

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0204.2
(664 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
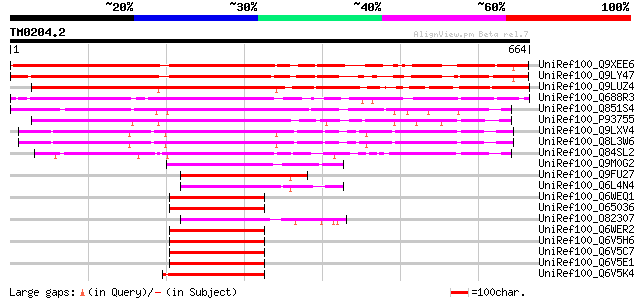
UniRef100_Q9XEE6 Hypothetical Cys-3-His zinc finger protein [Ara... 622 e-176
UniRef100_Q9LY47 Hypothetical protein F27K19_160 [Arabidopsis th... 599 e-170
UniRef100_Q9LUZ4 Zinc finger transcription factor-like protein [... 591 e-167
UniRef100_Q688R3 Putative finger transcription factor [Oryza sat... 489 e-136
UniRef100_Q851S4 Hypothetical protein OSJNBb0017F17.19 [Oryza sa... 459 e-127
UniRef100_P93755 Putative CCCH-type zinc finger protein [Arabido... 442 e-122
UniRef100_Q9LXV4 Zinc finger transcription factor-like protein [... 439 e-121
UniRef100_Q8L3W6 Zinc finger transcription factor-like protein [... 439 e-121
UniRef100_Q84SL2 CCCH-type zinc finger protein-like protein [Ory... 368 e-100
UniRef100_Q9M0G2 Hypothetical protein AT4g29190 [Arabidopsis tha... 201 7e-50
UniRef100_Q9FU27 CCCH-type zinc finger protein-like [Oryza sativa] 199 2e-49
UniRef100_Q6L4N4 Putative zinc finger transcription factor [Oryz... 193 2e-47
UniRef100_Q6WEQ1 Zing finger transcription factor PEI1 [Boechera... 189 2e-46
UniRef100_O65036 Zinc finger transcription factor [Arabidopsis t... 189 3e-46
UniRef100_O82307 Putative CCCH-type zinc finger protein [Arabido... 187 9e-46
UniRef100_Q6WER2 Zing finger transcription factor PEI1 [Arabidop... 187 1e-45
UniRef100_Q6V5H6 Cu2+ plastocyanin-like [Arabidopsis arenosa] 187 1e-45
UniRef100_Q6V5C7 Zn-finger transcription factor [Arabidopsis are... 187 1e-45
UniRef100_Q6V5E1 Zn-finger transcription factor [Olimarabidopsis... 186 2e-45
UniRef100_Q6V5K4 Zinc finger transcription factor [Brassica oler... 185 3e-45
>UniRef100_Q9XEE6 Hypothetical Cys-3-His zinc finger protein [Arabidopsis thaliana]
Length = 597
Score = 622 bits (1603), Expect = e-176
Identities = 344/686 (50%), Positives = 437/686 (63%), Gaps = 114/686 (16%)
Query: 1 MCSGSKSKLSSSEQVMESQ--KENCDGLHNFSILLELSASNDFEALKREVDEK-GLDVNE 57
MC G+KS L SS+ + E + ++ + + + LLE +A +D + KRE++E ++++E
Sbjct: 1 MC-GAKSNLCSSKTLTEVEFMRQKSEDGASATCLLEFAACDDLSSFKREIEENPSVEIDE 59
Query: 58 AGFWYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCA 117
+GFWY RR+GSKKMG E+RTPLM+A+++GS V+ Y++ TGR DVN+ C +K TALHCA
Sbjct: 60 SGFWYCRRVGSKKMGFEERTPLMVAAMYGSMEVLNYIIATGRSDVNRVCSDEKVTALHCA 119
Query: 118 VAGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGES 177
V+G S +E++K+LLDA + + +DA GNKP++L+A RKA+E+LL G
Sbjct: 120 VSGCSVSIVEIIKILLDASASPNCVDANGNKPVDLLAKDSRFVPNQSRKAVEVLLTGIHG 179
Query: 178 GQLIHQEMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRA 237
+ +E +L+ + +YP D SLPDIN G+YG+D+FRM+SFKVKPCSRA
Sbjct: 180 SVMEEEEEELKSVVT-----------KYPADASLPDINEGVYGTDDFRMFSFKVKPCSRA 228
Query: 238 YSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPA 297
YSHDWTECPFVHPGENARRRDPRKYPY+CVPCPEFRKG+C KGDSCEYAHGVFESWLHPA
Sbjct: 229 YSHDWTECPFVHPGENARRRDPRKYPYTCVPCPEFRKGSCPKGDSCEYAHGVFESWLHPA 288
Query: 298 QYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALS 357
QYRTRLCKDETGC R+VCFFAHR +ELRPV ASTGSAM SP+S S+ + +MSVMSPL L
Sbjct: 289 QYRTRLCKDETGCARRVCFFAHRRDELRPVNASTGSAMVSPRS-SNQSPEMSVMSPLTLG 347
Query: 358 SSSLPMSTVSTPPMSPLAGS--LSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDF 415
SS P SP+A LSP++G LWQN++N +LTPP LQL GSRLK+ LSARD
