

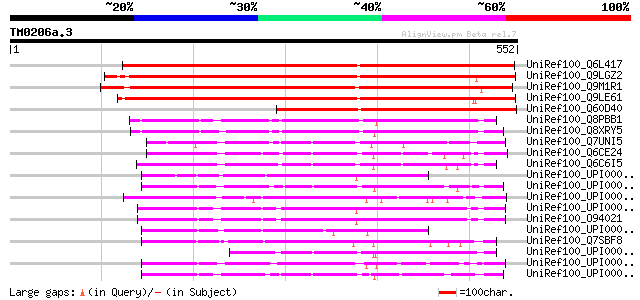
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0206a.3
(552 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6L417 Putative isopenicillin N epimerase [Solanum dem... 584 e-165
UniRef100_Q9LGZ2 ESTs AU091298 [Oryza sativa] 578 e-163
UniRef100_Q9M1R1 Hypothetical protein T17J13.90 [Arabidopsis tha... 567 e-160
UniRef100_Q9LE61 Similar to Streptomyces clavuligerus isopenicil... 541 e-152
UniRef100_Q60D40 Hypothetical protein [Solanum tuberosum] 378 e-103
UniRef100_Q8PBB1 Isopenicillin N epimerase [Xanthomonas campestris] 183 1e-44
UniRef100_Q8XRY5 PUTATIVE ISOPENICILLIN N EPIMERASE PROTEIN [Ral... 166 1e-39
UniRef100_Q7UNI5 Isopenicillin N-epimerase [Rhodopirellula baltica] 142 3e-32
UniRef100_Q6CE24 Similar to tr|O94021 Candida albicans Hypotheti... 137 1e-30
UniRef100_Q6C6I5 Similar to tr|O94021 Candida albicans CA49C10.0... 135 3e-30
UniRef100_UPI0000234B94 UPI0000234B94 UniRef100 entry 135 4e-30
UniRef100_UPI0000320766 UPI0000320766 UniRef100 entry 134 5e-30
UniRef100_UPI00003C1FB4 UPI00003C1FB4 UniRef100 entry 127 6e-28
UniRef100_UPI000004DFD4 UPI000004DFD4 UniRef100 entry 127 1e-27
UniRef100_O94021 Hypothetical protein Ca49C10.07c [Candida albic... 126 2e-27
UniRef100_UPI000021A892 UPI000021A892 UniRef100 entry 125 3e-27
UniRef100_Q7SBF8 Hypothetical protein [Neurospora crassa] 123 1e-26
UniRef100_UPI0000277927 UPI0000277927 UniRef100 entry 123 2e-26
UniRef100_UPI0000281D0D UPI0000281D0D UniRef100 entry 120 1e-25
UniRef100_UPI000025CD0A UPI000025CD0A UniRef100 entry 117 9e-25
>UniRef100_Q6L417 Putative isopenicillin N epimerase [Solanum demissum]
Length = 736
Score = 584 bits (1506), Expect = e-165
Identities = 281/427 (65%), Positives = 346/427 (80%), Gaps = 4/427 (0%)
Query: 124 PLSSSPFITPSEIQSEFSHHDPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFN 183
P +++ IT +EI +EFSHHD ++ARINNGSFG C S+ +AQ WQL++L+QPD FYFN
Sbjct: 12 PSTTAKPITETEIVAEFSHHDLTIARINNGSFGSCPKSIISAQQQWQLQFLQQPDYFYFN 71
Query: 184 HLKQGILRSRTIIKDLVNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLML 243
LK +L+SRT+I+ LVNA +DEISIVDNATTAAAIVLQ W+F F+PGD +ML
Sbjct: 72 TLKPSMLKSRTLIQSLVNAADVDEISIVDNATTAAAIVLQYITWSFFTSHFRPGDAAVML 131
Query: 244 HYAYGAVKKSMEAYVTRAGGTVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVI 303
HYAYG+VK S++AYV RAGG VIEV LPFP+ S+++IV EF AL+ GK G K+RLAVI
Sbjct: 132 HYAYGSVKSSVQAYVARAGGKVIEVHLPFPLNSNEEIVTEFDKALKMGKMNGGKIRLAVI 191
Query: 304 DHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFF 363
DH+TSMP VVIPVKELV++CR+EGVD +FVD AH+IG ++D+ +IGADFYTSNLHKWFF
Sbjct: 192 DHITSMPSVVIPVKELVQMCRDEGVDFIFVDGAHAIGNVEIDVVDIGADFYTSNLHKWFF 251
Query: 364 CPPSIAFLYSRKHPKGGSGSELHHPVVSHEYGNGLAVESAWIGTRDYSAQLVVPAVID-F 422
PS AFLY ++ K +LHHPVVS EYGNGLA+ESAWIGTRDYSAQLV+P V++ F
Sbjct: 252 TLPSAAFLYCKRSEK---VVDLHHPVVSVEYGNGLAIESAWIGTRDYSAQLVIPDVVELF 308
Query: 423 VNRFEGGIEGIKKRNHETVVEMGDMLVKAWGTHVGCPPHMCASMVMVGLPTCLGVQSDSD 482
VNRFEGGIEGI++RNH+ VVEM +MLVKAWGT +G PP MC+SM MVG+P CLG+ +SD
Sbjct: 309 VNRFEGGIEGIRRRNHDMVVEMAEMLVKAWGTELGTPPEMCSSMAMVGMPACLGISGNSD 368
Query: 483 ALKLRTHLREAFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDYYKFRDAVIQLVD 542
ALKLRTHLR F VEVPIYYR P +GEV P+TGYARISHQVYN ++DYY+FRDA+I+LV
Sbjct: 369 ALKLRTHLRVLFKVEVPIYYRAPLEGEVNPITGYARISHQVYNTIEDYYRFRDAIIKLVS 428
Query: 543 KGFTCAL 549
++ +
Sbjct: 429 DAYSAVV 435
>UniRef100_Q9LGZ2 ESTs AU091298 [Oryza sativa]
Length = 482
Score = 578 bits (1491), Expect = e-163
Identities = 290/455 (63%), Positives = 345/455 (75%), Gaps = 15/455 (3%)
Query: 104 NHNNNNNGCTTHIPKKPKLSPLSSSPFITPSEIQSEFSHHDPSVARINNGSFGCCSASVT 163
N N NG P P P S I+ ++I++EF HH+ VAR+NNGSFGCC +S+
Sbjct: 25 NGKGNGNGPAPRPP--PAKRPRS---VISAAQIRAEFEHHEAGVARVNNGSFGCCPSSLL 79
Query: 164 AAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNADHLDEISIVDNATTAAAIVLQ 223
AQ WQ ++ QPD+FYF+ L+ G+ RSR + LVNA + E+S+VDNATTAAAIVLQ
Sbjct: 80 DAQARWQRLFIAQPDDFYFHALQPGLRRSRAAVAGLVNAGDVAEVSLVDNATTAAAIVLQ 139
Query: 224 NTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGGTVIEVPLPFPVKSSDDIVRE 283
+ AW+F EG F GD VLMLHYAYGAVKKS+ AYV RAG TV+EVPLPFPV S+D I+ E
Sbjct: 140 HAAWSFAEGRFSRGDAVLMLHYAYGAVKKSIHAYVARAGATVVEVPLPFPVASADAIIAE 199
