

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0212.5
(129 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
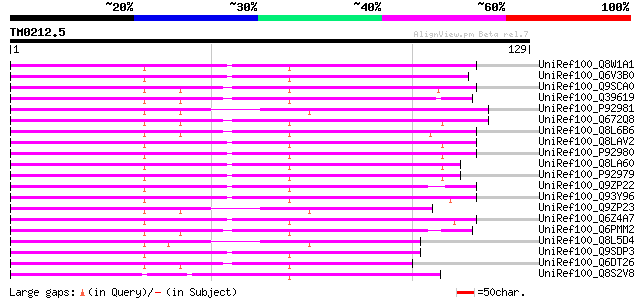
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8W1A1 Adenosine 5'-phosphosulfate reductase [Glycine ... 105 2e-22
UniRef100_Q6V3B0 Adenosine 5' phosphosulfate reductase [Populus ... 97 6e-20
UniRef100_Q9SCA0 Adenosine 5'-phosphosulphate reductase [Lemna m... 97 1e-19
UniRef100_Q39619 PAPS-reductase-like protein precursor [Catharan... 95 3e-19
UniRef100_P92981 5'-adenylylsulfate reductase 2, chloroplast pre... 93 1e-18
UniRef100_Q672Q8 Adenylyl-sulfate reductase precursor [Lycopersi... 93 2e-18
UniRef100_Q8L6B6 APS reductase [Solanum tuberosum] 92 3e-18
UniRef100_Q8LAV2 PRH26 protein [Arabidopsis thaliana] 91 4e-18
UniRef100_P92980 5'-adenylylsulfate reductase 3, chloroplast pre... 91 4e-18
UniRef100_Q8LA60 5-adenylylsulfate reductase [Arabidopsis thaliana] 90 1e-17
UniRef100_P92979 5'-adenylylsulfate reductase 1, chloroplast pre... 90 1e-17
UniRef100_Q9ZP22 APS reductase precursor [Brassica juncea] 90 1e-17
UniRef100_Q93Y96 Adenosine 5'-phosphosulfate reductase [Zea mays] 89 2e-17
UniRef100_Q9ZP23 APS reductase precursor [Brassica juncea] 88 4e-17
UniRef100_Q6Z4A7 Putative phosphoadenylyl-sulfate reductase [Ory... 88 5e-17
UniRef100_Q6PMM2 Adenosine 5'phosphosulfate reductase [Ceratopte... 87 8e-17
UniRef100_Q8L5D4 Adenosine 5' phosphosulfate reductase [Physcomi... 86 2e-16
UniRef100_Q9SDP3 APS-reductase [Allium cepa] 84 7e-16
UniRef100_Q6DT26 Adenosine 5-phosphosulfate reductase [Nicotiana... 82 3e-15
UniRef100_Q8S2V8 5'-adenylylsulfate reductase [Chlamydomonas rei... 71 6e-12
>UniRef100_Q8W1A1 Adenosine 5'-phosphosulfate reductase [Glycine max]
Length = 470
Score = 105 bits (262), Expect = 2e-22
Identities = 66/143 (46%), Positives = 78/143 (54%), Gaps = 28/143 (19%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 202 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGIGSLVKWNP 261
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 262 VANVNGLDIWSFLRT-MDVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKAK 320
Query: 94 DCGLHKGNVKQDVGAQLNGNGAA 116
+CGLHKGN+K + AQLNGNGA+
Sbjct: 321 ECGLHKGNIKHEDAAQLNGNGAS 343
>UniRef100_Q6V3B0 Adenosine 5' phosphosulfate reductase [Populus alba x Populus
tremula]
Length = 465
Score = 97.4 bits (241), Expect = 6e-20
Identities = 63/141 (44%), Positives = 75/141 (52%), Gaps = 28/141 (19%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +L+KWN
Sbjct: 195 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGAGSLIKWNP 254
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
+A + L M + ++ + + P VL QHEREG WWEDA AK
Sbjct: 255 MANVEGQDVWKFLRT-MDVPVNSLHSKGYISIGCEPCTRPVLPGQHEREGRWWWEDATAK 313
Query: 94 DCGLHKGNVKQDVGAQLNGNG 114
+CGLHKGN+KQ AQLNGNG
Sbjct: 314 ECGLHKGNLKQGDAAQLNGNG 334
