

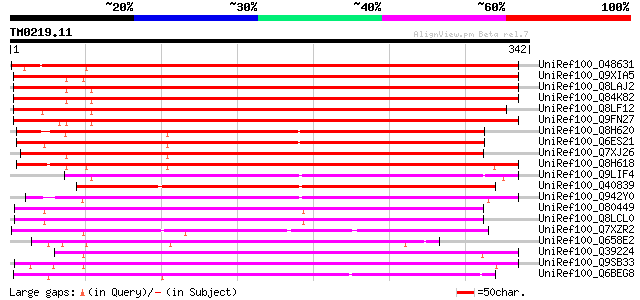
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0219.11
(342 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O48631 Ethylene-forming-enzyme-like dioxygenase [Prunu... 337 4e-91
UniRef100_Q9XIA5 Similar to ethylene-forming-enzyme-like dioxyge... 322 9e-87
UniRef100_Q8LAJ2 Ethylene-forming-enzyme-like dioxygenase-like p... 312 1e-83
UniRef100_Q84K82 Putative ethylene-forming dioxygenase [Arabidop... 311 1e-83
UniRef100_Q8LF12 Ethylene-forming-enzyme-like dioxygenase-like [... 301 1e-80
UniRef100_Q9FN27 Ethylene-forming-enzyme-like dioxygenase [Arabi... 298 1e-79
UniRef100_Q8H620 Putative iron/ascorbate-dependent oxidoreductas... 286 4e-76
UniRef100_Q6ES21 Putative iron/ascorbate-dependent oxidoreductas... 271 2e-71
UniRef100_Q7XJ26 Iron/ascorbate-dependent oxidoreductase [Hordeu... 262 9e-69
UniRef100_Q8H618 Putative iron/ascorbate-dependent oxidoreductas... 261 1e-68
UniRef100_Q9LIF4 Arabidopsis thaliana genomic DNA, chromosome 3,... 229 8e-59
UniRef100_Q40839 Ethylene-forming enzyme [Picea glauca] 224 3e-57
UniRef100_Q942Y0 Putative ethylene-forming enzyme [Oryza sativa] 220 4e-56
UniRef100_O80449 Putative anthocyanidin synthase [Arabidopsis th... 219 9e-56
UniRef100_Q8LCL0 Putative anthocyanidin synthase [Arabidopsis th... 218 2e-55
UniRef100_Q7XZR2 Salt-induced protein [Atriplex nummularia] 215 2e-54
UniRef100_Q658E2 Putative iron/ascorbate-dependent oxidoreductas... 213 5e-54
UniRef100_Q39224 SRG1 protein [Arabidopsis thaliana] 212 1e-53
UniRef100_Q9SB33 SRG1-like protein [Arabidopsis thaliana] 210 4e-53
UniRef100_Q6BEG8 Flavonol synthase [Eustoma grandiflorum] 207 3e-52
>UniRef100_O48631 Ethylene-forming-enzyme-like dioxygenase [Prunus armeniaca]
Length = 348
Score = 337 bits (863), Expect = 4e-91
Identities = 167/341 (48%), Positives = 236/341 (68%), Gaps = 8/341 (2%)
Query: 2 SKSVQEM--SMDSDEPPSAYVVERNSFGSKDSSTLIPIPIIDVSLLSSED-----EQGKL 54
S +VQE+ ++ P Y+ + + S+ L+ IP+ID+ LL + + KL
Sbjct: 9 SLTVQELLGKGKGEQVPEKYI-HKVGAPNASSAQLMDIPVIDLGLLLTPSSITAPQLEKL 67
Query: 55 RSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVS 114
RSAL++ GCFQ I HGM+ +LD++RE+ K FFALPVEEKQ+Y R VN+ +GYGND V S
Sbjct: 68 RSALTTWGCFQVINHGMTPEFLDEVREMTKQFFALPVEEKQQYLRQVNDIQGYGNDMVFS 127
Query: 115 KKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLN 174
++Q LDWS RL L V+P+E R+L W ++P F E+L +++ K++ + +L MARSLN
Sbjct: 128 EQQTLDWSDRLYLSVYPEEHRKLKFWAQDPKSFSETLHQYTMKLQVVTKTVLEAMARSLN 187
Query: 175 LEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLV 234
L+ F +GEQ + RFNFYPPCSRPD+VLGVKPH D + IT+LLQD++VEGLQ L
Sbjct: 188 LDVNCFRDLYGEQGKMDVRFNFYPPCSRPDVVLGVKPHADGTIITLLLQDKQVEGLQFLK 247
Query: 235 DDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEI 294
DD+W P +P+AL++N+GDQ +I+SNGIFKSP+HRV+TN ++ R+S+A F PE + +I
Sbjct: 248 DDQWFRAPIVPEALLINVGDQAEILSNGIFKSPVHRVVTNPDKERISLAAFCIPESDKDI 307
Query: 295 GPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQI 335
P E L+NE+ P LY+ V NY I + YQ+GK +E +I
Sbjct: 308 EPFESLVNESTPGLYKKVKNYVGIYFEYYQQGKRPIEAAKI 348
>UniRef100_Q9XIA5 Similar to ethylene-forming-enzyme-like dioxygenase [Arabidopsis
thaliana]
Length = 348
Score = 322 bits (825), Expect = 9e-87
Identities = 162/342 (47%), Positives = 224/342 (65%), Gaps = 9/342 (2%)
Query: 3 KSVQEMSMDSDEPPSAYVVERNSFG-SKDSSTLIP---IPIIDVSLLSS-----EDEQGK 53
K+VQE+ P Y+ G S+ + +P IP ID+SLL S ++E K
Sbjct: 7 KTVQEVVAAGQGLPERYLHAPTGEGESQPLNGAVPEMDIPAIDLSLLFSSSVDGQEEMKK 66
Query: 54 LRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVV 113
L SALS+ G Q + HG++ +LDKI ++ K FFALP EEK K AR +GYGND ++
Sbjct: 67 LHSALSTWGVVQVMNHGITEAFLDKIYKLTKQFFALPTEEKHKCARETGNIQGYGNDMIL 126