Query: 284 FRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHSIGCTD 343
FR+AL+ K+GG+KVRLAVIDH+TSMP VVIPVKELV ICREEGVD+VF+DAAHSIG
Sbjct: 200 FRAALDVAKAGGRKVRLAVIDHITSMPSVVIPVKELVAICREEGVDKVFIDAAHSIGQVP 259
Query: 344 VDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGSELHHPVVSHEYGNGLAVESA 403
VD+++IGADFYTSNLHKWFFCPP++AFL++RK S+LHHPVVSHEYGNGL +ES
Sbjct: 260 VDVRDIGADFYTSNLHKWFFCPPAVAFLHTRKDDP--IASQLHHPVVSHEYGNGLPMESG 317
Query: 404 WIGTRDYSAQLVVPAVIDFVNRFEGGIEGIKKRNHETVVEMGDMLVKAWGTHVGCPPHMC 463
WIGTRDYSAQLVVP IDFVNRFEGGIEGI+ RNHE V+EMG ML +AWGT +G PP +C
Sbjct: 318 WIGTRDYSAQLVVPESIDFVNRFEGGIEGIRSRNHEKVIEMGKMLAEAWGTFLGTPPELC 377
Query: 464 ASMVMVGLPTCLGVQSDSDALKLRTHLREAFGVEVPIYYRPPR--------DGEVEPVTG 515
SMVMVGLP CLGV+SD D +++RT LR+ F VEVPIYY R D + VTG
Sbjct: 378 GSMVMVGLPGCLGVESDDDVMRMRTMLRKDFMVEVPIYYNSRRVEAQEMAKDKNGDAVTG 437
Query: 516 YARISHQVYNKVDDYYKFRDAVIQLVDKGFTCALL 550
Y RISHQVYN +DY K RDAV +LV GFT + L
Sbjct: 438 YVRISHQVYNVTEDYEKLRDAVNKLVADGFTSSKL 472
>UniRef100_Q9M1R1 Hypothetical protein T17J13.90 [Arabidopsis thaliana]
Length = 454
Score = 567 bits (1461), Expect = e-160
Identities = 272/456 (59%), Positives = 346/456 (75%), Gaps = 15/456 (3%)
Query: 99 MASTHNHNNNNNGCTTHIPKKPKLSPLSSSPFITPSEIQSEFSHHDPSVARINNGSFGCC 158
M + N ++ PKKP+L+ L +T S+I SEF+HH VARINNGSFGCC
Sbjct: 1 MEAGERRNGDSMSHNHRAPKKPRLAGL-----LTESDIDSEFAHHQTGVARINNGSFGCC 55
Query: 159 SASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNADHLDEISIVDNATTAA 218
SV AQ +WQL+YLRQPD FYFN L++G+L SRT+I DL+NAD +DE+S+VDNATTAA
Sbjct: 56 PGSVLEAQREWQLRYLRQPDEFYFNGLRRGLLASRTVISDLINADDVDEVSLVDNATTAA 115
Query: 219 AIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGGTVIEVPLPFPVKSSD 278
AIVLQ F EG ++ D V+M H A+ +VKKS++AYV+R GG+ +EV LPFPV S++
Sbjct: 116 AIVLQKVGRCFSEGKYKKEDTVVMFHCAFQSVKKSIQAYVSRVGGSTVEVRLPFPVNSNE 175
Query: 279 DIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHS 338
+I+ +FR LE+G++ G+ VRLA+IDH+TSMPCV++PV+ELVKICREEGV++VFVDAAH+
Sbjct: 176 EIISKFREGLEKGRANGRTVRLAIIDHITSMPCVLMPVRELVKICREEGVEQVFVDAAHA 235
Query: 339 IGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGSELHHPVVSHEYGNGL 398
IG VD+KEIGAD+Y SNLHKWFFCPPSIAF Y +K GS S++HHPVVSHE+GNGL
Sbjct: 236 IGSVKVDVKEIGADYYVSNLHKWFFCPPSIAFFYCKKR---GSESDVHHPVVSHEFGNGL 292
Query: 399 AVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIEGIKKRNHETVVEMGDMLVKAWGTHVGC 458
+ESAWIGTRDYS+QLVVP+V++FVNRFEGG+EGI +NH+ V MG ML AWGT++G
Sbjct: 293 PIESAWIGTRDYSSQLVVPSVMEFVNRFEGGMEGIMMKNHDEAVRMGLMLADAWGTNLGS 352
Query: 459 PPHMCASMVMVGLPTCLGVQSDSDALKLRTHLREAFGVEVPIYYRPPRDGEVEP------ 512
PP MC MVM+GLP+ L V SD DA+KLR++LR + VEVP++Y RDGE
Sbjct: 353 PPEMCVGMVMIGLPSKLCVGSDEDAIKLRSYLRVHYSVEVPVFYLGLRDGEEGVKDKDSG 412
Query: 513 -VTGYARISHQVYNKVDDYYKFRDAVIQLVDKGFTC 547
+T Y RISHQVYNK +DY + RDA+ +LV TC
Sbjct: 413 LITAYVRISHQVYNKTEDYERLRDAITELVKDQMTC 448
>UniRef100_Q9LE61 Similar to Streptomyces clavuligerus isopenicillin epimerase [Oryza
sativa]
Length = 476
Score = 541 bits (1395), Expect = e-152
Identities = 269/445 (60%), Positives = 334/445 (74%), Gaps = 16/445 (3%)
Query: 118 KKPKLSPLSSSPFITPSEIQSEFSHHDPSVARINNGSFGCCSASVTAAQHDWQLKYLRQP 177
K+P+ + + IT +E+++EF+HHD +VAR+NNG+FGCC ASV AA+ WQ +L QP
Sbjct: 29 KRPRAG--AGAAAITDAEVRAEFAHHDRAVARLNNGTFGCCPASVLAARARWQRLFLSQP 86
Query: 178 DNFYFNHLKQGILRSRTIIKDLVNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPG 237
D FYF+HL+ G+ RSR + V A E+S+VDN TTAAAI++Q+ AW+F EG F G
Sbjct: 87 DAFYFHHLQPGLARSRAAVAAAVGAGDASEVSLVDNVTTAAAIIMQHVAWSFAEGDFARG 146
Query: 238 DVVLMLHYAYGAVKKSMEAYVTRAGGTVIEVPLPFPVKSSDDIVREFRSALERGKSGG-K 296
DVVLM Y Y ++K S+ AYV RAG TV+EVPLPFPV S D IV EFR+AL + GG +
Sbjct: 147 DVVLMFLYTYCSIKNSIHAYVARAGATVVEVPLPFPVSSPDAIVAEFRAALAVARDGGRR 206
Query: 297 KVRLAVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTS 356
+VRLAVIDH+T+MP V+IPVKELV ICREEGVD+VFVDAAH++G VD+++IGADFY S
Sbjct: 207 RVRLAVIDHITAMPTVLIPVKELVAICREEGVDKVFVDAAHAVGQVPVDVRDIGADFYAS 266
Query: 357 NLHKWFFCPPSIAFLYSRKHPKGGSGSELHHPVVSHEYGNGLAVESAWIGTRDYSAQLVV 416
NLHKWFFCP ++AF+++RK S+LHHPVVS EYGNGL +ESAWIG RDYSAQLVV
Sbjct: 267 NLHKWFFCPSAVAFIHTRKDDP--VSSKLHHPVVSSEYGNGLPMESAWIGVRDYSAQLVV 324
Query: 417 PAVIDFVNRFEGGIEGIKKRNHETVVEMGDMLVKAWGTHVGCPPHMCASMVMVGLPTCLG 476
