

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0207.16
(596 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
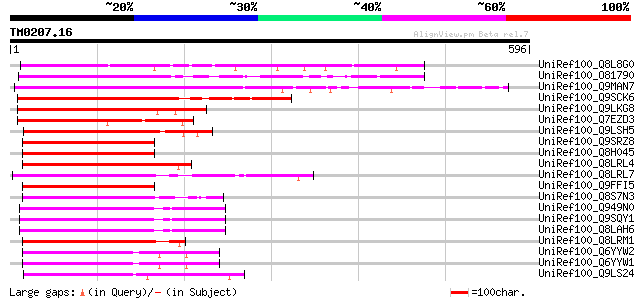
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L8G0 Nam-like protein 1 [Petunia hybrida] 301 3e-80
UniRef100_O81790 NAM / CUC2-like protein [Arabidopsis thaliana] 288 4e-76
UniRef100_Q9MAN7 CDS [Arabidopsis thaliana] 276 2e-72
UniRef100_Q9SCK6 Putative NAC2 protein [Arabidopsis thaliana] 273 1e-71
UniRef100_Q9LKG8 TIP [Arabidopsis thaliana] 258 5e-67
UniRef100_Q7EZD3 NAC2 protein-like [Oryza sativa] 216 2e-54
UniRef100_Q9LSH5 Similarity to NAM [Arabidopsis thaliana] 204 6e-51
UniRef100_Q9SRZ8 F12P19.8 protein [Arabidopsis thaliana] 203 1e-50
UniRef100_Q8H045 Hypothetical protein OJ1263H11.11 [Oryza sativa] 198 3e-49
UniRef100_Q8LRL4 Nam-like protein 11 [Petunia hybrida] 194 6e-48
UniRef100_Q8LRL7 Nam-like protein 8 [Petunia hybrida] 192 2e-47
UniRef100_Q9FFI5 NAM (No apical meristem)-like protein [Arabidop... 191 4e-47
UniRef100_Q8S7N3 Putative NAM (No apical meristem) protein [Oryz... 190 1e-46
UniRef100_Q949N0 Hypothetical protein At3g10500 [Arabidopsis tha... 189 3e-46
UniRef100_Q9SQY1 Hypothetical protein F13M14.22 [Arabidopsis tha... 189 3e-46
UniRef100_Q8LAH6 NAC, putative [Arabidopsis thaliana] 187 1e-45
UniRef100_Q8LRM1 Nam-like protein 4 [Petunia hybrida] 185 4e-45
UniRef100_Q6YYW2 Putative NAC2 protein [Oryza sativa] 184 5e-45
UniRef100_Q6YYW1 Putative NAC2 protein [Oryza sativa] 184 5e-45
UniRef100_Q9LS24 NAM-like [Arabidopsis thaliana] 184 8e-45
>UniRef100_Q8L8G0 Nam-like protein 1 [Petunia hybrida]
Length = 585
Score = 301 bits (772), Expect = 3e-80
Identities = 208/505 (41%), Positives = 278/505 (54%), Gaps = 50/505 (9%)
Query: 13 LNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDP 72
LNSLP G+RFRP+DEE+++ YLR KING +EV VIRE+D+CK EPWDLP SV+ + D
Sbjct: 24 LNSLPVGYRFRPTDEELVNHYLRLKINGADNEVSVIREVDICKLEPWDLPIWSVVESNDN 83
Query: 73 EWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKI--KSGSSLIGMKKTLVFYHGRAPK 130
EWFFFCP+DRKY NG RLNRAT GYWKATGKDR I K G+ IGMKKTLV+Y GRAP+
Sbjct: 84 EWFFFCPKDRKYQNGQRLNRATERGYWKATGKDRNIVTKKGAK-IGMKKTLVYYIGRAPE 142
Query: 131 GKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFK----KQDESIEVSISGEAEQTASTP 186
GKRT+WV+HEYR T + LDGT+PGQ +++CRLF+ KQDE +E S +AEQ A+
Sbjct: 143 GKRTHWVIHEYRATDKSLDGTHPGQGAFLLCRLFRKNDLKQDEHVESSNLDDAEQNAT-- 200
Query: 187 MAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNSDRYDA 246
AAK ++ S+ PLVA L + DK A E + P++ +S+ A
Sbjct: 201 -AAKSPTDDELSEAATPLVA--GQPLGDSDKSYAAKFPKGEPYGKQL--PLESHSNSCIA 255
Query: 247 QNAQPQNVKLAA---EENQPLNLDMYNDLKNDIFDDKLFSPALVHLPPVF--DYQASNEP 301
+ + Q V + + + L + D + D K+FSP + F Y +
Sbjct: 256 DDTEDQMVDITSIPPDLELEKALGNFCDQSSQPLDWKIFSPLHAQMQVEFGSSYLQAPMN 315
Query: 302 DDRSG------LQYGTNETAISDLFDS--LTWDQLSYEESGSQP---------MNFPLFN 344
+D S QYGTN I+D DS ++ D+ S +SG + +F
Sbjct: 316 NDMSAFQKDVQFQYGTNALDINDFLDSVIVSSDEFSCGDSGQEVGLHNFDTTIHSFTGAP 375
Query: 345 VKDSGSGSDSDFEITNM--------PSMQIPYPEEAIDMKIPLGTAPEFF-SPVGVPFDH 395
