

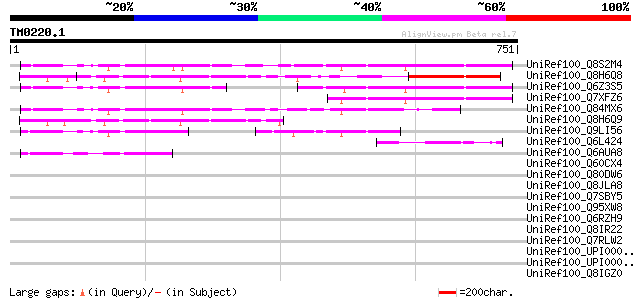
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0220.1
(751 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8S2M4 Hypothetical protein OSJNBa0090K04.9 [Oryza sat... 283 1e-74
UniRef100_Q8H6Q8 CTV.20 [Poncirus trifoliata] 276 2e-72
UniRef100_Q6Z3S5 Hypothetical protein OSJNBa0025J22.29 [Oryza sa... 225 3e-57
UniRef100_Q7XFZ6 Hypothetical protein [Oryza sativa] 224 7e-57
UniRef100_Q84MX6 Hypothetical protein OSJNBa0015G17.8 [Oryza sat... 189 2e-46
UniRef100_Q8H6Q9 CTV.19 [Poncirus trifoliata] 162 3e-38
UniRef100_Q9LI56 EST AU055734(S20025) corresponds to a region of... 96 3e-18
UniRef100_Q6L424 Hypothetical protein PGEC989P08.3 [Solanum demi... 82 4e-14
UniRef100_Q6AUA8 Hypothetical protein P0017D10.13 [Oryza sativa] 69 5e-10
UniRef100_Q60CX4 Hypothetical protein [Solanum tuberosum] 47 0.002
UniRef100_Q80DW6 A5L protein [Cowpox virus] 47 0.003
UniRef100_Q8JLA8 EVM107 [Ectromelia virus] 45 0.010
UniRef100_Q7SBY5 Hypothetical protein [Neurospora crassa] 43 0.030
UniRef100_Q95XW8 Hypothetical protein Y55B1BR.3 [Caenorhabditis ... 43 0.039
UniRef100_Q6RZH9 RPXV112 [Rabbitpox virus] 42 0.067
UniRef100_Q8IR22 CG32580-PA [Drosophila melanogaster] 42 0.067
UniRef100_Q7RLW2 Mannosyltransferase-related [Plasmodium yoelii ... 42 0.067
UniRef100_UPI000046BA1D UPI000046BA1D UniRef100 entry 42 0.088
UniRef100_UPI000046B980 UPI000046B980 UniRef100 entry 42 0.088
UniRef100_Q8IGZ0 RE01659p [Drosophila melanogaster] 42 0.088
>UniRef100_Q8S2M4 Hypothetical protein OSJNBa0090K04.9 [Oryza sativa]
Length = 1634
Score = 283 bits (724), Expect = 1e-74
Identities = 223/763 (29%), Positives = 346/763 (45%), Gaps = 126/763 (16%)
Query: 17 GETTLFLTDYDKANVIVPKTIQWSDITLPDKWSLEKAAPAQPVETPEFREIIQHQSGQVD 76
G TTL T+ + + I PKT++W +ITLP+KW L +A + ++ E +I+ G V+
Sbjct: 15 GLTTLLQTNPNNRHTI-PKTLKWDEITLPEKWVLSQAVEPKSMDQSEVESLIETLDGDVE 73
Query: 77 LIFDRRNSFSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTIHLAGYVN 136
+ F A +++F SR PS+S L
Sbjct: 74 ITF--------------------ASKQKAFLQSR---------PSVS-------LDSRPR 97
Query: 137 TTPVKTTY----DKSDEDHQSTLSPTYSSLETPGETSDFIGAITTE-FVIDKEGIRADFY 191
T P Y D SDE S L+ F+ A+ E F IDKE +R +
Sbjct: 98 TKPQNVVYATYEDNSDEPSISDFDINVIELDV-----GFVIALEEEEFEIDKELLRREIR 152
Query: 192 SDVRTKQREWFFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWFQSYAI----------- 240
+ + + + R KI+E ++ + + ++N+FFFDW+++ I
Sbjct: 153 LPKNRAKTKRYLEEVDESFRMKIREVWHNEMREQRRNIFFFDWYENSQIIHFEEFFKRKN 212
Query: 241 ---KKKISYPYQVDMIK-----WQHKDGKETLSNLPPKTPFLLK--GAQSTPVLASPFKT 290
K++ S + +IK W+ G + S PP L G ++ P+ + T
Sbjct: 213 MIKKEQKSEAEDLTVIKKVSTEWETASGNKVDSVHPPFESIQLSHNGGKACPLKSISKNT 272
Query: 291 REEEGEVTSKDIKDLMEQANYTNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSDLDIPK 350
E +V + I L+EQ NY N L++LG+ ++ + +
Sbjct: 273 YGETAKV--EHIGHLVEQQNYANISLRSLGQQTDRIETI------------------LME 312
Query: 351 GCQLEKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTPEA 410
G + +P K SN S+Q++ + + T +P +
Sbjct: 313 GYKTGRPEVKINIPSNSQSSSSQSV---------------------SPMFVPTIDPNIKF 351
Query: 411 VSNPAVSAPAPPKTRTSTRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTVIHNSRNQWRK 470
A P + + LNN++ K ++ D I VN + S
Sbjct: 352 GKQKAFG---PAISEELVNELALKLNNLKVNKNINEISDNEKYDI-VNKIFKPST--LTS 405
Query: 471 ETKLYYPRATAPDLLLEES---SNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATTY 527
T+ YYPR T DL EE N ++ + EWN+DG EY I + M M A
Sbjct: 406 TTRNYYPRPTYADLQFEEMPQIQNMTYYNGKEIVEWNLDGFTEYQIFTLCHQMIMYANAC 465
Query: 528 VTANSCPESLIVEVLVAGFCGQLKGWWDNYLTEDEKNQILTAIKTDEEGNPIIE-DGKF- 585
+ AN E ++V GF GQLKGWW+NYL E ++ +IL A+K D++G P+ + DG
Sbjct: 466 I-ANGNKEREAANMIVIGFSGQLKGWWNNYLNETQRQEILCAVKRDDQGRPLPDRDGNGN 524
Query: 586 ----ILDAVNSLIFTIAQHFVGDPSLIKDRFGDLLSNLKCKSLGDFRWYKDTFLTRVYTR 641
I DA+ +LI+ I HF G+ I ++ + L NLKCK++ DFRWYKDTFL+++YT
Sbjct: 525 PTGNISDALATLIYNIIYHFAGNYHDIYEKNREQLINLKCKTMSDFRWYKDTFLSKLYTL 584
Query: 642 EDSQQAFWKEKFLAGLPKSFGDKVREKLRSQNPGGEIPYHTLSYGQLIAIIQRVALKICQ 701
+ Q FWKEK+++GLP F +KVR LR + GG I YH L G++ IQ V ++C
Sbjct: 585 PEPNQDFWKEKYISGLPPLFAEKVRNSLRKEG-GGSINYHYLDIGKITQKIQLVGAELCN 643
Query: 702 DDRIQQQLTKEKSQNRRDLGTFCEQFGIQGCPKKPKPRKHDPP 744
D +I+ QL K++ +R++G FC QFG Q K + H P
Sbjct: 644 DLKIKDQLKKQRILGKREMGDFCYQFGFQDPYVYRKRKTHSKP 686
>UniRef100_Q8H6Q8 CTV.20 [Poncirus trifoliata]
Length = 3148
Score = 276 bits (705), Expect = 2e-72
Identities = 215/749 (28%), Positives = 347/749 (45%), Gaps = 159/749 (21%)
Query: 15 KSGETTLFLTDYDKANVIVPKTIQWSDITLPDKWSLEKAA----PAQPVETPEFREIIQH 70
K ET L TD +AN ++PK IQW DI LP++W LE AA P Q E + + Q+
Sbjct: 206 KRDETLLLQTDLSRANTVIPKPIQWKDINLPEEWILEGAAPPTIPKQLEPNTELQNVTQY 265
Query: 71 QSGQVDLIFDRRNS---------FSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPS 121
G+V L+F R S SIP R + S ++ S + + +S PS