>UniRef100_Q9SCA0 Adenosine 5'-phosphosulphate reductase [Lemna minor]
Length = 459
Score = 96.7 bits (239), Expect = 1e-19
Identities = 64/146 (43%), Positives = 77/146 (51%), Gaps = 32/146 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR L+G+ I GQ KDQSPGT +L+KWN
Sbjct: 199 CCRVRKVRPLRRALRGLRAWITGQRKDQSPGTRASVPTVQVDPSFEGFEGGTGSLIKWNP 258
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA + I+ M + ++ + + P VL QHEREG WWEDAKA
Sbjct: 259 VANVDGQDIWRFLRT--MAVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 316
Query: 93 KDCGLHKGNVKQD--VGAQLNGNGAA 116
K+CGLHKGN+KQD V NGNG A
Sbjct: 317 KECGLHKGNIKQDESVPPSSNGNGTA 342
>UniRef100_Q39619 PAPS-reductase-like protein precursor [Catharanthus roseus]
Length = 464
Score = 95.1 bits (235), Expect = 3e-19
Identities = 63/143 (44%), Positives = 76/143 (53%), Gaps = 31/143 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 197 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGMDGGVGSLVKWNP 256
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA + I+ M + ++ + + P VL QHEREG WWEDAKA
Sbjct: 257 VANVEGKDIWNFLRA--MDVPVNTLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 314
Query: 93 KDCGLHKGNVKQDVGAQLNGNGA 115
K+CGLHKGN+K++ NGNGA
Sbjct: 315 KECGLHKGNIKEET-LNNNGNGA 336
>UniRef100_P92981 5'-adenylylsulfate reductase 2, chloroplast precursor [Arabidopsis
thaliana]
Length = 454
Score = 93.2 bits (230), Expect = 1e-18
Identities = 62/157 (39%), Positives = 77/157 (48%), Gaps = 50/157 (31%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
+A + A ++ +R P+ AL VL QHERE
Sbjct: 254 LANVEGADVWNF------------LRTMDVPVNALHAQGYVSIGCEPCTRPVLPGQHERE 301
Query: 83 GNRWWEDAKAKDCGLHKGNVKQDVGAQLNGNGAAMQV 119
G WWEDAKAK+CGLHKGN+K++ GA + A ++
Sbjct: 302 GRWWWEDAKAKECGLHKGNIKEEDGAADSKPAAVQEI 338
>UniRef100_Q672Q8 Adenylyl-sulfate reductase precursor [Lycopersicon esculentum]
Length = 461
Score = 92.8 bits (229), Expect = 2e-18
Identities = 62/150 (41%), Positives = 75/150 (49%), Gaps = 33/150 (22%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 198 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPIVQVDPSFEGLDGGAGSLVKWNP 257
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA + I+ M + ++ + + P VL QHEREG WWEDAKA
Sbjct: 258 VANVDGKDIWNFLRA--MNVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 315
Query: 93 KDCGLHKGNVKQDV---GAQLNGNGAAMQV 119
K+CGLHKGN+K + AQ NG +
Sbjct: 316 KECGLHKGNIKDETVNGAAQTNGTATVADI 345
>UniRef100_Q8L6B6 APS reductase [Solanum tuberosum]
Length = 329
Score = 91.7 bits (226), Expect = 3e-18
Identities = 62/145 (42%), Positives = 75/145 (50%), Gaps = 31/145 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 78 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPIVQVDPSFEGLDGGAGSLVKWNP 137
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA + I+ M + ++ + + P VL QHEREG WWEDAKA
