

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0218.25
(334 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
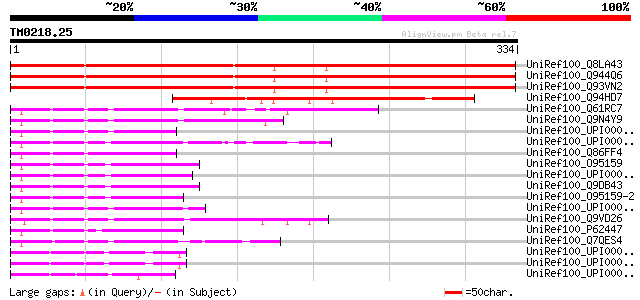
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LA43 Hypothetical protein [Arabidopsis thaliana] 489 e-137
UniRef100_Q944Q6 Hypothetical protein [Arabidopsis thaliana] 489 e-137
UniRef100_Q93VN2 At2g14841/At2g14841 [Arabidopsis thaliana] 486 e-136
UniRef100_Q94HD7 Hypothetical protein OSJNBa0045C13.14 [Oryza sa... 225 1e-57
UniRef100_Q61RC7 Hypothetical protein CBG06644 [Caenorhabditis b... 122 2e-26
UniRef100_Q9N4Y9 Hypothetical protein Y45G12B.2 [Caenorhabditis ... 116 7e-25
UniRef100_UPI0000431937 UPI0000431937 UniRef100 entry 96 1e-18
UniRef100_UPI000036EE80 UPI000036EE80 UniRef100 entry 96 1e-18
UniRef100_Q86FF4 Clone ZZD1216 mRNA sequence [Schistosoma japoni... 95 2e-18
UniRef100_O95159 Zinc finger protein-like 1 [Homo sapiens] 95 3e-18
UniRef100_UPI000045650B UPI000045650B UniRef100 entry 94 4e-18
UniRef100_Q9DB43 Zinc finger protein-like 1 [Mus musculus] 94 4e-18
UniRef100_O95159-2 Splice isoform 2 of O95159 [Homo sapiens] 94 4e-18
UniRef100_UPI00002F9C1C UPI00002F9C1C UniRef100 entry 94 5e-18
UniRef100_Q9VD26 CG5382-PB, isoform B [Drosophila melanogaster] 93 9e-18
UniRef100_P62447 Zinc finger protein-like 1 [Brachydanio rerio] 92 2e-17
UniRef100_Q7QES4 ENSANGP00000019860 [Anopheles gambiae str. PEST] 87 6e-16
UniRef100_UPI0000499912 UPI0000499912 UniRef100 entry 57 9e-07
UniRef100_UPI000049978E UPI000049978E UniRef100 entry 57 9e-07
UniRef100_UPI000049924F UPI000049924F UniRef100 entry 55 3e-06
>UniRef100_Q8LA43 Hypothetical protein [Arabidopsis thaliana]
Length = 343
Score = 489 bits (1260), Expect = e-137
Identities = 240/344 (69%), Positives = 274/344 (78%), Gaps = 13/344 (3%)
Query: 1 MVVCKCRKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQAV 60
MVVCKC+KAT+LYCFVHK PVCGECICFPEHQ CV+RTYSEWVIDGEYD PKCCQCQA
Sbjct: 1 MVVCKCKKATRLYCFVHKAPVCGECICFPEHQTCVVRTYSEWVIDGEYD-QPKCCQCQAT 59
Query: 61 LEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSR 120
+EG G Q TRLGCLH IHT+CLVS IKSFPPHTAPAGY CP+CST IWPP VKD+GSR
Sbjct: 60 FDEGAGHQVTRLGCLHAIHTSCLVSLIKSFPPHTAPAGYVCPSCSTPIWPPMMVKDAGSR 119
Query: 121 LHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSD------- 173
LH+ L+E I QTG+EKN+ GNHPVS S TESRSPPPAFAS+ LI + ++
Sbjct: 120 LHALLREVITQTGLEKNLLGNHPVSRS-TESRSPPPAFASDALINISSSSHTQEGNNLPD 178
Query: 174 --SPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSP--VGPATRKGAFNVERQNSEISYY 229
S A E K +V++IVEI+ SAG+++K SSP A RKG V+RQNSE YY