Sbjct: 266 SDGKVKLLFRRSMSSRFSNKESCSSIPTLERKFTKIPSVINLPYQSTKSQPRFSTSDIPS 325
Query: 122 LSHEGSTIHLAGYVNTTP----VKTTYDKSDEDHQSTL--SPTYSSLETPGETSDFIGAI 175
S+IH Y P + + +S E+ + +L SPT+S++ T + I I
Sbjct: 326 -----SSIHSVDYTTNVPHPIYTSSQHVQSQEEKEPSLPTSPTFSAV-----TENVINII 375
Query: 176 TTEFVIDKEGIRADFYSDVRTKQREWFFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWF 235
EF +DK + DFYSD+ +++ WFF ++ +RK+IQ+ +Y F+ + ++ FFDWF
Sbjct: 376 EKEFELDKTLLHNDFYSDLNKEKKIWFFKQFL-NQRKEIQQIYYEFVNFHKVHILFFDWF 434
Query: 236 QSYAIKKKISYPYQVD--------MIKWQHKDGKETL-SNLPPKTPFLL-KGAQSTPVLA 285
+ Y+ + I+YP++ + +W+ D +T+ S PP + G + A
Sbjct: 435 EIYSSENNITYPFKESNPITIRKKIPEWKLSDSDKTIESEHPPLRSLTIDHGEPPIQIRA 494
Query: 286 SPFKTREEEGEVTSKDIKDLMEQANYTNKYLQTLGESQSH-DDSSRKGKGKVKTEPSSSS 344
SP+K + + ++ +++Q N+ N L T+G+ + ++ +K V + S
Sbjct: 495 SPYKIPKPND--SDSNLSSIIQQNNFCNTNLNTIGKQLTRIENQFQKSTITVSSTSPIPS 552
Query: 345 DLDIPKGCQLEKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETP 404
D K +L++ +FK FQ+S S+ Q ES + + + EQL +
Sbjct: 553 KSDYDK--KLKEAIFKPFQVSKTSQKLVQ------ESKSDFAKAIREQLDRIE------- 597
Query: 405 EPTPEAVSNPAVSAPAPPKTR-----TSTRNTSTSLNNIEDGKVESDSEDQSVKSIPVNT 459
+ + SN AP P++ TS + +++E K E+ + P
Sbjct: 598 --SASSSSNKVQIAPNTPQSSKIGVLEQDDQTSIASSDLEAFKDEAPT--------PRAN 647
Query: 460 VIHNSRNQWRKETKLYYPRA-TAPDLLLEESSNFKSFSANNVYEWNIDGENEYGITKILQ 518
IH W +L P + PDL ++ ++LQ
Sbjct: 648 KIH-----W----ELAPPTVKSPPDLAIDNR------------------------PRVLQ 674
Query: 519 NMTMVATTYVTANSCPESLIVEVLVAGFCGQLKGWWDNYLTEDEKNQILTAIKTDEEGNP 578
MTM A Y T + + I E+L+AGF GQLKG
Sbjct: 675 QMTMAANAYKTQSGTSDMAIAEILIAGFTGQLKG-------------------------- 708
Query: 579 IIEDGKFILDAVNSLIFTIAQHFVGDPSLIKDRFGDLLSNLKCKSLGDFRWYKDTFLTRV 638
DPS ++D+ +LL NL+C+ L DF+ YK TF TR+
Sbjct: 709 -------------------------DPSHLRDKNAELLHNLRCRKLSDFQLYKTTFFTRL 743
Query: 639 YTREDSQQAFWKEKFLAGLPKSFGDKVREKLRSQNPGGEIPYHTLSYGQLIAIIQRVALK 698
+ R+D+ WKEKFLAGLP G+KVR +++ IPY L+YGQL++ + + LK
Sbjct: 744 FLRDDANHTTWKEKFLAGLPTLLGEKVRNSIKALY-DNRIPYDELTYGQLVSFVNKEGLK 802
Query: 699 ICQDDRIQQQLTKEKSQNRRDLGTFCEQF 727
ICQD ++Q++L +E Q++R+LG+FC+QF
Sbjct: 803 ICQDLKLQKRLKQELRQSKRELGSFCKQF 831
Score = 130 bits (328), Expect = 1e-28
Identities = 60/137 (43%), Positives = 95/137 (68%), Gaps = 1/137 (0%)
Query: 591 NSLIFTIAQHFVGDPSLIKDRFGDLLSNLKCKSLGDFRWYKDTFLTRVYTREDSQQAFWK 650
N+ + + Q+ GDPS ++D+ +LL NL+C+ L DF+ YK TF TR++ R+D+ WK
Sbjct: 1951 NTELQNVTQYSDGDPSHLRDKNAELLHNLRCRKLSDFQLYKTTFFTRLFLRDDANHTTWK 2010
Query: 651 EKFLAGLPKSFGDKVREKLRSQNPGGEIPYHTLSYGQLIAIIQRVALKICQDDRIQQQLT 710
EKFLA LP G+KVR +++ IPY L+YG+L++ + + LKICQD ++Q++L
Sbjct: 2011 EKFLACLPTLLGEKVRNFIKALY-DNRIPYDELTYGELVSFVNKEGLKICQDLKLQKRLK 2069
Query: 711 KEKSQNRRDLGTFCEQF 727
+E Q++R+LG+FC+QF
Sbjct: 2070 QEIRQSKRELGSFCKQF 2086
Score = 59.7 bits (143), Expect = 3e-07
Identities = 33/88 (37%), Positives = 43/88 (48%), Gaps = 4/88 (4%)
Query: 15 KSGETTLFLTDYDKANVIVPKTIQWSDITLPDKWSLEKAA----PAQPVETPEFREIIQH 70
K ET L TD +AN ++PK IQW DI LP++W LE AA P Q E + + Q+
Sbjct: 1901 KRDETLLLQTDLSRANTVIPKPIQWKDINLPEEWILEGAAPPTFPKQLEPNTELQNVTQY 1960
Query: 71 QSGQVDLIFDRRNSFSIPRSIRTSSDFQ 98
G + D+ R SDFQ
Sbjct: 1961 SDGDPSHLRDKNAELLHNLRCRKLSDFQ 1988
>UniRef100_Q6Z3S5 Hypothetical protein OSJNBa0025J22.29 [Oryza sativa]
Length = 1275
Score = 225 bits (574), Expect = 3e-57
Identities = 127/327 (38%), Positives = 181/327 (54%), Gaps = 13/327 (3%)
Query: 427 STRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTVIHNSRNQWRKETKLYYPRATAPDLLL 486
S R+ N IE +E + K VN + S T+ YYPR T DL
Sbjct: 444 SLRSLGQQTNRIETILMEGYKTGRPEKYDMVNKIFKPST--LTSTTRNYYPRPTYADLQF 501
Query: 487 EESSNFKS---FSANNVYEWNIDGENEYGITKILQNMTMVATTYVTANSCPESLIVEVLV 543
EE ++ ++ + EWN+DG EY I + M M A + AN E ++V
Sbjct: 502 EEMPQIQNMIYYNGKEIVEWNLDGFTEYQIFTLCHQMIMYANACI-ANGNKEREAANMIV 560
Query: 544 AGFCGQLKGWWDNYLTEDEKNQILTAIKTDEEGNPIIE-DGKF-----ILDAVNSLIFTI 597
GF GQLKGWW+NYL E ++ +IL A+K D++G P+ + DG I DA+ +LI+ I
Sbjct: 561 IGFSGQLKGWWNNYLNETQRQEILCAVKRDDQGRPLPDRDGNGNPTGNISDALATLIYNI 620
Query: 598 AQHFVGDPSLIKDRFGDLLSNLKCKSLGDFRWYKDTFLTRVYTREDSQQAFWKEKFLAGL 657
HF G+ I ++ + L NLKCK++ DFRWYKDTFL+++YT + Q FWKEK+++GL
Sbjct: 621 IYHFAGNYHDIYEKNREQLINLKCKTMSDFRWYKDTFLSKLYTLPEPNQDFWKEKYISGL 680
Query: 658 PKSFGDKVREKLRSQNPGGEIPYHTLSYGQLIAIIQRVALKICQDDRIQQQLTKEKSQNR 717
P F +KVR LR + GG I YH L G++ IQ V ++C D +I+ QL K++ +
Sbjct: 681 PPLFAEKVRNSLRKEG-GGSINYHYLDIGKITQKIQLVGAELCNDLKIKDQLKKQRILGK 739
Query: 718 RDLGTFCEQFGIQGCPKKPKPRKHDPP 744
R++G FC QFG Q K + H P
Sbjct: 740 REMGDFCYQFGFQDPYVHRKRKTHSKP 766
Score = 85.5 bits (210), Expect = 5e-15
Identities = 84/331 (25%), Positives = 140/331 (41%), Gaps = 70/331 (21%)
Query: 17 GETTLFLTDYDKANVIVPKTIQWSDITLPDKWSLEKAAPAQPVETPEFREIIQHQSGQVD 76
G TTL T+ + + I PKT++W +ITLP+KW L +A + ++ E +I+ G V+