P V+DFVNRF+GG+EGI++RNH+ VVEMG ML AWGT +G PP MC SM+MVGLP LG
Sbjct: 325 PDVVDFVNRFDGGVEGIRRRNHDKVVEMGTMLAAAWGTFLGTPPEMCGSMLMVGLPGSLG 384
Query: 477 VQSDSDALKLRTHLREAFGVEVPIYYR--------PP---RDGEVEPVTGYARISHQVYN 525
V S+ DA+ LRT LR+ F VEVP+YY PP +DG +PVTGY RISHQVYN
Sbjct: 385 VGSEDDAVGLRTMLRKQFKVEVPLYYNSKAAAADAPPEMVKDGNGDPVTGYVRISHQVYN 444
Query: 526 KVDDYYKFRDAVIQLVDKGFTCALL 550
++Y RDAV +LV GFTC L
Sbjct: 445 VREEYEALRDAVAKLVADGFTCRKL 469
>UniRef100_Q60D40 Hypothetical protein [Solanum tuberosum]
Length = 541
Score = 378 bits (971), Expect = e-103
Identities = 178/262 (67%), Positives = 215/262 (81%), Gaps = 4/262 (1%)
Query: 291 GKSGGKKVRLAVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIG 350
GK G K+RLAVIDH+TSMP VVIPVKELV++CR+EGVD +FVD AH+IG ++D+ +IG
Sbjct: 2 GKMNGGKIRLAVIDHITSMPSVVIPVKELVQMCRDEGVDFIFVDGAHAIGNVEIDVVDIG 61
Query: 351 ADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGSELHHPVVSHEYGNGLAVESAWIGTRDY 410
ADFYTSNLHKWFF PPS AFLY ++ K +LHHPVVS EYGNGLA+ESAWIGTRDY
Sbjct: 62 ADFYTSNLHKWFFTPPSAAFLYCKRSEKV---VDLHHPVVSVEYGNGLAIESAWIGTRDY 118
Query: 411 SAQLVVPAVID-FVNRFEGGIEGIKKRNHETVVEMGDMLVKAWGTHVGCPPHMCASMVMV 469
SAQLV+P V++ FVNRFEGGIEGI++RNH+ VVEM +MLVKAWGT +G PP MC+SM MV
Sbjct: 119 SAQLVIPDVVESFVNRFEGGIEGIRRRNHDMVVEMAEMLVKAWGTELGTPPEMCSSMAMV 178
Query: 470 GLPTCLGVQSDSDALKLRTHLREAFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDD 529
G+P C+G+ +SDALKLRTHLR +F VEVPIYYR P +GEV P+TGYARISHQVYN + D
Sbjct: 179 GMPACIGISGNSDALKLRTHLRVSFKVEVPIYYRAPLEGEVNPITGYARISHQVYNTIVD 238
Query: 530 YYKFRDAVIQLVDKGFTCALLS 551
Y++FRD +I+L K L+
Sbjct: 239 YFRFRDVIIKLTKKNMVTLFLN 260
>UniRef100_Q8PBB1 Isopenicillin N epimerase [Xanthomonas campestris]
Length = 415
Score = 183 bits (464), Expect = 1e-44
Identities = 133/407 (32%), Positives = 195/407 (47%), Gaps = 33/407 (8%)
Query: 131 ITPSEIQSEFSHHDPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGIL 190
++P+ ++ FS DP VA +N+G G C SV Q + + RQP F L +
Sbjct: 6 VSPTRFKAHFSL-DPGVACLNHGMLGACPISVQQRQSTLRAQLERQPAAFVLRALPALLD 64
Query: 191 RSRTIIKDLVNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAV 250
+R + +++ AD D + ++ N TTA + VL R F PGD +L +AY +
Sbjct: 65 TARNCLAEVIGADPQD-LLLLPNVTTAMSAVL-------RSRIFAPGDQILTTDHAYLSC 116
Query: 251 KKSMEAYVTRAGGTVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMP 310
+ +E G V+ P+ PV+ D IV + LER + + RLAV+DHVTS
Sbjct: 117 RNLLEFIARSTGAEVMVAPVKVPVQHPDAIVD---AVLERVTA---RTRLAVLDHVTSPT 170
Query: 311 CVVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAF 370
+V P+ LV+ G+D + VD AH+ G +D+ IGA +Y + HKW P F
Sbjct: 171 AIVFPIARLVERLAALGIDTL-VDGAHAPGMLPLDVIAIGAAYYAGDCHKWLCTPRGAGF 229
Query: 371 LYSRKHPKGGSGSELHHPVVSHEYGNG------LAVESAWIGTRDYSAQLVVPAVIDFV- 423
L+ R+ + G LH PV+S YG+ L +E W+GT D +A L +PA IDF+
Sbjct: 230 LHVRRDRQDG----LHPPVISRGYGDATPGRPRLHLEFDWLGTADPTALLCIPAAIDFLA 285
Query: 424 NRFEGGIEGIKKRNHETVVEMGDMLVKAWGTHVGCPPHMCASMVMVGLPTCLGVQSDSDA 483
GG+ + RNH V + L +A P M SMV LP D A
Sbjct: 286 TLLPGGLPAVFARNHALVTQAAARLAQALPLARLAPDAMVGSMVAFQLPDAASDAPDDAA 345
Query: 484 LKLRTHLREAFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDY 530
L+ L +A ++V + P R RIS QVYN +D+
Sbjct: 346 ASLQRWLYDAHRIDVAVTPWPHRTNRT------LRISAQVYNAAEDF 386
>UniRef100_Q8XRY5 PUTATIVE ISOPENICILLIN N EPIMERASE PROTEIN [Ralstonia solanacearum]
Length = 419
Score = 166 bits (421), Expect = 1e-39
Identities = 127/413 (30%), Positives = 191/413 (45%), Gaps = 33/413 (7%)
Query: 132 TPSEIQSEFSHHDPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILR 191
+P +S F DPSV +N+G G C A V Q+ + + RQP F L +
Sbjct: 10 SPKAFRSRFLL-DPSVTCLNHGMLGACPAEVLEQQNALRARIERQPAAFILRELTGLLDE 68
Query: 192 SRTIIKDLVNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVK 251
+R + L+ AD D ++++ N TTA + VL++ A+A PGD +L +AY +
Sbjct: 69 ARQALAGLIAADPAD-LALLPNVTTALSAVLRSRAFA-------PGDEILTTSHAYLSCT 120
Query: 252 KSMEAYVTRAGGTVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPC 311
++ G V+ +P PV +D +V + LER + RLAV+DHVTS
Sbjct: 121 NLLDFVARETGARVVTAIVPTPVTHADAVVD---AVLERVTP---RTRLAVLDHVTSPTG 174
Query: 312 VVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFL 371
+V P+ LV+ GVD + VD AH+ G +D++ IGA +Y N HKW P FL