VKDSGS S+S+ E+T P + + ++A D+K + + G P+
Sbjct: 376 VKDSGSCSESEAEVTQRLVEPDFFEPEILLGNFDQATDVKGGVNSIQTTVQDAAGAPY-F 434
Query: 396 SGDEQKSNVGLFQNQNAAQMTFSSDVSMGQVYNVVNDYEQPRNWNVA---TGGDTGIIIR 452
S D NVG QN Q FS+ QV N + E NV +G TGI +R
Sbjct: 435 SNDHVMGNVGFLQNTYIGQNDFSAGYGGIQVPNFLTVQESGGYNNVVSSDSGSGTGIKLR 494
Query: 453 ARQAQNELVNTNIN-QGSANRRIRL 476
RQ QN+ + QG+ANRRIRL
Sbjct: 495 TRQMQNQPDDRQSRMQGTANRRIRL 519
>UniRef100_O81790 NAM / CUC2-like protein [Arabidopsis thaliana]
Length = 534
Score = 288 bits (736), Expect = 4e-76
Identities = 199/475 (41%), Positives = 252/475 (52%), Gaps = 77/475 (16%)
Query: 11 LSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNR 70
+S+ SLP GFRFRP+DEE+++ YLR KING +V VI +IDVCKWEPWDLP LSVI+
Sbjct: 4 VSMESLPLGFRFRPTDEELVNHYLRLKINGRHSDVRVIPDIDVCKWEPWDLPALSVIKTD 63
Query: 71 DPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPK 130
DPEWFFFCP+DRKYPNG+R NRAT GYWKATGKDR IKS +LIGMKKTLVFY GRAPK
Sbjct: 64 DPEWFFFCPRDRKYPNGHRSNRATDSGYWKATGKDRSIKSKKTLIGMKKTLVFYRGRAPK 123
Query: 131 GKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAAK 190
G+RTNW+MHEYRPTL++LDGT+PGQ+PYV+CRLF K D+ + S EA TAS K
Sbjct: 124 GERTNWIMHEYRPTLKDLDGTSPGQSPYVLCRLFHKPDDRVNGVKSDEAAFTASN----K 179
Query: 191 YSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNSDRYDAQNAQ 250
YSP++ SD L+ E A + + S+ + Y N+
Sbjct: 180 YSPDDTSSD-----------LVQETPSSDAAVEKPSDYS----------GGCGYAHSNST 218
Query: 251 PQNVKL-AAEENQPLNLDMYNDLKNDIFDDKLFSPALVHLPPVFDYQASNEPDDRSGLQY 309
+ A EEN L+ D+ D K LP + A + D G Q+
Sbjct: 219 ADGTMIEAPEENLWLSCDL-EDQK-------------APLPCMDSIYAGDFSYDEIGFQF 264
Query: 310 --GTNE--TAISDLFDSL--TWDQLSYEESGSQPMNFPLFNVKDSGSGSDSDFEITNMPS 363
GT+E ++++L + + D S EES S+ N S +G S ++
Sbjct: 265 QDGTSEPDVSLTELLEEVFNNPDDFSCEESISRE------NPAVSPNGIFSSAKM----- 313
Query: 364 MQIPYPEEAIDMKIPLGTAPEFFSPVGVPFDHSGDEQKSNVGLFQNQNAAQMTFSSDVSM 423
+Q PE+A FF+ F D L + SD +
Sbjct: 314 LQSAAPEDA------------FFN----DFMAFTDTDAEMAQLQYGSEGGASGWPSDTN- 356
Query: 424 GQVYNVVNDYEQPRNWNVATG--GDTGIIIRARQAQNELVNTNINQGSANRRIRL 476
Y+ + EQ N N GI IRARQ QN INQG A RRIRL
Sbjct: 357 -SYYSDLVQQEQMINHNTENNLTEGRGIKIRARQPQNRQSTGLINQGIAPRRIRL 410
>UniRef100_Q9MAN7 CDS [Arabidopsis thaliana]
Length = 577
Score = 276 bits (705), Expect = 2e-72
Identities = 199/607 (32%), Positives = 295/607 (47%), Gaps = 83/607 (13%)
Query: 6 TAVPVLSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLS 65
T +LS+ +LP GFRFRP+DEE+I+ YLR KING EV VI EIDVCKWEPWDLP LS
Sbjct: 3 TEQALLSMEALPLGFRFRPTDEELINHYLRLKINGRDLEVRVIPEIDVCKWEPWDLPGLS 62
Query: 66 VIRNRDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYH 125
VI+ D EWFFFCP+DRKYP+G+R NRAT +GYWKATGKDR IKS +IGMKKTLVFY
Sbjct: 63 VIKTDDQEWFFFCPRDRKYPSGHRSNRATDIGYWKATGKDRTIKSKKMIIGMKKTLVFYR 122
Query: 126 GRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTAST 185
GRAP+G+RTNW+MHEYR T +ELDGT PGQNPYV+CRLF K +S + + E E+ T
Sbjct: 123 GRAPRGERTNWIMHEYRATDKELDGTGPGQNPYVLCRLFHKPSDSCDPAHCEEIEKVNFT 182
Query: 186 P-MAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNS-DR 243
