300 (27%), Positives = 132/300 (44%), Gaps = 46/300 (15%)
Query: 37 EGLDDIDLEDEDQSSGKK----------RTRWRVKDDLLLVQSWLNISKDPTVGTDQTAA 86
E DD EDE +GKK R W +D++L+ WLN SKDP VG +Q A
Sbjct: 309 EWSDDDPSEDELPIAGKKGKKGKKAKKPRRNWSSTEDVVLISGWLNTSKDPVVGNEQKGA 368
Query: 87 KFWDRIRDQFDEYRDFDTPPRTGKM-LKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKD 145
FW+RI ++ + G + K R+ K++ + F G Y + SG ++ D
Sbjct: 369 AFWERIAVYYNSSPKLKGVEKRGHICCKQRWSKVNDAVNKFVGSYLAASKQQTSGQNDDD 428
Query: 146 IMAEAHALFQVDHKKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSA 205
+++ AH +F D+ FT E+ WR + + KW QS ++ +K AD S
Sbjct: 429 VVSLAHQIFSKDYGCKFTCEHAWRERRYDQKWIAQSTHGKAKRRK---CEAD------SE 479
Query: 206 SIDCDEYEATPPTTRPKGKKAEKRKAKTTDTASSTLSFVPHPDVLAMGKAKMEMMANFRE 265
+ ++ EA RP G KA K AK A GKAK+ +
Sbjct: 480 PVGVEDKEA-----RPIGVKAAKAAAK------------------AKGKAKLSPVEGEET 516
Query: 266 IRNRELDLQQADQQLKQSELQLRQEELKFKKAENFRAYMDILNKNTSGMNDEELRTHNAL 325
+E+ Q+ ++K+ + +++ + K +N + + L T ++D E+ N L
Sbjct: 517 NALKEI---QSIWEIKEKDHAAKEKLIIIKDKKNRTKFFERLLGKTEPLSDIEIELKNKL 573
>UniRef100_Q9FHE1 Glutathione transferase-like [Arabidopsis thaliana]
Length = 590
Score = 99.8 bits (247), Expect = 1e-19
Identities = 65/209 (31%), Positives = 97/209 (46%), Gaps = 11/209 (5%)
Query: 37 EGLDDIDLEDEDQSSGKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQF 96
E DD +L K R +W +D +L+ +WLN SKDP V + A FW RI F
Sbjct: 252 EKSDDPNLVQNTTDRRKHRRKWSRAEDAILISAWLNTSKDPIVDNEHKACAFWKRIGAYF 311
Query: 97 DEYRDF-DTPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQ 155
+ + P R K R+ KL+ + F GCY++ SG SE D+ A+ ++
Sbjct: 312 NNSASLANLPKREPSHCKQRWSKLNDKVCKFVGCYDQALNQRSSGQSEDDVFQVAYQVYT 371
Query: 156 VDHKKDFTHENVWRMVKDEPKWKGQSMKTNSRG-------QKKSGAGADGTSTDP-SASI 207
++K +FT E+ WR ++ KW NS+G + +G +S++P S I
Sbjct: 372 NNYKSNFTLEHAWRELRHSKKWCSLYPFENSKGGGSSKRTKLNNGDRVYSSSSNPESVPI 431
Query: 208 DCDEYEATPPTTRPKGKKAEKRKAKTTDT 236
DE E P G K+ K+K K T
Sbjct: 432 ALDEEEQV--MDLPLGVKSSKQKEKKVAT 458
>UniRef100_Q7XEN8 Putative NAM-like protein [Oryza sativa]
Length = 370
Score = 99.8 bits (247), Expect = 1e-19
Identities = 78/278 (28%), Positives = 121/278 (43%), Gaps = 33/278 (11%)
Query: 51 SGKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGK 110
+ K+ + +D +LV +WLN + D G +Q +W RI F E ++F +
Sbjct: 93 NNKRSKNFNTAEDQMLVSAWLNTTLDAITGVEQHRDSYWARIHQYFHENKNFPSDQNQNS 152
Query: 111 MLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKKDFTHENVWRM 170