Query: 114 SKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSL 173
S QVLDW RL L +P++KR+L WP+ P F E+L E++ K + +++ + MARSL
Sbjct: 127 SDNQVLDWIDRLFLTTYPEDKRQLKFWPQVPVGFSETLDEYTMKQRVLIEKFFKAMARSL 186
Query: 174 NLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL 233
LEE FL +GE + + +RFNF+PPC RPD V+G+KPH D S IT+LL D++VEGLQ L
Sbjct: 187 ELEENCFLEMYGENAVMNSRFNFFPPCPRPDKVIGIKPHADGSAITLLLPDKDVEGLQFL 246
Query: 234 VDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENE 293
D KW P +PD +++ LGDQM+IMSNGI+KSP+HRV+TN E+ R+SVA F P + E
Sbjct: 247 KDGKWYKAPIVPDTILITLGDQMEIMSNGIYKSPVHRVVTNREKERISVATFCVPGLDKE 306
Query: 294 IGPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQI 335
I P +GL+ E RPRLY+ V Y D++Y+ YQ+G+ +E I
Sbjct: 307 IHPADGLVTEARPRLYKTVTKYVDLHYKYYQQGRRTIEAALI 348
>UniRef100_Q8LAJ2 Ethylene-forming-enzyme-like dioxygenase-like protein [Arabidopsis
thaliana]
Length = 348
Score = 312 bits (799), Expect = 1e-83
Identities = 158/342 (46%), Positives = 223/342 (65%), Gaps = 9/342 (2%)
Query: 3 KSVQEMSMDSDEPPSAYVVERNSFGS-KDSSTLIP---IPIIDVSLLSSEDEQG-----K 53
K+VQE+ + P Y+ G + + +P IP ID++LL S E G K
Sbjct: 7 KTVQEVVAAGEGLPERYLHAPTGDGEVQPLNAAVPEMDIPAIDLNLLLSSSEDGREELRK 66
Query: 54 LRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVV 113
L SALS+ G Q + HG++ +LDKI ++ K FFALP EEKQK AR ++ +GYGND ++
Sbjct: 67 LHSALSTWGVVQVMNHGITKAFLDKIYKLTKEFFALPTEEKQKCAREIDSIQGYGNDMIL 126
Query: 114 SKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSL 173
QVLDW RL + +P+++R+L+ WPE P F E+L E++ K + +++ + MARSL
Sbjct: 127 WDDQVLDWIDRLYITTYPEDQRQLNFWPEVPLGFRETLHEYTMKQRIVIEQFFKAMARSL 186
Query: 174 NLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL 233
LEE SFL +GE ++L RFN YPPC PD V+GVKPH D S IT+LL D++V GLQ
Sbjct: 187 ELEENSFLDMYGESATLDTRFNMYPPCPSPDKVIGVKPHADGSAITLLLPDKDVGGLQFQ 246
Query: 234 VDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENE 293
D KW P +PD +++N+GDQM+IMSNGI+KSP+HRV+TN E+ R+SVA F P + E
Sbjct: 247 KDGKWYKAPIVPDTILINVGDQMEIMSNGIYKSPVHRVVTNREKERISVATFCIPGADKE 306
Query: 294 IGPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQI 335
I PV L++E RPRLY+ V Y ++ ++ YQ+G+ +E I
Sbjct: 307 IQPVNELVSEARPRLYKTVKKYVELYFKYYQQGRRPIEAALI 348
>UniRef100_Q84K82 Putative ethylene-forming dioxygenase [Arabidopsis thaliana]
Length = 348
Score = 311 bits (798), Expect = 1e-83
Identities = 158/342 (46%), Positives = 223/342 (65%), Gaps = 9/342 (2%)
Query: 3 KSVQEMSMDSDEPPSAYVVERNSFGS-KDSSTLIP---IPIIDVSLLSSEDEQG-----K 53
K+VQE+ + P Y+ G + + +P IP ID++LL S E G K
Sbjct: 7 KTVQEVVAAGEGLPERYLHAPTGDGEVQPLNAAVPEMDIPAIDLNLLLSSSEAGQQELSK 66
Query: 54 LRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVV 113
L SALS+ G Q + HG++ +LDKI ++ K FFALP EEKQK AR ++ +GYGND ++
Sbjct: 67 LHSALSTWGVVQVMNHGITKAFLDKIYKLTKEFFALPTEEKQKCAREIDSIQGYGNDMIL 126
Query: 114 SKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSL 173
QVLDW RL + +P+++R+L+ WPE P F E+L E++ K + +++ + MARSL
Sbjct: 127 WDDQVLDWIDRLYITTYPEDQRQLNFWPEVPLGFRETLHEYTMKQRIVIEQFFKAMARSL 186
Query: 174 NLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL 233
LEE SFL +GE ++L RFN YPPC PD V+GVKPH D S IT+LL D++V GLQ
Sbjct: 187 ELEENSFLDMYGESATLDTRFNMYPPCPSPDKVIGVKPHADGSAITLLLPDKDVGGLQFQ 246
Query: 234 VDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENE 293
D KW P +PD +++N+GDQM+IMSNGI+KSP+HRV+TN E+ R+SVA F P + E
Sbjct: 247 KDGKWYKAPIVPDTILINVGDQMEIMSNGIYKSPVHRVVTNREKERISVATFCIPGADKE 306
Query: 294 IGPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQI 335
I PV L++E RPRLY+ V Y ++ ++ YQ+G+ +E I
Sbjct: 307 IQPVNELVSEARPRLYKTVKKYVELYFKYYQQGRRPIEAALI 348