Sbjct: 348 SS---------PMNSPMANGVPLSPRNGGLWQNRVN-SLTPPPLQLNGSRLKSTLSARDM 397
Query: 416 DLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNL 475
D+EME+ G R R+GDL P+NL++ GS DS
Sbjct: 398 DMEMELRFRGLDNR-------------------------RLGDLKPSNLEETFGSYDSAS 432
Query: 476 LSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVA 535
+ Q LQS SRH MNH PSSPVR+P GF+SS+A+A
Sbjct: 433 VMQ------------LQSPSRH----SQMNH--------YPSSPVRQPPPHGFESSAAMA 468
Query: 536 AAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVN 595
AAVMN+RS+AFAKRS SF V+S SDW SP+GKL+WG+
Sbjct: 469 AAVMNARSSAFAKRSLSF------------------KPAPVASNVSDWGSPNGKLEWGMQ 510
Query: 596 GDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKDDL------------ 643
DEL+KLR+SASFG N P A EPDVSWV+SLVK++
Sbjct: 511 RDELNKLRRSASFGIHGNNNNSVSRP-ARDYSDEPDVSWVNSLVKENAPERVNERVGNTV 569
Query: 644 ------SKEMLPPWVEQLYIEQEQMV 663
K LP W EQ+YI+ EQ +
Sbjct: 570 NGAASRDKFKLPSWAEQMYIDHEQQI 595
>UniRef100_Q9LY47 Hypothetical protein F27K19_160 [Arabidopsis thaliana]
Length = 586
Score = 599 bits (1545), Expect = e-170
Identities = 338/679 (49%), Positives = 429/679 (62%), Gaps = 116/679 (17%)
Query: 1 MCSGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGF 60
MCSG KS L SS + E + + +LLE +A +D ++ KREV+EKGLD++E+G
Sbjct: 7 MCSGPKSNLCSSRTLTEIESRQKE--EETMLLLEFAACDDLDSFKREVEEKGLDLDESGL 64
Query: 61 WYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAG 120
WY RR+GSKKMG E+RTPLM+A+++GS +V+ ++V TG+ DVN+ACG ++ T LHCAVAG
Sbjct: 65 WYCRRVGSKKMGLEERTPLMVAAMYGSIKVLTFIVSTGKSDVNRACGEERVTPLHCAVAG 124
Query: 121 GSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESGQL 180
S +EV+ VLLDA + +S+DA GN+P+++ + RRKA+ELLLRGG G L
Sbjct: 125 CSVNMIEVINVLLDASALVNSVDANGNQPLDVFVRVSRFVASPRRKAVELLLRGGGVGGL 184
Query: 181 IHQ--EMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAY 238
I + E +++++S +YP D SLPDIN G+YGSDEFRMYSFKVKPCSRAY
Sbjct: 185 IDEAVEEEIKIVS------------KYPADASLPDINEGVYGSDEFRMYSFKVKPCSRAY 232
Query: 239 SHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQ 298
SHDWTEC FVHPGENARRRDPRKYPY+CVPCPEFRKG+C KGDSCEYAHGVFESWLHPAQ
Sbjct: 233 SHDWTECAFVHPGENARRRDPRKYPYTCVPCPEFRKGSCPKGDSCEYAHGVFESWLHPAQ 292
Query: 299 YRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSS 358
Y+TRLCKDETGC RKVCFFAH+ EE+RPV ASTGSA+ +S S+ M +SPLA SS
Sbjct: 293 YKTRLCKDETGCARKVCFFAHKREEMRPVNASTGSAV--AQSPFSSLEMMPGLSPLAYSS 350
Query: 359 SSLPMSTVSTPPMSPLAGSL--SPKSGNLWQNKINLNLTPPSLQL-PGSRLKTALSARDF 415
VSTPP+SP+A + SP++G WQN++N LTPP+LQL GSRLK+ LSARD
Sbjct: 351 G------VSTPPVSPMANGVPSSPRNGGSWQNRVN-TLTPPALQLNGGSRLKSTLSARDI 403
Query: 416 DLEMEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSNL 475
D+EMEM + F N N+++ GS
Sbjct: 404 DMEMEM------------------------ELRLRGFGN--------NVEETFGS----- 426
Query: 476 LSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRK-PSSLGFDSSSAV 534
++ S SR+ M QNMN + PSSPVR+ PS GF+SS+A
Sbjct: 427 --------------YVSSPSRNSQMGQNMN-------QHYPSSPVRQPPSQHGFESSAAA 465
Query: 535 AAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGV 594
A AVM +RS AFAKRS SF P+ + S SDW SP+GKL+WG+
Sbjct: 466 AVAVMKARSTAFAKRSLSF--------------KPATQAA-PQSNLSDWGSPNGKLEWGM 510
Query: 595 NGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKDDL----------S 644
G+EL+K+R+S SFG N + A EPDVSWV+SLVKD
Sbjct: 511 KGEELNKMRRSVSFGIHGN----NNNNAARDYRDEPDVSWVNSLVKDSTVVSERSFGMNE 566
Query: 645 KEMLPPWVEQLYIEQEQMV 663