Sbjct: 164 GLTTLLQTNPNNRHTI-PKTLKWDEITLPEKWVLSQAVEPKSMDQSEVESLIETPDGDVE 222
Query: 77 LIFDRRNSFSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTIHLAGYVN 136
+ F A +++F S RPS+S L
Sbjct: 223 ITF--------------------ASKQKAFLQS---------RPSVS-------LDSRPR 246
Query: 137 TTPVKTTY----DKSDEDHQSTLSPTYSSLETPGETSDFIGAI-TTEFVIDKEGIRADFY 191
T P Y D SDE S L+ F+ AI EF IDK+ ++ +
Sbjct: 247 TKPQNVVYATYEDNSDEPSISDFDINVIELDV-----GFVIAIEEDEFEIDKDLLKRELR 301
Query: 192 SDVRTKQREWFFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWFQSYAIKKKISYPYQVD 251
+ + +F + R KI+E ++ + + ++N+FFFDW++S ++ + + +
Sbjct: 302 LQKNRPKMKRYFERVDEPFRLKIRELWHKEMREQRKNIFFFDWYESSQVRHFEEFFKRKN 361
Query: 252 MIK-------------------WQHKDGKETLSNLPPKTPFLL--KGAQSTPVLASPFKT 290
MIK W+ G + S PP L G ++ P+ + T
Sbjct: 362 MIKKEQKSEAEDLTVIKKVSTEWETASGNKVDSVHPPFESVQLSHNGGKACPLKSISKNT 421
Query: 291 REEEGEVTSKDIKDLMEQANYTNKYLQTLGE 321
E +V + I L+EQ NY N L++LG+
Sbjct: 422 YGETAKV--EHIGHLVEQQNYANISLRSLGQ 450
>UniRef100_Q7XFZ6 Hypothetical protein [Oryza sativa]
Length = 1204
Score = 224 bits (571), Expect = 7e-57
Identities = 118/282 (41%), Positives = 167/282 (58%), Gaps = 11/282 (3%)
Query: 472 TKLYYPRATAPDLLLEES---SNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATTYV 528
T+ YYPR T DL EE N ++ + EWN+DG EY I + M M A +
Sbjct: 15 TRNYYPRPTYADLQFEEMPQIQNMTYYNGKEIVEWNLDGFTEYQIFTLCHQMIMYANACI 74
Query: 529 TANSCPESLIVEVLVAGFCGQLKGWWDNYLTEDEKNQILTAIKTDEEGNPIIE-DGKF-- 585
AN E ++V GF GQLKGWW+NYL E ++ +IL A+K D++G P+ + DG
Sbjct: 75 -ANGNKEREAANMIVIGFSGQLKGWWNNYLNETQRQEILCAVKRDDQGRPLPDRDGNGNP 133
Query: 586 ---ILDAVNSLIFTIAQHFVGDPSLIKDRFGDLLSNLKCKSLGDFRWYKDTFLTRVYTRE 642
I DA+ +LI+ I HF G+ I ++ + L NLKCK++ DFRWYKDTFL+++YT
Sbjct: 134 TGNISDALATLIYNIIYHFAGNYHDIYEKNREQLINLKCKTMSDFRWYKDTFLSKLYTLP 193
Query: 643 DSQQAFWKEKFLAGLPKSFGDKVREKLRSQNPGGEIPYHTLSYGQLIAIIQRVALKICQD 702
+ Q FWKEK+++GLP F +KVR LR + GG I YH L G++ IQ V +++C D
Sbjct: 194 EPNQDFWKEKYISGLPPLFAEKVRNSLRKEG-GGSINYHYLDIGKITQKIQLVGVELCND 252
Query: 703 DRIQQQLTKEKSQNRRDLGTFCEQFGIQGCPKKPKPRKHDPP 744
+I+ QL K++ +R++G FC QFG Q K + H P
Sbjct: 253 LKIKDQLKKQRILGKREMGDFCYQFGFQDPYVYRKRKTHSKP 294
>UniRef100_Q84MX6 Hypothetical protein OSJNBa0015G17.8 [Oryza sativa]
Length = 890
Score = 189 bits (481), Expect = 2e-46
Identities = 177/683 (25%), Positives = 283/683 (40%), Gaps = 161/683 (23%)
Query: 17 GETTLFLTDYDKANVIVPKTIQWSDITLPDKWSLEKAAPAQPVETPEFREIIQHQSGQVD 76
G TTL T+ + + I PKT++W +ITLP+KW L +A + ++ E +I+ G V+
Sbjct: 15 GLTTLLQTNPNNRHTI-PKTLKWDEITLPEKWVLSQAVEPKSMDQSEVESLIETPDGDVE 73
Query: 77 LIFDRRNSFSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTIHLAGYVN 136
+ F A +++F SR PS+S L
Sbjct: 74 ITF--------------------ASKQKAFLQSR---------PSVS-------LDSRPR 97
Query: 137 TTPVKTTY----DKSDEDHQSTLSPTYSSLETPGETSDFIGAITT-EFVIDKEGIRADFY 191
T P Y D SDE S L+ F+ AI EF IDK ++ +
Sbjct: 98 TKPQNVVYATYEDNSDEPSISDFDINVIELDV-----GFVIAIEEDEFEIDKYLLKRELR 152
Query: 192 SDVRTKQREWFFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWFQSYAIKKKISYPYQVD 251
+ + +F + R KI+E ++ + + ++N+FFFDW++S ++ + + +
Sbjct: 153 LQKNRPKMKRYFERVDEPFRLKIRELWHKEMREQRKNIFFFDWYESSQVRHFEEFFKRKN 212
Query: 252 MIK-------------------WQHKDGKETLSNLPPKTPFLLK--GAQSTPVLASPFKT 290
MIK W+ G + S PP L G ++ P+ + T
Sbjct: 213 MIKKEQKSEAEDLTVIKKVSTEWETASGDKVDSVHPPFESIQLSHNGGKACPLKSISKNT 272
Query: 291 REEEGEVTSKDIKDLMEQANYTNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSDLDIPK 350
E +V + I L+EQ NY N L++LG+ ++ + +
Sbjct: 273 FGEIAKV--EHIGHLVEQQNYANISLRSLGQQTDRIETI------------------LME 312
Query: 351 GCQLEKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTPEA 410
G + +P K SN S+Q++ + + ++L Q
Sbjct: 313 GYKTGRPEVKINIPSNSQSSSSQSVSPM------FVPTIDPNIKLGKQ-----------K 355
Query: 411 VSNPAVSAPAPPKTRTSTRN--TSTSLNNIEDGKVESDSEDQSVKSIPVNTVIHNSRNQW 468
PA+S K N + ++N I D ++ D K +T+ +RN
Sbjct: 356 AFGPAISEELVNKLALKLNNLKVNKNINEISD----NEKYDMVNKIFKPSTLTSTTRN-- 409
Query: 469 RKETKLYYPRATAPDLLLEES---SNFKSFSANNVYEWNIDGENEYGITKILQNMTMVAT 525
YYPR T DL EE N ++ + EWN+DG EY I + M M A
Sbjct: 410 ------YYPRPTYADLQFEEMPQIQNMTYYNGKEIVEWNLDGFTEYQIFTLCHQMIMYAN 463
Query: 526 TYVTANSCPESLIVEVLVAGFCGQLKGWWDNYLTEDEKNQILTAIKTDEEGNPIIE-DGK 584
+ AN E ++V GF GQLKGWW+NYL E ++ +IL A+K D++G P+ + DG
Sbjct: 464 ACI-ANGNKEREAANMIVIGFSGQLKGWWNNYLNETQRQEILCAVKRDDQGRPLPDRDGN 522
Query: 585 FILDAVNSLIFTIAQHFVGDPSLIKDRFGDLLSNLKCKSLGDFRWYKDTFLTRVYTREDS 644
G+P+ DFRWYKDTFL+++YT +
Sbjct: 523 ------------------GNPT-------------------DFRWYKDTFLSKLYTLPEP 545
Query: 645 QQAFWKEKFLAGLPKSFGDKVRE 667
Q FWKEK+++ LP F +K ++
Sbjct: 546 NQDFWKEKYISSLPPLFAEKKKK 568
>UniRef100_Q8H6Q9 CTV.19 [Poncirus trifoliata]
Length = 433
Score = 162 bits (411), Expect = 3e-38
Identities = 122/430 (28%), Positives = 208/430 (48%), Gaps = 60/430 (13%)
