

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
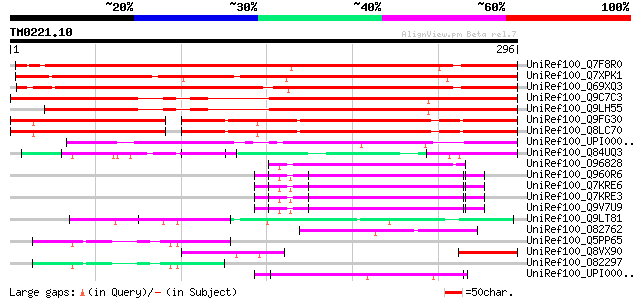
Query= TM0221.10
(296 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7F8R0 KH domain-containing protein-like [Oryza sativa] 349 6e-95
UniRef100_Q7XPK1 OSJNBa0087O24.9 protein [Oryza sativa] 347 2e-94
UniRef100_Q69XQ3 KH domain-containing protein / zinc finger prot... 320 2e-86
UniRef100_Q9C7C3 Hypothetical protein T21B14.5 [Arabidopsis thal... 294 2e-78
UniRef100_Q9LH55 Similarity to unknown protein [Arabidopsis thal... 261 1e-68
UniRef100_Q9FG30 Similarity to unknown protein [Arabidopsis thal... 201 3e-50
UniRef100_Q8LC70 Hypothetical protein [Arabidopsis thaliana] 201 3e-50
UniRef100_UPI0000343263 UPI0000343263 UniRef100 entry 113 7e-24
UniRef100_Q84UQ3 Putative zinc finger protein [Oryza sativa] 70 7e-11
UniRef100_O96828 EG:EG0003.2 protein [Drosophila melanogaster] 64 5e-09
UniRef100_Q960R6 LD38872p [Drosophila melanogaster] 64 5e-09
UniRef100_Q7KRE6 CG8912-PA, isoform A [Drosophila melanogaster] 64 5e-09
UniRef100_Q7KRE3 CG8912-PC, isoform C [Drosophila melanogaster] 64 5e-09
UniRef100_Q9V7U9 CG8912-PB, isoform B [Drosophila melanogaster] 64 5e-09
UniRef100_Q9LT81 Gb|AAC36178.1 [Arabidopsis thaliana] 63 1e-08
UniRef100_O82762 Hypothetical protein At2g25970 [Arabidopsis tha... 62 2e-08
UniRef100_Q5PP65 At2g35430 [Arabidopsis thaliana] 61 3e-08
UniRef100_Q8VX90 Similarity to the EST C99174 [Pinus pinaster] 61 3e-08
UniRef100_O82297 Hypothetical protein At2g35430 [Arabidopsis tha... 59 1e-07
UniRef100_UPI0000435F91 UPI0000435F91 UniRef100 entry 56 1e-06
>UniRef100_Q7F8R0 KH domain-containing protein-like [Oryza sativa]
Length = 300
Score = 349 bits (895), Expect = 6e-95
Identities = 183/298 (61%), Positives = 216/298 (72%), Gaps = 12/298 (4%)
Query: 4 RKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPG 63
RKRG+P+ G N GG K +E ES +GVGSKSKPCTKFFST+GCPFGE CHFLH+ PG
Sbjct: 6 RKRGKPD-GANGAGG--KRARESESFQTGVGSKSKPCTKFFSTSGCPFGEGCHFLHHFPG 62
Query: 64 GYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGS-TPAVKTRICNKFNTAEGCKFGDKCHF 122
GY AVA M NL A APP VP+G TP VKTR+CNK+NTAEGCK+GDKCHF
Sbjct: 63 GYQAVAKMTNLGGPAIAPPPGRMPMGNAVPDGPPTPTVKTRLCNKYNTAEGCKWGDKCHF 122
Query: 123 AHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPAT--GFGANATAKISVEASLA 180
AHGE ELGK + P+G P G P P +T FGA+ATAKISV+ASLA
Sbjct: 123 AHGERELGKPMLMDSSMPPPMGPRPTGHFAPPPMPSPAMSTPASFGASATAKISVDASLA 182
Query: 181 GAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQ 240
G IIG+GGVN+KQI R TGAKL+IR+HESD NL+NIELEG+FDQIK AS MV++L++++
Sbjct: 183 GGIIGRGGVNTKQISRVTGAKLAIRDHESDTNLKNIELEGTFDQIKNASAMVRELIVSIG 242
Query: 241 MSAPPKSNQ--GGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
APP+ + GG+ GG G +NFKTKLCENF KG+CTFG+RCHFAHG ELRK
Sbjct: 243 GGAPPQGKKPVGGSHRGGGPG----SNFKTKLCENFTKGSCTFGDRCHFAHGENELRK 296
Score = 44.3 bits (103), Expect = 0.004
Identities = 20/34 (58%), Positives = 23/34 (66%), Gaps = 1/34 (2%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIA 134
KT++C F T C FGD+CHFAHGE EL K A
Sbjct: 267 KTKLCENF-TKGSCTFGDRCHFAHGENELRKSAA 299
>UniRef100_Q7XPK1 OSJNBa0087O24.9 protein [Oryza sativa]
Length = 309
Score = 347 bits (890), Expect = 2e-94
Identities = 188/306 (61%), Positives = 215/306 (69%), Gaps = 20/306 (6%)
Query: 4 RKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPG 63
RKR RP+ G K+S+ E ES +G+ SK KPCTKFFST GCPFGE CHF H+VPG
Sbjct: 8 RKRSRPDTANGGAAGGKRSR-ETESFQTGLSSKLKPCTKFFSTIGCPFGEGCHFSHFVPG 66
Query: 64 GYNAVAHMMNLA-PSAQAPPRNVAAPPPPVPNGST-PAV--KTRICNKFNTAEGCKFGDK 119
GY AVA +NL P+ AP R AP G++ PA KTR+C K+NTAEGCKFGDK
Sbjct: 67 GYQAVAKTLNLGNPAVPAPAR---APMDHAAGGNSHPASSGKTRMCTKYNTAEGCKFGDK 123
Query: 120 CHFAHGEWELGKHIAPSFDDHRPIGHAP-AGRIGGRMEPPPG-----PATGFGANATAKI 173
CHFAHGE ELGK P++ H P GR GGR EPPP PA FGA+ATAKI
Sbjct: 124 CHFAHGERELGK---PAYMSHESAMAPPMGGRYGGRPEPPPPAAMGPPAGNFGASATAKI 180
Query: 174 SVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVK 233
SV+ASLAG IIGKGGVN+KQICR TG KLSIR+HESD NL+NIELEG+FDQIK+ASNMV
Sbjct: 181 SVDASLAGGIIGKGGVNTKQICRVTGVKLSIRDHESDSNLKNIELEGNFDQIKQASNMVG 240