Sbjct: 175 MVFPIAALVERLAARGVDTL-VDGAHAPGMLPLDVQAIGAAYYAGNCHKWLCSPRGAGFL 233
Query: 372 YSRKHPKGGSGSELHHPVVSHEYG------NGLAVESAWIGTRDYSAQLVVPAVIDFVN- 424
+ R+ G LH V+S YG L +E W+GT D + L + I F++
Sbjct: 234 HVRRDRHDG----LHPTVISRGYGATGTGRPRLHLEFDWLGTADPTPLLCIAHAIRFLDG 289
Query: 425 RFEGGIEGIKKRNHETVVEMGDMLVKAWGTHVGCPPHMCASMVMVGLPTCLGVQSDSDAL 484
GG+ + RNH +E + + P M SMV LP S A
Sbjct: 290 LLPGGLPELMARNHALAIEGAQRMAEGLPLKRLAPDSMVGSMVAFQLPETPEPASGDAAA 349
Query: 485 KLRTHLREAFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDYYKFRDAV 537
L+ L +A ++V P V R+S Q+YN +DD+ + D +
Sbjct: 350 SLQRWLYDAHRIDVAAAAWPAAHSRV------LRVSAQIYNAIDDFIRLGDVL 396
>UniRef100_Q7UNI5 Isopenicillin N-epimerase [Rhodopirellula baltica]
Length = 395
Score = 142 bits (357), Expect = 3e-32
Identities = 115/406 (28%), Positives = 184/406 (44%), Gaps = 44/406 (10%)
Query: 150 INNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLV----NADHL 205
+N+GSFG V Q WQ L + D F ++ +L +++ V NA
Sbjct: 18 LNHGSFGATPRCVIENQRHWQ--NLLEEDPIEFLAPERSLLPKLDFVRETVAKEINASSR 75
Query: 206 DEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGGTV 265
D + V NAT V+++ + GD +L+ ++ Y A ++ AG V
Sbjct: 76 DVV-FVRNATEGVNAVVRSLP-------LRAGDEILVTNHGYNACINAVSQAANIAGAAV 127
Query: 266 IEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKICRE 325
+PFP++S D++VR A+ER S K +IDHVTS +V+PV +L+++
Sbjct: 128 TTANIPFPIQSPDEVVR----AIERRIS--PKTTWMLIDHVTSPTGIVLPVAQLIELAHS 181
Query: 326 EGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGSEL 385
+ RV VD AH+ G +++ E+ D+YT+N HKW+ P FLY + S E+
Sbjct: 182 NNI-RVMVDGAHAPGMLPLNLNELKPDYYTANHHKWWCGPKVSGFLYVDEE----SQDEV 236
Query: 386 HHPVVSH-----EYG-NGLAVESAWIGTRDYSAQLVVPAVIDFVNRFE-----GGIEGIK 434
++SH YG + + W GT D S L +P IDF+ + G+
Sbjct: 237 LPSIISHGANMEGYGPSKFQSQFNWPGTFDPSPLLALPTAIDFLAGLHPADGPNRLAGLL 296
Query: 435 KRNHETVVEMGDMLVKAWGTHVGCPPHMCASMVMVGLPTCLGVQSDSDALKLRTHLREAF 494
+ NHE VE +++ P M S+ + +P + +RT LR +
Sbjct: 297 RHNHELAVEGRRVILNELKLAEPAPESMLGSLATIPVPAWTN-HTSVQIQAVRTALRTEY 355
Query: 495 GVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDYYKFRDAVIQL 540
E+P++ + RIS Q YN +D Y + DAV +L
Sbjct: 356 RFELPVF-------RFDATNVCLRISAQTYNSLDQYERLADAVTKL 394
>UniRef100_Q6CE24 Similar to tr|O94021 Candida albicans Hypothetical 48.1 kDa protein
[Yarrowia lipolytica]
Length = 430
Score = 137 bits (344), Expect = 1e-30
Identities = 117/412 (28%), Positives = 195/412 (46%), Gaps = 50/412 (12%)
Query: 150 INNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNADHLDEIS 209
+N+GSFG C V A++ K PD + L + I +R + ++++ L+ +
Sbjct: 33 LNHGSFGACPLPVREARNQILAKIEANPDLYMRTTLYEDIEVARATACEWIDSEPLNTV- 91
Query: 210 IVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGGTVIEVP 269
V NAT V+++ + GD ++ YGA +K+++ R G + V
Sbjct: 92 FVQNATVGFNTVIRSLP-------LKKGDTIIYCSTTYGACEKTLKFLELRHGIKHVSVD 144
Query: 270 LPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKICREEGVD 329
+ +P+ + ++IV +R A++ S + + D V+SMP ++P +LVK+CRE+ V
Sbjct: 145 IEYPM-NDEEIVDVYRKAIDAHPS----TVICLFDTVSSMPAAILPYNQLVKLCREKEV- 198
Query: 330 RVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLY-SRKHPKGGSGSELHHP 388
F+D AHSIG V M++ DFY +N+HKW + A LY + +H + +H
Sbjct: 199 LSFIDGAHSIGLIPVSMRKTQPDFYVTNVHKWGYGVRGGAILYVAEEHHR-----LIHTL 253
Query: 389 VVSHEY------------GNGLAVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIEGIKKR 436
VSH Y L +IGT+D+S + + A +F + GG I+K
Sbjct: 254 PVSHTYLDDSEDLEPEQEERRLVDRFTFIGTQDFSPYIAITAAFEFRKKI-GGEAQIRKY 312
Query: 437 NHETVVEMGDMLVKAWGTHVGCPPHMCASMVMVGLP---TCLGVQSDSDALKLRTHLRE- 492
++ V++GD+ W T V +MV V LP L S+ +L + +
Sbjct: 313 CNDLAVKVGDLAAGQWRTEV---LGHAGAMVTVRLPIPEEFLAAASEERKQQLFQLICDH 369
Query: 493 --AFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDYYKFRDAVIQLVD 542
G VP Y +G++ Y R S QVYN+++DY DAV + +D
Sbjct: 370 PLTRGTYVPPIY---HNGKM-----YVRFSAQVYNELEDYQVGIDAVNEALD 413
>UniRef100_Q6C6I5 Similar to tr|O94021 Candida albicans CA49C10.07C hypothetical 48.1
kDa protein [Yarrowia lipolytica]
Length = 419
Score = 135 bits (340), Expect = 3e-30
Identities = 114/415 (27%), Positives = 187/415 (44%), Gaps = 50/415 (12%)
Query: 139 EFSHHDPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKD 198
+F D +N+GSFG V + + K + D F L + +R ++