Query: 15 KSGETTLFLTDYDKANVIVPKTIQWSDITLPDKWSLEKAA----PAQPVETPEFREIIQH 70
K ET L TD +AN ++PK IQW DI LP++W LE AA P Q E + + Q+
Sbjct: 14 KRDETLLLQTDLSRANTVIPKPIQWKDINLPEEWILEGAAPPTIPKQLEPNTELQNVTQY 73
Query: 71 QSGQVDLIF---------DRRNSFSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPS 121
G+V L F D+ + SIP R + S ++ S + + +S PS
Sbjct: 74 SDGKVKLSFRRSMSSRFSDKESCSSIPTLERKFTKIPSVINLPYQSTKSQPRFSTSDIPS 133
Query: 122 LSHEGSTIHLAGYVNTTP----VKTTYDKSDEDHQSTL--SPTYSSLETPGETSDFIGAI 175
S+IH Y P + + +S E+ + +L SPT+S++ T + I I
Sbjct: 134 -----SSIHSVDYTTNVPHPIYTSSQHVQSQEEKEPSLPTSPTFSAV-----TENVINVI 183
Query: 176 TTEFVIDKEGIRADFYSDVRTKQREWFFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWF 235
EF +DK + DFYSD+ +++ WFF ++ +RK+IQ+ +Y F+ + ++ FFDWF
Sbjct: 184 EKEFELDKTLLHNDFYSDLNKEKKIWFFKQFL-NQRKEIQQIYYEFVNFHKVHILFFDWF 242
Query: 236 QSYAIKKKISYPYQVD--------MIKWQHKDGKETL-SNLPPKTPFLL-KGAQSTPVLA 285
+ Y+ + I+YP++ + +W+ D +T+ S PP + G + A
Sbjct: 243 KIYSSENNITYPFKESNPITIRKKIPEWKLSDSDKTIESEHPPLRSLTIDHGEPPIQIRA 302
Query: 286 SPFKTREEEGEVTSKDIKDLMEQANYTNKYLQTLGESQSH-DDSSRKGKGKVKTEPSSSS 344
SP+K + + ++ +++Q N+ N L T+G+ + ++ +K V + S
Sbjct: 303 SPYKIPKPND--SDSNLSSIIQQNNFCNTNLNTIGKQLTRIENQFQKSTITVSSTSPIPS 360
Query: 345 DLDIPKGCQLEKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLN------- 397
D K + ++P+FKHFQ+S S+ Q ES + + + EQL +
Sbjct: 361 KSDYDK--KFKEPIFKHFQVSKTSQKLVQ------ESKSDFAKAIREQLDRIESASSSSN 412
Query: 398 --QVIPETPE 405
Q+ P+TP+
Sbjct: 413 KVQIAPDTPQ 422
>UniRef100_Q9LI56 EST AU055734(S20025) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 719
Score = 96.3 bits (238), Expect = 3e-18
Identities = 69/236 (29%), Positives = 104/236 (43%), Gaps = 26/236 (11%)
Query: 365 SNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTPEAVSNPAVSAP----- 419
S++ RH + + KN KE + E L ++ +V E + S+ +P
Sbjct: 321 SSQVRHFEEFFKGKNMMKKEQKSEA-EDLTVIKKVSTEWETTSGNKSSSSQSVSPMFVPT 379
Query: 420 -------------APPKTRTSTRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTVIHNSRN 466
P + + LNN++ K ++ D K VN + S
Sbjct: 380 IDPNIKLGKQKAFGPAISEELVSELALKLNNLKVNKNINEISDNE-KYDMVNKIFKPST- 437
Query: 467 QWRKETKLYYPRATAPDLLLEES---SNFKSFSANNVYEWNIDGENEYGITKILQNMTMV 523
T+ YYPR T DL EE N ++ + EWN+DG EY I + M M
Sbjct: 438 -LTSTTRNYYPRPTYADLQFEEMPQIQNMTYYNGKEIVEWNLDGFTEYQIFTLCHQMIMY 496
Query: 524 ATTYVTANSCPESLIVEVLVAGFCGQLKGWWDNYLTEDEKNQILTAIKTDEEGNPI 579
A + AN E ++V GF GQLKGWW+NYL E ++ +IL A+K D++G P+
Sbjct: 497 ANACI-ANGNKEREAANMIVIGFSGQLKGWWNNYLNETQRQEILCAVKRDDQGRPL 551
Score = 73.2 bits (178), Expect = 3e-11
Identities = 64/254 (25%), Positives = 109/254 (42%), Gaps = 47/254 (18%)
Query: 17 GETTLFLTDYDKANVIVPKTIQWSDITLPDKWSLEKAAPAQPVETPEFREIIQHQSGQVD 76
G TTL T+ + PKT++W +ITLP+KW L +A + ++ E +I+ G V+
Sbjct: 138 GLTTLLQTNPNN-RCTTPKTLKWDEITLPEKWVLSQAVEPKSMDQSEVESLIETPDGDVE 196
Query: 77 LIFDRRNSFSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTIHLAGYVN 136
+ F A +++F S RPS+S L
Sbjct: 197 ITF--------------------ASKQKAFLQS---------RPSVS-------LDSRPR 220
Query: 137 TTPVKTTY----DKSDEDHQSTLSPTYSSLETPGETSDFIGAI-TTEFVIDKEGIRADFY 191
T P Y D SDE S L+ F+ AI EF IDK+ ++ +
Sbjct: 221 TKPQNVVYATYEDNSDEPSISDFDINVIELDV-----GFVIAIEEDEFEIDKDLLKKELR 275
Query: 192 SDVRTKQREWFFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWFQSYAIKKKISYPYQVD 251
+ + +F + R KI+E ++ + + ++N+FFFDW++S ++ + +
Sbjct: 276 LQKNRPKMKRYFERVDEPFRLKIRELWHKEMREQRKNIFFFDWYESSQVRHFEEFFKGKN 335
Query: 252 MIKWQHKDGKETLS 265
M+K + K E L+
Sbjct: 336 MMKKEQKSEAEDLT 349
>UniRef100_Q6L424 Hypothetical protein PGEC989P08.3 [Solanum demissum]
Length = 387
Score = 82.4 bits (202), Expect = 4e-14
Identities = 53/186 (28%), Positives = 82/186 (43%), Gaps = 65/186 (34%)
Query: 544 AGFCGQLKGWWDNYLTEDEKNQILTAIKTDEEGNPIIEDGKFILDAVNSLIFTIAQHFVG 603
+GF GQL+GWWDNY+T ++K ++ + E
Sbjct: 75 SGFTGQLRGWWDNYMTFEQKAAVINVTSSSEVRT-------------------------- 108
Query: 604 DPSLIKDRFGDLLSNLKCKSLGDFRWYKDTFLTRVYTREDSQQAFWKEKFLAGLPKSFGD 663
LL+ L+C++LG+FRWYKDT+++ V +++ K KF+ GLP F
Sbjct: 109 -----------LLNGLQCRNLGEFRWYKDTYMSSVIELPENKYGHLKAKFIDGLPPLFAL 157
Query: 664 KVREKLRSQNPGGEIPYHTLSYGQLIAIIQRVALKICQDDRIQQQLTKEKSQNRRDLGTF 723
+V++ L N G+IPY +Y +L KE+SQ +G F
Sbjct: 158 RVKKIL--SNEHGDIPYDDYTYDKL----------------------KERSQ----VGDF 189
Query: 724 CEQFGI 729
C QF +
Sbjct: 190 CTQFDL 195
>UniRef100_Q6AUA8 Hypothetical protein P0017D10.13 [Oryza sativa]
Length = 950
Score = 68.9 bits (167), Expect = 5e-10
Identities = 58/226 (25%), Positives = 96/226 (41%), Gaps = 41/226 (18%)
Query: 17 GETTLFLTDYDKANVIVPKTIQWSDITLPDKWSLEKAAPAQPVETPEFREIIQHQSGQVD 76
G TTL T+ + PKT++W +ITLP+KW L +A + ++ E +I+ G V+
Sbjct: 85 GLTTLLQTNPNN-RCTTPKTLKWDEITLPEKWVLSQAVEPKSMDQSEVESLIETPDGDVE 143
Query: 77 LIFDRRNSFSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTIHLAGYVN 136