Query: 234 DLLLTLQMSAPPKSNQGGAGG--PGGHGHHGS-NNFKTKLCENFAKGTCTFGERCHFAHG 290
+L+ T+ S P K G A G P G G G +N+KTKLCENF KGTCTFG+RCHFAHG
Sbjct: 241 ELIATISPSTPAKKPAGSAAGAAPAGRGGPGGRSNYKTKLCENFVKGTCTFGDRCHFAHG 300
Query: 291 PAELRK 296
E RK
Sbjct: 301 ENEQRK 306
Score = 40.8 bits (94), Expect = 0.044
Identities = 26/65 (40%), Positives = 31/65 (47%), Gaps = 5/65 (7%)
Query: 74 LAPSAQAP-PRNVAAPPPPVPNGSTPA---VKTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
++PS A P AA P G KT++C F C FGD+CHFAHGE E
Sbjct: 246 ISPSTPAKKPAGSAAGAAPAGRGGPGGRSNYKTKLCENFVKGT-CTFGDRCHFAHGENEQ 304
Query: 130 GKHIA 134
K A
Sbjct: 305 RKGAA 309
>UniRef100_Q69XQ3 KH domain-containing protein / zinc finger protein-like [Oryza
sativa]
Length = 295
Score = 320 bits (821), Expect = 2e-86
Identities = 171/297 (57%), Positives = 203/297 (67%), Gaps = 18/297 (6%)
Query: 5 KRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGG 64
KR PE +NG K+++ ES GVGSK KPCTKFFST+GCPFG SCHFLH PGG
Sbjct: 7 KRAAPE---GTNGAAKRARAS-ESSQVGVGSKLKPCTKFFSTSGCPFGSSCHFLHNFPGG 62
Query: 65 YNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGS-TPAVKTRICNKFNTAEGCKFGDKCHFA 123
Y A A M + +A A P P PNG T +VKTR+CNK+NTAEGCK+G KCHFA
Sbjct: 63 YQAAAKMTSHGGTAVAAPPGRMPLGPGAPNGPPTSSVKTRMCNKYNTAEGCKWGSKCHFA 122
Query: 124 HGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGP----ATGFGANATAKISVEASL 179
HGE ELGK + P+G P P PGP + FGA+ATAKISV+ASL
Sbjct: 123 HGERELGKPMLLDNSMPHPMGSMPF-----EAPPMPGPDIVPPSTFGASATAKISVDASL 177
Query: 180 AGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKDLLLTL 239
AG IIGKGG N+K I R TGAKL+IR++ES+PNL+NIELEG+FDQIK AS MV +L++ +
Sbjct: 178 AGGIIGKGGTNTKHISRMTGAKLAIRDNESNPNLKNIELEGTFDQIKHASAMVTELIVRI 237
Query: 240 QMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
+APP N G GG G +NFKTKLCENF KG+CTFG+RCHFAHG +ELRK
Sbjct: 238 SGNAPPAKNPGRGSHAGGPG----SNFKTKLCENFNKGSCTFGDRCHFAHGESELRK 290
>UniRef100_Q9C7C3 Hypothetical protein T21B14.5 [Arabidopsis thaliana]
Length = 248
Score = 294 bits (753), Expect = 2e-78
Identities = 164/301 (54%), Positives = 190/301 (62%), Gaps = 62/301 (20%)
Query: 1 MDFRKRGRPEPG-FNSNGG-IKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFL 58
MD RKRGRPE G FNSNGG KKSKQE+ES S+G+GSKSKPCTKFFST+GCPFGE+CHFL
Sbjct: 1 MDTRKRGRPEAGSFNSNGGGYKKSKQEMESYSTGLGSKSKPCTKFFSTSGCPFGENCHFL 60
Query: 59 HYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGD 118
HYVPGGYNAV+ M N+ PP+P S N + G +F
Sbjct: 61 HYVPGGYNAVSQMTNMG--------------PPIPQVSR--------NMQGSGNGGRF-- 96
Query: 119 KCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEAS 178
GR E PG + FG +ATA+ SV+AS
Sbjct: 97 ---------------------------------SGRGESGPGHVSNFGDSATARFSVDAS 123
Query: 179 LAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIKEASNMVKDLLLT 238
LAGAIIGKGGV+SKQICRQTG KLSI++HE DPNL+NI LEG+ +QI EAS MVKDL+
Sbjct: 124 LAGAIIGKGGVSSKQICRQTGVKLSIQDHERDPNLKNIVLEGTLEQISEASAMVKDLIGR 183
Query: 239 LQMSA--PPKSNQGGAGGPGGHGH-HGSNNFKTKLCENFAKGTCTFGERCHFAHGPAELR 295
L +A PP GG GG G G H +NFKTK+CE F+KG CTFG+RCHFAHG AELR
Sbjct: 184 LNSAAKKPPGGGLGGGGGMGSEGKPHPGSNFKTKICERFSKGNCTFGDRCHFAHGEAELR 243
Query: 296 K 296
K
Sbjct: 244 K 244
>UniRef100_Q9LH55 Similarity to unknown protein [Arabidopsis thaliana]
Length = 231
Score = 261 bits (668), Expect = 1e-68
Identities = 143/279 (51%), Positives = 171/279 (61%), Gaps = 60/279 (21%)
Query: 21 KSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQA 80
+S ++ES S+G+GSKSKPCTKFFST+GCPFGE+CHFLHYVPGGYNAV+ M N+
Sbjct: 6 RSLSKMESYSTGLGSKSKPCTKFFSTSGCPFGENCHFLHYVPGGYNAVSQMTNMG----- 60
Query: 81 PPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDH 140
PP+P S N + G +F
Sbjct: 61 ---------PPIPQVSR--------NMQGSGNGGRF------------------------ 79
Query: 141 RPIGHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGA 200
GR E PG + FG +ATA+ SV+ASLAGAIIGKGGV+SKQICRQTG
Sbjct: 80 -----------SGRGESGPGHVSNFGDSATARFSVDASLAGAIIGKGGVSSKQICRQTGV 128
Query: 201 KLSIREHESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQMSA--PPKSNQGGAGGPGGH 258
KLSI++HE DPNL+NI LEG+ +QI EAS MVKDL+ L +A PP GG GG G