Sbjct: 12 KFWDFDSQNTPLNHGSFGATPKDVEEKRFELIRKIEKNTDKFMRVDLYKMEDETRELVSK 71
Query: 199 LVNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYV 258
+N++ L+ + V NA+ V+++ + GDV++ YGA K+++
Sbjct: 72 WINSEKLNTV-FVPNASVGFNTVIRSLP-------LKEGDVIVHCSTLYGACDKTLQFME 123
Query: 259 TRAGGTVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKE 318
R G +V + +P S DIV +FR ++ K ++ + D V+SMP ++P E
Sbjct: 124 NRYGVKSAKVDITYPEDSDKDIVEKFRKVIKENP----KTKMVIFDTVSSMPGCLLPFNE 179
Query: 319 LVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFL-YSRKHP 377
L ++C++ V F+D AH IG ++++KE DF+ SNLHKW + P A L ++KH
Sbjct: 180 LTQLCKDLDV-LSFIDGAHGIGLVELNLKENEPDFFVSNLHKWGYVPRGAAVLVVAKKH- 237
Query: 378 KGGSGSELHHPVVSHEY-------------GNGLAVESAWIGTRDYSAQLVVPAVIDFVN 424
+++H VSH Y L ++GT D+S L PA + F
Sbjct: 238 ----HNKIHTLPVSHTYLDDEFEAASELDKSRRLVDRFTFVGTTDFSTHLSTPAAVKFRE 293
Query: 425 RFEGGIEGIKKRNHETVVEMGDMLVKAWGTHV--GCPPHMCASMVMVGLPTC---LGVQS 479
+ GG E I+ E ++G +GT V + +MV + LP L S
Sbjct: 294 QI-GGEEAIRNYCFELAKKVGTFAADFFGTEVLENAAGTLTTAMVNIRLPVSEKWLNEAS 352
Query: 480 DSDALKL----RTHLREAFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDY 530
D L T+ E F VP + +G++ Y R+S QVYN++ DY
Sbjct: 353 AEDKEHLLQVINTYPLENFDTFVPPVF---HNGKL-----YIRLSCQVYNELSDY 399
>UniRef100_UPI0000234B94 UPI0000234B94 UniRef100 entry
Length = 470
Score = 135 bits (339), Expect = 4e-30
Identities = 101/336 (30%), Positives = 157/336 (46%), Gaps = 30/336 (8%)
Query: 144 DPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGIL-RSRTIIKDLVNA 202
DP+ +N+GSFG + V Q Q +PD F +++ G++ SR + L+N
Sbjct: 18 DPNYNNLNHGSFGTYPSQVLEKQQSIQKSLESRPDIF-IRYIQPGLIDTSRAALAPLLNV 76
Query: 203 DHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAG 262
+ ++ +V NATT VL N A T DV+ YGAV++++ A G
Sbjct: 77 P-VSDLVLVKNATTGVNTVLHNLALT---RTLTADDVIFYFDTVYGAVERALFALKESWG 132
Query: 263 GTVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKI 322
+ +V FP++ +V+ FR AL+ + G +LAV + V S P + P +E+ +
Sbjct: 133 VKLRKVKYVFPLEEGG-MVKRFREALKSVRKEGLTPKLAVFETVVSNPGIRFPFEEITRA 191
Query: 323 CREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKH------ 376
C+EEGV + +D AH++G +D+ +G DF+TSN HKW + P S A LY +
Sbjct: 192 CKEEGVLSL-IDGAHAVGMIKLDLAALGVDFFTSNCHKWLYTPRSCAVLYVPERNQKFIR 250
Query: 377 --------------PKGGSGSELHHP--VVSHEYGNGLAVESAWIGTRDYSAQLVVPAVI 420
P SG + P + + + + GT D SA VPA +
Sbjct: 251 TSLPTSWGYVPPQVPPSESGEDKDIPPSTLPNTGKSPFVALFEFTGTTDDSAYACVPAAL 310
Query: 421 DFVNRFEGGIEGIKKRNHETVVEMGDMLVKAWGTHV 456
+F + GG E I E G++L A GT V
Sbjct: 311 NFRDEVCGGEERIYAYLERLAGEAGELLASALGTDV 346
>UniRef100_UPI0000320766 UPI0000320766 UniRef100 entry
Length = 396
Score = 134 bits (338), Expect = 5e-30
Identities = 116/407 (28%), Positives = 188/407 (45%), Gaps = 43/407 (10%)
Query: 144 DPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNAD 203
DP +N+GSFG SV Q + P F+ + + + + +NAD
Sbjct: 16 DPETCFLNHGSFGATPVSVLEEQSRLRNLIESDPVRFFERDYMPMMKGAVGKLSEFINAD 75
Query: 204 HLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGG 263
+ + V N T VL++ +PGD +++ +++Y A +++ R+
Sbjct: 76 P-EGLVFVKNTTEGVNTVLRSL-------DLKPGDEIIVTNHSYQACWNAVDFVTERSDA 127
Query: 264 TVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKIC 323
+ V +PF VKS +++V L K GK V LA+ID VTS + +P ++LV+
Sbjct: 128 KTVVVDIPFRVKSEEEVVD-----LIMDKVTGKTV-LAMIDTVTSATGLRMPFEKLVRNI 181
Query: 324 REEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGS 383
+ GVD V +DAAH G +D+ + A + T N HKW P AFL+ R+ K
Sbjct: 182 QGAGVD-VLLDAAHGPGIVPLDLIAMDAAYCTGNCHKWICTPKGSAFLHIREDRK----- 235
Query: 384 ELHHPV-VSHEYG------NGLAVESAWIGTRDYSAQLVVPAVIDFV-NRFEGGIEGIKK 435
L P+ +SH + E W GT+D +A L +P I+F+ + +GG + I +
Sbjct: 236 SLVRPLSISHGHSFEGTDQEKFEYEFGWPGTQDPTAWLCIPHAIEFLGSLLDGGWQDIME 295
Query: 436 RNHETVVEMGDMLVKAWGTHVGCPPHMCASMVMVGLPTCLGVQSDSDALKL-----RTHL 490
N ++ +L A GT P M +S+ V +P + D + L L
Sbjct: 296 HNRALAIQGRKILCGALGTSPPVPDSMVSSIAAVEMPG----EGDVGPMSLEGDPYHNFL 351
Query: 491 REAFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDYYKFRDAV 537
+ FG++VP++ P R Y RIS Q+YN V++Y D +
Sbjct: 352 LDEFGIQVPVF--PWRHHNKR----YIRISAQLYNHVEEYEFLADCL 392
>UniRef100_UPI00003C1FB4 UPI00003C1FB4 UniRef100 entry
Length = 465
Score = 127 bits (320), Expect = 6e-28