>UniRef100_Q8LF12 Ethylene-forming-enzyme-like dioxygenase-like [Arabidopsis
thaliana]
Length = 349
Score = 301 bits (772), Expect = 1e-80
Identities = 156/335 (46%), Positives = 217/335 (64%), Gaps = 10/335 (2%)
Query: 3 KSVQEMSMDSDEPPSAYV----VERNSFGSKDSSTLIPIPIIDVSLLSSEDEQG-----K 53
K+VQE+ + P Y+ V+ N + ++ IP ID+SLL S + G K
Sbjct: 7 KTVQEVVAAGEGIPERYLQPPAVDDNGQHLNAAVPVMDIPAIDLSLLLSPSDDGREELSK 66
Query: 54 LRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVV 113
L SALS+ G Q I HG++ LDKI ++ K F ALP EEKQKYAR + +GYGND ++
Sbjct: 67 LHSALSTWGVVQVINHGITKALLDKIYKLTKEFCALPSEEKQKYAREIGSIQGYGNDMIL 126
Query: 114 SKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSL 173
QVLDW RL + +P+++R+L WP+ P F E+L E++ K + + + + MA SL
Sbjct: 127 WDDQVLDWIDRLYITTYPEDQRQLKFWPDVPVGFRETLHEYTMKQHLVFNQVFKAMAISL 186
Query: 174 NLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL 233
LEE FL GE +++ RFN YPPC RPD V+GV+PH D+S T+LL D+ VEGLQ L
Sbjct: 187 ELEENCFLDMCGENATMDTRFNMYPPCPRPDKVIGVRPHADKSAFTLLLPDKNVEGLQFL 246
Query: 234 VDDKWVNVPTI-PDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPEN 292
D KW P + D +++N+GDQM+IMSNGI+KSP+HRV+TNTE+ R+SVA F P +
Sbjct: 247 KDGKWYKAPVVASDTILINVGDQMEIMSNGIYKSPVHRVVTNTEKERISVATFCIPGADK 306
Query: 293 EIGPVEGLINETRPRLYRNVNNYGDINYRCYQEGK 327
EI PV+GL++E RPRLY+ V NY D+ + Y +G+
Sbjct: 307 EIQPVDGLVSEARPRLYKPVKNYVDLLNKYYIQGQ 341
>UniRef100_Q9FN27 Ethylene-forming-enzyme-like dioxygenase [Arabidopsis thaliana]
Length = 349
Score = 298 bits (763), Expect = 1e-79
Identities = 151/343 (44%), Positives = 218/343 (63%), Gaps = 10/343 (2%)
Query: 3 KSVQEMSMDSDEPPSAYVVERNSFGSKDS--STLIP---IPIIDVSLLSSEDEQG----- 52
K+VQE+ ++ P Y+ G D + L+P I IID++LL S + G
Sbjct: 7 KTVQEVVAAGEKLPERYLYTPTGDGEGDQPFNGLLPEMKISIIDLNLLFSSSDDGREELS 66
Query: 53 KLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRV 112
KL SA+S+ G Q + HG+S LDKI E+ K FF LP +EKQKYAR ++ +G+GND +
Sbjct: 67 KLHSAISTWGVVQVMNHGISEALLDKIHELTKQFFVLPTKEKQKYAREISSFQGFGNDMI 126
Query: 113 VSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARS 172
+S QVLDW RL L +P+++R+L WPENPS F E+L E++ K + +++ + +ARS
Sbjct: 127 LSDDQVLDWVDRLYLITYPEDQRQLKFWPENPSGFRETLHEYTMKQQLVVEKFFKALARS 186
Query: 173 LNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQV 232
L LE+ FL GE ++L RFN YPPC RPD VLG+KPH+D S T++L D+ VEGLQ
Sbjct: 187 LELEDNCFLEMHGENATLETRFNIYPPCPRPDKVLGLKPHSDGSAFTLILPDKNVEGLQF 246
Query: 233 LVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPEN 292
L D KW +P +++N+GD M++MSNGI+KSP+HRV+ N ++ R+ VA F + +
Sbjct: 247 LKDGKWYKASILPHTILINVGDTMEVMSNGIYKSPVHRVVLNGKKERIYVATFCNADEDK 306
Query: 293 EIGPVEGLINETRPRLYRNVNNYGDINYRCYQEGKIALETVQI 335
EI P+ GL++E RPRLY+ V + YQ+G+ +E I
Sbjct: 307 EIQPLNGLVSEARPRLYKAVKKSEKNFFDYYQQGRRPIEAAMI 349
>UniRef100_Q8H620 Putative iron/ascorbate-dependent oxidoreductase [Oryza sativa]
Length = 350
Score = 286 bits (733), Expect = 4e-76
Identities = 144/321 (44%), Positives = 212/321 (65%), Gaps = 18/321 (5%)
Query: 5 VQEMSMDS-DEPPSAYVVERNSFGSKDSSTLI-------PIPIIDVSLLSSEDEQGKLRS 56
VQE++ +EPPS YV+ G KD S + PIP++D+S L+ DE KLR+
Sbjct: 12 VQELAAAGVEEPPSRYVL-----GEKDRSDELVAAELPEPIPVVDLSRLAGADEAAKLRA 66
Query: 57 ALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVN----EAEGYGNDRV 112
AL + G F HG+ ++ +D + +A+ FF P+E K+K++ ++ + EGYG DRV
Sbjct: 67 ALQNWGFFLLTNHGVETSLMDDVLNLAREFFNQPIERKRKFSNLIDGKNFQVEGYGTDRV 126
Query: 113 VSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARS 172
V++ Q+LDWS RL LRV PKE+R L+ WP++P F + L E++++ K + D +++ M++
Sbjct: 127 VTQDQILDWSDRLFLRVEPKEERNLAFWPDHPESFRDVLNEYASRTKRIRDDIVQAMSKL 186
Query: 173 LNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQV 232