Sbjct: 129 KLSIQDHERDPNLKNIVLEGTLEQISEASAMVKDLIGRLNSAAKKPPGGGLGGGGGMGSE 188
Query: 259 GH-HGSNNFKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
G H +NFKTK+CE F+KG CTFG+RCHFAHG AELRK
Sbjct: 189 GKPHPGSNFKTKICERFSKGNCTFGDRCHFAHGEAELRK 227
>UniRef100_Q9FG30 Similarity to unknown protein [Arabidopsis thaliana]
Length = 240
Score = 201 bits (510), Expect = 3e-50
Identities = 107/206 (51%), Positives = 138/206 (66%), Gaps = 17/206 (8%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPI----------GHAPAGR 150
K++ C KF + GC FGD CHF H G + A + RP P GR
Sbjct: 38 KSKPCTKFFSTSGCPFGDNCHFLHYV-PGGYNAAAQMTNLRPPVSQVSRNMQGSGGPGGR 96
Query: 151 IGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESD 210
GR +P GP + FGA+ T+KISV+ASLAGAIIGKGG++SKQICR+TGAKLSI++HE D
Sbjct: 97 FSGRGDPGSGPVSIFGAS-TSKISVDASLAGAIIGKGGIHSKQICRETGAKLSIKDHERD 155
Query: 211 PNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKL 270
PNL+ IELEG+F+QI AS MV++L+ L P+ G GGP G H GS N+KTK+
Sbjct: 156 PNLKIIELEGTFEQINVASGMVRELIGRLGSVKKPQ----GIGGPEGKPHPGS-NYKTKI 210
Query: 271 CENFAKGTCTFGERCHFAHGPAELRK 296
C+ ++KG CT+G+RCHFAHG +ELR+
Sbjct: 211 CDRYSKGNCTYGDRCHFAHGESELRR 236
Score = 133 bits (334), Expect = 6e-30
Identities = 62/93 (66%), Positives = 71/93 (75%), Gaps = 2/93 (2%)
Query: 1 MDFRKRGRPEPG--FNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFL 58
MD RKRGRPE NSNGG K+SKQE+ES+S+G+GSKSKPCTKFFST+GCPFG++CHFL
Sbjct: 1 MDARKRGRPEAAASHNSNGGFKRSKQEMESISTGLGSKSKPCTKFFSTSGCPFGDNCHFL 60
Query: 59 HYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPP 91
HYVPGGYNA A M NL P RN+ P
Sbjct: 61 HYVPGGYNAAAQMTNLRPPVSQVSRNMQGSGGP 93
>UniRef100_Q8LC70 Hypothetical protein [Arabidopsis thaliana]
Length = 240
Score = 201 bits (510), Expect = 3e-50
Identities = 107/206 (51%), Positives = 138/206 (66%), Gaps = 17/206 (8%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPI----------GHAPAGR 150
K++ C KF + GC FGD CHF H G + A + RP P GR
Sbjct: 38 KSKPCTKFFSTSGCPFGDNCHFLHYV-PGGYNAAAQMKNLRPPVSQVSRNMQGSGGPGGR 96
Query: 151 IGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESD 210
GR +P GP + FGA+ T+KISV+ASLAGAIIGKGG++SKQICR+TGAKLSI++HE D
Sbjct: 97 FSGRGDPGSGPVSIFGAS-TSKISVDASLAGAIIGKGGIHSKQICRETGAKLSIKDHERD 155
Query: 211 PNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKL 270
PNL+ IELEG+F+QI AS MV++L+ L P+ G GGP G H GS N+KTK+
Sbjct: 156 PNLKIIELEGTFEQINVASGMVRELIGRLGSVKKPQ----GIGGPEGKPHPGS-NYKTKI 210
Query: 271 CENFAKGTCTFGERCHFAHGPAELRK 296
C+ ++KG CT+G+RCHFAHG +ELR+
Sbjct: 211 CDRYSKGNCTYGDRCHFAHGESELRR 236
Score = 133 bits (334), Expect = 6e-30
Identities = 62/93 (66%), Positives = 71/93 (75%), Gaps = 2/93 (2%)
Query: 1 MDFRKRGRPEPG--FNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFL 58
MD RKRGRPE NSNGG K+SKQE+ES+S+G+GSKSKPCTKFFST+GCPFG++CHFL
Sbjct: 1 MDARKRGRPEAAASHNSNGGFKRSKQEMESISTGLGSKSKPCTKFFSTSGCPFGDNCHFL 60
Query: 59 HYVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPP 91
HYVPGGYNA A M NL P RN+ P
Sbjct: 61 HYVPGGYNAAAQMKNLRPPVSQVSRNMQGSGGP 93
>UniRef100_UPI0000343263 UPI0000343263 UniRef100 entry
Length = 246
Score = 113 bits (282), Expect = 7e-24
Identities = 86/271 (31%), Positives = 121/271 (43%), Gaps = 53/271 (19%)
Query: 34 GSKSKPCTKFFSTAGCPFGESCHFLH-YVPGGYNAVAHMMNLAPSAQAPPRNVAAPPPPV 92
G K C KFFS GC +G+ CH+LH Y PG P+ AP V
Sbjct: 17 GKSDKLCVKFFSIHGCAYGDECHYLHTYRPG---------LPVPARPAPLPYVYTTNAYG 67
Query: 93 PNGSTPAVKTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIG 152
+ +KTR+C F + EGC+FGD+C FAHG+ EL DD
Sbjct: 68 AAQANEKMKTRLCRNFQSPEGCRFGDRCSFAHGKGELRS------DDLNG---------D 112
Query: 153 GRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI--REHESD 210
G ME T G + V S GAI+GK G + Q+ +GAK+S+ E+ +
Sbjct: 113 GTME---NLETHQGFGKAVLVPVPQSQVGAIVGKAGSSIAQVSAASGAKVSMLSAEYTNS 169
Query: 211 PNLRNIELEGSFDQIKEASNMVKDLLLTLQM----SAPPKSNQGGAGGPGGHGHHGSNNF 266
R L GS ++ A ++ L ++ + PP+
Sbjct: 170 DGDRLCRLVGSPFDVQRAKELIYHRLAVGKVKKRDTKPPRP------------------C 211
Query: 267 KTKLCENFAK-GTCTFGERCHFAHGPAELRK 296
KTK+C + K G+C +G++CHFAHG AEL+K
Sbjct: 212 KTKICALWQKDGSCMYGDKCHFAHGIAELQK 242