Identities = 112/455 (24%), Positives = 207/455 (44%), Gaps = 56/455 (12%)
Query: 125 LSSSPFITPSEIQSEFSHHDPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNH 184
L S PF + DPS +NNGSFG C V + + R+PD F
Sbjct: 18 LLSKPFPGFGHALKPYYGFDPSYVPLNNGSFGACPIYVLDIIKELLDEAERRPDVFLRKQ 77
Query: 185 LKQGILRSRTIIKDLVNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLH 244
+ + +R + +V D D + V+NAT+ +L+ GT++ D +L+
Sbjct: 78 YQSLLDHARKEVAGIVRCDVADLV-FVNNATSGVNAILRGL-----NGTWKKRDAILVYD 131
Query: 245 YAYGAVKKSMEAYVTRAGGT----VIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRL 300
YGA K+ + Y+ + + ++ +PL +P+ + D+++ + + + +S G K+R+
Sbjct: 132 TVYGACGKTAQ-YIVDSNPSFELQLVTLPLSYPL-THDEVLAKTKQTILDAESKGIKIRV 189
Query: 301 AVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHK 360
++D ++S+P V++P +++ + R+ G+ + +D AH++G +++++ DF+ SN HK
Sbjct: 190 GIVDAISSVPGVIVPWEKITTLFRQHGILSL-IDGAHAVGQIPLNLRQADPDFFISNCHK 248
Query: 361 WFFCPPSIAFLYS-RKHPKGGSGSELHH----PVVSHEYGNGLAVESA---------WIG 406
W AFLY+ +++ + H P V L +A W G
Sbjct: 249 WLSAHRGCAFLYAPKRNQQFAQAIPTSHFYLSPNVPKSNAPDLIPTNAPSNWVATWEWTG 308
Query: 407 TRDYSAQLVVPAVIDFVNRFEGGIEGIKKRNHETVVEMGDMLVKAWG-----THVGCP-- 459
T D S L VPA I+F ++ GG + + + N + G+++ G + P
Sbjct: 309 TIDLSNYLSVPAAIEF-RKWMGGEDAVMQHNAQLARRAGEIVSSRLGKGSEVMEIETPTE 367
Query: 460 -PHMCASMVMVGLPTCL------------GVQSDSDALKLRTHLREAFGVEVPIYYRPPR 506
+ ASMV + +P L VQ D A KL+T L + + +RP
Sbjct: 368 SERLTASMVNISVPITLPKSSSNLSAEEVNVQLDVLATKLQTRLANEH--DTFVMFRPHA 425
Query: 507 DGEVEPVTGYARISHQVYNKVDDYYKFRDAVIQLV 541
+ + R+S QV+ + D+ D + Q++
Sbjct: 426 N------KIWIRLSAQVWLEEKDFEWVADKIAQMI 454
>UniRef100_UPI000004DFD4 UPI000004DFD4 UniRef100 entry
Length = 421
Score = 127 bits (318), Expect = 1e-27
Identities = 110/413 (26%), Positives = 190/413 (45%), Gaps = 36/413 (8%)
Query: 140 FSHHDPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDL 199
F++ D V +N+GS+G V D +K D F +K + S + +
Sbjct: 17 FTNMDDEVFPVNHGSYGLTPTPVLEKYLDLIVKNASYTDKFMKYGIKDSYVESLKAVGRV 76
Query: 200 VNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVT 259
+N D+ ++ VDNAT+ +L R + GD +++ YGA +++
Sbjct: 77 LNCDY-HNLAFVDNATSGVNTIL-------RSYPLKKGDKLVIQSTVYGACGNTVKFLHD 128
Query: 260 RAGGTVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKEL 319
R G I V L +P+ + ++I+ +F +K +L + D ++SMP VV P +++
Sbjct: 129 RYGVEFIVVDLNYPI-TDEEILSKFERVFVE-----EKPKLCMFDTISSMPGVVFPYEKM 182
Query: 320 VKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLY-SRKH-- 376
K+C++ V + +D AH IGC D+ + DFY +NLHKWF+ P A LY KH
Sbjct: 183 TKLCKKYSVLSL-IDGAHGIGCIPQDLGNLKPDFYVTNLHKWFYIPFGCAVLYIDPKHHN 241
Query: 377 -----PKGGSGSELHHPVVSHEYGNGLAVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIE 431
P S E H + + N L GT+++++ V+P I F + G
Sbjct: 242 VVHTLPISHSYLEDHVKLSDGDQKNRLIDRFFLYGTKNFASIQVIPEAIKFRSEVCDGET 301
Query: 432 GIKKRNHETVVEMGDMLVKAWGTHV--GCPPHMCASMVMVGLPTCLGVQSDSDALKLRTH 489
I H ++G+++ + WGT + ++MV V +PT + + + H
Sbjct: 302 KIYDYCHGLAKQVGELVSRKWGTSYLDQKGSTLISTMVTVEVPTQDYPEIVKNWSVIDHH 361
Query: 490 L-REAFGVEVPIYYRP--PRDGEVEPVTGYARISHQVYNKVDDYYKFRDAVIQ 539
+ + F + Y P +G++ +AR S QVYN++ DY D V++
Sbjct: 362 VYNKMFDRKA---YTPCIAHNGKL-----FARFSCQVYNELSDYDYASDVVLE 406
>UniRef100_O94021 Hypothetical protein Ca49C10.07c [Candida albicans]
Length = 421
Score = 126 bits (316), Expect = 2e-27
Identities = 109/413 (26%), Positives = 190/413 (45%), Gaps = 36/413 (8%)
Query: 140 FSHHDPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDL 199
F++ D V +N+GS+G V D +K D F +K + S + +
Sbjct: 17 FTNMDDEVFPVNHGSYGLTPTPVLEKYLDLIVKNASYTDKFMKYGIKDSYVESLKAVGKV 76
Query: 200 VNADHLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVT 259
+N D+ ++ VDNAT+ +L R + D +++ YGA +++
Sbjct: 77 LNCDY-HNLAFVDNATSGVNTIL-------RSYPLKKVDKLVIQSTVYGACGNTVKFLHD 128
Query: 260 RAGGTVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKEL 319
R G I V L +P+ + ++I+ +F +K +L + D ++SMP V+ P +++
Sbjct: 129 RYGVEFIVVDLNYPI-TDEEILSKFERVFVE-----EKPKLCMFDTISSMPGVIFPYEKM 182
Query: 320 VKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLY-SRKH-- 376
K+C++ V + +D AH IGC D+ + DFY +NLHKWF+ P A LY KH
Sbjct: 183 TKLCKKYSVLSL-IDGAHGIGCIPQDLGNLKPDFYVTNLHKWFYIPFGCAVLYIDPKHHN 241
Query: 377 -----PKGGSGSELHHPVVSHEYGNGLAVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIE 431