+ F S SR S S+ R + +S
Sbjct: 144 ITF-------------ASKQKAFLQSRPSVSLDSRPRTKSQ------------------- 171
Query: 137 TTPVKTTY-DKSDEDHQSTLSPTYSSLETPGETSDFIGAI-TTEFVIDKEGIRADFYSDV 194
V TY D DE S + L+ DF+ A+ EF IDKE +R +
Sbjct: 172 -NVVYATYEDNFDEPSISDFNINVIELD-----DDFVIALEEEEFEIDKELLRREIRLPK 225
Query: 195 RTKQREWFFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWFQSYAI 240
+ + + + R KI+E ++ + + ++N+FFFDW+++ I
Sbjct: 226 NRTKTKRYLEEVDKSFRMKIREVWHNEMREQRRNIFFFDWYENSQI 271
>UniRef100_Q60CX4 Hypothetical protein [Solanum tuberosum]
Length = 187
Score = 47.4 bits (111), Expect = 0.002
Identities = 21/57 (36%), Positives = 35/57 (60%), Gaps = 1/57 (1%)
Query: 513 ITKILQNMTMVATTYVTANSCPESLIVEVLVAGFCGQLKGWWDNYLTEDEKNQILTA 569
+T ++ M M AT + N+ + I ++++ GF QL+GWWDNY+T D K ++ A
Sbjct: 124 LTILVHRMLMYATICKSVNNTDRT-ICKMIILGFTCQLRGWWDNYMTTDAKAAVINA 179
>UniRef100_Q80DW6 A5L protein [Cowpox virus]
Length = 283
Score = 46.6 bits (109), Expect = 0.003
Identities = 44/180 (24%), Positives = 87/180 (47%), Gaps = 16/180 (8%)
Query: 310 NYTNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSDLDIPKGCQLEKPLFKHFQISNRSR 369
++ NK+ Q L ES + S + K +P + D I G + ++ ++Q R +
Sbjct: 2 DFFNKFSQGLAESSTPKSSIYYSEEK---DPDTKKDEAIEIGLKSQE---SYYQRQLREQ 55
Query: 370 HSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTPEAVSNPAVSAPAPPKTRTSTR 429
+ + A + ++ +Q + + + Q +P P PTP + P+ +AP P K R T
Sbjct: 56 LARDNMMA---ASRQPIQPLQPTIHITPQPVP-VPTPTPAPILLPSSTAPTP-KPRQQT- 109
Query: 430 NTSTSLNNIEDGKVESDSEDQSVKSIPVNT---VIHNSRNQWRKETKLYYPRATAPDLLL 486
NTS+ ++N+ D + +D++ QS + +T + + +++ K+ K P +T P L
Sbjct: 110 NTSSDMSNLFDW-LSADTDTQSNSLLTASTPSNAVQDIISKFNKDQKTTTPPSTQPSQTL 168
>UniRef100_Q8JLA8 EVM107 [Ectromelia virus]
Length = 281
Score = 44.7 bits (104), Expect = 0.010
Identities = 45/181 (24%), Positives = 85/181 (46%), Gaps = 20/181 (11%)
Query: 310 NYTNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSDLDIPKGCQLEKPLFKHFQISNRSR 369
++ NK+ Q L E+ + S + K +P + D I G + ++ ++Q R +
Sbjct: 2 DFFNKFSQGLVEASTPKSSIYYSEEK---DPDTKKDEAIEIGLKSQE---SYYQRQLREQ 55
Query: 370 HSAQTLRAKNESDKELLQKVVEQLQLLNQVIPE-TPEPTPEAVSNPAVSAPAPPKTRTST 428
+ + N ++ ++ LQ + P+ P TP A+ P+ +AP PK R T
Sbjct: 56 LARDNMMVAN-------RRPIQPLQPTIHITPQPVPTSTPSAILLPSSTAPT-PKPRQQT 107
Query: 429 RNTSTSLNNIEDGKVESDSEDQSVKSIPVNT---VIHNSRNQWRKETKLYYPRATAPDLL 485
NTS+ ++N+ D + SD++ QS +P T + + +++ K+ K P +T P
Sbjct: 108 -NTSSDMSNLFDW-LSSDTDTQSNSLLPALTPSNAVQDIISKFNKDQKTTTPPSTQPSQT 165
Query: 486 L 486
L
Sbjct: 166 L 166
>UniRef100_Q7SBY5 Hypothetical protein [Neurospora crassa]
Length = 1081
Score = 43.1 bits (100), Expect = 0.030
Identities = 54/283 (19%), Positives = 108/283 (38%), Gaps = 14/283 (4%)
Query: 259 DGKETLSNLPPKTPFLLKGAQSTPVLASPFKTREEEGEVTSKDIKDLMEQANYTNKYLQT 318
D E + K P G P A+ K +EEE +++ K+ + +
Sbjct: 155 DAPEVPAGDAQKEPVASPGEDKVPEPAAEEKPKEEEKPAAAEEAKEDKPAEPKADVATEA 214
Query: 319 LGESQSHDDSSRKGKGKVKTEPSSSSDLDIPKGCQLEKPLFKHFQISNRSRHSAQTLRAK 378
E++S D + K K + ++ + P P + + + AK
Sbjct: 215 KSEAKSDDGPAAKEKEEAAAPAATETPATEPASKPPASPAKQQEDVEMADAPATVEEPAK 274
Query: 379 NESDKEL-LQKVVEQLQLLNQVIPETPEPTP-------EAVSNPAVSAPAPPKTRTSTRN 430
+E K Q + + + + P PTP + P+ +AP P T ++ +
Sbjct: 275 SEQAKPAEATPTATQQETQDTAVSDVPPPTPAEDEKKDKESPQPSPAAPVAPVTADTSMS 334
Query: 431 TSTSLNNIEDGKVESDSEDQSV-KSIPVNTVIH--NSRNQWRKETKL---YYPRATAPDL 484
+ ++ + E DSED+ V K V+ V +++N+ ++E K+ P A++
Sbjct: 335 DAPPPSSKPYREREEDSEDEPVAKRAKVSPVADQASAKNEAKREDKMDVDSKPAASSAST 394
Query: 485 LLEESSNFKSFSANNVYEWNIDGENEYGITKILQNMTMVATTY 527
+ + S KS + +++ I I ++L + Y
Sbjct: 395 SVHDRSEPKSLADDSINHLPISDYMNRQIRQVLAGVKKTKAGY 437
>UniRef100_Q95XW8 Hypothetical protein Y55B1BR.3 [Caenorhabditis elegans]
Length = 679
Score = 42.7 bits (99), Expect = 0.039
Identities = 53/219 (24%), Positives = 90/219 (40%), Gaps = 17/219 (7%)
Query: 240 IKKKISYPYQVDMIKWQHKDGKETLSNLPPKTPFLLKGAQSTPVLASPFKTREEEGEVTS 299
+KKK S K D +E + P K +P +S +EE E S
Sbjct: 378 VKKKKSSKINKRKAKESSSDEEEEVEESPKKKT-------KSPRKSSKKSAAKEESEEES 430
Query: 300 KDIKDLMEQANYTNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSDLDIPKGCQLEKPLF 359
D ++ E+ +Y+ K + +S SS+K KV++E S ++ + + E P+
Sbjct: 431 SDNEE-EEEVDYSPKK-----KVKSPKKSSKKPAAKVESEEPSDNEEEEEE--VEESPIK 482
Query: 360 KHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTP--EAVSNPAVS 417
K SR SA + + S E ++V E + + ++ + + E N
Sbjct: 483 KDKTPRKYSRKSAAKVESTESSGNEEEEEVEESPKKKGKTPRKSSKKSAAVEESDNEEED 542
Query: 418 APAPPKTRTSTRNTSTSLNNIEDGKVESDSEDQSVKSIP 456
PK RTS R +S E+ + SD+E + V+ P
Sbjct: 543 VEESPKKRTSPRKSSKKRAAKEESEESSDNEVEEVEDSP 581
>UniRef100_Q6RZH9 RPXV112 [Rabbitpox virus]
Length = 283
Score = 42.0 bits (97), Expect = 0.067
Identities = 42/180 (23%), Positives = 85/180 (46%), Gaps = 16/180 (8%)
Query: 310 NYTNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSDLDIPKGCQLEKPLFKHFQISNRSR 369