P + SP++ S+ L D + + + ++ ++ S +
Sbjct: 183 PTTTTRCSPDDTSSEMVQETATSGVHALDRSDDTERCLSDKGNNDVKPDVSVINNTSVNH 242
Query: 244 YDAQNAQPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHLPPVFDYQASNEPDD 303
+ A+ +N+ E PL D+ L I +K + P + + S D
Sbjct: 243 AETSRAKDRNLGKTLVEENPLLRDV-PTLHGPILSEKSYYPGQSSIGFATSHMDSMYSSD 301
Query: 304 RSGLQYGTN--------ETAISDLFDSLTWDQLSYEESGSQPMNFPLFN----------- 344
YG + + +++D+ D + ++ ES + +F L N
Sbjct: 302 FGNCDYGLHFQDGASEQDASLTDVLDEVFH---NHNESSNDRKDFVLPNMMHWPGNTRLL 358
Query: 345 ------VKDSGSGSDSDFEITNMPSMQIP-------YPEEAIDMKIPLGTAPEFFSPVGV 391
+KDS + D E++ +P E+ +D K A E S G
Sbjct: 359 STEYPFLKDSVAFVDGSAEVSGSQQF-VPDILASRWVSEQNVDSK----EAVEILSSTG- 412
Query: 392 PFDHSGDEQKSNVGLFQNQNAAQMTFSSDVSMGQVYNVVNDYEQPR------NWNVATGG 445
+ N Q SS ++ VN EQ + N++
Sbjct: 413 --------SSRTLTPLHNNVFGQYASSSYAAIDPFNYNVNQPEQSSFEQSHVDRNISPSN 464
Query: 446 DTGIIIRARQAQNELVNTNINQGSANRRIRLLKSAHGSSRVVKGGRRAQEDRNSKPITAV 505
R+R+ Q +L ++ ++QG+A RRIRL Q ++ P+T
Sbjct: 465 IFEFKARSRENQRDL-DSVVDQGTAPRRIRL-----------------QIEQPLTPVTNK 506
Query: 506 EKKASESLAADEDDTLTNHVKAMPKTPKSTNNRKIYQRGSNAGSSTKVLKDLLLLRRVPC 565
+++ +++ +ED+ + K + + P + + + QR T++ K L+ L
Sbjct: 507 KERDADNY-EEEDEVQSAMSKVVEEEPANLSAQGTAQR--RIRLQTRLRKPLITLNN--- 560
Query: 566 ISKASSN 572
+K +SN
Sbjct: 561 -TKRNSN 566
>UniRef100_Q9SCK6 Putative NAC2 protein [Arabidopsis thaliana]
Length = 469
Score = 273 bits (697), Expect = 1e-71
Identities = 151/320 (47%), Positives = 206/320 (64%), Gaps = 26/320 (8%)
Query: 10 VLSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRN 69
VLS++SLP G RFRP+DEE+I +YLR+KING+ D+V IREID+CKWEPWDLPD SVI+
Sbjct: 7 VLSMDSLPVGLRFRPTDEELIRYYLRRKINGHDDDVKAIREIDICKWEPWDLPDFSVIKT 66
Query: 70 RDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSG-SSLIGMKKTLVFYHGRA 128
+D EW +FCP DRKYP+G+R NRAT+ GYWKATGKDRKIKSG +++IG+K+TLVF+ GRA
Sbjct: 67 KDSEWLYFCPLDRKYPSGSRQNRATVAGYWKATGKDRKIKSGKTNIIGVKRTLVFHAGRA 126
Query: 129 PKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESI--EVSISGEAEQTASTP 186
P+G RTNW++HEYR T +L GTNPGQ+P+VIC+LFKK++ + E S S E E+ A +
Sbjct: 127 PRGTRTNWIIHEYRATEDDLSGTNPGQSPFVICKLFKKEELVLGEEDSKSDEVEEPAVSS 186
Query: 187 MAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNSDRYD- 245
+ + E VS + TED K I S+++ D +SD D
Sbjct: 187 PTVEVTKSE-----------VSEVIKTEDVKRHDI-------AESSLVISGDSHSDACDE 228
Query: 246 AQNAQPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHLPPVFDYQASNEPDDRS 305
A A+ + K E + L+ +++ L + + +L S P ++ +N +
Sbjct: 229 ATTAELVDFKW-YPELESLDFTLFSPLHSQV-QSELGSSYNTFQPGSSNFSGNNNNSFQI 286
Query: 306 GLQYGTNE--TAISDLFDSL 323
QYGTNE T ISD DS+
Sbjct: 287 QTQYGTNEVDTYISDFLDSI 306
>UniRef100_Q9LKG8 TIP [Arabidopsis thaliana]
Length = 451
Score = 258 bits (658), Expect = 5e-67
Identities = 129/230 (56%), Positives = 165/230 (71%), Gaps = 13/230 (5%)
Query: 10 VLSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRN 69
VLSL SLP GFRF P+DEE++ +YLR KING+ ++V VIREID+CKWEPWDLPD SV++
Sbjct: 7 VLSLASLPVGFRFSPTDEELVRYYLRLKINGHDNDVRVIREIDICKWEPWDLPDFSVVKT 66