P S E H + + N L GT+++++ V+P I F + GG
Sbjct: 242 VVHTLPISHSYLEDHVKLSDGDQKNRLIDRFFLYGTKNFASIQVIPEAIKFRSEVCGGET 301
Query: 432 GIKKRNHETVVEMGDMLVKAWGTHV--GCPPHMCASMVMVGLPTCLGVQSDSDALKLRTH 489
I H ++G+++ + WGT + ++MV V +PT + + + H
Sbjct: 302 KIYDYCHGLAKQVGELVSRKWGTSYLDQKGSTLISTMVTVEVPTQDYPEIVKNWSVIDHH 361
Query: 490 L-REAFGVEVPIYYRP--PRDGEVEPVTGYARISHQVYNKVDDYYKFRDAVIQ 539
+ + F + Y P +G++ +AR S QVYN++ DY D V++
Sbjct: 362 VYNKMFDRKA---YTPCIAHNGKL-----FARFSCQVYNELSDYDYASDVVLE 406
>UniRef100_UPI000021A892 UPI000021A892 UniRef100 entry
Length = 449
Score = 125 bits (314), Expect = 3e-27
Identities = 95/322 (29%), Positives = 155/322 (47%), Gaps = 19/322 (5%)
Query: 144 DPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNAD 203
DP +N+GSFG V AA+ +Q K +PD + + Q + SR + L+ D
Sbjct: 36 DPKFRNMNHGSFGVIPRPVHAARRYYQDKSEERPDVWIRYNWSQLLEGSRAAVAPLLGVD 95
Query: 204 HLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGG 263
D I+ V NAT VL+N W D +L + Y A K+++ + + G
Sbjct: 96 K-DTIAFVPNATVGVNTVLRNLVW-----NDDKKDEILYFNTIYAACGKTVQYMIEISRG 149
Query: 264 TVI--EVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVK 321
V VPL +P+ + D++V F+ ++ ++ GK+ R AVID V+S+P V +P + LV+
Sbjct: 150 HVSGRSVPLEYPL-TDDELVALFKKGIQDCRAAGKRPRAAVIDTVSSIPAVRLPFEALVQ 208
Query: 322 ICREEGVDRVFVDAAHSIGCTDVDMKEIGA----DFYTSNLHKWFFCPPSIAFLYSRKHP 377
+C +EG+ + VD A +G +D+K +G DF+ +N HKW + P A L+ KH
Sbjct: 209 VCHDEGILSI-VDGAQGVGM--IDLKHLGTQVKPDFFITNCHKWLYTPRGCAVLHVPKHN 265
Query: 378 KGGSGSELHHP---VVSHEYGNGLAVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIEGIK 434
+ S L V S E A+ T D S + V + ++ GG + +
Sbjct: 266 QALMRSTLPTSWGWVPSGEGDPDFIDNFAFASTLDNSNYMAVQHAVQWIQEALGGEDAVI 325
Query: 435 KRNHETVVEMGDMLVKAWGTHV 456
+ + G+M+ + GT V
Sbjct: 326 EYMMSLNKKGGNMVAEMLGTKV 347
>UniRef100_Q7SBF8 Hypothetical protein [Neurospora crassa]
Length = 1356
Score = 123 bits (309), Expect = 1e-26
Identities = 120/430 (27%), Positives = 186/430 (42%), Gaps = 59/430 (13%)
Query: 144 DPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNAD 203
DP+ +N+GSFG +++ +Q +P F + SR + +L+
Sbjct: 37 DPAYRNLNHGSFGTIPSAIQQKLRSYQTAAEARPCPFLRYQTPVLLDESRAAVANLLKVP 96
Query: 204 HLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGG 263
++ + V NAT VL+N W+ +G D +L YGA K+++ + G
Sbjct: 97 -VETVVFVANATMGVNTVLRNIVWS-ADGK----DEILYFDTIYGACGKTIDYVIEDKRG 150
Query: 264 TVIE--VPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVK 321
V +PL +P + DD+V FR A+++ + GK+ RLAVID V+SMP V P +++VK
Sbjct: 151 IVSSRCIPLIYPAED-DDVVAAFRDAIKKSREEGKRPRLAVIDVVSSMPGVRFPFEDIVK 209
Query: 322 ICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLY---SRKHPK 378
IC+EE + VD A IG D+ + E DF SN HKW F P A Y +H
Sbjct: 210 ICKEEEIISC-VDGAQGIGMVDLKITETDPDFLISNCHKWLFTPRGCAVFYVPVRNQHLI 268
Query: 379 GGSGSELHH--PVVSHEY------GNGLAVES--AWIGTRDYSAQLVVPAVIDFVNRFEG 428
+ H P V + + GN A S ++GT D S V I + G
Sbjct: 269 RSTLPTSHGFVPQVGNRFNPLVPAGNKSAFVSNFEFVGTVDNSPFFCVKDAIKWREEVLG 328
Query: 429 GIEGIKKRNHETVVEMGDMLVKAWGTHV---GCPPHMCASMVMVGLPTCLG--------- 476
G E I + + E G + + GT V + +MV + LP +G
Sbjct: 329 GEERIMEYMTKLAREGGQKVAEILGTRVLENSTGTLIRCAMVNIALPFVVGEDPKAPVKL 388
Query: 477 ---VQSDSDALKLRTH-------------LREAFGVEVPIYYRPPRDGEVEPVTGYARIS 520
+ D + L H L++ F VP+ + R +AR+S
Sbjct: 389 TEKEEKDVEGLYEIPHEEANMAFKWMYNVLQDEFNTFVPMTFHRRR--------FWARLS 440
Query: 521 HQVYNKVDDY 530
QVY ++ D+
Sbjct: 441 AQVYLEMSDF 450
>UniRef100_UPI0000277927 UPI0000277927 UniRef100 entry
Length = 293
Score = 123 bits (308), Expect = 2e-26
Identities = 84/299 (28%), Positives = 150/299 (50%), Gaps = 25/299 (8%)
Query: 240 VLMLHYAYGAVKKSMEAYVTRAGGTVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVR 299
+L+ + Y A +++ R+G + VPLPFP+++ ++ + S KK +
Sbjct: 1 ILVPDHTYQACWNTVDFITKRSGAKTVIVPLPFPIENPEEATKAILSHTT------KKTK 54
Query: 300 LAVIDHVTSMPCVVIPVKELVKICREEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLH 359
LA+ID VTS + +P +++V + +G+D V +DAAH G +++K++ + T N H
Sbjct: 55 LALIDTVTSPTGIRLPFEQIVFELQSKGID-VLLDAAHGPGIVPLNLKKLKPAYATGNAH 113
Query: 360 KWFFCPPSIAFLYSRKHPKGGSGSELHHPVVSHEY---GN---GLAVESAWIGTRDYSAQ 413
KW P AFLY R+ + SE++ +SH + GN + +E W GT+D +
Sbjct: 114 KWLCTPKGAAFLYIREDKR----SEINPLSISHGFSVDGNVDEKIRLEFDWTGTQDITPW 169