Sbjct: 179 GYSVAGNGEYSKSAVSEIVEIDVPASAGSYMKSSSPGLAAAAARKGVPAVDRQNSETLYY 238
Query: 230 ADDEDGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQ 289
ADDEDGNRKKY++RGP HKFLRALLPFWSSALPTLPVTAPPRKDA+ A +GSEGR RHQ
Sbjct: 239 ADDEDGNRKKYSRRGPLRHKFLRALLPFWSSALPTLPVTAPPRKDAAKADDGSEGRVRHQ 298
Query: 290 RSSRMDPRKILLLIAIMACMATMGILYYRLVQRGPGEELPNEEQ 333
RSS+MD RKIL+ IA++ACMATMGILYYRL + G+ELP+EEQ
Sbjct: 299 RSSKMDIRKILIFIALIACMATMGILYYRLALQAIGQELPDEEQ 342
>UniRef100_Q944Q6 Hypothetical protein [Arabidopsis thaliana]
Length = 343
Score = 489 bits (1259), Expect = e-137
Identities = 240/344 (69%), Positives = 273/344 (78%), Gaps = 13/344 (3%)
Query: 1 MVVCKCRKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQAV 60
MVVCKC+KAT+LYCFVHK PVCGECICFPEHQ CV+RTYSEWVIDGEYD PKCCQCQA
Sbjct: 1 MVVCKCKKATRLYCFVHKAPVCGECICFPEHQTCVVRTYSEWVIDGEYD-QPKCCQCQAT 59
Query: 61 LEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSR 120
+EG G Q TRLGCLH IHT+CLVS IKSFPPHTAPAGY CP CST IWPP VKD+GSR
Sbjct: 60 FDEGAGHQVTRLGCLHAIHTSCLVSLIKSFPPHTAPAGYVCPACSTPIWPPMMVKDAGSR 119
Query: 121 LHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSD------- 173
LH+ L+E I QTG+EKN+ GNHPVS S TESRSPPPAFAS+ LI + ++
Sbjct: 120 LHALLREVITQTGLEKNLLGNHPVSRS-TESRSPPPAFASDALINISSSSHTQEGNNLPD 178
Query: 174 --SPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSP--VGPATRKGAFNVERQNSEISYY 229
S A E K +V++IVEI+ SAG+++K SSP A RKG V+RQNSE YY
Sbjct: 179 GYSVAGNGEYSKSAVSEIVEIDVPASAGSYMKSSSPGLAAAAARKGVPAVDRQNSETLYY 238
Query: 230 ADDEDGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQ 289
ADDEDGNRKKY++RGP HKFLRALLPFWSSALPTLPVTAPPRKDA+ A +GSEGR RHQ
Sbjct: 239 ADDEDGNRKKYSRRGPLRHKFLRALLPFWSSALPTLPVTAPPRKDAAKADDGSEGRVRHQ 298
Query: 290 RSSRMDPRKILLLIAIMACMATMGILYYRLVQRGPGEELPNEEQ 333
RSS+MD RKIL+ IA++ACMATMGILYYRL + G+ELP+EEQ
Sbjct: 299 RSSKMDIRKILIFIALIACMATMGILYYRLALQAIGQELPDEEQ 342
>UniRef100_Q93VN2 At2g14841/At2g14841 [Arabidopsis thaliana]
Length = 343
Score = 486 bits (1252), Expect = e-136
Identities = 239/344 (69%), Positives = 272/344 (78%), Gaps = 13/344 (3%)
Query: 1 MVVCKCRKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQAV 60
MVVCKC+KAT+LYCFVHK PVCGECICFPEHQ CV+RTYSEWVIDGEYD PKCCQCQA
Sbjct: 1 MVVCKCKKATRLYCFVHKAPVCGECICFPEHQTCVVRTYSEWVIDGEYD-QPKCCQCQAT 59
Query: 61 LEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSGSR 120
+EG G Q TRLGCLH IHT+CLVS IKSFPPHTAPAGY CP CST IWPP VKD+GSR
Sbjct: 60 FDEGAGHQVTRLGCLHAIHTSCLVSLIKSFPPHTAPAGYVCPACSTPIWPPMMVKDAGSR 119
Query: 121 LHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSD------- 173