+ + W EQ+Y E+EQ V
Sbjct: 567 RVRIMSWAEQMYREKEQTV 585
>UniRef100_Q9LUZ4 Zinc finger transcription factor-like protein [Arabidopsis
thaliana]
Length = 607
Score = 591 bits (1523), Expect = e-167
Identities = 327/647 (50%), Positives = 412/647 (63%), Gaps = 61/647 (9%)
Query: 29 FSILLELSASNDFEALKREVDEKGLD-VNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGS 87
FS+LLE SA ND K V+E+GL+ ++ +G WYGRR+GSKKMG E+RTPLMIA+LFGS
Sbjct: 11 FSLLLESSACNDLSGFKSLVEEEGLESIDGSGLWYGRRLGSKKMGFEERTPLMIAALFGS 70
Query: 88 TRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGN 147
VV Y++ TG VDVN++CGSD ATALHCAV+G S SLE+V +LL ++ DS DA GN
Sbjct: 71 KEVVDYIISTGLVDVNRSCGSDGATALHCAVSGLSANSLEIVTLLLKGSANPDSCDAYGN 130
Query: 148 KPMNLIAPAFNSSSKSRRKAMELLLRGGESGQLIHQEMDLEL---ISAPFSSKEGSDKKE 204
KP ++I P + +R K +E LL+G + ++ + + E + S GS++KE
Sbjct: 131 KPGDVIFPCLSPVFSARMKVLERLLKGNDDLNEVNGQEESEPEVEVEVEVSPPRGSERKE 190
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
YPVD +LPDI NG+YG+DEFRMY+FK+KPCSRAYSHDWTECPFVHPGENARRRDPRKY Y
Sbjct: 191 YPVDPTLPDIKNGVYGTDEFRMYAFKIKPCSRAYSHDWTECPFVHPGENARRRDPRKYHY 250
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
SCVPCPEFRKG+C +GD+CEYAHG+FE WLHPAQYRTRLCKDET C R+VCFFAH+PEEL
Sbjct: 251 SCVPCPEFRKGSCSRGDTCEYAHGIFECWLHPAQYRTRLCKDETNCSRRVCFFAHKPEEL 310
Query: 325 RPVYASTGSAMPSPKS-----YSSNAVDMSVMSPLALSSSSLPMSTVSTPPMSPLAGSLS 379
RP+Y STGS +PSP+S SS A DM +SP LP+ +TPP+SP S
Sbjct: 311 RPLYPSTGSGVPSPRSSFSSCNSSTAFDMGPISP-------LPIGATTTPPLSPNGVSSP 363
Query: 380 PKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTRQQQQQQQQQQ 439
G W N N+TPP+LQLPGSRLK+AL+AR+ D EM L SPT
Sbjct: 364 IGGGKTWMNW--PNITPPALQLPGSRLKSALNAREIDFSEEMQSLTSPTTWNNTPMSSPF 421
Query: 440 LIEEIARISSPSFRNRIGDLTPTN-LDDLLGSVDSNLLSQLHGLSGPSTPTHLQSQSRHQ 498
+ + R++ G ++P N L D+ G+ D+ GL Q R
Sbjct: 422 SGKGMNRLAG-------GAMSPVNSLSDMFGTEDNT-----SGL-----------QIRRS 458
Query: 499 MMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVMNSRSAAFAK-RSQSFIDRG 557
++ + + +++ SSPV S DSS AV+ SR+A FAK RSQSFI+R
Sbjct: 459 VINPQL------HSNSLSSSPVGANSLFSMDSS-----AVLASRAAEFAKQRSQSFIERN 507
Query: 558 AAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAP 617
HH +SS ++ +DW S GKLDW V GDEL KLRKS SF R GM
Sbjct: 508 NGLNHHPAISS------MTTTCLNDWGSLDGKLDWSVQGDELQKLRKSTSFRLRAGGM-E 560
Query: 618 TGSPMAHSEHAEPDVSWVHSLVKDDLSKEMLPPWVEQLYIEQEQMVA 664
+ P + EPDVSWV LVK+ + P W+EQ Y+E EQ VA
Sbjct: 561 SRLPNEGTGLEEPDVSWVEPLVKEPQETRLAPVWMEQSYMETEQTVA 607
>UniRef100_Q688R3 Putative finger transcription factor [Oryza sativa]
Length = 601
Score = 489 bits (1259), Expect = e-136
Identities = 296/676 (43%), Positives = 389/676 (56%), Gaps = 87/676 (12%)
Query: 1 MCSGSKSKLSSSEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGF 60
MCSG + S+ + + QKE + S+LLEL+A++D A++R V+E+ + + AG
Sbjct: 1 MCSGPRKP--STPPLPQQQKEAT--VMAASLLLELAAADDVAAVRRVVEEEKVSLGVAGL 56
Query: 61 WYGRRI-GSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVA 119
WYG G ++G E+RT M+A+L+GST V+ Y+V + +A +D AT LH A A
Sbjct: 57 WYGPSASGVARLGMERRTAAMVAALYGSTGVLGYVVAAAPAEAARASETDGATPLHMAAA 116
Query: 120 GGSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESGQ 179
GG+ ++ ++LL AG+ D+L A G + +L+ A + KA+ LLL+ S
Sbjct: 117 GGAANAVAATRLLLAAGASVDALSASGLRAGDLLPRA-----TAAEKAIRLLLK---SPA 168
Query: 180 LIHQEMDLELISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYS 239