>UniRef100_Q84UQ3 Putative zinc finger protein [Oryza sativa]
Length = 367
Score = 70.1 bits (170), Expect = 7e-11
Identities = 44/118 (37%), Positives = 57/118 (48%), Gaps = 21/118 (17%)
Query: 31 SGVGSKSKPCTKFFSTAGCPFGESCHFLH----------------YVPGGYNAVAHMMNL 74
SG K + C KF++ GCP+G++C FLH V GG ++
Sbjct: 166 SGRAYKGRHCKKFYTDEGCPYGDACTFLHDEQSKARESVAISLSPSVGGGGGGGSYNSAA 225
Query: 75 APSAQAPPRNVAAPPPPVPNGSTPAVKTRICNKFNTAEGCKFGDKCHFAHGEWELGKH 132
A +A A + AA P+ S KTRICNK+ C FG KCHFAHG EL K+
Sbjct: 226 AAAASA---SAAAGNGPMQKPSN--WKTRICNKWEMTGYCPFGSKCHFAHGAAELHKY 278
Score = 54.3 bits (129), Expect = 4e-06
Identities = 28/58 (48%), Positives = 32/58 (54%), Gaps = 5/58 (8%)
Query: 244 PPKSNQGGAGGPGGHGHH-----GSNNFKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
P K ++GG GG GG G FKTKLC F GTC + C+FAHG ELRK
Sbjct: 61 PGKKSRGGGGGEGGGNTSKSRAIGKMFFKTKLCCKFRAGTCPYVTNCNFAHGMEELRK 118
Score = 50.4 bits (119), Expect = 5e-05
Identities = 43/140 (30%), Positives = 54/140 (37%), Gaps = 22/140 (15%)
Query: 8 RPEPGFNSNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFGESCHFLHYV--- 61
+P PG S GG S S +G K+K C KF CP+ +C+F H +
Sbjct: 58 QPPPGKKSRGGGGGEGGGNTSKSRAIGKMFFKTKLCCKF-RAGTCPYVTNCNFAHGMEEL 116
Query: 62 ----PGGYNAVA-----------HMMNLAPSAQAPPRNVAAPPPPVPNGSTPAVKTRICN 106
P VA H + + S+ A GS A K R C
Sbjct: 117 RKPPPNWQEIVAAHEEATEAREEHQIPIMTSSGPTAGGDAGCGGGGGGGSGRAYKGRHCK 176
Query: 107 KFNTAEGCKFGDKCHFAHGE 126
KF T EGC +GD C F H E
Sbjct: 177 KFYTDEGCPYGDACTFLHDE 196
Score = 47.8 bits (112), Expect = 4e-04
Identities = 54/212 (25%), Positives = 75/212 (34%), Gaps = 39/212 (18%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIAPSFDDHRPIGHAPAGRIGGRMEPPPG 160
KT++C KF A C + C+FAHG EL K P H A + P
Sbjct: 89 KTKLCCKFR-AGTCPYVTNCNFAHGMEELRK--PPPNWQEIVAAHEEATEAREEHQIPIM 145
Query: 161 PATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEG 220
++G A A G GG S + + K + L
Sbjct: 146 TSSGPTAGGDAGCG----------GGGGGGSGRAYKGRHCKKFYTDEGCPYGDACTFLHD 195
Query: 221 SFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGP---------------GGHGHHGSNN 265
+ +E+ + +S P GG GG G +N
Sbjct: 196 EQSKARES----------VAISLSPSVGGGGGGGSYNSAAAAAASASAAAGNGPMQKPSN 245
Query: 266 FKTKLCENFAK-GTCTFGERCHFAHGPAELRK 296
+KT++C + G C FG +CHFAHG AEL K
Sbjct: 246 WKTRICNKWEMTGYCPFGSKCHFAHGAAELHK 277
Score = 33.9 bits (76), Expect = 5.3
Identities = 40/166 (24%), Positives = 59/166 (35%), Gaps = 21/166 (12%)
Query: 138 DDHRPIGHAPAGRIG----GRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQ 193
DD+ +A G G G +PPPG + G + S A IGK +K
Sbjct: 36 DDYNSQWNADGGGGGSSRAGSEQPPPGKKSRGGGGGEGGGNTSKSRA---IGKMFFKTKL 92
Query: 194 ICRQTGAKLSI---------REHESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAP 244
C+ E P E+ + ++ EA + ++T S P
Sbjct: 93 CCKFRAGTCPYVTNCNFAHGMEELRKPPPNWQEIVAAHEEATEAREEHQIPIMT--SSGP 150
Query: 245 PKSNQGGAGGPGGHGHHGSNNFKTKLCENF-AKGTCTFGERCHFAH 289
G GG GG G +K + C+ F C +G+ C F H
Sbjct: 151 TAGGDAGCGGGGGGG--SGRAYKGRHCKKFYTDEGCPYGDACTFLH 194
>UniRef100_O96828 EG:EG0003.2 protein [Drosophila melanogaster]
Length = 806
Score = 63.9 bits (154), Expect = 5e-09
Identities = 42/123 (34%), Positives = 62/123 (50%), Gaps = 12/123 (9%)
Query: 152 GGRME-------PPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI 204
GG++E PPPGP+ G +I + + G +IGKGG KQ+ +TGAK+ I
Sbjct: 204 GGQVEVLLTINMPPPGPS---GYPPYQEIMIPGAKVGLVIGKGGDTIKQLQEKTGAKMII 260
Query: 205 REHESDPNL-RNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGS 263
+ + L + + + G +I+ A MV DL+ + +GG GG GG G G
Sbjct: 261 IQDGPNQELIKPLRISGEAQKIEHAKQMVLDLIAQKDAQGQQQGGRGGGGGGGGPG-MGF 319
Query: 264 NNF 266
NNF
Sbjct: 320 NNF 322
Score = 45.1 bits (105), Expect = 0.002
Identities = 27/84 (32%), Positives = 43/84 (51%), Gaps = 1/84 (1%)
Query: 183 IIGKGGVNSKQICRQTGAKLSI-REHESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQM 241
+IG+GG K I +Q+GA + R + PN + + +G+ DQ++ A M+ + +
Sbjct: 450 VIGRGGETIKLINQQSGAHTEMDRNASNPPNEKLFKSKGTTDQVEAARQMISEKINMELN 509
Query: 242 SAPPKSNQGGAGGPGGHGHHGSNN 265
K GG GG GG+ G NN
Sbjct: 510 VISRKPIGGGPGGGGGNSGGGQNN 533
Score = 35.0 bits (79), Expect = 2.4