LH+ L+E I QTG+EKN+ GNHPVS S TESRSPPPAFAS+ LI + ++
Sbjct: 120 LHALLREVITQTGLEKNLLGNHPVSRS-TESRSPPPAFASDALINISSSSHTQEGNNLPD 178
Query: 174 --SPATGSEPPKLSVTDIVEIEGANSAGNFVKGSSP--VGPATRKGAFNVERQNSEISYY 229
S A E K +V++IVEI+ SAG+++K SSP A RKG V+RQNSE YY
Sbjct: 179 GYSVAGNGEYSKSAVSEIVEIDVPASAGSYMKSSSPGLAAAAARKGVPAVDRQNSETLYY 238
Query: 230 ADDEDGNRKKYTKRGPFYHKFLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQ 289
ADDEDGNRKKY++RGP HKFLRALLPFWSSALPTLPVTAPPRKDA+ A +GSEGR RHQ
Sbjct: 239 ADDEDGNRKKYSRRGPLRHKFLRALLPFWSSALPTLPVTAPPRKDAAKADDGSEGRVRHQ 298
Query: 290 RSSRMDPRKILLLIAIMACMATMGILYYRLVQRGPGEELPNEEQ 333
RSS+MD KIL+ IA++ACMATMGILYYRL + G+ELP+EEQ
Sbjct: 299 RSSKMDIGKILIFIALIACMATMGILYYRLALQAIGQELPDEEQ 342
>UniRef100_Q94HD7 Hypothetical protein OSJNBa0045C13.14 [Oryza sativa]
Length = 383
Score = 225 bits (573), Expect = 1e-57
Identities = 130/237 (54%), Positives = 157/237 (65%), Gaps = 43/237 (18%)
Query: 108 IWPPKSVKDSGSRLHSKLKEAIMQ------TGMEKNIFGNHPVSLSVTESRSPPPAFASE 161
IWPP ++KD+GSRLHSKLKEAI Q TG+EKN+FGNH V++ ++R+PP AFAS+
Sbjct: 116 IWPPSTIKDTGSRLHSKLKEAIAQISINVKTGLEKNVFGNHFVTMPKADTRTPP-AFASD 174
Query: 162 PLI------GRENHGNS----------------DSPATGSEPPKLSVTDIVEIEGAN--- 196
PL RE++G + S GS P +IVEI+G +
Sbjct: 175 PLKRVSISGDRESNGANIINSAIDANVQSGGMYSSATVGSGTPSHVEPEIVEIDGPSPIT 234
Query: 197 -----SAGNFVKGSSPVGPA--TRKGAFNVERQNSEISYYADDEDGNRKKYTKRGPFYHK 249
NF++ SP GP+ TRKGA VERQNSEISYYADDED NRKKYTKRG F HK
Sbjct: 235 TQFPEQESNFIRSPSPHGPSAMTRKGANYVERQNSEISYYADDEDANRKKYTKRGTFRHK 294
Query: 250 FLRALLPFWSSALPTLPVTAPPRKDASNATEGSEGRTRHQRSSRMDPRKILLLIAIM 306
FLR LLPFWSSALPTLPVTAPPRK+ ++ EGR+RHQ+SSRMDP KILL +AI+
Sbjct: 295 FLRMLLPFWSSALPTLPVTAPPRKE----SDAPEGRSRHQKSSRMDPTKILLALAII 347
>UniRef100_Q61RC7 Hypothetical protein CBG06644 [Caenorhabditis briggsae]
Length = 319
Score = 122 bits (305), Expect = 2e-26
Identities = 83/254 (32%), Positives = 115/254 (44%), Gaps = 28/254 (11%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+C+ H+V VC C+ H CV+++Y W+ D +YD P C C
Sbjct: 1 MGLCKCPKRKVTNLFCYEHRVNVCEFCLV-DNHPNCVVQSYLNWLTDQDYD--PNCSLCH 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSG 118
L +G+ T RL CLH++H C SFPP TAPAGY CP CS ++PP +
Sbjct: 58 TTLTQGE---TIRLNCLHLLHWRCFDDWAASFPPTTAPAGYRCPCCSQEVFPP---INEV 111
Query: 119 SRLHSKLKEAIMQTGMEKNIFG-------NHPVSLSVTESRSPPPAFASEPLIGRENHGN 171
S L KL+E + Q+ +N G N PV ++ PPP P + ++
Sbjct: 112 SPLIEKLREQLKQSNWARNALGLPVLPELNRPVK-NIAPIPPPPP-----PQVKHVSY-- 163
Query: 172 SDSPATGSEP--PKLSVTDIVEIEGANSAGNFVKGSSPVGPATRKGAFNVERQNSEISYY 229