+ + S P KKEYP D++LPD+ +GL+ +DEFRMYSFKVKPCSRAYS
Sbjct: 169 VSPSSSPKKSASPPSPPPPQEAKKEYPPDLTLPDLKSGLFSTDEFRMYSFKVKPCSRAYS 228
Query: 240 HDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKG-ACQKGDSCEYAHGVFESWLHPAQ 298
HDWTECPFVHPGENARRRDPR+Y YSCVPCPEFRKG +C+KGD+CEYAHGVFE WLHPAQ
Sbjct: 229 HDWTECPFVHPGENARRRDPRRYSYSCVPCPEFRKGGSCRKGDACEYAHGVFECWLHPAQ 288
Query: 299 YRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSS 358
YRTRLCKDE GC R++CFFAH+P+ELR V S S P +V SP +
Sbjct: 289 YRTRLCKDEVGCARRICFFAHKPDELRAVNPSAVSVGMQP----------TVSSPRSSPP 338
Query: 359 SSLPMSTVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLE 418
+ L M+ + MSP W + P SRLKTAL AR+ D +
Sbjct: 339 NGLDMAAAAAAMMSPA-----------WPSS------------PASRLKTALGARELDFD 375
Query: 419 MEMLGLGSPTRQQQQQQQQQQLIEEIARISSP-----SFRNRIGDLTPT----NLDDLLG 469
+EML L Q QQ+L ++++ SP + N + +P + DLLG
Sbjct: 376 LEMLAL---------DQYQQKLFDKVSGAPSPRASWGAAANGLATASPARAVPDYTDLLG 426
Query: 470 SVDSNLLSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFD 529
SVD +LSQLH LS S Q +M P+SP+ ++ F
Sbjct: 427 SVDPAMLSQLHALSLKQAGDMPAYSSMADTTQMHM-----------PTSPMVGGANTAFG 475
Query: 530 SSSAVAAAVMNSRSAAFAKRSQSFIDRGAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGK 589
++A A+M+SR++AFAKRSQSFIDRG + SP+ S SDW SP GK
Sbjct: 476 LDHSMAKAIMSSRASAFAKRSQSFIDRGGRAPAARSLMSPATTGA--PSILSDWGSPDGK 533
Query: 590 LDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKD-DLSKEML 648
LDWGV GDEL KLRKSASF FR P + H+ AEPDVSWV+SLVKD + ++
Sbjct: 534 LDWGVQGDELHKLRKSASFAFRGQSAMPVAT---HAAAAEPDVSWVNSLVKDGHAAGDIF 590
Query: 649 PPWVEQLYIEQEQMVA 664
W EQEQMVA
Sbjct: 591 AQWP-----EQEQMVA 601
>UniRef100_Q851S4 Hypothetical protein OSJNBb0017F17.19 [Oryza sativa]
Length = 749
Score = 459 bits (1181), Expect = e-127
Identities = 290/695 (41%), Positives = 390/695 (55%), Gaps = 87/695 (12%)
Query: 1 MCSGSKSKLSSSEQVMESQKE-NCDGLHNFSILLELSASNDFEALKREVDEKG-LDVNEA 58
+ + +K +++ V E K D F+ LLEL+A +D E L+R ++ +EA
Sbjct: 23 LAAATKKSAAAAAAVAEMAKTLTVDTDDAFAGLLELAADDDAEGLRRALERAPPAAADEA 82
Query: 59 GFWYGRRIGSKKMGSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAV 118
G WYGRR E RTPLM+A+ +GS V++ L+ VDVN+ CGSD TALHCA
Sbjct: 83 GLWYGRR-----KVLEHRTPLMVAATYGSLAVLRLLLSLPSVDVNRRCGSDGTTALHCAA 137
Query: 119 AGGSELSLEVVKVLLDAGSDADSLDACGNKPMNLIAPAFNSSSKSRRKAMELLLRGGESG 178
+GGS +E VK+LL AG+DAD+ DA G +P ++I+ + A++ LL ++G
Sbjct: 138 SGGSPSCVEAVKLLLAAGADADATDASGYRPADVISVP--PKMFDAKIALQDLLGCPKAG 195
Query: 179 QLIHQEMD------LELISAPFSSKEGS--------------------DKKEYPVDVSLP 212
+ + + L +S+P + S +KKEYPVD SLP
Sbjct: 196 HGVLRVVTRAANSMLSPVSSPTAEDARSPSAAVMMTTKFADLPRVVTSEKKEYPVDPSLP 255
Query: 213 DINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEF 272
DI N +Y SDEFRMYSFK++PCSRAYSHDWTECPFVHPGENARRRDPRKY YSCVPCP+F
Sbjct: 256 DIKNSIYASDEFRMYSFKIRPCSRAYSHDWTECPFVHPGENARRRDPRKYHYSCVPCPDF 315
Query: 273 RKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTG 332
RKG C++GD CEYAHGVFE WLHPAQYRTRLCKD T C R+VCFFAH +ELRP+Y STG
Sbjct: 316 RKGVCRRGDMCEYAHGVFECWLHPAQYRTRLCKDGTSCNRRVCFFAHTTDELRPLYVSTG 375
Query: 333 SAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVS--TPPMSPLAGSLSPKSGNLWQNKI 390
SA+PSP++ ++ ++M+ L S S + +S TPPMSP + P G WQ
Sbjct: 376 SAVPSPRASATATMEMAAAMGLMPGSPSSVSAVMSPFTPPMSPSGNGMPPSLG--WQQPN 433
Query: 391 NLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSPTRQQQQQQQQQQLIEEI--ARI- 447