L L+E F + + +L ARFN+YPPC RPDLV GV+PH+D S T+LL D +V GLQ+
Sbjct: 187 LGLDEDYFFDRLNKAPAL-ARFNYYPPCPRPDLVFGVRPHSDGSLFTILLVDEDVGGLQI 245
Query: 233 LVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPEN 292
D KW NV P+ L++NLGD M+++ NGIF+SP+HRV+TN ER R+S+AMF E
Sbjct: 246 QRDGKWYNVQVTPNTLLINLGDTMEVLCNGIFRSPVHRVVTNAERERISLAMFYSVNDEK 305
Query: 293 EIGPVEGLINETRPRLYRNVN 313
+IGP GL++E RP YR V+
Sbjct: 306 DIGPAAGLLDENRPARYRKVS 326
>UniRef100_Q6ES21 Putative iron/ascorbate-dependent oxidoreductase [Oryza sativa]
Length = 351
Score = 271 bits (693), Expect = 2e-71
Identities = 134/317 (42%), Positives = 207/317 (65%), Gaps = 9/317 (2%)
Query: 5 VQEMSMDS-DEPPSAYVV---ERNSFGSKDSSTLIPIPIIDVSLLSSEDEQGKLRSALSS 60
VQE++ +EPPS Y++ +R+ + P+P++D+S L +E KLR AL +
Sbjct: 12 VQELAAAGVEEPPSRYLLREKDRSDVKLVAAELPEPLPVVDLSRLDGAEEATKLRVALQN 71
Query: 61 AGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVN----EAEGYGNDRVVSKK 116
G F HG+ ++ +D + +++ FF P+E KQK++ ++ + +GYG DRVV++
Sbjct: 72 WGFFLLTNHGVEASLMDSVMNLSREFFNQPIERKQKFSNLIDGKNFQIQGYGTDRVVTQD 131
Query: 117 QVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLE 176
Q+LDWS RL LRV PKE++ L+ WP++P F + L ++++ K + D +++ MA+ L L+
Sbjct: 132 QILDWSDRLHLRVEPKEEQDLAFWPDHPESFRDVLNKYASGTKRIRDDIIQAMAKLLELD 191
Query: 177 EGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDD 236
E FL + E + ARFN+YPPC RPDLV G++PH+D + +T+LL D++V GLQV D
Sbjct: 192 EDYFLDRLNEAPAF-ARFNYYPPCPRPDLVFGIRPHSDGTLLTILLVDKDVSGLQVQRDG 250
Query: 237 KWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGP 296
KW NV P L++NLGD M++M NGIF+SP+HRV+TN E+ R+S+AM E +I P
Sbjct: 251 KWSNVEATPHTLLINLGDTMEVMCNGIFRSPVHRVVTNAEKERISLAMLYSVNDEKDIEP 310
Query: 297 VEGLINETRPRLYRNVN 313
GL++E RP YR V+
Sbjct: 311 AAGLLDENRPARYRKVS 327
>UniRef100_Q7XJ26 Iron/ascorbate-dependent oxidoreductase [Hordeum vulgare]
Length = 352
Score = 262 bits (670), Expect = 9e-69
Identities = 136/314 (43%), Positives = 205/314 (64%), Gaps = 9/314 (2%)
Query: 8 MSMDSDEPPSAYVV-ERNSFGSKDSSTLIP--IPIIDVSLLSSEDEQGKLRSALSSAGCF 64
+S EPPS Y+V E++ G ++ +P IP+ID+S L DE KLR+AL + G F
Sbjct: 14 VSAGVQEPPSRYLVHEQDRHGDLLAAHEMPEPIPLIDLSRLMDADEADKLRAALQTWGFF 73
Query: 65 QAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVN------EAEGYGNDRVVSKKQV 118
A HG+ + ++ + ++ FF P EEKQK + V+ + EGYG+D+V S+ QV
Sbjct: 74 LATNHGIEDSLMEAMMSASREFFRQPSEEKQKCSNLVDGNGKHYQVEGYGSDKVESEDQV 133
Query: 119 LDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEG 178
L+W+ RL LRV P+++R + WP +P F + L E+++K K + D +LR++A+ L ++E
Sbjct: 134 LNWNDRLHLRVEPEDERNFAKWPSHPESFRDVLNEYASKTKKIRDLVLRSIAKLLEIDED 193
Query: 179 SFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKW 238
F++Q ++S AR +YPPC RPDLVLG+ PH+D + +T+L D +V GLQV D KW
Sbjct: 194 YFVNQISNKASGFARLYYYPPCPRPDLVLGLTPHSDGNLLTILFVDDDVGGLQVQRDGKW 253
Query: 239 VNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVE 298
NVP P LV+NL D ++IM+NGIF+SP+HRV+TNTE+ R+S+A+F + E + P
Sbjct: 254 YNVPAKPHTLVINLADCLEIMNNGIFRSPVHRVVTNTEKERLSLAVFYAVDEETVLEPAP 313
Query: 299 GLINETRPRLYRNV 312
GL++E RP YR +
Sbjct: 314 GLLDEKRPPRYRKM 327
>UniRef100_Q8H618 Putative iron/ascorbate-dependent oxidoreductase [Oryza sativa]
Length = 354
Score = 261 bits (668), Expect = 1e-68
Identities = 145/344 (42%), Positives = 216/344 (62%), Gaps = 14/344 (4%)
Query: 5 VQEMSMDSDEPPSAYVVERNSFGSKDSSTLIP--IPIIDVSLLSSED---EQGKLRSALS 59
VQE++ EPPS Y+ + G + + IP IP ID+ LS D E KLRSAL
Sbjct: 12 VQELAAGVQEPPSRYLQDLAG-GDQLAGAEIPEPIPTIDLGRLSGSDGADEAAKLRSALQ 70
Query: 60 SAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVN----EAEGYGNDRVVSK 115
+ G F HG+ ++ +D + E A+ FF PVEEK+K + ++ + EGYGND V +K