Identities = 28/105 (26%), Positives = 47/105 (44%), Gaps = 7/105 (6%)
Query: 144 GHAPAGRIGGRMEPPPGPA-----TGFGANATAKISVEASLAGAIIGKGGVNSKQICRQT 198
G GR GG PG G G +T ++ V G +IGKGG ++I +
Sbjct: 300 GQQQGGRGGGGGGGGPGMGFNNFNNGNGGEST-EVFVPKIAVGVVIGKGGDMIRKIQTEC 358
Query: 199 GAKLSIREHESDP-NLRNIELEGSFDQIKEASNMVKDLLLTLQMS 242
G KL + ++D R ++G+ Q+ +A + L+ + +S
Sbjct: 359 GCKLQFIQGKNDEMGDRRCVIQGTRQQVDDAKRTIDGLIENVMVS 403
>UniRef100_Q960R6 LD38872p [Drosophila melanogaster]
Length = 661
Score = 63.9 bits (154), Expect = 5e-09
Identities = 42/123 (34%), Positives = 62/123 (50%), Gaps = 12/123 (9%)
Query: 152 GGRME-------PPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI 204
GG++E PPPGP+ G +I + + G +IGKGG KQ+ +TGAK+ I
Sbjct: 55 GGQVEVLLTINMPPPGPS---GYPPYQEIMIPGAKVGLVIGKGGDTIKQLQEKTGAKMII 111
Query: 205 REHESDPNL-RNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGS 263
+ + L + + + G +I+ A MV DL+ + +GG GG GG G G
Sbjct: 112 IQDGPNQELIKPLRISGEAQKIEHAKQMVLDLIAQKDAQGQQQGGRGGGGGGGGPG-MGF 170
Query: 264 NNF 266
NNF
Sbjct: 171 NNF 173
Score = 50.4 bits (119), Expect = 5e-05
Identities = 31/92 (33%), Positives = 47/92 (50%), Gaps = 1/92 (1%)
Query: 175 VEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNLRNIELEGSFDQIKEASNMVK 233
V AS G +IG+GG K I +Q+GA + R + PN + + +G+ DQ++ A M+
Sbjct: 297 VPASKCGIVIGRGGETIKLINQQSGAHTEMDRNASNPPNEKLFKSKGTTDQVEAARQMIS 356
Query: 234 DLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNN 265
+ + K GG GG GG+ G NN
Sbjct: 357 EKINMELNVISRKPIGGGPGGGGGNSGGGQNN 388
Score = 49.7 bits (117), Expect = 9e-05
Identities = 40/141 (28%), Positives = 65/141 (45%), Gaps = 10/141 (7%)
Query: 144 GHAPAGRIGGRMEPPPGPA-----TGFGANATAKISVEASLAGAIIGKGGVNSKQICRQT 198
G GR GG PG G G +T ++ V G +IGKGG ++I +
Sbjct: 151 GQQQGGRGGGGGGGGPGMGFNNFNNGNGGEST-EVFVPKIAVGVVIGKGGDMIRKIQTEC 209
Query: 199 GAKLSIREHESDP-NLRNIELEGSFDQIKEASNMVKDLL-LTLQMSAPPKSNQGGAGGPG 256
G KL + ++D R ++G+ Q+ +A + L+ +Q + ++ GG GGPG
Sbjct: 210 GCKLQFIQGKNDEMGDRRCVIQGTRQQVDDAKRTIDGLIENVMQRNGMNRNGNGGGGGPG 269
Query: 257 GHGHHGSNNFKTKLCENFAKG 277
G G++N+ N A+G
Sbjct: 270 GDS--GNSNYGYGYGVNHAQG 288
>UniRef100_Q7KRE6 CG8912-PA, isoform A [Drosophila melanogaster]
Length = 716
Score = 63.9 bits (154), Expect = 5e-09
Identities = 42/123 (34%), Positives = 62/123 (50%), Gaps = 12/123 (9%)
Query: 152 GGRME-------PPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI 204
GG++E PPPGP+ G +I + + G +IGKGG KQ+ +TGAK+ I
Sbjct: 111 GGQVEVLLTINMPPPGPS---GYPPYQEIMIPGAKVGLVIGKGGDTIKQLQEKTGAKMII 167
Query: 205 REHESDPNL-RNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGS 263
+ + L + + + G +I+ A MV DL+ + +GG GG GG G G
Sbjct: 168 IQDGPNQELIKPLRISGEAQKIEHAKQMVLDLIAQKDAQGQQQGGRGGGGGGGGPG-MGF 226
Query: 264 NNF 266
NNF
Sbjct: 227 NNF 229
Score = 50.4 bits (119), Expect = 5e-05
Identities = 31/92 (33%), Positives = 47/92 (50%), Gaps = 1/92 (1%)
Query: 175 VEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNLRNIELEGSFDQIKEASNMVK 233
V AS G +IG+GG K I +Q+GA + R + PN + + +G+ DQ++ A M+
Sbjct: 353 VPASKCGIVIGRGGETIKLINQQSGAHTEMDRNASNPPNEKLFKSKGTTDQVEAARQMIS 412
Query: 234 DLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNN 265
+ + K GG GG GG+ G NN
Sbjct: 413 EKINMELNVISRKPIGGGPGGGGGNSGGGQNN 444
Score = 49.7 bits (117), Expect = 9e-05
Identities = 40/141 (28%), Positives = 65/141 (45%), Gaps = 10/141 (7%)
Query: 144 GHAPAGRIGGRMEPPPGPA-----TGFGANATAKISVEASLAGAIIGKGGVNSKQICRQT 198
G GR GG PG G G +T ++ V G +IGKGG ++I +
Sbjct: 207 GQQQGGRGGGGGGGGPGMGFNNFNNGNGGEST-EVFVPKIAVGVVIGKGGDMIRKIQTEC 265
Query: 199 GAKLSIREHESDP-NLRNIELEGSFDQIKEASNMVKDLL-LTLQMSAPPKSNQGGAGGPG 256
G KL + ++D R ++G+ Q+ +A + L+ +Q + ++ GG GGPG
Sbjct: 266 GCKLQFIQGKNDEMGDRRCVIQGTRQQVDDAKRTIDGLIENVMQRNGMNRNGNGGGGGPG 325
Query: 257 GHGHHGSNNFKTKLCENFAKG 277
G G++N+ N A+G
Sbjct: 326 GDS--GNSNYGYGYGVNHAQG 344
>UniRef100_Q7KRE3 CG8912-PC, isoform C [Drosophila melanogaster]
Length = 797
Score = 63.9 bits (154), Expect = 5e-09
Identities = 42/123 (34%), Positives = 62/123 (50%), Gaps = 12/123 (9%)
Query: 152 GGRME-------PPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI 204