L P L SRL+T+LSARD + L Q QLI ++ +RI
Sbjct: 434 VPTLHLPGSSLQSSRLRTSLSARDMPADDYSL----------MQDIDSQLINDLCYSRIG 483
Query: 448 -SSPSFRNRIGDLTPTNLDDLLGS--VDSNLLSQLHGLSGPSTPTHL-----QSQSRHQM 499
S+ + +R L P+NLDDL + V S S G +P+H Q Q + Q
Sbjct: 484 SSTGNHTSRTKSLNPSNLDDLFSAEMVSSPRYSNA-DQGGMFSPSHKAAFLNQFQQQQQA 542
Query: 500 MQQNMNHL---RSSYPSNIPSSPVRKPSSLGFDSSSAV------AAAVMNSR-SAAFAKR 549
+ +N + +S +PS +SLG S + + + MNS +AA A+R
Sbjct: 543 LLSPINTVFSPKSVDNQQLPSHSSLLQASLGISSPGRMSPRCVESGSPMNSHLAAALAQR 602
Query: 550 SQSFIDRGAAGTHHLGMSSPSNP---SCRVSSGFSDWNSPSGKLDWGVNGDELSKLRKSA 606
+ + + LG S+ +SS +S W SPSG DWGVNG+EL KLR+S+
Sbjct: 603 EKQQQTMRSLSSRDLGPSAARASGVVGSPLSSSWSKWGSPSGTPDWGVNGEELGKLRRSS 662
Query: 607 SFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKD 641
SF R+ G +PD+SWVH+LVK+
Sbjct: 663 SFELRSGG-------------DDPDLSWVHTLVKE 684
>UniRef100_P93755 Putative CCCH-type zinc finger protein [Arabidopsis thaliana]
Length = 716
Score = 442 bits (1136), Expect = e-122
Identities = 275/667 (41%), Positives = 371/667 (55%), Gaps = 100/667 (14%)
Query: 29 FSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKMGSEKRTPLMIASLFGST 88
F+ LLEL+A+ND E ++ ++ V+EAG WYGR+ GSK M ++ RTPLM+A+ +GS
Sbjct: 45 FASLLELAANNDVEGVRLSIERDPSCVDEAGLWYGRQKGSKAMVNDYRTPLMVAATYGSI 104
Query: 89 RVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACGNK 148
V+K +V DVN+ACG+D+ TALHCA +GG+ +++VVK+LL AG+D + LDA G +
Sbjct: 105 DVIKLIVSLTDADVNRACGNDQTTALHCAASGGAVNAIQVVKLLLAAGADLNLLDAEGQR 164
Query: 149 PMNLIAP---------------AFNSSSKSRRKAMELLLRGGESGQLIHQEMDLE----- 188
++I + + SS + R + S H
Sbjct: 165 AGDVIVVPPKLEGVKLMLQELLSADGSSTAERNLRVVTNVPNRSSSPCHSPTGENGGSGS 224
Query: 189 --LISAPFSSKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECP 246
+ +PF K KKEYPVD SLPDI N +Y +DEFRMYSFKV+PCSRAYSHDWTECP
Sbjct: 225 GSPLGSPFKLKSTEFKKEYPVDPSLPDIKNSIYATDEFRMYSFKVRPCSRAYSHDWTECP 284
Query: 247 FVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKD 306
FVHPGENARRRDPRK+ YSCVPCP+FRKGAC++GD CEYAHGVFE WLHPAQYRTRLCKD
Sbjct: 285 FVHPGENARRRDPRKFHYSCVPCPDFRKGACRRGDMCEYAHGVFECWLHPAQYRTRLCKD 344
Query: 307 ETGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVM--SPLALSSSSLPMS 364
TGC R+VCFFAH PEELRP+YASTGSA+PSP+S + A +S++ SP +S S
Sbjct: 345 GTGCARRVCFFAHTPEELRPLYASTGSAVPSPRSNADYAAALSLLPGSPSGVSVMS---- 400
Query: 365 TVSTPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPG-----SRLKTALSARDFDL-E 418
P+SP A N+ + N+ P+L LPG SRL+++L+ARD E
Sbjct: 401 -----PLSPSAAGNGMSHSNMAWPQPNV----PALHLPGSNLQSSRLRSSLNARDIPTDE 451
Query: 419 MEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRNRIGDLTPTNLDDLLGSVDSN--LL 476
ML +QQQL+ E + S S R+ + P+NL+DL + S+
Sbjct: 452 FNMLA----------DYEQQQLLNEYSNALSRS--GRMKSMPPSNLEDLFSAEGSSSPRF 499
Query: 477 SQLHGLSGPSTPTHL-----QSQSRHQMMQQNMNHLRSSY--PSNIPSS----------- 518
+ S +PTH Q Q + Q Q ++ + +S+ P ++ S
Sbjct: 500 TDSALASAVFSPTHKSAVFNQFQQQQQQQQSMLSPINTSFSSPKSVDHSLFSGGGRMSPR 559
Query: 519 PVRKPSSLGFDSSSAVAAAVMNSRSAAFAKRSQ----SFIDRGAAGTHHLGMSSPSNPSC 574
V +P S S +A V + ++ Q S R + SP N
Sbjct: 560 NVVEPISPMSARVSMLAQCVKQQQQQQQQQQQQHQFRSLSSRELRTNSSPIVGSPVNN-- 617
Query: 575 RVSSGFSDWNSPSGKLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSW 634
++ S W S +G+ DWG++ + L KLR S+SF + EPDVSW
Sbjct: 618 --NTWSSKWGSSNGQPDWGMSSEALGKLRSSSSF-----------------DGDEPDVSW 658
Query: 635 VHSLVKD 641