DSPA P S T +E ++A V S + A +
Sbjct: 164 DDSPAQKEIPIHHNRSETPATHLEMEDTASYSVSNSDVTFARKKNFASESSSDTRPLLRQ 223
Query: 230 ADDEDGNRKKYTKR 243
D D KY +R
Sbjct: 224 DRDADNEENKYKRR 237
>UniRef100_Q9N4Y9 Hypothetical protein Y45G12B.2 [Caenorhabditis elegans]
Length = 309
Score = 116 bits (291), Expect = 7e-25
Identities = 69/186 (37%), Positives = 98/186 (52%), Gaps = 15/186 (8%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+C+ H+V VC C+ H CV+++Y W+ D +YD P C C+
Sbjct: 1 MGLCKCPKRKVTNLFCYEHRVNVCEFCLV-DNHPNCVVQSYLTWLTDQDYD--PNCSLCK 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSG 118
L EGD T RL CLH++H C +FP TAPAGY CP CS ++PP +
Sbjct: 58 TTLAEGD---TIRLNCLHLLHWKCFDEWAANFPATTAPAGYRCPCCSQEVFPP---INEV 111
Query: 119 SRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSP-PPAFASEPLIGRE---NHGNSDS 174
S L KL+E + Q+ + G + SP PP + P++ +E ++ S +
Sbjct: 112 SPLIEKLREQLKQSNWARAALGLPTLPELNRPVPSPAPPQLKNAPVMHKEVPVHNNRSST 171
Query: 175 PATGSE 180
PAT E
Sbjct: 172 PATHLE 177
>UniRef100_UPI0000431937 UPI0000431937 UniRef100 entry
Length = 110
Score = 96.3 bits (238), Expect = 1e-18
Identities = 47/112 (41%), Positives = 63/112 (55%), Gaps = 8/112 (7%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC R+ T L+CF H+V VC C+ H C++++Y W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRRVTNLFCFEHRVNVCEHCMV-TNHPKCIVQSYILWLHDHDYN--PICTLCS 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWP 110
L EGD RL C H+ H CL + + P TAPAGY CPTC I+P
Sbjct: 58 VNLSEGD---CVRLTCYHMYHWTCLDKYARELPATTAPAGYTCPTCKERIFP 106
>UniRef100_UPI000036EE80 UPI000036EE80 UniRef100 entry
Length = 310
Score = 96.3 bits (238), Expect = 1e-18
Identities = 69/214 (32%), Positives = 105/214 (48%), Gaps = 27/214 (12%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRKVTNLFCFEHRVNVCEHCLV-ANHAKCIVQSYLQWLQDSDYN--PNCRLCN 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSG 118
L +TTRL C + H CL P +TAPAGY CP+C+ I+PP ++
Sbjct: 58 IPLAS---RETTRLVCYDLFHWACLNERAAQLPRNTAPAGYQCPSCNGPIFPPTNL---A 111
Query: 119 SRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSDSPATG 178
+ S L+E + + G P+ + E SP P EPL + S A+
Sbjct: 112 GPVASALREKLATVNWARAGLG-LPL---IDEVVSPEP----EPLNTSDFSDWSSFNASS 163
Query: 179 SEPPKLSVTDIVEIEGANSAGNFVKGSSPVGPAT 212
+ P+ E++GA++A F +P PA+
Sbjct: 164 TPGPE-------EVDGASAAPAFY-SQAPRPPAS 189
>UniRef100_Q86FF4 Clone ZZD1216 mRNA sequence [Schistosoma japonicum]
Length = 337
Score = 95.1 bits (235), Expect = 2e-18
Identities = 43/112 (38%), Positives = 63/112 (55%), Gaps = 5/112 (4%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC R+ T L+CF H+V VC C+ H C++++Y W+ D +++ P C C