GG++E PPPGP+ G +I + + G +IGKGG KQ+ +TGAK+ I
Sbjct: 191 GGQVEVLLTINMPPPGPS---GYPPYQEIMIPGAKVGLVIGKGGDTIKQLQEKTGAKMII 247
Query: 205 REHESDPNL-RNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGS 263
+ + L + + + G +I+ A MV DL+ + +GG GG GG G G
Sbjct: 248 IQDGPNQELIKPLRISGEAQKIEHAKQMVLDLIAQKDAQGQQQGGRGGGGGGGGPG-MGF 306
Query: 264 NNF 266
NNF
Sbjct: 307 NNF 309
Score = 50.4 bits (119), Expect = 5e-05
Identities = 31/92 (33%), Positives = 47/92 (50%), Gaps = 1/92 (1%)
Query: 175 VEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNLRNIELEGSFDQIKEASNMVK 233
V AS G +IG+GG K I +Q+GA + R + PN + + +G+ DQ++ A M+
Sbjct: 433 VPASKCGIVIGRGGETIKLINQQSGAHTEMDRNASNPPNEKLFKSKGTTDQVEAARQMIS 492
Query: 234 DLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNN 265
+ + K GG GG GG+ G NN
Sbjct: 493 EKINMELNVISRKPIGGGPGGGGGNSGGGQNN 524
Score = 49.7 bits (117), Expect = 9e-05
Identities = 40/141 (28%), Positives = 65/141 (45%), Gaps = 10/141 (7%)
Query: 144 GHAPAGRIGGRMEPPPGPA-----TGFGANATAKISVEASLAGAIIGKGGVNSKQICRQT 198
G GR GG PG G G +T ++ V G +IGKGG ++I +
Sbjct: 287 GQQQGGRGGGGGGGGPGMGFNNFNNGNGGEST-EVFVPKIAVGVVIGKGGDMIRKIQTEC 345
Query: 199 GAKLSIREHESDP-NLRNIELEGSFDQIKEASNMVKDLL-LTLQMSAPPKSNQGGAGGPG 256
G KL + ++D R ++G+ Q+ +A + L+ +Q + ++ GG GGPG
Sbjct: 346 GCKLQFIQGKNDEMGDRRCVIQGTRQQVDDAKRTIDGLIENVMQRNGMNRNGNGGGGGPG 405
Query: 257 GHGHHGSNNFKTKLCENFAKG 277
G G++N+ N A+G
Sbjct: 406 GDS--GNSNYGYGYGVNHAQG 424
>UniRef100_Q9V7U9 CG8912-PB, isoform B [Drosophila melanogaster]
Length = 796
Score = 63.9 bits (154), Expect = 5e-09
Identities = 42/123 (34%), Positives = 62/123 (50%), Gaps = 12/123 (9%)
Query: 152 GGRME-------PPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI 204
GG++E PPPGP+ G +I + + G +IGKGG KQ+ +TGAK+ I
Sbjct: 191 GGQVEVLLTINMPPPGPS---GYPPYQEIMIPGAKVGLVIGKGGDTIKQLQEKTGAKMII 247
Query: 205 REHESDPNL-RNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGS 263
+ + L + + + G +I+ A MV DL+ + +GG GG GG G G
Sbjct: 248 IQDGPNQELIKPLRISGEAQKIEHAKQMVLDLIAQKDAQGQQQGGRGGGGGGGGPG-MGF 306
Query: 264 NNF 266
NNF
Sbjct: 307 NNF 309
Score = 50.4 bits (119), Expect = 5e-05
Identities = 31/92 (33%), Positives = 47/92 (50%), Gaps = 1/92 (1%)
Query: 175 VEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPNLRNIELEGSFDQIKEASNMVK 233
V AS G +IG+GG K I +Q+GA + R + PN + + +G+ DQ++ A M+
Sbjct: 433 VPASKCGIVIGRGGETIKLINQQSGAHTEMDRNASNPPNEKLFKSKGTTDQVEAARQMIS 492
Query: 234 DLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNN 265
+ + K GG GG GG+ G NN
Sbjct: 493 EKINMELNVISRKPIGGGPGGGGGNSGGGQNN 524
Score = 49.7 bits (117), Expect = 9e-05
Identities = 40/141 (28%), Positives = 65/141 (45%), Gaps = 10/141 (7%)
Query: 144 GHAPAGRIGGRMEPPPGPA-----TGFGANATAKISVEASLAGAIIGKGGVNSKQICRQT 198
G GR GG PG G G +T ++ V G +IGKGG ++I +
Sbjct: 287 GQQQGGRGGGGGGGGPGMGFNNFNNGNGGEST-EVFVPKIAVGVVIGKGGDMIRKIQTEC 345
Query: 199 GAKLSIREHESDP-NLRNIELEGSFDQIKEASNMVKDLL-LTLQMSAPPKSNQGGAGGPG 256
G KL + ++D R ++G+ Q+ +A + L+ +Q + ++ GG GGPG
Sbjct: 346 GCKLQFIQGKNDEMGDRRCVIQGTRQQVDDAKRTIDGLIENVMQRNGMNRNGNGGGGGPG 405
Query: 257 GHGHHGSNNFKTKLCENFAKG 277
G G++N+ N A+G
Sbjct: 406 GDS--GNSNYGYGYGVNHAQG 424
>UniRef100_Q9LT81 Gb|AAC36178.1 [Arabidopsis thaliana]
Length = 386
Score = 62.8 bits (151), Expect = 1e-08
Identities = 38/115 (33%), Positives = 55/115 (47%), Gaps = 21/115 (18%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHY------VPGGYNAVAHMMNLAPSAQAPPRNVAAP- 88
+ K C KF CP+G+ C+F+H G + ++++ +A P + A+
Sbjct: 185 RMKLCRKFCFGEECPYGDRCNFIHEDLSKFREDSGKLRESSVISVGATAADQPSDTASNL 244
Query: 89 -----------PPPVPNGS---TPAVKTRICNKFNTAEGCKFGDKCHFAHGEWEL 129
P P+ NG T KTR+C KF+ C FGDKCHFAHG+ EL
Sbjct: 245 IEVNRQGSIPVPAPMNNGGVVKTVYWKTRLCMKFDITGQCPFGDKCHFAHGQAEL 299
Score = 60.8 bits (146), Expect = 4e-08
Identities = 65/233 (27%), Positives = 92/233 (38%), Gaps = 27/233 (11%)
Query: 76 PSAQAPPRNVAAPPPPVPNGSTPAV-KTRICNKFNTAEGCKFGDKCHFAHGEWELGKHIA 134
PS+ P + PPPV G+ KTR+C KF A C+ G+ C+FAHG +L +
Sbjct: 80 PSSSNPWMVPSLNPPPVNKGTANIFYKTRMCAKFR-AGTCRNGELCNFAHGIEDLRQ--- 135