V SLVK+
Sbjct: 659 VQSLVKE 665
>UniRef100_Q9LXV4 Zinc finger transcription factor-like protein [Arabidopsis
thaliana]
Length = 706
Score = 439 bits (1130), Expect = e-121
Identities = 275/678 (40%), Positives = 389/678 (56%), Gaps = 85/678 (12%)
Query: 12 SEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKM 71
+E + + + + H+FS LLE +A ND E +R++ + +N+ G WY R+ ++M
Sbjct: 22 NESLAKPMNDAAEWEHSFSALLEFAADNDVEGFRRQLSDVSC-INQMGLWYRRQRFVRRM 80
Query: 72 GSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKV 131
E+RTPLM+ASL+GS VVK+++ ++N +CG DK+TALHCA +G S SL+VVK+
Sbjct: 81 VLEQRTPLMVASLYGSLDVVKFILSFPEAELNLSCGPDKSTALHCAASGASVNSLDVVKL 140
Query: 132 LLDAGSDADSLDACGNKPMN-----------------------LIAPAFNSSSKSRRKAM 168
LL G+D + DA GN+P++ +I+ ++SS S +
Sbjct: 141 LLSVGADPNIPDAHGNRPVDVLVVSPHAPGLRTILEEILKKDEIISEDLHASSSSLGSSF 200
Query: 169 ELLLRGGESGQLIHQEMDLELISAPFSSKE-----GSDKKEYPVDVSLPDINNGLYGSDE 223
L ++G + + L+ +S+P S+KKEYP+D SLPDI +G+Y +DE
Sbjct: 201 RSLSSSPDNGSSL---LSLDSVSSPTKPHGTDVTFASEKKEYPIDPSLPDIKSGIYSTDE 257
Query: 224 FRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSC 283
FRM+SFK++PCSRAYSHDWTECPF HPGENARRRDPRK+ Y+CVPCP+F+KG+C++GD C
Sbjct: 258 FRMFSFKIRPCSRAYSHDWTECPFAHPGENARRRDPRKFHYTCVPCPDFKKGSCKQGDMC 317
Query: 284 EYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPK---S 340
EYAHGVFE WLHPAQYRTRLCKD GC R+VCFFAH EELRP+Y STGS +PSP+ +
Sbjct: 318 EYAHGVFECWLHPAQYRTRLCKDGMGCNRRVCFFAHANEELRPLYPSTGSGLPSPRASSA 377
Query: 341 YSSNAVDM-SVMSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNLW--QNKINLNLTPP 397
S++ +DM SV++ L S S+ S TPP+SP P S W QN LNL
Sbjct: 378 VSASTMDMASVLNMLPGSPSAAQHS--FTPPISPSGNGSMPHSSMGWPQQNIPALNLPGS 435
Query: 398 SLQLPGSRLKTALSARDFDLE-MEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRN-- 454
++QL SRL+++L+ARD E + ML + + Q+QL + + SP F N
Sbjct: 436 NIQL--SRLRSSLNARDIPSEQLSML---------HEFEMQRQLAGD---MHSPRFMNHS 481
Query: 455 -RIGDLTPTNLDDLLGS-VDSNLLSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYP 512
R L P+NL++L + V S S +S +P+H + +Q N + S
Sbjct: 482 ARPKTLNPSNLEELFSAEVASPRFSDQLAVSSVLSPSH--KSALLNQLQNNKQSMLSPIK 539
Query: 513 SNIPSSP--VRKPSSLGFDSSSAVAAAV--MNSR-SAAFAKRSQSFIDRGAAGTHHLGMS 567
+N+ SSP V + S L SS + MN+R RS S D G++ L +
Sbjct: 540 TNLMSSPKNVEQHSLLQQASSPRGGEPISPMNARMKQQLHSRSLSSRDFGSSLPRDLMPT 599
Query: 568 SPSNPSCRVSSGFSDWNSPSG-KLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSE 626
+P S +S W+ G K+DW V DEL +LRKS S +A++
Sbjct: 600 DSGSP----LSPWSSWDQTHGSKVDWSVQSDELGRLRKSHS--------------LANNP 641
Query: 627 HAEPDVSWVHSLVKDDLS 644
+ E DVSW ++KD S
Sbjct: 642 NREADVSWAQQMLKDSSS 659
>UniRef100_Q8L3W6 Zinc finger transcription factor-like protein [Arabidopsis
thaliana]
Length = 706
Score = 439 bits (1129), Expect = e-121
Identities = 275/678 (40%), Positives = 389/678 (56%), Gaps = 85/678 (12%)
Query: 12 SEQVMESQKENCDGLHNFSILLELSASNDFEALKREVDEKGLDVNEAGFWYGRRIGSKKM 71
+E + + + + H+FS LLE +A ND E +R++ + +N+ G WY R+ ++M
Sbjct: 22 NESLAKPMNDVAEWEHSFSALLEFAADNDVEGFRRQLSDVSC-INQMGLWYRRQRFVRRM 80
Query: 72 GSEKRTPLMIASLFGSTRVVKYLVETGRVDVNKACGSDKATALHCAVAGGSELSLEVVKV 131
E+RTPLM+ASL+GS VVK+++ ++N +CG DK+TALHCA +G S SL+VVK+
Sbjct: 81 VLEQRTPLMVASLYGSLDVVKFILSFPEAELNLSCGPDKSTALHCAASGASVNSLDVVKL 140
Query: 132 LLDAGSDADSLDACGNKPMN-----------------------LIAPAFNSSSKSRRKAM 168
LL G+D + DA GN+P++ +I+ ++SS S +
