

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
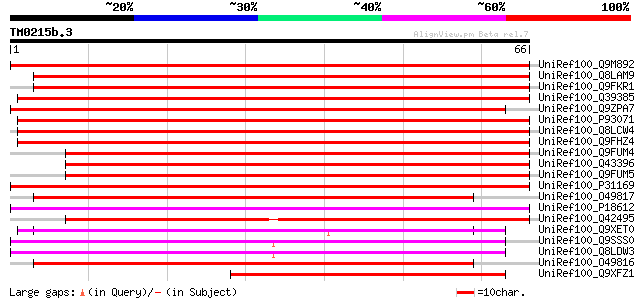
Query= TM0215b.3
(66 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M892 F16B3.11 protein [Arabidopsis thaliana] 86 3e-16
UniRef100_Q8LAM9 Pollen coat-like protein [Arabidopsis thaliana] 85 4e-16
UniRef100_Q9FKR1 Pollen coat protein-like [Arabidopsis thaliana] 84 1e-15
UniRef100_Q39385 Pollen coat protein [Brassica oleracea] 82 4e-15
UniRef100_Q9ZPA7 ABA-inducible protein [Fagus sylvatica] 82 5e-15
UniRef100_P93071 Pollen coat protein homolog [Brassica campestris] 80 1e-14
UniRef100_Q8LCW4 ABA-inducible protein-like protein [Arabidopsis... 79 4e-14
UniRef100_Q9FHZ4 ABA-inducible protein-like [Arabidopsis thaliana] 77 1e-13
UniRef100_Q9FUM4 BN28b [Brassica napus] 53 2e-06
UniRef100_Q43396 Kin1 protein [Brassica napus] 52 3e-06
UniRef100_Q9FUM5 BN28a [Brassica napus] 52 4e-06
UniRef100_P31169 Stress-induced KIN2 protein [Arabidopsis thaliana] 51 9e-06
UniRef100_O49817 Late embryogenesis abundant protein 2 [Cicer ar... 48 7e-05
UniRef100_P18612 Stress-induced KIN1 protein [Arabidopsis thaliana] 48 7e-05
UniRef100_Q42495 Kin1 protein [Brassica campestris] 47 2e-04
UniRef100_Q9XET0 Seed maturation protein PM30 [Glycine max] 47 2e-04
UniRef100_Q9SSS0 F6D8.10 [Arabidopsis thaliana] 46 2e-04
UniRef100_Q8LDW3 Hypothetical protein [Arabidopsis thaliana] 46 2e-04
UniRef100_O49816 Late embryogenesis abundant protein 1 [Cicer ar... 46 2e-04
UniRef100_Q9XFZ1 Cold shock protein [Camelina sativa] 46 3e-04
>UniRef100_Q9M892 F16B3.11 protein [Arabidopsis thaliana]
Length = 68
Score = 85.5 bits (210), Expect = 3e-16
Identities = 43/66 (65%), Positives = 48/66 (72%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
MD QN S+QAGQATGQ +EKA MM A++AA SAQ S Q+ GQQMK KAQGAAD VK
Sbjct: 1 MDNKQNASYQAGQATGQTKEKAGGMMDKAKDAAASAQDSLQQTGQQMKEKAQGAADVVKD 60
Query: 61 AIGANK 66
G NK
Sbjct: 61 KTGMNK 66
>UniRef100_Q8LAM9 Pollen coat-like protein [Arabidopsis thaliana]
Length = 67
Score = 85.1 bits (209), Expect = 4e-16
Identities = 43/63 (68%), Positives = 50/63 (79%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIG 63
SQN+S QAGQA GQ QEKAS MM A NAAQSA++S QE GQQ+K KAQGA ++VK+A G
Sbjct: 5 SQNISFQAGQAKGQTQEKASTMMDKASNAAQSAKESLQETGQQIKEKAQGATESVKNATG 64
Query: 64 ANK 66
NK
Sbjct: 65 MNK 67
>UniRef100_Q9FKR1 Pollen coat protein-like [Arabidopsis thaliana]
Length = 67
Score = 83.6 bits (205), Expect = 1e-15
Identities = 42/63 (66%), Positives = 50/63 (78%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIG 63
SQN+S QAGQA GQ QEKAS MM A NAAQSA++S +E GQQ+K KAQGA ++VK+A G
Sbjct: 5 SQNISFQAGQAKGQTQEKASTMMDKASNAAQSAKESLKETGQQIKEKAQGATESVKNATG 64
Query: 64 ANK 66
NK
Sbjct: 65 MNK 67
>UniRef100_Q39385 Pollen coat protein [Brassica oleracea]