Sbjct: 71 NWGLFLVSNHGVETSLIDAVIEAAREFFRQPVEEKKKLSNLIDGKRFQIEGYGNDPVQTK 130
Query: 116 KQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNL 175
Q+LDWS RL L+V P+ R L+ WP +P F + L E++ K+K++ + +L +A+ L L
Sbjct: 131 DQILDWSDRLHLKVEPECDRNLAFWPTHPKSFRDILHEYTLKIKTVKNDILLALAKLLEL 190
Query: 176 EEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVD 235
+E L+QF +++ ARFN+Y PC RPDLVLG+KPH+D +TVLL D+EV GLQVL D
Sbjct: 191 DEDCLLNQFSDRAITTARFNYYSPCPRPDLVLGLKPHSDLCALTVLLTDKEVGGLQVLRD 250
Query: 236 DKWVNVPTIPD-ALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEI 294
W +VP + D +L++N+G ++IM+NG F++P+HRV+TN ER RMSVAMF + E EI
Sbjct: 251 GTWYSVPAVRDYSLLINIGVTLEIMTNGTFRAPLHRVVTNAERERMSVAMFYAVDGEKEI 310
Query: 295 GPVEGLIN-ETRPRLYRNVNNYGDI--NYRCYQEGKIALETVQI 335
PV L+ + + YR + + +Y + G +++++I
Sbjct: 311 EPVAELLGLKQQSARYRGIKGKDLLIGHYEHFSRGGRVVDSLKI 354
>UniRef100_Q9LIF4 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone:MHC9
[Arabidopsis thaliana]
Length = 364
Score = 229 bits (584), Expect = 8e-59
Identities = 129/310 (41%), Positives = 182/310 (58%), Gaps = 13/310 (4%)
Query: 37 IPIIDVSLLSSEDEQG------KLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALP 90
IP+ID+S LS D KL A G FQ I HG+ ++ I EVA FF +P
Sbjct: 55 IPVIDLSKLSKPDNDDFFFEILKLSQACEDWGFFQVINHGIEVEVVEDIEEVASEFFDMP 114
Query: 91 VEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGES 150
+EEK+KY +GYG + S+ Q LDW +L V P + R LWP P+ F ES
Sbjct: 115 LEEKKKYPMEPGTVQGYGQAFIFSEDQKLDWCNMFALGVHPPQIRNPKLWPSKPARFSES 174
Query: 151 LVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVK 210
L +S +++ + LL+ +A SL L+E F FGE V R N+YPPCS PDLVLG+
Sbjct: 175 LEGYSKEIRELCKRLLKYIAISLGLKEERFEEMFGEAVQAV-RMNYYPPCSSPDLVLGLS 233
Query: 211 PHTDRSGITVLLQDR-EVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMH 269
PH+D S +TVL Q + GLQ+L D+ WV V +P+ALV+N+GD ++++SNG +KS H
Sbjct: 234 PHSDGSALTVLQQSKNSCVGLQILKDNTWVPVKPLPNALVINIGDTIEVLSNGKYKSVEH 293
Query: 270 RVLTNTERLRMSVAMFNEPEPENEIGPVEGLI-NETRPRLYRNVNNYGDINYRCYQ---E 325
R +TN E+ R+++ F P E EI P+ L+ +ET P YR+ N+GD +Y +
Sbjct: 294 RAVTNREKERLTIVTFYAPNYEVEIEPMSELVDDETNPCKYRSY-NHGDYSYHYVSNKLQ 352
Query: 326 GKIALETVQI 335
GK +L+ +I
Sbjct: 353 GKKSLDFAKI 362
>UniRef100_Q40839 Ethylene-forming enzyme [Picea glauca]
Length = 298
Score = 224 bits (571), Expect = 3e-57
Identities = 115/276 (41%), Positives = 169/276 (60%), Gaps = 3/276 (1%)
Query: 45 LSSEDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEA 104
L E+E K+ A G FQ + HG+ + +D I + K FF LP EEK+KYA + +
Sbjct: 8 LQKEEEMAKIDKACEEWGFFQVVNHGVPHSLMDDITRMGKEFFRLPAEEKEKYA--IRDF 65
Query: 105 EGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDH 164
+GYG VVS++Q DW L L + P + R LS+WP PS+F + + ++ +++S+
Sbjct: 66 QGYGQIFVVSEEQKRDWGDLLGLIISPPQSRNLSVWPSVPSEFRQIVEAYNMEIRSLAVK 125
Query: 165 LLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQD 224
+L +A +L+L+ F FG + R N+YP C RPDLVLG+ PH D SGIT+LLQD
Sbjct: 126 ILSLIAENLHLKPDYFEQSFGNTYQKM-RMNYYPACPRPDLVLGLSPHADGSGITLLLQD 184
Query: 225 REVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAM 284
+VEGL V DD WV V IP ALV+N+G+ +++++NG +KS HR +TN + R+S+ +
Sbjct: 185 EKVEGLHVRKDDIWVAVQPIPYALVINIGNLLEVITNGRYKSIQHRAVTNKHKSRLSIDV 244
Query: 285 FNEPEPENEIGPVEGLINETRPRLYRNVNNYGDINY 320
F P + EIGP LI+E+ P L+R + I Y
Sbjct: 245 FYSPGFDAEIGPAPELIDESHPCLFRKFIHEDHIKY 280
>UniRef100_Q942Y0 Putative ethylene-forming enzyme [Oryza sativa]
Length = 366
Score = 220 bits (561), Expect = 4e-56
Identities = 131/330 (39%), Positives = 182/330 (54%), Gaps = 13/330 (3%)
Query: 11 DSDEPPSAYVVERNSFGSKDSSTLIPIPIIDVSLLS--SEDEQGKLRSALSSAGCFQAIG 68
D D+ P VV+ D IP+IDV L SEDE LR A G FQ +