Sbjct: 141 LLSVGADPNIPDAHGNRPVDVLVVSPHAPGLRTILEEILKKDEIISEDLHASSSSLGSSF 200
Query: 169 ELLLRGGESGQLIHQEMDLELISAPFSSKE-----GSDKKEYPVDVSLPDINNGLYGSDE 223
L ++G + + L+ +S+P S+KKEYP+D SLPDI +G+Y +DE
Sbjct: 201 RSLSSSPDNGSSL---LSLDSVSSPTKPHGTDVTFASEKKEYPIDPSLPDIKSGIYSTDE 257
Query: 224 FRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQKGDSC 283
FRM+SFK++PCSRAYSHDWTECPF HPGENARRRDPRK+ Y+CVPCP+F+KG+C++GD C
Sbjct: 258 FRMFSFKIRPCSRAYSHDWTECPFAHPGENARRRDPRKFHYTCVPCPDFKKGSCKQGDMC 317
Query: 284 EYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSPK---S 340
EYAHGVFE WLHPAQYRTRLCKD GC R+VCFFAH EELRP+Y STGS +PSP+ +
Sbjct: 318 EYAHGVFECWLHPAQYRTRLCKDGMGCNRRVCFFAHANEELRPLYPSTGSGLPSPRASSA 377
Query: 341 YSSNAVDM-SVMSPLALSSSSLPMSTVSTPPMSPLAGSLSPKSGNLW--QNKINLNLTPP 397
S++ +DM SV++ L S S+ S TPP+SP P S W QN LNL
Sbjct: 378 VSASTMDMASVLNMLPGSPSAAQHS--FTPPISPSGNGSMPHSSMGWPQQNIPALNLPGS 435
Query: 398 SLQLPGSRLKTALSARDFDLE-MEMLGLGSPTRQQQQQQQQQQLIEEIARISSPSFRN-- 454
++QL SRL+++L+ARD E + ML + + Q+QL + + SP F N
Sbjct: 436 NIQL--SRLRSSLNARDIPSEQLSML---------HEFEMQRQLAGD---MHSPRFMNHS 481
Query: 455 -RIGDLTPTNLDDLLGS-VDSNLLSQLHGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYP 512
R L P+NL++L + V S S +S +P+H + +Q N + S
Sbjct: 482 ARPKTLNPSNLEELFSAEVASPRFSDQLAVSSVLSPSH--KSALLNQLQNNKQSMLSPIK 539
Query: 513 SNIPSSP--VRKPSSLGFDSSSAVAAAV--MNSR-SAAFAKRSQSFIDRGAAGTHHLGMS 567
+N+ SSP V + S L SS + MN+R RS S D G++ L +
Sbjct: 540 TNLMSSPKNVEQHSLLQQASSPRGGEPISPMNARMKQQLHSRSLSSRDFGSSLPRDLMPT 599
Query: 568 SPSNPSCRVSSGFSDWNSPSG-KLDWGVNGDELSKLRKSASFGFRNNGMAPTGSPMAHSE 626
+P S +S W+ G K+DW V DEL +LRKS S +A++
Sbjct: 600 DSGSP----LSPWSSWDQTHGSKVDWSVQSDELGRLRKSHS--------------LANNP 641
Query: 627 HAEPDVSWVHSLVKDDLS 644
+ E DVSW ++KD S
Sbjct: 642 NREADVSWAQQMLKDSSS 659
>UniRef100_Q84SL2 CCCH-type zinc finger protein-like protein [Oryza sativa]
Length = 657
Score = 368 bits (945), Expect = e-100
Identities = 252/643 (39%), Positives = 346/643 (53%), Gaps = 94/643 (14%)
Query: 32 LLELSASNDFEALKREVDEKGLDVNE----AGFWYGRRIGSKKMGSEKRTPLMIASLFGS 87
LLEL+A++D + L+ + E G + E G WYGR E RTPLM+A+ +GS
Sbjct: 27 LLELAAADDVDGLRGALAEGGEEAAELADGVGLWYGR-----SKAYEARTPLMVAATYGS 81
Query: 88 TRVVKYLVETGR-VDVNKACGSDKATALHCAVAGGSELSLEVVKVLLDAGSDADSLDACG 146
VV LV G VDVN+ G+D ATALHCA +GGS ++ VVK+LL AG+D + D+ G
Sbjct: 82 AGVVSLLVGLGGCVDVNRRPGADGATALHCAASGGSRNAVAVVKLLLAAGADPATPDSAG 141
Query: 147 NKPMNLI-APAFNSSSKS-------RRKAMELL--LRGGESGQLIHQEMDLELISAPFSS 196
P ++I AP + + RR+A+ + + G S + D E +P SS
Sbjct: 142 RFPADVILAPPASPDALGDLEVLLGRRRALAVATSVASGSSSPPLSSSPD-EGNRSP-SS 199
Query: 197 KEGS--------DKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFV 248
+ S KKEYPVD +LPDI + +Y SDEFRM++FKV+PCSRAYSHDWTECPFV
Sbjct: 200 RSSSLSPITVDRGKKEYPVDPTLPDIKSSVYASDEFRMFAFKVRPCSRAYSHDWTECPFV 259
Query: 249 HPGENARRRDPRKYPYSCVPCPEFRK-GACQKGDSCEYAHGVFESWLHPAQYRTRLCKDE 307
HPGENARRRDPRK+PY+ VPCP FR+ G C GDSCE++HGVFESWLHP+QYRTRLCK+
Sbjct: 260 HPGENARRRDPRKHPYTAVPCPNFRRPGGCPSGDSCEFSHGVFESWLHPSQYRTRLCKEG 319
Query: 308 TGCGRKVCFFAHRPEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVS 367
C R++CFFAH +ELR V ++G+ + SP+ +S+++DM+ + L L S P +
Sbjct: 320 AACARRICFFAHDEDELRHVPHNSGAGLLSPR--ASSSIDMTAAAALGLLPGS-PTRHFA 376
Query: 368 TPPMSPLAGSLSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARD-------FDLEME 420
PP+SP AGS + W L GSRL+++ +ARD LE E