Query: 135 PSFDDHRPIGHAPAG--------RIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGK 186
P + +G PAG R R P P KI + L
Sbjct: 136 PPSNWQEIVGPPPAGQDRERERERERERERPSLAPVVNNNWEDDQKIILRMKLCRKFCFG 195
Query: 187 GGVNSKQICRQTGAKLSIREHESDPNLRNIELEG----SFDQIKEASNMVKDLLLTLQMS 242
C LS + E LR + + DQ + ++ + ++ +
Sbjct: 196 EECPYGDRCNFIHEDLS-KFREDSGKLRESSVISVGATAADQPSDTASNLIEVNRQGSIP 254
Query: 243 APPKSNQGGAGGPGGHGHHGSNNFKTKLCENF-AKGTCTFGERCHFAHGPAEL 294
P N GG + +KT+LC F G C FG++CHFAHG AEL
Sbjct: 255 VPAPMNNGGVVK--------TVYWKTRLCMKFDITGQCPFGDKCHFAHGQAEL 299
Score = 43.9 bits (102), Expect = 0.005
Identities = 28/86 (32%), Positives = 39/86 (44%), Gaps = 10/86 (11%)
Query: 4 RKRGRPEPGFNSNGGIKKSKQELESLSSGVGSKSKPCTKFFSTAGCPFGESCHFLHYVPG 63
R+ P P +NGG+ K+ V K++ C KF T CPFG+ CHF H
Sbjct: 249 RQGSIPVPAPMNNGGVVKT----------VYWKTRLCMKFDITGQCPFGDKCHFAHGQAE 298
Query: 64 GYNAVAHMMNLAPSAQAPPRNVAAPP 89
+N+V + A +A A A P
Sbjct: 299 LHNSVGRVEGEAMNAVASVNKQAVVP 324
Score = 41.6 bits (96), Expect = 0.026
Identities = 16/31 (51%), Positives = 22/31 (70%)
Query: 266 FKTKLCENFAKGTCTFGERCHFAHGPAELRK 296
+KT++C F GTC GE C+FAHG +LR+
Sbjct: 105 YKTRMCAKFRAGTCRNGELCNFAHGIEDLRQ 135
Score = 37.4 bits (85), Expect = 0.48
Identities = 29/118 (24%), Positives = 43/118 (35%), Gaps = 44/118 (37%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVPGGYNAVAHMMNLAPSAQAPPRN---VAAPPP-- 90
K++ C KF C GE C+F H + + PP N + PPP
Sbjct: 106 KTRMCAKF-RAGTCRNGELCNFAHGIE--------------DLRQPPSNWQEIVGPPPAG 150
Query: 91 -------------------PVPNGSTP-----AVKTRICNKFNTAEGCKFGDKCHFAH 124
PV N + ++ ++C KF E C +GD+C+F H
Sbjct: 151 QDRERERERERERERPSLAPVVNNNWEDDQKIILRMKLCRKFCFGEECPYGDRCNFIH 208
>UniRef100_O82762 Hypothetical protein At2g25970 [Arabidopsis thaliana]
Length = 632
Score = 61.6 bits (148), Expect = 2e-08
Identities = 38/107 (35%), Positives = 58/107 (53%), Gaps = 5/107 (4%)
Query: 170 TAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSI-REHESDPN--LRNIELEGSFDQIK 226
T KI + G IIGKGG K + Q+GAK+ + R+ ++DPN R ++L G+ DQI
Sbjct: 135 TKKIDIPNMRVGVIIGKGGETIKYLQLQSGAKIQVTRDMDADPNCATRTVDLTGTPDQIS 194
Query: 227 EASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGHHGSNNFKTKLCEN 273
+A ++ D+L + A + GG GG G G++ F K+ N
Sbjct: 195 KAEQLITDVL--QEAEAGNTAGSGGGGGRRMGGQAGADQFVMKIPNN 239
Score = 35.0 bits (79), Expect = 2.4
Identities = 18/60 (30%), Positives = 33/60 (55%), Gaps = 4/60 (6%)
Query: 181 GAIIGKGGVNSKQICRQTGAKLSIREHESDPN----LRNIELEGSFDQIKEASNMVKDLL 236
G IIGKGG K + +TGA++ + P R ++++G +QI+ A +V +++
Sbjct: 242 GLIIGKGGETIKSMQAKTGARIQVIPLHLPPGDPTPERTLQIDGITEQIEHAKQLVNEII 301
>UniRef100_Q5PP65 At2g35430 [Arabidopsis thaliana]
Length = 252
Score = 61.2 bits (147), Expect = 3e-08
Identities = 50/146 (34%), Positives = 59/146 (40%), Gaps = 50/146 (34%)
Query: 14 NSNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFG-ESCHFLHYVPGGYNAVA 69
NS+G SK+ S SS +G K+K C KF CP+ SCHF H
Sbjct: 48 NSDGAESPSKKTRSSSSSEIGKSFFKTKLCFKF-RAGTCPYSASSCHFAHS--------- 97
Query: 70 HMMNLAPSAQAPPRNVAAPPPPV--------------------PNGS------TPAVKTR 103
A + PP PPPP P G+ +P KTR
Sbjct: 98 -----AEELRLPP-----PPPPNWQETVTEASRNRESFAVSLGPRGNVAQTLKSPNWKTR 147
Query: 104 ICNKFNTAEGCKFGDKCHFAHGEWEL 129
ICNK+ T C FG CHFAHG EL
Sbjct: 148 ICNKWQTTGYCPFGSHCHFAHGPSEL 173
Score = 43.9 bits (102), Expect = 0.005
Identities = 18/33 (54%), Positives = 23/33 (69%), Gaps = 1/33 (3%)
Query: 263 SNNFKTKLCENF-AKGTCTFGERCHFAHGPAEL 294
S N+KT++C + G C FG CHFAHGP+EL
Sbjct: 141 SPNWKTRICNKWQTTGYCPFGSHCHFAHGPSEL 173
Score = 41.2 bits (95), Expect = 0.033
Identities = 19/35 (54%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Query: 262 GSNNFKTKLCENFAKGTCTF-GERCHFAHGPAELR 295
G + FKTKLC F GTC + CHFAH ELR
Sbjct: 68 GKSFFKTKLCFKFRAGTCPYSASSCHFAHSAEELR 102
>UniRef100_Q8VX90 Similarity to the EST C99174 [Pinus pinaster]
Length = 80
Score = 61.2 bits (147), Expect = 3e-08
Identities = 31/66 (46%), Positives = 39/66 (58%), Gaps = 6/66 (9%)
Query: 101 KTRICNKFNTAEGCKFGDKCHFAHGEWELGK--HIAPSFDDHRPIG----HAPAGRIGGR 154
KTR+C+ F T GC+FGDKCHFAHGE ELGK +A + D G P G + G+