Sbjct: 138 VANVDGKDIWNFLRA--MNVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 195
Query: 93 KDCGLHKGNVK-QDVGAQLNGNGAA 116
K+CGLHKGN+K + V + NG A
Sbjct: 196 KECGLHKGNIKDESVDGAVQTNGTA 220
>UniRef100_Q8LAV2 PRH26 protein [Arabidopsis thaliana]
Length = 458
Score = 91.3 bits (225), Expect = 4e-18
Identities = 62/145 (42%), Positives = 75/145 (50%), Gaps = 30/145 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 254 VANVEGNDVWNFLRT-MDVPVNTLHAAGYVSIGCEPCTRAVLPGQHEREGRWWWEDAKAK 312
Query: 94 DCGLHKGNVKQDV--GAQLNGNGAA 116
+CGLHKGN+K++ A N NG A
Sbjct: 313 ECGLHKGNIKENTNGNATANVNGTA 337
>UniRef100_P92980 5'-adenylylsulfate reductase 3, chloroplast precursor [Arabidopsis
thaliana]
Length = 458
Score = 91.3 bits (225), Expect = 4e-18
Identities = 62/145 (42%), Positives = 75/145 (50%), Gaps = 30/145 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 194 CCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 253
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 254 VANVEGNDVWNFLRT-MDVPVNTLHAAGYVSIGCEPCTRAVLPGQHEREGRWWWEDAKAK 312
Query: 94 DCGLHKGNVKQDV--GAQLNGNGAA 116
+CGLHKGN+K++ A N NG A
Sbjct: 313 ECGLHKGNIKENTNGNATANVNGTA 337
>UniRef100_Q8LA60 5-adenylylsulfate reductase [Arabidopsis thaliana]
Length = 465
Score = 90.1 bits (222), Expect = 1e-17
Identities = 62/140 (44%), Positives = 75/140 (53%), Gaps = 29/140 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 202 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 261
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 262 VANVEGNDVWNFLRT-MDVPVNTLHAAGYISIGCEPCTKAVLPGQHEREGRWWWEDAKAK 320
Query: 94 DCGLHKGNVKQDV-GAQLNG 112
+CGLHKGNVK++ A++NG
Sbjct: 321 ECGLHKGNVKENSDDAKVNG 340
>UniRef100_P92979 5'-adenylylsulfate reductase 1, chloroplast precursor [Arabidopsis
thaliana]
Length = 465
Score = 90.1 bits (222), Expect = 1e-17
Identities = 62/140 (44%), Positives = 75/140 (53%), Gaps = 29/140 (20%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 202 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGVGSLVKWNP 261
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 262 VANVEGNDVWNFLRT-MDVPVNTLHAAGYISIGCEPCTKAVLPGQHEREGRWWWEDAKAK 320
Query: 94 DCGLHKGNVKQDV-GAQLNG 112
+CGLHKGNVK++ A++NG
Sbjct: 321 ECGLHKGNVKENSDDAKVNG 340
>UniRef100_Q9ZP22 APS reductase precursor [Brassica juncea]
Length = 464
Score = 89.7 bits (221), Expect = 1e-17
Identities = 61/143 (42%), Positives = 73/143 (50%), Gaps = 32/143 (22%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 199 CCRIRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPVFEGLDGGAGSLVKWNP 258
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 259 VANVEGNDVWNFLRT-MDVPVNTLHAAGYVSIGCEPCTRAVLPGQHEREGRWWWEDAKAK 317
Query: 94 DCGLHKGNVKQDVGAQLNGNGAA 116
+CGLHKGN+K+ NGN A
Sbjct: 318 ECGLHKGNIKE----SSNGNATA 336
>UniRef100_Q93Y96 Adenosine 5'-phosphosulfate reductase [Zea mays]