Query: 70 RDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSG-SSLIGMKKTLVFYHGRA 128
D EW FFCP DRKYP+G+R+NRAT+ GYWKATGKDRKIKSG + +IG+K+TLVFY GRA
Sbjct: 67 TDSEWLFFCPLDRKYPSGSRMNRATVAGYWKATGKDRKIKSGKTKIIGVKRTLVFYTGRA 126
Query: 129 PKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQD-----ESIEVSISGEAEQTA 183
PKG RT W+MHEYR T ++LDGT GQNP+V+C+LFKKQD E S S E E
Sbjct: 127 PKGTRTCWIMHEYRATEKDLDGTKSGQNPFVVCKLFKKQDIVNGAAEPEESKSCEVEPAV 186
Query: 184 STPMA------AKYSPEEVQSDTPLPL-VAVSSSLLTEDDKHQAIIPETS 226
S+P ++ SP +++ P VA SS ++ + + +PE +
Sbjct: 187 SSPTVVDEVEMSEVSPVFPKTEETNPCDVAESSLVIPSECRSGYSVPEVT 236
>UniRef100_Q7EZD3 NAC2 protein-like [Oryza sativa]
Length = 729
Score = 216 bits (550), Expect = 2e-54
Identities = 112/212 (52%), Positives = 143/212 (66%), Gaps = 12/212 (5%)
Query: 10 VLSLNSLPTGFRFRPSDEEIIDFYLRQKINGN-GDEVWVIREIDVCKWEPWDLPDLSVIR 68
V+ L LP GFRF P+DEE++ YL+ KI G E VI EIDVCK EPWDLPD S+IR
Sbjct: 3 VMELKKLPLGFRFHPTDEELVRHYLKGKITGQIRSEADVIPEIDVCKCEPWDLPDKSLIR 62
Query: 69 NRDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKS-----GSSLIGMKKTLVF 123
+ DPEWFFF P+DRKYPNG+R NRAT GYWKATGKDR I+S +IGMKKTLVF
Sbjct: 63 SDDPEWFFFAPKDRKYPNGSRSNRATEAGYWKATGKDRVIRSKGDKKKQQVIGMKKTLVF 122
Query: 124 YHGRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTA 183
+ GRAPKG+RT W+MHEYR T E + Q YV+ RLF+KQ+E IE E +++
Sbjct: 123 HRGRAPKGERTGWIMHEYRTTEPEFESGE--QGGYVLYRLFRKQEEKIERPSPDEVDRSG 180
Query: 184 STPMAAKYSPEEVQ----SDTPLPLVAVSSSL 211
+P ++ +P+ ++ +TPL + S+L
Sbjct: 181 YSPTPSRSTPDNMEPIEDGNTPLNRESPESAL 212
>UniRef100_Q9LSH5 Similarity to NAM [Arabidopsis thaliana]
Length = 246
Score = 204 bits (519), Expect = 6e-51
Identities = 110/234 (47%), Positives = 150/234 (64%), Gaps = 21/234 (8%)
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE++++YL++KING E+ +I E+D+ K EPWDL + S + +RDPEW+
Sbjct: 6 LPPGFRFHPTDEELVNYYLKRKINGQEIELDIIPEVDLYKCEPWDLAEKSFLPSRDPEWY 65
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRTN 135
FF P+DRKYPNG R NRAT GYWK+TGKDR++ S S IGMKKTLV+Y GRAP+G RT+
Sbjct: 66 FFGPRDRKYPNGFRTNRATRGGYWKSTGKDRRVTSQSRAIGMKKTLVYYKGRAPQGIRTD 125
Query: 136 WVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAA--KYSP 193
WVMHEYR ++ D + Q+ Y +CR+FKK E+ E+E+ T + S
Sbjct: 126 WVMHEYRLDDKDCDDPSSLQDSYALCRVFKKNGICSEL----ESERQLQTGQCSFTTASM 181
Query: 194 EEVQS----------DTPLPLVAVSSSLLTE-----DDKHQAIIPETSEETTSN 232
EE+ S +T P V VSS+ + E DD I + + +T+SN
Sbjct: 182 EEINSNNNNNYNNDYETMSPEVGVSSACVEEVVDDKDDSWMQFITDDAWDTSSN 235
>UniRef100_Q9SRZ8 F12P19.8 protein [Arabidopsis thaliana]
Length = 631
Score = 203 bits (516), Expect = 1e-50
Identities = 92/152 (60%), Positives = 117/152 (76%)
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
S+P GFRF P+DEE++ +YL++KING E+ +I EID+ K EPWDLP S++ ++D EW
Sbjct: 5 SMPPGFRFHPTDEELVIYYLKRKINGRTIELEIIPEIDLYKCEPWDLPGKSLLPSKDLEW 64
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
FFF P+DRKYPNG+R NRAT GYWKATGKDRK+ S S ++G KKTLV+Y GRAP G RT
Sbjct: 65 FFFSPRDRKYPNGSRTNRATKAGYWKATGKDRKVTSHSRMVGTKKTLVYYRGRAPHGSRT 124
Query: 135 NWVMHEYRPTLQELDGTNPGQNPYVICRLFKK 166