Sbjct: 1 MGLCKCERRRVTNLFCFEHRVNVCEFCLV-ANHNKCIVKSYLRWIKDSDFN--PSCSLCH 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWP 110
+ E + RL CL V H +CL +++ S PP T PAG+ CP C I P
Sbjct: 58 ELFETDPTKKCVRLVCLDVFHWDCLNNYVLSLPPTTTPAGFLCPLCDQPIIP 109
>UniRef100_O95159 Zinc finger protein-like 1 [Homo sapiens]
Length = 310
Score = 94.7 bits (234), Expect = 3e-18
Identities = 50/128 (39%), Positives = 70/128 (54%), Gaps = 9/128 (7%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRKVTNLFCFEHRVNVCEHCLV-ANHAKCIVQSYLQWLQDSDYN--PNCRLCN 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDS- 117
L +TTRL C + H CL P +TAPAGY CP+C+ I+PP ++
Sbjct: 58 IPLAS---RETTRLVCYDLFHWACLNERAAQLPRNTAPAGYQCPSCNGPIFPPTNLAGPV 114
Query: 118 GSRLHSKL 125
S L KL
Sbjct: 115 ASALREKL 122
>UniRef100_UPI000045650B UPI000045650B UniRef100 entry
Length = 117
Score = 94.4 bits (233), Expect = 4e-18
Identities = 48/122 (39%), Positives = 67/122 (54%), Gaps = 8/122 (6%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRKVTNLFCFEHRVNVCEHCLV-ANHAKCIVQSYLQWLQDSDYN--PNCRLCN 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSG 118
L +TTRL C + H CL P +TAPAGY CP+C+ I+PP + G
Sbjct: 58 IPLAS---RETTRLVCYDLFHWACLNERAAQLPRNTAPAGYQCPSCNGPIFPPTNGWPRG 114
Query: 119 SR 120
R
Sbjct: 115 LR 116
>UniRef100_Q9DB43 Zinc finger protein-like 1 [Mus musculus]
Length = 310
Score = 94.4 bits (233), Expect = 4e-18
Identities = 49/128 (38%), Positives = 70/128 (54%), Gaps = 9/128 (7%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRKVTNLFCFEHRVNVCEHCLV-ANHAKCIVQSYLQWLQDSDYN--PNCRLCN 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDS- 117
L +TTRL C + H C+ P +TAPAGY CP+C+ I+PP ++
Sbjct: 58 TPLAS---RETTRLVCYDLFHWACINERAAQLPRNTAPAGYQCPSCNGPIFPPANLAGPV 114
Query: 118 GSRLHSKL 125
S L KL
Sbjct: 115 ASALREKL 122
>UniRef100_O95159-2 Splice isoform 2 of O95159 [Homo sapiens]
Length = 118
Score = 94.4 bits (233), Expect = 4e-18
Identities = 46/116 (39%), Positives = 66/116 (56%), Gaps = 8/116 (6%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRKVTNLFCFEHRVNVCEHCLV-ANHAKCIVQSYLQWLQDSDYN--PNCRLCN 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSV 114
L +TTRL C + H CL P +TAPAGY CP+C+ I+PP ++
Sbjct: 58 IPLAS---RETTRLVCYDLFHWACLNERAAQLPRNTAPAGYQCPSCNGPIFPPTNL 110
>UniRef100_UPI00002F9C1C UPI00002F9C1C UniRef100 entry
Length = 323
Score = 94.0 bits (232), Expect = 5e-18