Length = 302
Score = 89.4 bits (220), Expect = 2e-17
Identities = 62/147 (42%), Positives = 74/147 (50%), Gaps = 32/147 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 111 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRASIPIVQVDPSFEGLDGGAGSLVKWNP 170
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA L M + ++ + + P VL QHEREG WWE AKA+
Sbjct: 171 VANVDGKDIWTFLRT-MDVPVNTLHAQGYVSIGCEPCTRPVLPGQHEREGRWWWEGAKAE 229
Query: 94 DCGLHKGNVKQDVGA----QLNGNGAA 116
+CGLHKGN+ +D A NGNG+A
Sbjct: 230 ECGLHKGNIDKDAQAAAPRSANGNGSA 256
>UniRef100_Q9ZP23 APS reductase precursor [Brassica juncea]
Length = 454
Score = 88.2 bits (217), Expect = 4e-17
Identities = 58/143 (40%), Positives = 70/143 (48%), Gaps = 50/143 (34%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 192 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRSEIPIVQVDPVFEGLDGGVGSLVKWNP 251
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
+A + ++ +R P+ AL VL QHERE
Sbjct: 252 LANVEGGDVWNF------------LRTMDVPVNALHAQGYVSIGCEPCTRPVLPGQHERE 299
Query: 83 GNRWWEDAKAKDCGLHKGNVKQD 105
G WWEDAKAK+CGLHKGN+K++
Sbjct: 300 GRWWWEDAKAKECGLHKGNIKKE 322
>UniRef100_Q6Z4A7 Putative phosphoadenylyl-sulfate reductase [Oryza sativa]
Length = 475
Score = 87.8 bits (216), Expect = 5e-17
Identities = 62/152 (40%), Positives = 73/152 (47%), Gaps = 37/152 (24%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR RKVR LRR L+G+ I GQ KDQSPGT +LVKWN
Sbjct: 202 CCRARKVRPLRRALRGLRAWITGQRKDQSPGTRAAIPVVQVDPSFEGLAGGAGSLVKWNP 261
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
VA L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 262 VANVDGKDVWTFLRA-MDVPVNALHAQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKAK 320
Query: 94 DCGLHKGNVKQDVGAQ---------LNGNGAA 116
+CGLHKGN+ GA NGNG+A
Sbjct: 321 ECGLHKGNIDDQGGAAAAAAHKAGGANGNGSA 352
>UniRef100_Q6PMM2 Adenosine 5'phosphosulfate reductase [Ceratopteris richardii]
Length = 456
Score = 87.0 bits (214), Expect = 8e-17
Identities = 57/143 (39%), Positives = 75/143 (51%), Gaps = 33/143 (23%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCR+RKVR LRR LKG+ I GQ +DQSPGT +L+KWN
Sbjct: 194 CCRIRKVRPLRRALKGLKAWITGQRRDQSPGTRSNIPIVQIDPSFEGMDGGPGSLIKWNP 253
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA ++ I+ M + ++ + + P VL HEREG WWEDAKA
Sbjct: 254 VANVTGTDIWSFLRT--MNVPVNVLHSQGYISIGCEPCTRAVLPGMHEREGRWWWEDAKA 311
Query: 93 KDCGLHKGNVKQDVGAQLNGNGA 115
K+CGLHKGN++ A+ + NGA
Sbjct: 312 KECGLHKGNIET---AETSNNGA 331
>UniRef100_Q8L5D4 Adenosine 5' phosphosulfate reductase [Physcomitrella patens]
Length = 465
Score = 85.9 bits (211), Expect = 2e-16
Identities = 58/140 (41%), Positives = 67/140 (47%), Gaps = 50/140 (35%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWN- 38
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 203 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRANVPVVQVDPAFEGLDGGPGSLVKWNP 262