+WVMHEYR QE D + Q+ Y +CR+FKK
Sbjct: 125 DWVMHEYRLEEQECDSKSGIQDAYALCRVFKK 156
>UniRef100_Q8H045 Hypothetical protein OJ1263H11.11 [Oryza sativa]
Length = 650
Score = 198 bits (504), Expect = 3e-49
Identities = 90/152 (59%), Positives = 115/152 (75%)
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SLP GFRF P+DEE+I +YL++KING E+ +I E+D+ K EPWDLP+ S + ++D EW
Sbjct: 5 SLPPGFRFHPTDEELIIYYLKRKINGRQIELEIIPEVDLYKCEPWDLPEKSFLPSKDLEW 64
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF P+DRKYPNG+R NRAT GYWKATGKDRK+ S +GMKKTLV+Y GRAP G RT
Sbjct: 65 YFFSPRDRKYPNGSRTNRATKAGYWKATGKDRKVNSQRRAVGMKKTLVYYRGRAPHGSRT 124
Query: 135 NWVMHEYRPTLQELDGTNPGQNPYVICRLFKK 166
+WVMHEYR +E + Q+ Y +CR+FKK
Sbjct: 125 DWVMHEYRLDERECETDTGLQDAYALCRVFKK 156
>UniRef100_Q8LRL4 Nam-like protein 11 [Petunia hybrida]
Length = 337
Score = 194 bits (493), Expect = 6e-48
Identities = 94/199 (47%), Positives = 129/199 (64%), Gaps = 4/199 (2%)
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SLP GFRF P+DEE++ +YL++K +G E+ VI ID+ K++PW+LP+ S + RD EW
Sbjct: 5 SLPPGFRFHPTDEELVGYYLKRKTDGLEIELEVIPVIDLYKFDPWELPEKSFLPKRDLEW 64
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
FFFCP+D+KYPNG+R NRAT GYWKATGKDRK+ +++G +KTLVFY GRAP G RT
Sbjct: 65 FFFCPRDKKYPNGSRTNRATKSGYWKATGKDRKVVCHPTVVGYRKTLVFYRGRAPLGDRT 124
Query: 135 NWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAAKYS-- 192
+WVMHEYR GT Q P+ +CR+ K+ D S++ +Q + ++
Sbjct: 125 DWVMHEYRLCDDVSQGTPIFQGPFALCRVIKRNDVSLKTKDVQGVKQVGCSSATGGFTSV 184
Query: 193 --PEEVQSDTPLPLVAVSS 209
P + DTP VS+
Sbjct: 185 NEPILLSDDTPTQSTYVSN 203
>UniRef100_Q8LRL7 Nam-like protein 8 [Petunia hybrida]
Length = 392
Score = 192 bits (489), Expect = 2e-47
Identities = 126/354 (35%), Positives = 176/354 (49%), Gaps = 43/354 (12%)
Query: 4 VETAVPVLSLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPD 63
V T P + SL GFRF P+DEE++ +YLR+KI G + EIDV K EPW+L
Sbjct: 11 VATPAPAPAPTSLAPGFRFHPTDEELVRYYLRRKIIGKPFRFQAVTEIDVYKSEPWELAG 70
Query: 64 LSVIRNRDPEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVF 123
S ++NRD EW+FF P D+KY NG+RLNRAT GYWKATGKDR ++ S IGMKKTLVF
Sbjct: 71 YSSLKNRDLEWYFFSPVDKKYGNGSRLNRATGQGYWKATGKDRHVRHKSQDIGMKKTLVF 130
Query: 124 YHGRAPKGKRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTA 183
+ GRAP GKRTNWVMHEYR T +EL+ Q+ +V+CR+F K
Sbjct: 131 HSGRAPDGKRTNWVMHEYRLTDEELEKAGIVQDSFVLCRIFHKSG--------------L 176
Query: 184 STPMAAKYSPEEVQSDTPLPLVAVSSSLLTEDDKHQAIIP--ETSEETTSNIITPVDRNS 241
P + +Y+P + E D A++P E ++ + V+ N
Sbjct: 177 GPPNSDRYAP----------------FVEEEWDDDAAVVPGEEAEDDVANGDEARVEGND 220
Query: 242 DRYDAQNAQPQNVKLAAEENQPLNLDMYNDLKNDIFDDKLFSPALVHLPPVFDYQASNEP 301
DA+ P++ + E P NL + K + +D F P D +S+
Sbjct: 221 LDQDARQKTPRHCENLIE---PQNLPLV--CKRESSEDHEFLSLSQSKRPKHDDPSSSRA 275
Query: 302 DDRSGLQYGTNETAISDLFDSLTWDQLSY------EESGSQPMNFPLFNVKDSG 349
+ ++E + + + L + E SQP N P F+ + G
Sbjct: 276 NGSEDSTISSHEPPTMMMMTNSSPTLLGFPLLESTEPKESQPSNPPAFDSANLG 329
>UniRef100_Q9FFI5 NAM (No apical meristem)-like protein [Arabidopsis thaliana]
Length = 476
Score = 191 bits (486), Expect = 4e-47