++ NK+ Q L ES + S + K +P + D I G + ++ ++Q R +
Sbjct: 2 DFFNKFSQGLAESSTPKSSIYYSE---KKDPDTKKDEAIEIGLKSQE---SYYQRQLREQ 55
Query: 370 HSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTPEAVSNPAVSAPAPPKTRTSTR 429
+ + A + ++ +Q + + + Q +P P PT + P+ +AP P K R T
Sbjct: 56 LARDNMMA---ASRQPIQPLQPTIHITPQPVP-VPTPTSAPILLPSSTAPTP-KPRQQT- 109
Query: 430 NTSTSLNNIEDGKVESDSEDQSVKSIPVNT---VIHNSRNQWRKETKLYYPRATAPDLLL 486
NTS+ ++N+ D + +D++ + +P T + + +++ K+ K P +T P L
Sbjct: 110 NTSSDMSNLFDW-LSADTDAPASSLLPALTPSNAVQDIISKFNKDQKTTTPPSTQPSQTL 168
>UniRef100_Q8IR22 CG32580-PA [Drosophila melanogaster]
Length = 16223
Score = 42.0 bits (97), Expect = 0.067
Identities = 85/437 (19%), Positives = 159/437 (35%), Gaps = 46/437 (10%)
Query: 86 SIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTI---HLAGYVNTTPVKT 142
S+P+SI T++ + S + S + P L+ E S+ A TT
Sbjct: 6770 SVPQSIATANSLLTGSSAEEQTAQEETSERSKSLPQLTTEESSSFQESSAEENQTTNQSV 6829
Query: 143 TYDKSDEDHQSTLSPTYSSLETPGETSDFIGAITTEFVIDKEGIRADFYSDVRTKQREWF 202
DK+ ++ TLS S ++ T+ + + E +E D S + E
Sbjct: 6830 NEDKTSQEDTRTLS--ISVPQSIATTNSLLTGSSAEEQTAQEETSEDSKSLPQLTTEE-- 6885
Query: 203 FAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWFQSYAIKKKISYPYQVDMIKWQHKDGKE 262
+ +Q ++ Q + T + QS + K I V+ K +D +
Sbjct: 6886 SSSFQESSAEENQTTEVPWTPSTSPS-------QSSSKTKNIFSSQSVNEDKTSQEDTRT 6938
Query: 263 TLSNLPPK---TPFLLKGAQSTPVLASPFKTREEEG--EVTSKDIKDLMEQANYTNKYLQ 317
++P T LL G+ + A + + ++T+++ E + N+ +
Sbjct: 6939 LSISVPQSIATTNSLLTGSSAEEQTAQEETSEHSKSLPQLTTEESSSFQESSAEENQMTE 6998
Query: 318 ---TLGESQSHDDSSRK--------GKGKVKTEPSSSSDLDIPKGCQLEKPLFKHFQISN 366
TL S S S K + K E + + + +P+ L ++
Sbjct: 6999 VPWTLSTSLSQSSSKTKNIFSSQSVNEDKTSQEDTRTLSISVPQSIATTNSL-----LTG 7053
Query: 367 RSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTPEAVSNPAVSAPAPPKTRT 426
S +E K L Q E+ + E + T P + +P ++ +
Sbjct: 7054 SSAEEQTAQEETSEDSKSLPQLTTEESSSFQESSAEENQTT----EVPWTPSTSPSQSSS 7109
Query: 427 STRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTVIHN------SRNQWRKETKLYYPRAT 480
T+N +S + ED + D+ S+ S+P + N S + + + +
Sbjct: 7110 KTKNIFSSQSVNEDKTSQEDTRTLSI-SVPQSIATTNSLLTGSSAEEQTAQEETSEDSKS 7168
Query: 481 APDLLLEESSNFKSFSA 497
P L EESS+F+ SA
Sbjct: 7169 LPQLTTEESSSFQKSSA 7185
Score = 39.3 bits (90), Expect = 0.44
Identities = 86/451 (19%), Positives = 164/451 (36%), Gaps = 42/451 (9%)
Query: 81 RRNSFSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTI---HLAGYVNT 137
R S S+P+SI T++ + S + +S + P L+ E S+ A T
Sbjct: 6840 RTLSISVPQSIATTNSLLTGSSAEEQTAQEETSEDSKSLPQLTTEESSSFQESSAEENQT 6899
Query: 138 TPVKTTYDKSDEDHQSTLSPTYSSL-----ETPGETSDFIGAITTEFVIDKEGIRADFYS 192
T V T S S +SS +T E + + + + + +
Sbjct: 6900 TEVPWTPSTSPSQSSSKTKNIFSSQSVNEDKTSQEDTRTLSISVPQSIATTNSLLTGSSA 6959
Query: 193 DVRTKQREWFFAKYQGKKRKKIQEKFYGFLEQT--QQNLFFFDW------FQSYAIKKKI 244
+ +T Q E +++ + E+ F E + + + W QS + K I
Sbjct: 6960 EEQTAQEE--TSEHSKSLPQLTTEESSSFQESSAEENQMTEVPWTLSTSLSQSSSKTKNI 7017
Query: 245 SYPYQVDMIKWQHKDGKETLSNLPPK---TPFLLKGAQSTPVLASPFKTREEEG--EVTS 299
V+ K +D + ++P T LL G+ + A + + + ++T+
Sbjct: 7018 FSSQSVNEDKTSQEDTRTLSISVPQSIATTNSLLTGSSAEEQTAQEETSEDSKSLPQLTT 7077
Query: 300 KDIKDLME---QANYTNKYLQTLGESQSHDDSSRK--------GKGKVKTEPSSSSDLDI 348
++ E + N T + T S S S K + K E + + + +
Sbjct: 7078 EESSSFQESSAEENQTTEVPWTPSTSPSQSSSKTKNIFSSQSVNEDKTSQEDTRTLSISV 7137
Query: 349 PKGCQLEKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPT- 407
P+ L ++ S +E K L Q E+ + E + T
Sbjct: 7138 PQSIATTNSL-----LTGSSAEEQTAQEETSEDSKSLPQLTTEESSSFQKSSAEENQMTE 7192
Query: 408 -PEAVSNPAVSAPAPPKTRTSTRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTVIHNSRN 466
P +S + + PK S+++ + + ED + S S Q + + ++
Sbjct: 7193 VPWTLSTSLSQSSSEPKNIFSSQSLNEDNISQEDTRTLSISVPQPIATANSLLTGSSAEE 7252
Query: 467 QWRKETKLYYPRATAPDLLLEESSNFKSFSA 497
Q +E + + + + P L EESS+F+ SA
Sbjct: 7253 QTAQEERSEHSK-SLPQLTTEESSSFQESSA 7282
Score = 38.5 bits (88), Expect = 0.74
Identities = 88/446 (19%), Positives = 156/446 (34%), Gaps = 42/446 (9%)
Query: 86 SIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTIHL---AGYVNTTPVKT 142
S+P+SI T+ + S + S + P L+ E S+ L A TT V
Sbjct: 4693 SVPQSIATAHSLLTGSSAEEQTAQEETSEHSKSLPQLTTEESSSFLESSAEENQTTEVPW 4752
Query: 143 TYDKSDEDHQSTLSPTYSSL-----ETPGETSDFIGAITTEFVIDKEGIRADFYSDVRTK 197
T S S +SS +T E + + + + + ++ +T
Sbjct: 4753 TLSTSPSQSSSKTKNIFSSQSVNEDKTSQEDTRTLPISVPQSIATTNSLLTGSSAEEQTA 4812
Query: 198 QREWFFAKYQGKKRKKIQEKFYGFLEQT--QQNLFFFDWFQSYAIKKKISYPYQVDMIKW 255
Q E +++ + E+ FLE + + W S ++ + +
Sbjct: 4813 QEE--TSEHSKSLPQLTTEESSSFLESSAEENQTTEVPWTLSTSLSQSSLQAKNIFSSPS 4870
Query: 256 QHKD--GKETLSNLPPKTPFLLKGAQSTPVLAS-PFKTREEEGEVTSKDIKDL------- 305
++D +E LP P + A S +S +T +EE SK + L
Sbjct: 4871 LNEDNISQEDTRTLPISVPQSIATANSLLTGSSAEEQTAQEETSEHSKSLPQLTTEESSS 4930