Query: 414 LVVPAVIDFVNRF-EGGIEGIKKRNHETVVEMGDMLVKAWGTHVGCPPHMCASMVMVGLP 472
L +P I+ VN EGG I + N +E +++++ T CP M + V LP
Sbjct: 170 LCIPKSIEHVNSLVEGGWAEIMEHNTNLAIEARNIILEVVETPKLCPDSMIVGLSAVALP 229
Query: 473 -TCLGVQSDSDALKLRTHLREAFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDY 530
+ ++ + L T L E + ++VP++ P + Y R + +YN +++Y
Sbjct: 230 GEGVSTKTVLEPDPLHTLLYEKYNIQVPVFSWPHHNRR------YLRTASHLYNSLEEY 282
>UniRef100_UPI0000281D0D UPI0000281D0D UniRef100 entry
Length = 378
Score = 120 bits (301), Expect = 1e-25
Identities = 107/410 (26%), Positives = 174/410 (42%), Gaps = 56/410 (13%)
Query: 144 DPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNAD 203
D +N+GS+G C V WQ QP F + + SR + V +
Sbjct: 11 DSETIFLNHGSYGACPRPVFEEYQKWQRMLEIQPVRFQTETVYSALKDSRVALGKFVGCE 70
Query: 204 HLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGG 263
E+ N TTA + VL N + D +LM + YGA+ ++ A R G
Sbjct: 71 E-GELLFFQNPTTAISNVLYNLE-------LKSEDEILMTDHEYGALVRAWNALGRRTGA 122
Query: 264 TVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKIC 323
+I+ +P + + +D + F K +K ++ I +TS ++ P+K+++K
Sbjct: 123 NIIQQNIPTNLVTEEDFINAF------WKGVTEKTKVIFISQITSPTALIFPIKKIIKRA 176
Query: 324 REEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGS 383
+E G+ + VD AH G D+ + ++G DFYT LHKW P +FL+ +K
Sbjct: 177 KEIGILTI-VDGAHVPGQLDLKINDLGCDFYTGALHKWLCAPKGTSFLFVKKE------- 228
Query: 384 ELHH----PVVSHEYGNG-------LAVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIEG 432
HH PV+ NG + W GTRD S+ L +P I+F + I+
Sbjct: 229 --HHSWMKPVIYSWGKNGDDPGPTEFLQDFQWQGTRDMSSFLTIPKAIEF---YHECIKP 283
Query: 433 IKKRNHETVVEMGDMLVKAWGTHVGCPPHMCASMV--MVGLPTCLGVQSDSDALKLRTHL 490
++++ + +E A GT P A + MV P + + D + L H
Sbjct: 284 LQEKCRQINLEAYSKFQVALGT---SPLSEGAGWIGQMVSHPLPIDMPEDLKKILLDDH- 339
Query: 491 REAFGVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDYYKFRDAVIQL 540
+E+PI+ E + ++ RIS Q+YN D A+ L
Sbjct: 340 ----KIEIPIF-------EWKGLS-LIRISIQIYNTKKDVESLMSALASL 377
>UniRef100_UPI000025CD0A UPI000025CD0A UniRef100 entry
Length = 377
Score = 117 bits (293), Expect = 9e-25
Identities = 101/403 (25%), Positives = 173/403 (42%), Gaps = 47/403 (11%)
Query: 144 DPSVARINNGSFGCCSASVTAAQHDWQLKYLRQPDNFYFNHLKQGILRSRTIIKDLVNAD 203
+ ++ +N+GSFG C SV +WQ QP F+ + + + + SRT + +L+ D
Sbjct: 8 EKNIIFLNHGSFGACPKSVFKNYQNWQRLLENQPVQFFSHDVYKYLEESRTHLSNLITCD 67
Query: 204 HLDEISIVDNATTAAAIVLQNTAWAFREGTFQPGDVVLMLHYAYGAVKKSMEAYVTRAGG 263
D+I + N TTA V+ R VL ++ YGA+ ++ E + G
Sbjct: 68 S-DDIIFIPNPTTAINTVI-------RSLMIDKNYEVLTSNHEYGAMIRAWEWFSKNKGY 119
Query: 264 TVIEVPLPFPVKSSDDIVREFRSALERGKSGGKKVRLAVIDHVTSMPCVVIPVKELVKIC 323
T+ + L P + D +F L G + KK ++ I H+TS ++ PV+E+
Sbjct: 120 TLTQKHLRIPFTTRD----KFLDNLWEGIT--KKTKIIFISHITSPTAIIFPVEEICHRA 173
Query: 324 REEGVDRVFVDAAHSIGCTDVDMKEIGADFYTSNLHKWFFCPPSIAFLYSRKHPKGGSGS 383
R EG+ +D AH G +D+ + D Y HKW P +F+Y +K +
Sbjct: 174 RNEGI-LTIIDGAHVPGHIQLDISALDPDIYIGACHKWLSAPKGSSFMYVKKDTQ----- 227
Query: 384 ELHHPVVSHEYG-------NGLAVESAWIGTRDYSAQLVVPAVIDFVNRFEGGIEGIKKR 436
++ P++ +G + E+ + GTRD+S L VP I+F N + +K R
Sbjct: 228 DMIDPLII-SWGKEVDPSPSTFINENQYQGTRDFSPYLAVPTAINFQN--DNNWNLVKAR 284
Query: 437 NHETVVEMGDMLVKAWGTHVGCP--PHMCASMVMVGLPTCLGVQSDSDALKLRTHLREAF 494
+ + L T + CP A M + +P +D+ ++ L +
Sbjct: 285 CKDLTKKTLISLQNILKTDLLCPINDDWLAQMASIQIPV-------NDSTAIKNKLLNNY 337
Query: 495 GVEVPIYYRPPRDGEVEPVTGYARISHQVYNKVDDYYKFRDAV 537
+E+PI+ ++ R S YN D DA+
Sbjct: 338 NIEIPIFKWDNKN--------MLRFSFNAYNDESDSIALIDAI 372
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 968,069,146
Number of Sequences: 2790947
Number of extensions: 41616876
Number of successful extensions: 140074
Number of sequences better than 10.0: 911
Number of HSP's better than 10.0 without gapping: 128
Number of HSP's successfully gapped in prelim test: 784
Number of HSP's that attempted gapping in prelim test: 139092
Number of HSP's gapped (non-prelim): 1088
length of query: 552
length of database: 848,049,833
effective HSP length: 132
effective length of query: 420
effective length of database: 479,644,829
effective search space: 201450828180
effective search space used: 201450828180
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0206a.3