Identities = 91/153 (59%), Positives = 113/153 (73%), Gaps = 1/153 (0%)
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SLP GFRF P+DEE+I +YL++KING E+ +I E+D+ K EPWDLP S+I ++D EW
Sbjct: 5 SLPPGFRFHPTDEELITYYLKRKINGQEIELEIIPEVDLYKCEPWDLPGKSLIPSKDQEW 64
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
FFF P+DRKYPNG+R NRAT GYWKATGKDR++ IG KKTLV+Y GRAP G RT
Sbjct: 65 FFFSPRDRKYPNGSRTNRATKGGYWKATGKDRRVSWRDRAIGTKKTLVYYRGRAPHGIRT 124
Query: 135 NWVMHEYRPTLQELDGTNPG-QNPYVICRLFKK 166
WVMHEYR E + + G Q+ Y +CR+FKK
Sbjct: 125 GWVMHEYRLDESECEPSAFGMQDAYALCRVFKK 157
>UniRef100_Q8S7N3 Putative NAM (No apical meristem) protein [Oryza sativa]
Length = 326
Score = 190 bits (482), Expect = 1e-46
Identities = 102/231 (44%), Positives = 137/231 (59%), Gaps = 24/231 (10%)
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
+LP GFRF P+D+E++ +YL++K++ E+ VI ID+ K+EPW+LP+ S + RD EW
Sbjct: 2 TLPPGFRFHPTDDELVGYYLKRKVDSLKIELEVIPVIDLYKFEPWELPEKSFLPKRDLEW 61
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
FFFCP+DRKYPNG+R NRAT GYWKATGKDRKI + G++KTLVFY GRAP G+RT
Sbjct: 62 FFFCPRDRKYPNGSRTNRATSTGYWKATGKDRKIACAGEVFGLRKTLVFYKGRAPGGERT 121
Query: 135 NWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAAKYSPE 194
+WVMHEYR G + Y +CR+ K+ E + GE + M +K
Sbjct: 122 DWVMHEYRLCQDLAHGVSNFIGAYALCRVIKRH----EAGLHGEPPAAKAKGMISK---- 173
Query: 195 EVQSDTPLPLVAVSSSLLTEDDKHQAIIPETSEETTSNIITPVDRNSDRYD 245
V+ SSSL+T + HQ +S S TP + S D
Sbjct: 174 ----------VSSSSSLVTVE--HQL----SSRGNASPSFTPTNNGSPLVD 208
>UniRef100_Q949N0 Hypothetical protein At3g10500 [Arabidopsis thaliana]
Length = 549
Score = 189 bits (479), Expect = 3e-46
Identities = 102/239 (42%), Positives = 142/239 (58%), Gaps = 12/239 (5%)
Query: 12 SLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRD 71
S+ SL GFRF P+DEE++ +YL++KI + I DV K EPWDLPD S +++RD
Sbjct: 5 SVTSLAPGFRFHPTDEELVRYYLKRKICNKPFKFDAISVTDVYKSEPWDLPDKSRLKSRD 64
Query: 72 PEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKG 131
EW+FF D+KY NG++ NRAT +GYWK TGKDR+I +GS ++GMKKTLV++ GRAP+G
Sbjct: 65 LEWYFFSMLDKKYRNGSKTNRATEMGYWKTTGKDREILNGSKVVGMKKTLVYHKGRAPRG 124
Query: 132 KRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAAKY 191
+RTNWVMHEYR Q+LD T Q+ +V+CR+F+K SG + A +
Sbjct: 125 ERTNWVMHEYRLVDQDLDKTGVHQDAFVLCRIFQK---------SGSGPKNGE-QYGAPF 174
Query: 192 SPEEVQSDTPLPLVAVSSSLLTEDD--KHQAIIPETSEETTSNIITPVDRNSDRYDAQN 248
EE + + + V L +ED H I + SE P+ N ++ N
Sbjct: 175 VEEEWEEEDDMTFVPDQEDLGSEDHVYVHMDDIDQKSENFVVYDAIPIPLNFIHGESSN 233
>UniRef100_Q9SQY1 Hypothetical protein F13M14.22 [Arabidopsis thaliana]
Length = 558
Score = 189 bits (479), Expect = 3e-46
Identities = 102/239 (42%), Positives = 142/239 (58%), Gaps = 12/239 (5%)
Query: 12 SLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRD 71
S+ SL GFRF P+DEE++ +YL++KI + I DV K EPWDLPD S +++RD
Sbjct: 5 SVTSLAPGFRFHPTDEELVRYYLKRKICNKPFKFDAISVTDVYKSEPWDLPDKSRLKSRD 64
Query: 72 PEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKG 131
EW+FF D+KY NG++ NRAT +GYWK TGKDR+I +GS ++GMKKTLV++ GRAP+G