Sbjct: 45 DGDDRPDHAVVD-------DERAQERIPVIDVGELQRGSEDELDNLRLACEQWGFFQVVN 97
Query: 69 HGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLR 128
HG+ ++++ + A+ FF LP+EEK+KY +GYG+ V S Q LDW L+L
Sbjct: 98 HGVEEETMEEMEKAAREFFMLPLEEKEKYPMEPGGIQGYGHAFVFSDDQKLDWCNMLALG 157
Query: 129 VFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQS 188
V P RR +LWP P++F ++L ++S +++ + LL +A +L L FGE
Sbjct: 158 VEPAFIRRPNLWPTTPANFSKTLEKYSVEIRELCVRLLEHIAAALGLAPARLNGMFGEAV 217
Query: 189 SLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVL-VDDKWVNVPTIPDA 247
V R NFYPPC RP+LVLG+ PH+D S +TVL QD GLQVL WV V +P A
Sbjct: 218 QAV-RMNFYPPCPRPELVLGLSPHSDGSAVTVLQQDAAFAGLQVLRGGGGWVAVHPVPGA 276
Query: 248 LVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPR 307
LVVN+GD +++++NG +KS HR + + E RMSV F P + E+GP+ L+ + PR
Sbjct: 277 LVVNVGDTLEVLTNGRYKSVEHRAVASGEHDRMSVVTFYAPAYDVELGPLPELVADGEPR 336
Query: 308 LYRNVNN--YGDINYRCYQEGKIALETVQI 335
YR N+ Y +GK LE +I
Sbjct: 337 RYRTYNHGEYSRHYVTSRLQGKKTLEFAKI 366
>UniRef100_O80449 Putative anthocyanidin synthase [Arabidopsis thaliana]
Length = 353
Score = 219 bits (558), Expect = 9e-56
Identities = 119/316 (37%), Positives = 181/316 (56%), Gaps = 7/316 (2%)
Query: 4 SVQEMSMDS-DEPPSAYVV---ERNSFGSKDSSTLIPIPIIDVSLLSSEDEQGKL-RSAL 58
SVQ +S P+ YV +R F + S I IP++D++ + + E +L RSA
Sbjct: 11 SVQSLSQTGVPTVPNRYVKPAHQRPVFNTTQSDAGIEIPVLDMNDVWGKPEGLRLVRSAC 70
Query: 59 SSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQV 118
G FQ + HG++ + ++++R + FF LP+EEK+KYA + + EGYG+ V K
Sbjct: 71 EEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEKRKYANSPDTYEGYGSRLGVVKDAK 130
Query: 119 LDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEG 178
LDWS L P R S WP P E + ++ +V+ + + L T++ SL L+
Sbjct: 131 LDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELIEKYGEEVRKLCERLTETLSESLGLKPN 190
Query: 179 SFLSQFGEQSSLVA--RFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDD 236
+ G + A R NFYP C +P L LG+ H+D GIT+LL D +V GLQV D
Sbjct: 191 KLMQALGGGDKVGASLRTNFYPKCPQPQLTLGLSSHSDPGGITILLPDEKVAGLQVRRGD 250
Query: 237 KWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGP 296
WV + ++P+AL+VN+GDQ+QI+SNGI+KS H+V+ N+ R+S+A F P + +GP
Sbjct: 251 GWVTIKSVPNALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERVSLAFFYNPRSDIPVGP 310
Query: 297 VEGLINETRPRLYRNV 312
+E L+ RP LY+ +
Sbjct: 311 IEELVTANRPALYKPI 326
>UniRef100_Q8LCL0 Putative anthocyanidin synthase [Arabidopsis thaliana]
Length = 353
Score = 218 bits (555), Expect = 2e-55
Identities = 118/316 (37%), Positives = 181/316 (56%), Gaps = 7/316 (2%)
Query: 4 SVQEMSMDS-DEPPSAYVV---ERNSFGSKDSSTLIPIPIIDVSLLSSEDEQGKL-RSAL 58
SVQ +S P+ YV +R F + S I IP++D++ + + E +L RSA
Sbjct: 11 SVQSLSQTGVPTVPNRYVKPAHQRPVFNTTQSDAGIEIPVLDMNDVWGKPEGLRLVRSAC 70
Query: 59 SSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSKKQV 118
G FQ + HG++ + ++++R + FF LP+EEK+KYA + + EGYG+ V +
Sbjct: 71 EEWGFFQMVNHGVTHSLMERVRGAWREFFELPLEEKRKYANSPDTYEGYGSRLGVVRDAK 130
Query: 119 LDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEG 178
LDWS L P R S WP P E + ++ +V+ + + L T++ SL L+
Sbjct: 131 LDWSDYFFLNYLPSSIRNPSKWPSQPPKIRELIEKYGEEVRKLCERLTETLSESLGLKPN 190
Query: 179 SFLSQFGEQSSLVA--RFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDD 236
+ G + A R NFYP C +P L LG+ H+D GIT+LL D +V GLQV D
Sbjct: 191 KLMQALGGGDKVGASLRTNFYPKCPQPQLTLGLSSHSDPGGITILLPDEKVAGLQVRRGD 250
Query: 237 KWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGP 296
WV + ++P+AL+VN+GDQ+QI+SNGI+KS H+V+ N+ R+S+A F P + +GP
Sbjct: 251 GWVTIKSVPNALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERVSLAFFYNPRSDIPVGP 310
Query: 297 VEGLINETRPRLYRNV 312
+E L+ RP LY+ +