Length = 67
Score = 82.0 bits (201), Expect = 4e-15
Identities = 40/65 (61%), Positives = 50/65 (76%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSA 61
+ SQ++S AGQA GQ QEKASN+M A NAAQSA++S QE GQQ+K KAQGA++A+K
Sbjct: 3 NNSQSMSFNAGQAKGQTQEKASNLMDKASNAAQSAKESLQEGGQQLKQKAQGASEAIKEK 62
Query: 62 IGANK 66
G NK
Sbjct: 63 TGLNK 67
>UniRef100_Q9ZPA7 ABA-inducible protein [Fagus sylvatica]
Length = 68
Score = 81.6 bits (200), Expect = 5e-15
Identities = 40/63 (63%), Positives = 49/63 (77%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
MD+SQN QAGQA GQ QEKASN+M A NA QSA+++C EAG+Q+ AKAQGAA+ K
Sbjct: 1 MDKSQNTCFQAGQAKGQTQEKASNLMEKAGNAGQSAKETCLEAGKQVPAKAQGAANTAKD 60
Query: 61 AIG 63
A+G
Sbjct: 61 AVG 63
>UniRef100_P93071 Pollen coat protein homolog [Brassica campestris]
Length = 67
Score = 80.1 bits (196), Expect = 1e-14
Identities = 40/65 (61%), Positives = 47/65 (71%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSA 61
+ SQN S AGQA GQ QEKASN+M A NAAQSA++S E GQQ+K KAQGA +A+K
Sbjct: 3 NNSQNWSFNAGQAKGQTQEKASNLMDKASNAAQSAKESLSEGGQQLKQKAQGATEAIKEK 62
Query: 62 IGANK 66
G NK
Sbjct: 63 TGLNK 67
>UniRef100_Q8LCW4 ABA-inducible protein-like protein [Arabidopsis thaliana]
Length = 67
Score = 78.6 bits (192), Expect = 4e-14
Identities = 38/65 (58%), Positives = 49/65 (74%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSA 61
+ SQ++S AGQA GQ QEKASN+M A NAAQSA++S QE GQQ+K KAQGA++ +K
Sbjct: 3 NNSQSMSFNAGQAKGQTQEKASNLMDKASNAAQSAKESIQEGGQQLKQKAQGASETIKEK 62
Query: 62 IGANK 66
G +K
Sbjct: 63 TGISK 67
>UniRef100_Q9FHZ4 ABA-inducible protein-like [Arabidopsis thaliana]
Length = 67
Score = 77.0 bits (188), Expect = 1e-13
Identities = 37/65 (56%), Positives = 49/65 (74%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSA 61
+ SQ++S AGQA GQ QEKASN++ A NAAQSA++S QE GQQ+K KAQGA++ +K
Sbjct: 3 NNSQSMSFNAGQAKGQTQEKASNLIDKASNAAQSAKESIQEGGQQLKQKAQGASETIKEK 62
Query: 62 IGANK 66
G +K
Sbjct: 63 TGISK 67
>UniRef100_Q9FUM4 BN28b [Brassica napus]
Length = 65
Score = 52.8 bits (125), Expect = 2e-06
Identities = 27/59 (45%), Positives = 37/59 (61%)
Query: 8 SHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIGANK 66
S QAGQ G+A+EK++ +M ++AA SA S Q AGQ++ A GA + VK G NK
Sbjct: 7 SFQAGQTAGRAEEKSNVLMDKVKDAATSAGASAQTAGQKISETAGGAVNVVKEKTGINK 65
>UniRef100_Q43396 Kin1 protein [Brassica napus]
Length = 65
Score = 52.4 bits (124), Expect = 3e-06
Identities = 27/59 (45%), Positives = 38/59 (63%)
Query: 8 SHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIGANK 66
S QAGQA+G+A+EK + +M ++AA +A S Q AGQ++ A GA + VK G NK
Sbjct: 7 SFQAGQASGRAEEKGNVLMDKVKDAATAAGASAQTAGQKITEAAGGAVNLVKEKTGMNK 65
>UniRef100_Q9FUM5 BN28a [Brassica napus]
Length = 65
Score = 52.0 bits (123), Expect = 4e-06