Sbjct: 65 LEWYFFSMLDKKYRNGSKTNRATEMGYWKTTGKDREILNGSKVVGMKKTLVYHKGRAPRG 124
Query: 132 KRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAAKY 191
+RTNWVMHEYR Q+LD T Q+ +V+CR+F+K SG + A +
Sbjct: 125 ERTNWVMHEYRLVDQDLDKTGVHQDAFVLCRIFQK---------SGSGPKNGE-QYGAPF 174
Query: 192 SPEEVQSDTPLPLVAVSSSLLTEDD--KHQAIIPETSEETTSNIITPVDRNSDRYDAQN 248
EE + + + V L +ED H I + SE P+ N ++ N
Sbjct: 175 VEEEWEEEDDMTFVPDQEDLGSEDHVYVHMDDIDQKSENFVVYDAIPIPLNFIHGESSN 233
>UniRef100_Q8LAH6 NAC, putative [Arabidopsis thaliana]
Length = 549
Score = 187 bits (474), Expect = 1e-45
Identities = 101/239 (42%), Positives = 142/239 (59%), Gaps = 12/239 (5%)
Query: 12 SLNSLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRD 71
S+ SL GFRF P+DEE++ +YL++KI + I DV K EPWDLPD S +++RD
Sbjct: 5 SVTSLAPGFRFHPTDEELVRYYLKRKICNKPFKFDAISVTDVYKSEPWDLPDKSRLKSRD 64
Query: 72 PEWFFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKG 131
EW+FF ++KY NG++ NRAT +GYWK TGKDR+I +GS ++GMKKTLV++ GRAP+G
Sbjct: 65 LEWYFFSMLNKKYRNGSKTNRATEMGYWKTTGKDREILNGSKVVGMKKTLVYHKGRAPRG 124
Query: 132 KRTNWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAAKY 191
+RTNWVMHEYR Q+LD T Q+ +V+CR+F+K SG + A +
Sbjct: 125 ERTNWVMHEYRLVDQDLDKTGVHQDAFVLCRIFQK---------SGSGPKNGE-QYGAPF 174
Query: 192 SPEEVQSDTPLPLVAVSSSLLTEDD--KHQAIIPETSEETTSNIITPVDRNSDRYDAQN 248
EE + + + V L +ED H I + SE P+ N ++ N
Sbjct: 175 VEEEWEEEDDMTFVPDQEDLGSEDHVYVHMDDIDQKSENFVVYDAIPIPLNFIHGESSN 233
>UniRef100_Q8LRM1 Nam-like protein 4 [Petunia hybrida]
Length = 406
Score = 185 bits (469), Expect = 4e-45
Identities = 95/191 (49%), Positives = 119/191 (61%), Gaps = 17/191 (8%)
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SL GFRF P+DEE++ +YLR+K + EIDV K EPW+L + + ++ RD EW
Sbjct: 34 SLAPGFRFHPTDEELVRYYLRRKACAKPFRFQAVSEIDVYKSEPWELAEFASLKTRDLEW 93
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF P DRKY NG+RLNRAT GYWKATGKDR ++ S IGMKKTLVF+ GRAP GKRT
Sbjct: 94 YFFSPVDRKYGNGSRLNRATGKGYWKATGKDRPVRHKSQTIGMKKTLVFHSGRAPDGKRT 153
Query: 135 NWVMHEYRPTLQELDGTNPGQNPYVICRLFKKQDESIEVSISGEAEQTASTPMAAKYSP- 193
NWVMHEYR +EL+ Q +V+CR+F+K P +Y+P
Sbjct: 154 NWVMHEYRLADEELERAGVVQIAFVLCRIFQKSG--------------LGPPNGDRYAPF 199
Query: 194 --EEVQSDTPL 202
EE DTPL
Sbjct: 200 IEEEWDDDTPL 210
>UniRef100_Q6YYW2 Putative NAC2 protein [Oryza sativa]
Length = 656
Score = 184 bits (468), Expect = 5e-45
Identities = 99/247 (40%), Positives = 145/247 (58%), Gaps = 25/247 (10%)
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SL GFRF P+DEE++ +YL++K++G +V I E+D+ K EPWDLP S +R+RD +W
Sbjct: 20 SLAPGFRFHPTDEELVSYYLKRKVHGRPLKVDAIAEVDLYKVEPWDLPARSRLRSRDSQW 79
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF DRK+ N R NRAT GYWK TGKDR++++G + +GMKKTLVF+ GRAPKG+RT
Sbjct: 80 YFFSRLDRKHANRARTNRATAGGYWKTTGKDREVRNGPTTVGMKKTLVFHAGRAPKGERT 139
Query: 135 NWVMHEYRPTLQELDG--TNPGQNPYVICRLFKKQDES-----------IEVSISGEAEQ 181
NWVMHEYR LDG T P Q+ +V+CR+F+K +E + E
Sbjct: 140 NWVMHEYR-----LDGQTTIPPQDSFVVCRIFQKAGPGPQNGAQYGAPFVEEEWEEDDED 194
Query: 182 TASTPMAAKYSPEEVQSDTP-------LPLVAVSSSLLTEDDKHQAIIPETSEETTSNII 234
P+ K + ++ + + L + + +L+ +++ +P + SN