Sbjct: 311 IEELVTANRPALYKPI 326
>UniRef100_Q7XZR2 Salt-induced protein [Atriplex nummularia]
Length = 356
Score = 215 bits (547), Expect = 2e-54
Identities = 127/320 (39%), Positives = 191/320 (59%), Gaps = 11/320 (3%)
Query: 2 SKSVQEMSMD-SDEPPSAYVVERNSFGSKDSSTLIPIPI-IDVSLLSS--EDEQGKLRSA 57
S+ VQ+++ DE P ++ + +KD L + ID SLLSS +DE K RSA
Sbjct: 15 SRLVQKIAQTVEDEIPEQFLHKDGFPEAKDVPELWTRSLLIDFSLLSSGDQDEIVKFRSA 74
Query: 58 LSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVS--K 115
LS+ GCF HG+ ++L+++ EV++ FF+LP EEK KY + +GY D V
Sbjct: 75 LSNWGCFLVKNHGIEGSFLEEVIEVSRKFFSLPFEEKMKYYTD-DIFQGYDTDAVCQGLD 133
Query: 116 KQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNL 175
+Q ++W+ RL L ++PK K L WP+NPS+F E + E+S ++ ++ L + A+SLN+
Sbjct: 134 QQKMNWNDRLFLTMYPKSKGNLKAWPQNPSNFSEIVDEYSKELINLTKGLYKAAAKSLNV 193
Query: 176 EEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVD 235
E SF + ++ +++RF YP C RP VLG KPH D S T++L D+E GL+V D
Sbjct: 194 REDSFCLE--KEGVILSRFTLYPKCPRPKNVLGSKPHLDGSVFTIVLADQE--GLEVQKD 249
Query: 236 DKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIG 295
+W VP IP AL VN GD ++++NGI+KS MHRV T++ + R+SV F + E+
Sbjct: 250 GQWFKVPVIPGALFVNFGDFGEVLTNGIYKSTMHRVATHSAKDRLSVTAFCTMDATQELT 309
Query: 296 PVEGLINETRPRLYRNVNNY 315
P+ L+ RP LY+ Y
Sbjct: 310 PLTELVTANRPPLYKKFKVY 329
>UniRef100_Q658E2 Putative iron/ascorbate-dependent oxidoreductase [Oryza sativa]
Length = 367
Score = 213 bits (543), Expect = 5e-54
Identities = 125/303 (41%), Positives = 178/303 (58%), Gaps = 35/303 (11%)
Query: 15 PPSAYVVERN-----SFGSKDSST--LIPIPIIDVSLLSSED---------EQGKLRSAL 58
PPS YV+ + G+ ++ L IP IDVS L++E E KLRSAL
Sbjct: 22 PPSRYVLREKDRPVAAAGAVQAAQRELAAIPTIDVSRLAAESGDDVVDDGGEAAKLRSAL 81
Query: 59 SSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEA-------------- 104
S G F GHGM +LD+I + FF LP EEK++Y+ V A
Sbjct: 82 QSWGLFAVTGHGMPEPFLDEILAATREFFHLPPEEKERYSNVVAAADADGVGAGGERFQP 141
Query: 105 EGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDH 164
EGYG DRV + +Q+LDW RL L+V P+E+RRL WPE+P+ L E++ + + +
Sbjct: 142 EGYGIDRVDTDEQILDWCDRLYLQVQPEEERRLEFWPEHPAALRGLLEEYTRRSEQVFRR 201
Query: 165 LLRTMARSLNLEEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQD 224
+L ARSL E F + GE+ + ARF +YPPC RP+LV G+KPHTD S +TVLL D
Sbjct: 202 VLAATARSLGFGEEFFGDKVGEKVTTYARFTYYPPCPRPELVYGLKPHTDNSVLTVLLLD 261
Query: 225 REVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIM----SNGIFKSPMHRVLTNTERLRM 280
+ V GLQ+L D +W+++P + + L+V GD+++++ +F +P+HRV+T +ER RM
Sbjct: 262 KHVGGLQLLKDGRWLDIPVLTNELLVVAGDEIEVIVIKNHEQVFMAPVHRVVT-SERERM 320
Query: 281 SVA 283
SVA
Sbjct: 321 SVA 323
>UniRef100_Q39224 SRG1 protein [Arabidopsis thaliana]
Length = 358
Score = 212 bits (539), Expect = 1e-53
Identities = 116/313 (37%), Positives = 187/313 (59%), Gaps = 7/313 (2%)
Query: 30 DSSTLIPIPIIDVSLLSS----EDEQGKLRSALSSAGCFQAIGHGMSSTYLDKIREVAKH 85
D I IPIID+ L S + E KL A G FQ + HG+ S++LDK++ +
Sbjct: 46 DFDVKIEIPIIDMKRLCSSTTMDSEVEKLDFACKEWGFFQLVNHGIDSSFLDKVKSEIQD 105
Query: 86 FFALPVEEKQKYARAVNEAEGYGNDRVVSKKQVLDWSYRLSLRVFPKEKRRLSLWPENPS 145
FF LP+EEK+K+ + +E EG+G VVS+ Q LDW+ V P E R+ L+P+ P
Sbjct: 106 FFNLPMEEKKKFWQRPDEIEGFGQAFVVSEDQKLDWADLFFHTVQPVELRKPHLFPKLPL 165
Query: 146 DFGESLVEFSTKVKSMMDHLLRTMARSLNLEEGSFLSQFGEQSSLVA-RFNFYPPCSRPD 204
F ++L +S++V+S+ L+ MAR+L ++ F + S+ + R N+YPPC +PD
Sbjct: 166 PFRDTLEMYSSEVQSVAKILIAKMARALEIKPEELEKLFDDVDSVQSMRMNYYPPCPQPD 225
Query: 205 LVLGVKPHTDRSGITVLLQDREVEGLQVLVDDKWVNVPTIPDALVVNLGDQMQIMSNGIF 264
V+G+ PH+D G+TVL+Q +VEGLQ+ D KWV V +P+A +VN+GD ++I++NG +
Sbjct: 226 QVIGLTPHSDSVGLTVLMQVNDVEGLQIKKDGKWVPVKPLPNAFIVNIGDVLEIITNGTY 285
Query: 265 KSPMHRVLTNTERLRMSVAMFNEPEPENEIGPVEGLINETRPRLYR--NVNNYGDINYRC 322