Identities = 27/59 (45%), Positives = 37/59 (61%)
Query: 8 SHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIGANK 66
S QAGQA G+A+EK + +M ++AA +A S Q AGQ++ A GA + VK G NK
Sbjct: 7 SFQAGQAAGRAEEKGNVLMDKVKDAATAAGASAQTAGQKITEAAGGAVNLVKEKTGMNK 65
>UniRef100_P31169 Stress-induced KIN2 protein [Arabidopsis thaliana]
Length = 66
Score = 50.8 bits (120), Expect = 9e-06
Identities = 25/66 (37%), Positives = 41/66 (61%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
M ++ + QAGQA G+A+EK++ ++ A++AA +A S Q+AG+ + A G + VK
Sbjct: 1 MSETNKNAFQAGQAAGKAEEKSNVLLDKAKDAAAAAGASAQQAGKSISDAAVGGVNFVKD 60
Query: 61 AIGANK 66
G NK
Sbjct: 61 KTGLNK 66
>UniRef100_O49817 Late embryogenesis abundant protein 2 [Cicer arietinum]
Length = 155
Score = 47.8 bits (112), Expect = 7e-05
Identities = 22/56 (39%), Positives = 35/56 (62%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S + S++AG+A G+ +EK + M+GN + AQ+A++ Q+A Q K K A A K
Sbjct: 3 SHDQSYRAGEAKGRTEEKTNQMIGNIEDKAQAAKEKAQQAAQTAKDKTSQTAQAAK 58
Score = 43.9 bits (102), Expect = 0.001
Identities = 24/64 (37%), Positives = 35/64 (54%), Gaps = 4/64 (6%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQS----CQEAGQQMKAKAQGAADAVK 59
+Q +A TG+A+EK S M + + AQS + + Q+ G++ K AQGA DAVK
Sbjct: 76 TQATKEKAQDTTGRAKEKGSEMGQSTKETAQSGKDNSAGFLQQTGEKAKGMAQGATDAVK 135
Query: 60 SAIG 63
G
Sbjct: 136 QTFG 139
>UniRef100_P18612 Stress-induced KIN1 protein [Arabidopsis thaliana]
Length = 66
Score = 47.8 bits (112), Expect = 7e-05
Identities = 23/66 (34%), Positives = 38/66 (56%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKS 60
M ++ + QAGQ G+A+EK++ ++ A++AA A Q+AG+ + A G + VK
Sbjct: 1 MSETNKNAFQAGQTAGKAEEKSNVLLDKAKDAAAGAGAGAQQAGKSVSDAAAGGVNFVKD 60
Query: 61 AIGANK 66
G NK
Sbjct: 61 KTGLNK 66
>UniRef100_Q42495 Kin1 protein [Brassica campestris]
Length = 64
Score = 46.6 bits (109), Expect = 2e-04
Identities = 26/59 (44%), Positives = 37/59 (62%), Gaps = 1/59 (1%)
Query: 8 SHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIGANK 66
S QAGQA G+A+EK + +M ++AA +A S + AGQ++ A GA + VK G NK
Sbjct: 7 SFQAGQAAGRAEEKGNVLMDKVKDAA-TAGASAETAGQKITEAAGGAVNLVKEKTGMNK 64
>UniRef100_Q9XET0 Seed maturation protein PM30 [Glycine max]
Length = 140
Score = 46.6 bits (109), Expect = 2e-04
Identities = 25/66 (37%), Positives = 38/66 (56%), Gaps = 4/66 (6%)
Query: 2 DQSQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQS----CQEAGQQMKAKAQGAADA 57
D SQ + Q TG AQ+K S M + + +AQS + + Q+ G+++K AQGA +A
Sbjct: 63 DTSQAAKEKTQQNTGAAQQKTSEMGQSTKESAQSGKDNTQGFLQQTGEKVKGAAQGATEA 122
Query: 58 VKSAIG 63
VK +G
Sbjct: 123 VKQTLG 128
Score = 46.6 bits (109), Expect = 2e-04
Identities = 23/56 (41%), Positives = 31/56 (55%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S S++AGQ G+ +EK + MGN AQ+A++ QE Q K K Q A A K