Identities = 49/131 (37%), Positives = 71/131 (53%), Gaps = 11/131 (8%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC RK T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKRKVTNLFCFEHRVNVCEHCLV-SNHNKCIVQSYLQWLQDSDYN--PNCSLCN 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSG 118
L D T RL C V H +CL + P HTAPAGY CP C ++P ++
Sbjct: 58 DPLTAQD---TVRLVCYDVFHWSCLNNLASRLPLHTAPAGYQCPVCQGPVFPSPNL---A 111
Query: 119 SRLHSKLKEAI 129
S + +L+E +
Sbjct: 112 SPIADQLREQL 122
>UniRef100_Q9VD26 CG5382-PB, isoform B [Drosophila melanogaster]
Length = 299
Score = 93.2 bits (230), Expect = 9e-18
Identities = 65/221 (29%), Positives = 107/221 (48%), Gaps = 20/221 (9%)
Query: 1 MVVCKCRK--ATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC K T +CF H+V VC C+ H C++++Y +W+ D +Y C C
Sbjct: 1 MGLCKCPKRLVTNQFCFEHRVNVCEHCMV-QSHPKCIVQSYLQWLRDSDYI--SNCTLCG 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSG 118
LE+GD RL C HV H +CL + + P +TAP G+ CP CS I+P ++
Sbjct: 58 TTLEQGD---CVRLVCYHVFHWDCLNARQAALPANTAPRGHQCPACSVEIFPNANLV--- 111
Query: 119 SRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTES-RSPPPAFASEPLIG---RENHGNSDS 174
S + LK + Q +N G +S + S ++ P AS+ + + +H +S
Sbjct: 112 SPVADALKSFLSQVNWGRNGLGLALLSEEQSSSLKAIKPKVASQAAVSNMTKVHHIHSGG 171
Query: 175 PATGSEP---PKLSVTDIVEIEGAN--SAGNFVKGSSPVGP 210
++P +S ++ ++ N SAG++ P+ P
Sbjct: 172 ERERTKPNGHDAVSPHSVLLMDAFNPPSAGDYASSRRPLLP 212
>UniRef100_P62447 Zinc finger protein-like 1 [Brachydanio rerio]
Length = 317
Score = 92.0 bits (227), Expect = 2e-17
Identities = 43/116 (37%), Positives = 66/116 (56%), Gaps = 8/116 (6%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC +K T L+CF H+V VC C+ H C++++Y +W+ D +Y+ P C C
Sbjct: 1 MGLCKCPKKKVTNLFCFKHRVNVCEHCLV-SNHNKCIVQSYLQWLQDSDYN--PNCSLC- 56
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSV 114
++ D T RL C + H +CL P +TAP GY CPTC ++PP+++
Sbjct: 57 --IQPLDSQDTVRLVCYDLFHWSCLNELASHQPLNTAPDGYQCPTCQGPVFPPRNL 110
>UniRef100_Q7QES4 ENSANGP00000019860 [Anopheles gambiae str. PEST]
Length = 295
Score = 87.0 bits (214), Expect = 6e-16
Identities = 54/180 (30%), Positives = 88/180 (48%), Gaps = 21/180 (11%)
Query: 1 MVVCKC--RKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQ 58
M +CKC R+ T +CF H+V VC C+ H C +++Y +W+ D ++D C C
Sbjct: 1 MGLCKCPKRQVTTQFCFEHRVNVCENCMVV-NHTKCTVQSYIQWLKDSDFD--SNCTLCG 57
Query: 59 AVLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWPPKSVKDSG 118
+ E D RL C HV H CL + + P +TAP G+ CP CS ++PP
Sbjct: 58 SPRESDD---CVRLICYHVFHWKCLNARQQLLPANTAPGGHTCPICSEPMFPP------- 107
Query: 119 SRLHSKLKEAIMQTGMEKNIFGNHPVSLSVTESRSPPPAFASEPLIGRENHGNSDSPATG 178
S L S + + +++ + + +G + + L + S S A E ++G+ G + G