Query: 39 LVAISKAIIYGISLGP*MCL*IH*IRKDTFPLEAL----------------VLREQHERE 82
L +S ++ +R P+ AL VL QHERE
Sbjct: 263 LSNVSGTAVWSF------------LRTMDVPVNALHFKGYVSIGCEPCTRAVLPGQHERE 310
Query: 83 GNRWWEDAKAKDCGLHKGNV 102
G WWEDAKAK+CGLHKGN+
Sbjct: 311 GRWWWEDAKAKECGLHKGNL 330
>UniRef100_Q9SDP3 APS-reductase [Allium cepa]
Length = 442
Score = 84.0 bits (206), Expect = 7e-16
Identities = 56/129 (43%), Positives = 67/129 (51%), Gaps = 28/129 (21%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 176 CCRVRKVRPLRRALKGLKAWITGQRKDQSPGTRAHIPVVQVDPVFEGLEGGVGSLVKWNP 235
Query: 40 VAISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKAK 93
+A + L M + ++ + + P VL QHEREG WWEDAKAK
Sbjct: 236 LANVEGNNVWNFLRT-MDVPVNSLHTKGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKAK 294
Query: 94 DCGLHKGNV 102
+CGLHKGN+
Sbjct: 295 ECGLHKGNI 303
>UniRef100_Q6DT26 Adenosine 5-phosphosulfate reductase [Nicotiana tabacum]
Length = 161
Score = 82.0 bits (201), Expect = 3e-15
Identities = 55/128 (42%), Positives = 65/128 (49%), Gaps = 30/128 (23%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGT---------------------NLVKWNL 39
CCRVRKVR LRR LKG+ I GQ KDQSPGT +LVKWN
Sbjct: 36 CCRVRKVRPLRRALKGLRAWITGQRKDQSPGTRSEIPVVQVDPSFEGLGGGAGSLVKWNP 95
Query: 40 VA-ISKAIIYGISLGP*MCL*IH*IRKDTF------PLEALVLREQHEREGNRWWEDAKA 92
VA + I+ M + ++ + + P VL QHEREG WWEDAKA
Sbjct: 96 VANVDGKDIWNFLRA--MNVPVNSLHSQGYVSIGCEPCTRPVLPGQHEREGRWWWEDAKA 153
Query: 93 KDCGLHKG 100
K+CG HKG
Sbjct: 154 KECGFHKG 161
>UniRef100_Q8S2V8 5'-adenylylsulfate reductase [Chlamydomonas reinhardtii]
Length = 413
Score = 70.9 bits (172), Expect = 6e-12
Identities = 48/135 (35%), Positives = 64/135 (46%), Gaps = 30/135 (22%)
Query: 1 CCRVRKVRLLRRELKGVTT*IIGQ*KDQSPGTNLVKWNLVAISKAIIYGISLGP*MCL*I 60
CCR+RKV+ LR++LK I GQ KDQSPGT + +V + + G++ GP +
Sbjct: 153 CCRIRKVKPLRKQLKVYKAWITGQRKDQSPGTR-TEVPVVQVDP-VFEGVTGGPGSLIKY 210
Query: 61 H*IRKDTF----------------------------PLEALVLREQHEREGNRWWEDAKA 92
+ + T P VL Q EREG WWEDA A
Sbjct: 211 NPLSNMTSAEVWNFLRVMKVPTNKLHNCGYISIGCEPCTRPVLPNQAEREGRWWWEDAAA 270
Query: 93 KDCGLHKGNVKQDVG 107
K+CGLH GN+K+ G
Sbjct: 271 KECGLHSGNIKKADG 285
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.344 0.153 0.534
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 189,746,835
Number of Sequences: 2790947
Number of extensions: 6635039
Number of successful extensions: 22227
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 55
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 22105
Number of HSP's gapped (non-prelim): 108
length of query: 129
length of database: 848,049,833
effective HSP length: 105
effective length of query: 24
effective length of database: 555,000,398
effective search space: 13320009552
effective search space used: 13320009552
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0212.5