+ C +G + + + F G Y ++ +SG +D + +A L++ KK F+ + W +
Sbjct: 153 LNNC-WGTIQEQVNKFCGYYEQILNGPQSGMVVQDYITQAFTLYKSAEKKTFSLMHCWEI 211
Query: 171 VKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATP------PTTRPKGK 224
+ PKW + QK+ A D PSAS + E+ +P P RP GK
Sbjct: 212 LHHHPKWNDRLF------QKRQKAHVD-PLVRPSASTNSSEFHYSPDINTSDPLVRPPGK 264
Query: 225 KAEKRKAKTTDTASSTLSFVPHPDVLA---MGKAKMEMMANFREIRNRELDLQQADQQLK 281
K EK K +T S+ S P V+A M K EM A RE RN L
Sbjct: 265 KVEKAKCPRGNT--SSCSSESSPVVIALNNMWSEKKEMSAQSREERNDRL---------- 312
Query: 282 QSELQLRQEELKFKK----AENFRAYMDILNKNTSGMN 315
+ L +E LK +K E I+N + S M+
Sbjct: 313 VEVISLEKERLKVEKRKLDIETHEREERIMNMDLSAMD 350
>UniRef100_Q9LHC6 Gb|AAF34841.1 [Arabidopsis thaliana]
Length = 211
Score = 94.4 bits (233), Expect = 4e-18
Identities = 49/142 (34%), Positives = 76/142 (53%), Gaps = 2/142 (1%)
Query: 52 GKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDT-PPRTGK 110
G + +W K+D++LV++WLN +KD +G DQ F +I D D P R
Sbjct: 65 GLSQKKWFPKEDIVLVRAWLNTNKDHVIGNDQQCQSFLKQIASYVDISPQLDDLPKREHA 124
Query: 111 MLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKKDFTHENVWRM 170
K R+ K++K + F GCY TT SG SE D+M A+ ++ D KK F E+ WR
Sbjct: 125 KCKQRWSKVNKSVTKFVGCYKTATTHKTSGQSEDDVMKLAYEIYYNDTKKKFNLEHTWRK 184
Query: 171 VKDEPKW-KGQSMKTNSRGQKK 191
++ + KW + ++K N ++K
Sbjct: 185 LRYDQKWCEAATIKGNKNAKRK 206
>UniRef100_Q8H957 A protein [Oryza sativa]
Length = 352
Score = 91.7 bits (226), Expect = 3e-17
Identities = 69/282 (24%), Positives = 124/282 (43%), Gaps = 32/282 (11%)
Query: 42 IDLEDEDQSSGKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRD 101
ID ED + S + RW +++ L +WLN SKD G D+ FW + D+F++ +
Sbjct: 88 IDEEDNNDDSRAAKKRWTHEEEERLASAWLNASKDSIHGNDKKGDTFWKEVTDEFNK-KG 146
Query: 102 FDTPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKKD 161
R LK + +L I F ++ VT SG+S+ + EA L+ K
Sbjct: 147 NGKRRREINQLKVHWSRLKSAISEFNDYWSTVTQMHTSGYSDDMLEKEAQRLYANRFGKP 206
Query: 162 FTHENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATP-PTTR 220
F + W+++KDEPKW Q + D E +A P +R
Sbjct: 207 FALVHWWKILKDEPKWCAQF----------------------ESEKDKSEMDAVPEQQSR 244
Query: 221 PKGKKAEKRKAKTTDTASSTLSFVPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQL 280
P G++A K + + + ++ +G +++ + R +A Q+
Sbjct: 245 PIGREAAKSERNGKRKKENVME-----GIVLLGDNVQKIIKVHEDRRVDREKATEAQIQI 299
Query: 281 KQSELQLRQEELKFKKAENFRAYMDILNKNTSGMNDEELRTH 322
+ L +E+ K+A+ F Y +L+K+TS M+++++ +H