Sbjct: 108 SNLVSPVAD-VLRVRLGQVNWGRNELGLPLVSSSS-----AGELMVGQNGAGTMTGYSDG 161
>UniRef100_UPI0000499912 UPI0000499912 UniRef100 entry
Length = 260
Score = 56.6 bits (135), Expect = 9e-07
Identities = 39/121 (32%), Positives = 54/121 (44%), Gaps = 15/121 (12%)
Query: 1 MVVC-KCRKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQA 59
M +C +C+K T LYC HK +C ECI F H C + Y E+V + + C C
Sbjct: 1 MGICFECKKPTDLYCVHHKKHICIECI-FKNHINCTVCKYREYVEENREE-VNNCIICST 58
Query: 60 VLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWP----PKSVK 115
L +T RL C V H +CL+S + CP C+ I+ P +K
Sbjct: 59 ELA---SKETVRLPCFCVFHKDCLISLFDQ-----SIEEVKCPKCNQPIFTQQDIPNQIK 110
Query: 116 D 116
D
Sbjct: 111 D 111
>UniRef100_UPI000049978E UPI000049978E UniRef100 entry
Length = 260
Score = 56.6 bits (135), Expect = 9e-07
Identities = 39/121 (32%), Positives = 54/121 (44%), Gaps = 15/121 (12%)
Query: 1 MVVC-KCRKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQA 59
M +C +C+K T LYC HK +C ECI F H C + Y E+V + + C C
Sbjct: 1 MGICFECKKPTDLYCVHHKKHICIECI-FKNHINCTVCKYREYVEENREE-VNNCIICST 58
Query: 60 VLEEGDGSQTTRLGCLHVIHTNCLVSHIKSFPPHTAPAGYACPTCSTSIWP----PKSVK 115
L +T RL C V H +CL+S + CP C+ I+ P +K
Sbjct: 59 ELA---SKETVRLPCFCVFHKDCLISLFDQ-----SIEEVKCPKCNQPIFTQQDIPNQIK 110
Query: 116 D 116
D
Sbjct: 111 D 111
>UniRef100_UPI000049924F UPI000049924F UniRef100 entry
Length = 223
Score = 54.7 bits (130), Expect = 3e-06
Identities = 36/113 (31%), Positives = 52/113 (45%), Gaps = 8/113 (7%)
Query: 1 MVVCK-CRKATKLYCFVHKVPVCGECICFPEHQICVIRTYSEWVIDGEYDWPPKCCQCQA 59
M +C C++ T LYC H+ +C C+ H+ C I Y ++V DG+ C C
Sbjct: 1 MGICHICKEQTTLYCVDHEKHICLNCL-LKSHKECTICQYVDYV-DGKVKKDNSCHLCGE 58
Query: 60 VLEEGDGSQTTRLGCLHVIHTNCL---VSHIKSFPPHTAPAGYACPTCSTSIW 109
+ EGD RL CLH H CL +S+I + + CP C I+
Sbjct: 59 EIFEGD--LIVRLPCLHKFHQECLLKSLSNIHLNESESCENQFKCPECLIDIF 109
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.135 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 611,920,549
Number of Sequences: 2790947
Number of extensions: 26464726
Number of successful extensions: 69969
Number of sequences better than 10.0: 128
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 97
Number of HSP's that attempted gapping in prelim test: 69817
Number of HSP's gapped (non-prelim): 176
length of query: 334
length of database: 848,049,833
effective HSP length: 128
effective length of query: 206
effective length of database: 490,808,617
effective search space: 101106575102
effective search space used: 101106575102
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0218.25