Sbjct: 5 KTRLCSNFGTDTGCRFGDKCHFAHGEKELGKVNAVAHNLKDDLATGPFGSRFPVGGLDGK 64
Query: 155 MEPPPG 160
+ PG
Sbjct: 65 LGGRPG 70
Score = 48.5 bits (114), Expect = 2e-04
Identities = 20/35 (57%), Positives = 26/35 (74%), Gaps = 1/35 (2%)
Query: 263 SNNFKTKLCENFAKGT-CTFGERCHFAHGPAELRK 296
++N+KT+LC NF T C FG++CHFAHG EL K
Sbjct: 1 ASNYKTRLCSNFGTDTGCRFGDKCHFAHGEKELGK 35
Score = 37.4 bits (85), Expect = 0.48
Identities = 17/37 (45%), Positives = 22/37 (58%), Gaps = 2/37 (5%)
Query: 36 KSKPCTKFFSTAGCPFGESCHFLHYVP--GGYNAVAH 70
K++ C+ F + GC FG+ CHF H G NAVAH
Sbjct: 5 KTRLCSNFGTDTGCRFGDKCHFAHGEKELGKVNAVAH 41
>UniRef100_O82297 Hypothetical protein At2g35430 [Arabidopsis thaliana]
Length = 180
Score = 59.3 bits (142), Expect = 1e-07
Identities = 48/142 (33%), Positives = 57/142 (39%), Gaps = 50/142 (35%)
Query: 14 NSNGGIKKSKQELESLSSGVGS---KSKPCTKFFSTAGCPFG-ESCHFLHYVPGGYNAVA 69
NS+G SK+ S SS +G K+K C KF CP+ SCHF H
Sbjct: 48 NSDGAESPSKKTRSSSSSEIGKSFFKTKLCFKF-RAGTCPYSASSCHFAHS--------- 97
Query: 70 HMMNLAPSAQAPPRNVAAPPPPV--------------------PNGS------TPAVKTR 103
A + PP PPPP P G+ +P KTR
Sbjct: 98 -----AEELRLPP-----PPPPNWQETVTEASRNRESFAVSLGPRGNVAQTLKSPNWKTR 147
Query: 104 ICNKFNTAEGCKFGDKCHFAHG 125
ICNK+ T C FG CHFAHG
Sbjct: 148 ICNKWQTTGYCPFGSHCHFAHG 169
Score = 41.2 bits (95), Expect = 0.033
Identities = 19/35 (54%), Positives = 21/35 (59%), Gaps = 1/35 (2%)
Query: 262 GSNNFKTKLCENFAKGTCTF-GERCHFAHGPAELR 295
G + FKTKLC F GTC + CHFAH ELR
Sbjct: 68 GKSFFKTKLCFKFRAGTCPYSASSCHFAHSAEELR 102
Score = 40.4 bits (93), Expect = 0.057
Identities = 16/31 (51%), Positives = 21/31 (67%), Gaps = 1/31 (3%)
Query: 263 SNNFKTKLCENF-AKGTCTFGERCHFAHGPA 292
S N+KT++C + G C FG CHFAHGP+
Sbjct: 141 SPNWKTRICNKWQTTGYCPFGSHCHFAHGPS 171
>UniRef100_UPI0000435F91 UPI0000435F91 UniRef100 entry
Length = 654
Score = 56.2 bits (134), Expect = 1e-06
Identities = 42/125 (33%), Positives = 54/125 (42%), Gaps = 20/125 (16%)
Query: 144 GHAPAGRIGGRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLS 203
G P G GG PPG F S+ A G +IG+GG N K I +QTGA +
Sbjct: 383 GRGPGGNWGG----PPGSEMTF--------SIPAHKCGLVIGRGGENVKAINQQTGAFVE 430
Query: 204 IREH---ESDPNLRNIELEGSFDQIKEASNMVKDLLLTLQMSAPPKSNQGGAGGPGGHGH 260
I DPN + + GS QI A +++D ++ P G GGPG G
Sbjct: 431 ISRQPPPNGDPNFKLFTIRGSPQQIDHAKQLIED-----KIEGPLCPVGPGPGGPGPAGP 485
Query: 261 HGSNN 265
G N
Sbjct: 486 MGPYN 490
Score = 49.3 bits (116), Expect = 1e-04
Identities = 35/121 (28%), Positives = 58/121 (47%), Gaps = 6/121 (4%)
Query: 153 GRMEPPPGPATGFGANATAKISVEASLAGAIIGKGGVNSKQICRQTGAK-LSIREHESDP 211
GR PP + G+ ++ + A AG IIGKGG KQ+ + G K + I++ P
Sbjct: 170 GRGTPPSFHESTNGSGHMQEMVIPAGKAGLIIGKGGETIKQLQERAGVKMILIQDASQGP 229
Query: 212 NL-RNIELEGSFDQIKEASNMVKDLLLTLQMSAPPK----SNQGGAGGPGGHGHHGSNNF 266
N+ + + + G ++++A MV+++L + S GG GG GG G G
Sbjct: 230 NMDKPLRIIGDPYKVQQAREMVQEILRERDHPGFERNEYGSRMGGGGGGGGGGGGGGGGI 289
Query: 267 K 267
+
Sbjct: 290 E 290
Score = 35.4 bits (80), Expect = 1.8
Identities = 23/92 (25%), Positives = 44/92 (47%), Gaps = 3/92 (3%)
Query: 167 ANATAKISVEASLAGAIIGKGGVNSKQICRQTGAKLSIREHESDPNLRNIELEGSFDQIK 226
++ T + V + G IIG+GG +I + +G K+ I R++ + G + I+
Sbjct: 98 SSMTEEYRVPDGMVGLIIGRGGEQINKIQQDSGCKVQIAPDSGGLPDRSVSITGGPEAIQ 157
Query: 227 EASNMVKDLLLTLQMSAPPKSNQGGAGGPGGH 258
+A M+ D +++ PP ++ G GH
Sbjct: 158 KAKMMLDD-IVSRGRGTPPSFHESTNG--SGH 186
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.135 0.424
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 596,250,347
Number of Sequences: 2790947
Number of extensions: 28271677
Number of successful extensions: 149590
Number of sequences better than 10.0: 817
Number of HSP's better than 10.0 without gapping: 489
Number of HSP's successfully gapped in prelim test: 337
Number of HSP's that attempted gapping in prelim test: 146331
Number of HSP's gapped (non-prelim): 3228
length of query: 296
length of database: 848,049,833
effective HSP length: 126
effective length of query: 170
effective length of database: 496,390,511
effective search space: 84386386870
effective search space used: 84386386870
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0221.10