Sbjct: 300 SNATLLAAKEQ---KEAKMFDVYNTLLSKDTSNMSEDQMASH 338
>UniRef100_Q6AVB6 Hypothetical protein OJ1212_C10.14 [Oryza sativa]
Length = 377
Score = 91.7 bits (226), Expect = 3e-17
Identities = 80/318 (25%), Positives = 145/318 (45%), Gaps = 52/318 (16%)
Query: 25 TQCEATPDDTQHEGLDDIDLEDEDQSSGKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQT 84
T A+P + QH ++++ + +Q K+ + K+D LV +WLN+SKD G +Q+
Sbjct: 24 TMMGASPPEEQHSTVEEVVVVRRNQ---KRTKNFSGKEDEGLVSAWLNVSKDVVQGIEQS 80
Query: 85 AAKFWDRIRDQFDEYRDFDTPPRTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEK 144
+W RI D F +DF T R+ L R+ + +++ F GC +++ +S S +
Sbjct: 81 RCAYWKRIYDYFHANKDF-TSDRSQNSLMHRWSTIQENVTKFEGCLSRIGDRKQSWVSSQ 139
Query: 145 DIMAEAHALFQV--DHKKDFTHENVWRMVKDEPKWKGQSMKTNSRGQKK----SGAGADG 198
D + A AL++ ++ + F + W +++++ KW +S + S ++K S G
Sbjct: 140 DKIMHACALYKAEDENHRPFHMMHCWNLLRNQQKWIDRSSQLPSHKKQKITNNSSLGMST 199
Query: 199 TSTDPSASID--------------------CD-----EYEATPP----TTRPKGKKAEKR 229
T+TD +A+ C+ E PP + RP+ +K EK
Sbjct: 200 TTTDDTAAAPPPPPGFELSKTPESINAPPRCELSERPESRNAPPRCELSERPENRKGEKE 259
Query: 230 KAKTTDTASSTLSFVPHPDVLAMGK----AKMEMMANFREIRNRELD-----LQQADQQL 280
K + + FV D L K + E+ + R I++ L+ L+QA
Sbjct: 260 KLR----RGGDVVFVEDLDDLLAKKKEADVEKELKKDERYIQSHALEQEKVALEQAKVAN 315
Query: 281 KQSELQLRQEELKFKKAE 298
+ L++R +EL+ K E
Sbjct: 316 ETKNLEMRSKELELKTKE 333
>UniRef100_Q9XEP3 Hypothetical protein [Sorghum bicolor]
Length = 1475
Score = 91.7 bits (226), Expect = 3e-17
Identities = 67/284 (23%), Positives = 144/284 (50%), Gaps = 22/284 (7%)
Query: 49 QSSGKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTP--P 106
+ + ++ ++ +++D ++ +WLN+SKDP G +Q + FW RIR F++++D T
Sbjct: 1186 KGTSHRQKKFDIEEDKVICSAWLNVSKDPINGANQGRSTFWGRIRAFFEKHKDKKTAVCV 1245
Query: 107 RTGKMLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALF--QVDHKKDFTH 164
RT + R+ + +D+ F CY K+ SG + +D++ A + + K FT
Sbjct: 1246 RTESSIMHRWLTIQRDVNKFCSCYEKIERRNASGATIQDMINAALETYIGADEENKPFTL 1305
Query: 165 ENVWRMVKDEPKWKGQSMKTNSRGQKKSGAGADGTSTDPSASIDCDEYEATPPTT----R 220
+ + +K+E KWK ++ ++ KK + T ++ AS + + E P + R
Sbjct: 1306 MHCYLKLKEEDKWKSLRIEL-AQKNKKQKTTKESTPSNVEASNNDEIPEVAAPDSEERKR 1364
Query: 221 PKGKKAEKRKAKTTDTASSTLSFVPHPDVLAMGKAKMEMMANFREIRNRELDLQQADQQL 280
PKG+K + ++A+ D + + L M + K + R L++++A ++
Sbjct: 1365 PKGQK-QVKQARRDDASLAVLE--------KMLEKKEARELERDKAREASLEIERASLEI 1415
Query: 281 KQSELQLRQEEL----KFKKAENFRAYMDILNKNTSGMNDEELR 320