Sbjct: 195 VGLLPVEEKDNSDDQEKEISGAMEKGYLQMSDLVQNLVDQNENGTIALPVSDNSNNSNHS 254
Query: 235 TPVDRNS 241
VD NS
Sbjct: 255 EDVDGNS 261
>UniRef100_Q6YYW1 Putative NAC2 protein [Oryza sativa]
Length = 618
Score = 184 bits (468), Expect = 5e-45
Identities = 99/247 (40%), Positives = 145/247 (58%), Gaps = 25/247 (10%)
Query: 15 SLPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEW 74
SL GFRF P+DEE++ +YL++K++G +V I E+D+ K EPWDLP S +R+RD +W
Sbjct: 20 SLAPGFRFHPTDEELVSYYLKRKVHGRPLKVDAIAEVDLYKVEPWDLPARSRLRSRDSQW 79
Query: 75 FFFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKSGSSLIGMKKTLVFYHGRAPKGKRT 134
+FF DRK+ N R NRAT GYWK TGKDR++++G + +GMKKTLVF+ GRAPKG+RT
Sbjct: 80 YFFSRLDRKHANRARTNRATAGGYWKTTGKDREVRNGPTTVGMKKTLVFHAGRAPKGERT 139
Query: 135 NWVMHEYRPTLQELDG--TNPGQNPYVICRLFKKQDES-----------IEVSISGEAEQ 181
NWVMHEYR LDG T P Q+ +V+CR+F+K +E + E
Sbjct: 140 NWVMHEYR-----LDGQTTIPPQDSFVVCRIFQKAGPGPQNGAQYGAPFVEEEWEEDDED 194
Query: 182 TASTPMAAKYSPEEVQSDTP-------LPLVAVSSSLLTEDDKHQAIIPETSEETTSNII 234
P+ K + ++ + + L + + +L+ +++ +P + SN
Sbjct: 195 VGLLPVEEKDNSDDQEKEISGAMEKGYLQMSDLVQNLVDQNENGTIALPVSDNSNNSNHS 254
Query: 235 TPVDRNS 241
VD NS
Sbjct: 255 EDVDGNS 261
>UniRef100_Q9LS24 NAM-like [Arabidopsis thaliana]
Length = 292
Score = 184 bits (466), Expect = 8e-45
Identities = 109/266 (40%), Positives = 154/266 (56%), Gaps = 14/266 (5%)
Query: 16 LPTGFRFRPSDEEIIDFYLRQKINGNGDEVWVIREIDVCKWEPWDLPDLSVIRNRDPEWF 75
LP GFRF P+DEE+I++YL++K+ G E+ VI ID+ ++PW+LPD S + NRD EW+
Sbjct: 6 LPPGFRFHPTDEELIEYYLKRKVEGLEIELEVIPVIDLYSFDPWELPDKSFLPNRDMEWY 65
Query: 76 FFCPQDRKYPNGNRLNRATILGYWKATGKDRKIKS-GSSLIGMKKTLVFYHGRAPKGKRT 134
FFC +D+KYPNG R NR T GYWKATGKDRKI S SS+IG +KTLVFY GRAP G R+
Sbjct: 66 FFCSRDKKYPNGFRTNRGTKAGYWKATGKDRKITSRSSSIIGYRKTLVFYKGRAPLGDRS 125
Query: 135 NWVMHEYRPTLQELDGTNPGQN---PYVICRLFKKQDESIEVSISG-EAEQTASTPMAAK 190
NW+MHEYR L + D + QN +V+CR+ K + I +EQT + ++
Sbjct: 126 NWIMHEYR--LCDDDTSQGSQNLKGAFVLCRVAMKNEIKTNTKIRKIPSEQTIGSGESSG 183
Query: 191 YSPE--EVQSDTPLPLVAVSSSLLTEDDKHQA-IIPETSEETTSNIITPVDRNSDRYDAQ 247
S D +P + ++ + TE D I PE +++ + D S +
Sbjct: 184 LSSRVTSPSRDETMPFHSFANPVSTETDSSNIWISPEFILDSSKDYPQIQDVASQCFQQD 243
Query: 248 NAQP----QNVKLAAEENQPLNLDMY 269
P QN++ A + N+D +
Sbjct: 244 FDFPIIGNQNMEFPASTSLDQNMDEF 269
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.131 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,038,737,439
Number of Sequences: 2790947
Number of extensions: 46846298
Number of successful extensions: 86836
Number of sequences better than 10.0: 332
Number of HSP's better than 10.0 without gapping: 241
Number of HSP's successfully gapped in prelim test: 91
Number of HSP's that attempted gapping in prelim test: 86085
Number of HSP's gapped (non-prelim): 374
length of query: 596
length of database: 848,049,833
effective HSP length: 133
effective length of query: 463
effective length of database: 476,853,882
effective search space: 220783347366
effective search space used: 220783347366
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0207.16