Query: 306 ----MEQANYTNKYLQTLGESQSHDDSSRK---GKGKVKTEPSSSSDLDIPKGCQLEKPL 358
+ N T + TL S S S K + + +S D P +
Sbjct: 4931 FQESSAEENQTTEVPWTLSTSLSQSSSQAKNIFSSQSLNEDKTSQEDTRTPSISVPQSIA 4990
Query: 359 FKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTPEAVSNPAVSA 418
H ++ S +E K L Q E+ + E+ + P +
Sbjct: 4991 TAHSLLTGSSAEEQTAQEETSEHSKSLPQLTTEE----SSSFLESSAEENQTTEVPWTLS 5046
Query: 419 PAPPKTRTSTRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTVIHNS-------RNQWRKE 471
+P ++ + T+N +S + ED + D+ + S+P + NS Q +E
Sbjct: 5047 TSPSQSSSKTKNIFSSQSVNEDKTSQEDTRTLPI-SVPQSIATTNSLLTGSSAEEQTAQE 5105
Query: 472 TKLYYPRATAPDLLLEESSNFKSFSA 497
+ + + P L EESS+F SA
Sbjct: 5106 ETSEHSK-SLPQLTTEESSSFLESSA 5130
Score = 36.2 bits (82), Expect = 3.7
Identities = 92/457 (20%), Positives = 156/457 (34%), Gaps = 54/457 (11%)
Query: 81 RRNSFSIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTIHL---AGYVNT 137
R S S+P+SI T+ + S + S + P L+ E S+ L A T
Sbjct: 4979 RTPSISVPQSIATAHSLLTGSSAEEQTAQEETSEHSKSLPQLTTEESSSFLESSAEENQT 5038
Query: 138 TPVKTTYDKSDEDHQSTLSPTYSSL-----ETPGETSDFIGAITTEFVIDKEGIRADFYS 192
T V T S S +SS +T E + + + + + +
Sbjct: 5039 TEVPWTLSTSPSQSSSKTKNIFSSQSVNEDKTSQEDTRTLPISVPQSIATTNSLLTGSSA 5098
Query: 193 DVRTKQREWFFAKYQGKKRKKIQEKFYGFLEQT--QQNLFFFDW------FQSYAIKKKI 244
+ +T Q E +++ + E+ FLE + + W QS + K I
Sbjct: 5099 EEQTAQEE--TSEHSKSLPQLTTEESSSFLESSAEENQTTEVPWTLSTSPSQSSSKTKNI 5156
Query: 245 SYPYQVDMIKWQHKDGKETLSNLPPKTPFLLKGAQSTPVLASPFKTREEEGEVTSKDIKD 304
V+ + K +E LP P + A S +L + E TSK K
Sbjct: 5157 FSSQSVN----EDKTSQEDTRTLPISVPQSIATANS--LLTGSSAEEQTAQEETSKHSKS 5210
Query: 305 LME--------------QANYTNKYLQTLGESQSHDDSSRK--------GKGKVKTEPSS 342
L + + N T + TL S S S K + K E +
Sbjct: 5211 LPQLTTEESSSFQESSAEENQTTEVPWTLSTSPSQSSSKTKNIFSSQSVNEDKTSQEDTR 5270
Query: 343 SSDLDIPKGCQLEKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPE 402
+ + +P+ L ++ S +E K L Q E+ + E
Sbjct: 5271 TLPISVPQSIATTNSL-----LTGSSAEEQTAQEETSEHSKSLPQLTTEESSSFLESSAE 5325
Query: 403 TPEPT--PEAVSNPAVSAPAPPKTRTSTRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTV 460
+ T P +S + K S+ + + + ED + S QS+ +
Sbjct: 5326 ENQTTEVPWTLSTSLSQSSLQAKNIFSSPSLNEDNISQEDTRTLPISVPQSIATANSLLT 5385
Query: 461 IHNSRNQWRKETKLYYPRATAPDLLLEESSNFKSFSA 497
++ Q +E + + + P L EESS+F+ SA
Sbjct: 5386 GSSAEEQTAQEETSEHSK-SLPQLTTEESSSFQESSA 5421
Score = 35.0 bits (79), Expect = 8.2
Identities = 86/446 (19%), Positives = 160/446 (35%), Gaps = 42/446 (9%)
Query: 86 SIPRSIRTSSDFQSAMSRRSFSMSRRNQPESSARPSLSHEGSTI---HLAGYVNTTPVKT 142
S+P+SI T++ + S + S + P L+ E S+ A TT V
Sbjct: 8186 SVPQSIATANSLLTGSSAEEQTAQEETSEHSKSLPQLTTEESSSFQESSAEENQTTEVPW 8245
Query: 143 TYDKSDEDHQSTLSPTYSSL-----ETPGETSDFIGAITTEFVIDKEGIRADFYSDVRTK 197
T S S +SS +T E + + + + + ++ +T
Sbjct: 8246 TLSTSLSQSSSKTKNIFSSQSVNEDKTSQEDTRTLSISVPQSIATTNSLLTGSSAEEQTA 8305
Query: 198 QREWFFAKYQGKKRKKIQEKFYGFLEQT--QQNLFFFDW------FQSYAIKKKISYPYQ 249
Q E +++ + E+ F E + + + W QS +I K I
Sbjct: 8306 QEE--TSEHSKSLPQLTTEESSSFQESSAEENQMTEVPWTVLTSLSQSSSITKNIFSSQS 8363
Query: 250 VDMIKWQHKDGKETLSNLPPK---TPFLLKGAQSTPVLASPFKTREEEG--EVTSKDIKD 304
V+ K +D + ++P T LL G+ + A + + ++T+++
Sbjct: 8364 VNEDKTSQEDTRTLSISVPQSFATTNRLLTGSSAEEQTAQEETSEHSKSLPQLTTEERSS 8423
Query: 305 LMEQANYTNKYLQ---TLGESQSHDDSSRK--------GKGKVKTEPSSSSDLDIPKGCQ 353
L E + N+ + TL S S S K + K E + + + +P+
Sbjct: 8424 LQESSAEENQMTEVPWTLSTSLSQSSSITKNIFSSQSVNEDKTSQEDTRTLSISVPQSFA 8483
Query: 354 LEKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPT--PEAV 411
L ++ S +E K L Q E+ + E + T P V
Sbjct: 8484 TANSL-----LTGSSAEEQTAQEETSEHSKSLPQLTTEESSSFQESSAEENQMTEVPWTV 8538
Query: 412 SNPAVSAPAPPKTRTSTRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTVIHNSRNQWRKE 471
+ + K S+++ + + ED + S S QS + ++ Q +E
Sbjct: 8539 LTSLSQSSSITKNIFSSQSVNEDKTSQEDTRTLSISVPQSFATANSLLTGSSAEEQTAQE 8598
Query: 472 TKLYYPRATAPDLLLEESSNFKSFSA 497
+ + + P L EESS+F+ SA
Sbjct: 8599 ETSEHSK-SLPQLTTEESSSFQESSA 8623
>UniRef100_Q7RLW2 Mannosyltransferase-related [Plasmodium yoelii yoelii]
Length = 2013
Score = 42.0 bits (97), Expect = 0.067
Identities = 81/392 (20%), Positives = 148/392 (37%), Gaps = 71/392 (18%)
Query: 145 DKSDEDHQS-----TLSPTYSSLETPGETS-----DFIGAITTEFVIDKEGIRADFYSDV 194
+ +DED+++ + T E P + S D +G+ + + +KEG +F +
Sbjct: 1343 NNNDEDNENGESLNNMEETLKKEEKPFDLSKKALEDILGSAISNKIAEKEGNSMEFSKII 1402
Query: 195 RTKQRE------------W----FFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWFQSY 238
+ ++ + W Y+ KK ++ +K Q +N+ F +
Sbjct: 1403 KDEKNKNTIDSILSMLTPWEQKILALLYEQKKMREQVQKIKEKFSQRIKNINIF--IKKI 1460
Query: 239 AIKK-----------KISYPYQVDMIKWQHKDGKETLSNLPPKTP----FLLKGAQSTPV 283
I K K++ Y+ DM+K D + L K L + + V
Sbjct: 1461 LIDKIEVEKHFNRIVKVTEQYKEDMLKTSDVDSNFKFAGLMKKLEELSISLSEKTRDFNV 1520
Query: 284 LASPFKTREEEGEVTSKDIKDLMEQANY---TNKYLQTLGESQSHDDSSRKGKGKVKTEP 340