+++ L++ ++ L K +AE + DI+ + + +++E+L+
Sbjct: 1416 EKAALEIEKKRLSNEEKKIEAELMKEEKDIMLADKTSLDEEQLQ 1459
>UniRef100_Q9LHC8 Gb|AAF34841.1 [Arabidopsis thaliana]
Length = 171
Score = 91.3 bits (225), Expect = 3e-17
Identities = 48/142 (33%), Positives = 75/142 (52%), Gaps = 2/142 (1%)
Query: 52 GKKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFD-TPPRTGK 110
G + +W K+D++LV++WLN +KD +G DQ F +I D P R
Sbjct: 25 GLSQKKWFPKEDIVLVRAWLNTNKDHVIGNDQQCQSFLKQIASYVAISPQLDYLPKREHA 84
Query: 111 MLKCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKKDFTHENVWRM 170
K R+ K++K + F GCY TT SG SE D+M A+ ++ D KK F E+ WR
Sbjct: 85 KCKQRWSKVNKSVTKFVGCYKTATTHKTSGQSEDDVMKLAYEIYYNDTKKKFNLEHTWRK 144
Query: 171 VKDEPKW-KGQSMKTNSRGQKK 191
++ + KW + ++K N ++K
Sbjct: 145 LRYDQKWCEAATIKGNKNAKRK 166
>UniRef100_Q6K3A7 Cytokinin inducibl protein-like [Oryza sativa]
Length = 320
Score = 90.1 bits (222), Expect = 8e-17
Identities = 51/186 (27%), Positives = 89/186 (47%), Gaps = 7/186 (3%)
Query: 53 KKRTRWRVKDDLLLVQSWLNISKDPTVGTDQTAAKFWDRIRDQFDEYRDFDTPPRTGKML 112
K+ + + K+D+ L SW +IS DP +G +Q +W RI + + RDF++ T L
Sbjct: 31 KRSSNFNRKEDIQLCISWQSISSDPIIGNEQPGKAYWQRIAEHYHANRDFESDRNTFS-L 89
Query: 113 KCRFGKLSKDIQFFTGCYNKVTTPWKSGHSEKDIMAEAHALFQVDHKKD--FTHENVWRM 170
+ R+G + K++ F GCY ++ SG ++++ EA AL+ + K+ F + W
Sbjct: 90 EHRWGNIQKEVSKFQGCYEQIERRHPSGIPHQELVLEAEALYSSNAPKNRAFQFNHCWLK 149
Query: 171 VKDEPKWKGQSMKTNSRGQKKS----GAGADGTSTDPSASIDCDEYEATPPTTRPKGKKA 226
+++ PK++ R +K S AG + D + + RP G+K
Sbjct: 150 LRNSPKFQTLKSHKRPRSRKSSTPIESAGEEDEEGDDARKSTTPDLSQPSAKKRPMGRKE 209
Query: 227 EKRKAK 232
K K K
Sbjct: 210 AKEKLK 215
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.129 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 596,504,314
Number of Sequences: 2790947
Number of extensions: 25952111
Number of successful extensions: 74978
Number of sequences better than 10.0: 243
Number of HSP's better than 10.0 without gapping: 73
Number of HSP's successfully gapped in prelim test: 170
Number of HSP's that attempted gapping in prelim test: 74686
Number of HSP's gapped (non-prelim): 340
length of query: 336
length of database: 848,049,833
effective HSP length: 128
effective length of query: 208
effective length of database: 490,808,617
effective search space: 102088192336
effective search space used: 102088192336
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0223.4