Sbjct: 3 SHRQSYEAGQTKGRTEEKTNQTMGNIGEKAQAAKEKTQEMAQAAKEKTQQTAQAAK 58
>UniRef100_Q9SSS0 F6D8.10 [Arabidopsis thaliana]
Length = 114
Score = 46.2 bits (108), Expect = 2e-04
Identities = 30/89 (33%), Positives = 37/89 (40%), Gaps = 26/89 (29%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNA--------------------------AQ 34
M SQ LSH AG+ TGQ Q K + N +A
Sbjct: 1 MSSSQQLSHSAGEVTGQVQLKKEEYLNNVSHAMNQNADHHTHSQSQLHSEHDQNNPSLIS 60
Query: 35 SAQQSCQEAGQQMKAKAQGAADAVKSAIG 63
A Q+ G Q+K AQGAADAVK+ +G
Sbjct: 61 QASSVIQQTGGQVKNMAQGAADAVKNTLG 89
>UniRef100_Q8LDW3 Hypothetical protein [Arabidopsis thaliana]
Length = 114
Score = 46.2 bits (108), Expect = 2e-04
Identities = 30/89 (33%), Positives = 37/89 (40%), Gaps = 26/89 (29%)
Query: 1 MDQSQNLSHQAGQATGQAQEKASNMMGNARNA--------------------------AQ 34
M SQ LSH AG+ TGQ Q K + N +A
Sbjct: 1 MSSSQQLSHSAGEVTGQVQLKKEEYLNNVSHAMNQNADHHTHSQSQLHSEHDQNNPSLIS 60
Query: 35 SAQQSCQEAGQQMKAKAQGAADAVKSAIG 63
A Q+ G Q+K AQGAADAVK+ +G
Sbjct: 61 QASSVIQQTGGQVKNMAQGAADAVKNTLG 89
>UniRef100_O49816 Late embryogenesis abundant protein 1 [Cicer arietinum]
Length = 177
Score = 46.2 bits (108), Expect = 2e-04
Identities = 21/56 (37%), Positives = 34/56 (60%)
Query: 4 SQNLSHQAGQATGQAQEKASNMMGNARNAAQSAQQSCQEAGQQMKAKAQGAADAVK 59
S + S++AG+ G+ +EK + M+GN + AQ+A++ Q+A Q K K A A K
Sbjct: 3 SHDQSYKAGETMGRTEEKTNQMIGNIEDKAQAAKEKAQQAAQTAKDKTSQTAQAAK 58
Score = 43.5 bits (101), Expect = 0.001
Identities = 23/58 (39%), Positives = 34/58 (57%), Gaps = 4/58 (6%)
Query: 10 QAGQATGQAQEKASNMMGNARNAAQSAQQSC----QEAGQQMKAKAQGAADAVKSAIG 63
+A TG+A+EK S M + + AQS + + Q+ G+++K AQGA DAVK G
Sbjct: 104 KAQDTTGRAREKGSEMGQSTKETAQSGKDNSAGFLQQTGEKVKGMAQGATDAVKQTFG 161
>UniRef100_Q9XFZ1 Cold shock protein [Camelina sativa]
Length = 37
Score = 45.8 bits (107), Expect = 3e-04
Identities = 22/35 (62%), Positives = 27/35 (76%)
Query: 29 ARNAAQSAQQSCQEAGQQMKAKAQGAADAVKSAIG 63
A++AA SAQ+S Q+ GQQ+K KAQGAAD VK G
Sbjct: 3 AKDAAGSAQESAQQTGQQVKEKAQGAADVVKEKTG 37
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.300 0.108 0.271
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 68,076,146
Number of Sequences: 2790947
Number of extensions: 1413506
Number of successful extensions: 7135
Number of sequences better than 10.0: 140
Number of HSP's better than 10.0 without gapping: 105
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 6631
Number of HSP's gapped (non-prelim): 474
length of query: 66
length of database: 848,049,833
effective HSP length: 42
effective length of query: 24
effective length of database: 730,830,059
effective search space: 17539921416
effective search space used: 17539921416
T: 11
A: 40
X1: 17 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (21.8 bits)
S2: 68 (30.8 bits)
Lotus: description of TM0215b.3