LA +E E IK+L E+ KY + L E HD +S K K
Sbjct: 1521 LAEFHLNLRKENEKKDTLIKELEEENKLFYLVPKYNELLNELNCHDGNSIAAKELKKEFL 1580
Query: 341 SSSSDLDIPKGCQLEKPLF--------KHF------------QISNRSRHSAQTLRAKNE 380
L I K EK + KH + ++++ L +N+
Sbjct: 1581 LMYGRLMITKKKLQEKEVIENDIRNLNKHIYTELIESISIIKNLESQNKFLEYKLNLENK 1640
Query: 381 SDKELLQKVVEQLQLLNQVIPETPEPTPEAVSNPAVSAPAPPKTRTSTRNTSTSLNNIED 440
E ++K++ + + LN+VI E +A + ++ + T+ NT TS+NNI+
Sbjct: 1641 KKVEKVKKLLNEKEKLNKVISEMERMLEDANID---TSTLKNEILTNFNNTYTSINNIDS 1697
Query: 441 GKVESDSEDQSVKSIPVNTVIHNSRNQWRKET 472
+ + E + K N V + + +K+T
Sbjct: 1698 EEEKPPKELDTNKK--ENKVKKTKKKKKKKQT 1727
>UniRef100_UPI000046BA1D UPI000046BA1D UniRef100 entry
Length = 576
Score = 41.6 bits (96), Expect = 0.088
Identities = 69/307 (22%), Positives = 122/307 (39%), Gaps = 46/307 (14%)
Query: 289 KTREEEGEVTSKDIKDLMEQANYTNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSD-LD 347
K EG V +KD + + T+ ++Q ++++ K K SS+ D +D
Sbjct: 251 KQPNNEGSVGNKDNVHINPKNEDTSSDSTNKEKTQQKENATDKNSDKASVSDSSTKDVID 310
Query: 348 IPKGCQLEKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPT 407
KG K I S ++ K S+ + K E+ + +V P
Sbjct: 311 NTKG----KTEGVQSGIGEDSNTVDGNIKTKEPSEND---KQKEESSSIVKVPSVAPSNV 363
Query: 408 PEAVSNPAVSAPAPPKTRTSTRNTSTSLNNIE-DGKVESD---SEDQSVKSIPVNTVIHN 463
PE A + PP + +T ST+LN+ E +GK+ +D ++++ K T++
Sbjct: 364 PEQKKADANTLEFPPTSHNNT--NSTNLNDTESNGKISNDVIPKKNENNKPNNNQTILEE 421
Query: 464 SRNQWRKETKLYYPRATAPDLLLEESSNFKSFSANNVYEWNIDGENEYGITKILQNMTMV 523
+RN + +T + K + NN +E D E E + ++M ++
Sbjct: 422 ARNDPKVKT------------------STKDTAVNNNFEMRYDNEEE-NENSVDESMNIM 462
Query: 524 ATTYVTANSCPESLIVEVLVAGFCGQLKGWWDNYLTEDEKNQI------LTAIKTDEEGN 577
+ NS E +E A Q +N E+EK +I T+ K +++
Sbjct: 463 ESYTGDQNSDQEK--IEFADAKDIPQ-----NNKTNENEKEKISKKSRKRTSKKKEKKNE 515
Query: 578 PIIEDGK 584
PI D K
Sbjct: 516 PITIDSK 522
>UniRef100_UPI000046B980 UPI000046B980 UniRef100 entry
Length = 1661
Score = 41.6 bits (96), Expect = 0.088
Identities = 79/388 (20%), Positives = 144/388 (36%), Gaps = 70/388 (18%)
Query: 145 DKSDEDHQSTLSPTYSSLETPGETS-----DFIGAITTEFVIDKEGIRADFYSDVRTKQR 199
D + + + T E P + S D +G+ + + +KEG +F ++ ++
Sbjct: 1029 DNEKGESLNNMEETLKKEEKPYDLSKKALEDILGSAISNKIAEKEGDTMEFPKIIKDEKN 1088
Query: 200 E------------W----FFAKYQGKKRKKIQEKFYGFLEQTQQNLFFFDWFQSYAIKK- 242
+ W Y+ KK ++ +K Q +N+ F + I K
Sbjct: 1089 KNTIDSILSMLTPWEQKILALLYEQKKMREQVQKIKEKFSQRIKNINIF--IKKILIDKI 1146
Query: 243 ----------KISYPYQVDMIKWQHKDGKETLSNLPPKTP----FLLKGAQSTPVLASPF 288
K++ Y+ DM+K D + L K L + + VLA
Sbjct: 1147 EVEKHFNRIVKVTEQYKEDMLKTSDVDSNFKFAGLMKKLEELSISLSEKNRDFNVLAEFH 1206
Query: 289 KTREEEGEVTSKDIKDLMEQANY---TNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSD 345
+E E IK+L E+ KY + L E H+ +S K K
Sbjct: 1207 LNLRKENEKKDILIKELQEENKLFYLVPKYNELLNELNCHEGNSIAAKELKKEFLLMYGR 1266
Query: 346 LDIPKGCQLEKPLF--------KHF------------QISNRSRHSAQTLRAKNESDKEL 385
L I K EK + KH + ++++ L +N+ E
Sbjct: 1267 LMITKKKLQEKEVIENDIRNLNKHIYTELIESISIIKNLESQNKFLEYKLNLENKKKIEK 1326
Query: 386 LQKVVEQLQLLNQVIPETPEPTPEAVSNPAVSAPAPPKTRTSTRNTSTSLNNIEDGKVES 445
++K++ + + LN+VI E +A + ++ + T+ NT TS+NNI+
Sbjct: 1327 VKKLLNEKEKLNKVISEMERMLEDANID---TSTLKNEILTNFNNTYTSINNID------ 1377
Query: 446 DSEDQSVKSIPVNTVIHNSRNQWRKETK 473
ED+S K + N + ++ +K+ K
Sbjct: 1378 SEEDKSPKELDTNKKENKAKKTKKKKKK 1405
>UniRef100_Q8IGZ0 RE01659p [Drosophila melanogaster]
Length = 569
Score = 41.6 bits (96), Expect = 0.088
Identities = 48/170 (28%), Positives = 70/170 (40%), Gaps = 25/170 (14%)
Query: 295 GEVTSKDIKDLMEQANYTNKYLQTLGESQSHDDSSRKGKGKVKTEPSSSSDLDIPKGCQL 354
G+V ++ K++ NY NK+ E D SS TE SSS GC
Sbjct: 93 GDVIHEESKEIENYVNYKNKHKSQDPEKSDDDGSS------TATETESSS------GCGS 140
Query: 355 EKPLFKHFQISNRSRHSAQTLRAKNESDKELLQKVVEQLQLLNQVIPETPEPTPEAVSNP 414
I++ S+ S L A E KEL + + L + P+ P P P S P
Sbjct: 141 SAGNSNSSSINDSSQQSTVPLDALEEVFKELKTNLEHRETL--KAAPQEPPPVPPKKS-P 197
Query: 415 AVSAPAP---PKTRTSTRNTSTSLNNIEDGKVESDSEDQSVKSIPVNTVI 461
P P PK R + + + TS D + + DS++Q I +T+I
Sbjct: 198 HTIPPKPQIKPKPRVTQKFSRTS-----DIRRDEDSDNQ--HKINTDTII 240
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.131 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,308,938,579
Number of Sequences: 2790947
Number of extensions: 57253750
Number of successful extensions: 170454
Number of sequences better than 10.0: 268
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 256
Number of HSP's that attempted gapping in prelim test: 169934
Number of HSP's gapped (non-prelim): 664
length of query: 751
length of database: 848,049,833
effective HSP length: 135
effective length of query: 616
effective length of database: 471,271,988
effective search space: 290303544608
effective search space used: 290303544608
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0220.1