+S HR + N+E+ R+S+A F+ E+GP + L+ + ++ + Y D +
Sbjct: 286 RSIEHRGVVNSEKERLSIATFHNVGMYKEVGPAKSLVERQKVARFKRLTMKEYNDGLFSR 345
Query: 323 YQEGKIALETVQI 335
+GK L+ ++I
Sbjct: 346 TLDGKAYLDALRI 358
>UniRef100_Q9SB33 SRG1-like protein [Arabidopsis thaliana]
Length = 356
Score = 210 bits (535), Expect = 4e-53
Identities = 126/342 (36%), Positives = 196/342 (56%), Gaps = 10/342 (2%)
Query: 4 SVQEMSMDS--DEPPSAYVVERNSFG--SKDSSTLIPIPIIDVSLLSS----EDEQGKLR 55
SVQEM + P YV + DS IPIID+SLL S + E KL
Sbjct: 15 SVQEMVKEKMITTVPPRYVRSDQDVAEIAVDSGLRNQIPIIDMSLLCSSTSMDSEIDKLD 74
Query: 56 SALSSAGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYARAVNEAEGYGNDRVVSK 115
SA G FQ + HGM S++L+K++ + FF LP+EEK+ + +E EG+G VVS+
Sbjct: 75 SACKEWGFFQLVNHGMESSFLNKVKSEVQDFFNLPMEEKKNLWQQPDEIEGFGQVFVVSE 134
Query: 116 KQVLDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNL 175
+Q LDW+ L + P R+ L+P+ P F ++L +S +VKS+ LL +A +L +
Sbjct: 135 EQKLDWADMFFLTMQPVRLRKPHLFPKLPLPFRDTLDMYSAEVKSIAKILLGKIAVALKI 194
Query: 176 EEGSFLSQFGEQSSLVARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVD 235
+ F ++ R N+YP C PD V+G+ PH+D +G+T+LLQ EVEGLQ+ +
Sbjct: 195 KPEEMDKLFDDELGQRIRLNYYPRCPEPDKVIGLTPHSDSTGLTILLQANEVEGLQIKKN 254
Query: 236 DKWVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIG 295
KWV+V +P+ALVVN+GD ++I++NG ++S HR + N+E+ R+SVA F+ EIG
Sbjct: 255 AKWVSVKPLPNALVVNVGDILEIITNGTYRSIEHRGVVNSEKERLSVAAFHNIGLGKEIG 314
Query: 296 PVEGLINETRPRLYRNVNNYGDIN--YRCYQEGKIALETVQI 335
P+ L+ + +++V N + +GK L+ +++
Sbjct: 315 PMRSLVERHKAAFFKSVTTEEYFNGLFSRELDGKAYLDVMRL 356
>UniRef100_Q6BEG8 Flavonol synthase [Eustoma grandiflorum]
Length = 335
Score = 207 bits (527), Expect = 3e-52
Identities = 115/323 (35%), Positives = 178/323 (54%), Gaps = 7/323 (2%)
Query: 3 KSVQEMSMDSDEPPSAYVVERN--SFGSKDSSTLIPIPIIDVSLLSSEDEQGKLRSALSS 60
+ + +S D P+ Y+ N S S ++ +P++D+S + G + A
Sbjct: 7 QEIASLSKVIDTIPAEYIRSENEQSVISTVHGVVLEVPVVDLSDSDEKKIVGLVSEASKE 66
Query: 61 AGCFQAIGHGMSSTYLDKIREVAKHFFALPVEEKQKYAR--AVNEAEGYGNDRVVSKKQV 118
G FQ + HG+ + + K++EV KHFF LP EEK+ A+ EGYG
Sbjct: 67 WGIFQVVNHGIPNEVIRKLQEVGKHFFELPQEEKELIAKPEGSQSIEGYGTRLQKEVDGK 126
Query: 119 LDWSYRLSLRVFPKEKRRLSLWPENPSDFGESLVEFSTKVKSMMDHLLRTMARSLNLEEG 178
W L +++P WP+NP + E+ E++ +++ ++D+L + ++ L+LE
Sbjct: 127 KGWVDHLFHKIWPPSSINYQFWPKNPPAYREANEEYAKRLQLVVDNLFKYLSLGLDLEPN 186
Query: 179 SFLSQFGEQSSL-VARFNFYPPCSRPDLVLGVKPHTDRSGITVLLQDREVEGLQVLVDDK 237
SF G + + + N+YPPC RPDL LGV HTD S ITVL+ + EV+GLQV D
Sbjct: 187 SFKDGAGGDDLVYLMKINYYPPCPRPDLALGVVAHTDMSAITVLVPN-EVQGLQVYKDGH 245
Query: 238 WVNVPTIPDALVVNLGDQMQIMSNGIFKSPMHRVLTNTERLRMSVAMFNEPEPENEIGPV 297
W + IP+AL+V++GDQ++IMSNG +KS HR N E+ RMS +F EP P++E+GP+
Sbjct: 246 WYDCKYIPNALIVHIGDQVEIMSNGKYKSVYHRTTVNKEKTRMSWPVFLEPPPDHEVGPI 305
Query: 298 EGLINETRPRLYRNVNNYGDINY 320
L+NE P ++ Y D Y
Sbjct: 306 PKLVNEENPAKFK-TKKYKDYAY 327
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.133 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 551,030,476
Number of Sequences: 2790947
Number of extensions: 22275579
Number of successful extensions: 64736
Number of sequences better than 10.0: 1259
Number of HSP's better than 10.0 without gapping: 1068
Number of HSP's successfully gapped in prelim test: 191
Number of HSP's that attempted gapping in prelim test: 61803
Number of HSP's gapped (non-prelim): 1369
length of query: 342
length of database: 848,049,833
effective HSP length: 128
effective length of query: 214
effective length of database: 490,808,617
effective search space: 105033044038
effective search space used: 105033044038
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0219.11