Sbjct: 377 PPPVSPSAGSNGGAAAAHW--------------LQGSRLRSSFNARDAAVDDLGMLLEWE 422
Query: 421 MLGLGSPTRQQQQQQQQQQLIEEIARISSP-SFRNRIGDLTPTNLDDLLGSVDSNLLSQL 479
LG+ + Q R+S+ S R I P+NL+D+ S D + +
Sbjct: 423 SQYLGALCLPPSSRPQ--------PRLSAGLSIRPTI---APSNLEDMYAS-DMAMSPRF 470
Query: 480 HGLSGPSTPTHLQSQSRHQMMQQNMNHLRSSYPSNIPSSPVRKPSSLGFDSSSAVAAAVM 539
P+ H H+ N H + S + ++ + P +L S + M
Sbjct: 471 -----PNDQGHSVYSPAHKSALLNKLHQQKGLLSPVNTNRMYSPRALDPSSLAHSPFGGM 525
Query: 540 NSRSAAFAKRSQSFIDR-GAAGTHHLGMSSPSNPSCRVSSGFSDWNSPSGKLDWGVNGDE 598
+ RS + + R GA T + SP N SS + SP GK+DWGV+ +E
Sbjct: 526 SPRSPRTMEPTSPLSARVGAPATQRPSVGSPRN-----SSAWGTVGSPMGKVDWGVDSEE 580
Query: 599 LSKLRKSASFGFRNNGMAPTGSPMAHSEHAEPDVSWVHSLVKD 641
L +LR+ A GF + E DVSWV SLV +
Sbjct: 581 LVRLRRPAQPGFGED---------------ETDVSWVQSLVSN 608
>UniRef100_Q9M0G2 Hypothetical protein AT4g29190 [Arabidopsis thaliana]
Length = 356
Score = 201 bits (510), Expect = 7e-50
Identities = 107/229 (46%), Positives = 138/229 (59%), Gaps = 16/229 (6%)
Query: 201 DKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPR 260
D + YP D+ PD Y D FRMY FKV+ C+R SHDWTECP+ HPGE ARRRDPR
Sbjct: 58 DPESYP-DLLGPDSPIDAYSCDHFRMYDFKVRRCARGRSHDWTECPYAHPGEKARRRDPR 116
Query: 261 KYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHR 320
KY YS CP+FRKG C+KGDSCE+AHGVFE WLHPA+YRT+ CKD C RK+CFFAH
Sbjct: 117 KYHYSGTACPDFRKGGCKKGDSCEFAHGVFECWLHPARYRTQPCKDGGNCLRKICFFAHS 176
Query: 321 PEELRPVYASTGSAMPSPKSYSSNAVDMSVMSPLALSSSSLPMSTVS-TPPMSPLAGS-L 378
P++LR ++ + + S V SP+ + L +S VS +PPMSP A S
Sbjct: 177 PDQLRFLHTRSPDRVDS----------FDVSSPIRARAFQLSISPVSGSPPMSPRADSES 226
Query: 379 SPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSP 427
SP + +L ++ + ++ +P R S + F + G GSP
Sbjct: 227 SPMTQSLSRSLGSCSIND---VVPSFRNLQFNSVKSFPRNNPLFGFGSP 272
>UniRef100_Q9FU27 CCCH-type zinc finger protein-like [Oryza sativa]
Length = 386
Score = 199 bits (507), Expect = 2e-49
Identities = 97/171 (56%), Positives = 115/171 (66%), Gaps = 9/171 (5%)
Query: 219 YGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQ 278
Y DEFRMY FKV+ C+R SHDWTECPF HPGE ARRRDPRKY YS CP+FRKG C+
Sbjct: 72 YACDEFRMYEFKVRRCARGRSHDWTECPFAHPGEKARRRDPRKYHYSGTACPDFRKGGCK 131
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSP 338
+GD+CEYAHGVFE WLHPA+YRT+ CKD T C R+VCFFAH P++LR + A S
Sbjct: 132 RGDACEYAHGVFECWLHPARYRTQPCKDGTACRRRVCFFAHTPDQLRVLPAQQSSPRSVA 191
Query: 339 KSYSSNAVDMSVMSPLALSS-------SSLPMSTVSTPPMSPLAGS--LSP 380
S + + D S + A S SS P ST+ +PP SP + S LSP
Sbjct: 192 SSPLAESYDGSPLRRQAFESYLTKTIMSSSPTSTLMSPPKSPPSESPPLSP 242
>UniRef100_Q6L4N4 Putative zinc finger transcription factor [Oryza sativa]
Length = 402
Score = 193 bits (490), Expect = 2e-47
Identities = 103/230 (44%), Positives = 127/230 (54%), Gaps = 36/230 (15%)
Query: 219 YGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQ 278
Y DEFRMY FKV+ C+R SHDWTECPF HPGE ARRRDPR+Y YS CP+FRKG C+
Sbjct: 73 YACDEFRMYEFKVRRCARGRSHDWTECPFAHPGEKARRRDPRRYCYSGTACPDFRKGGCK 132
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSP 338
+GD+CE+AHGVFE WLHPA+YRT+ CKD T C R+VCFFAH P++LR + S SP
Sbjct: 133 RGDACEFAHGVFECWLHPARYRTQPCKDGTACRRRVCFFAHTPDQLRVLPPSQQQGSNSP 192
Query: 339 KSYSSNAVDMSVMSPLALS-------------------SSSLPMSTVSTPPMSPLAGS-- 377
+ + SPLA S SS P ST+ +PP SP + S
Sbjct: 193 RGCGGGGAG-AAASPLAESYDGSPLRRQAFESYLTKSIMSSSPTSTLVSPPRSPPSESPP 251
Query: 378 LSPKSGNLWQNKINLNLTPPSLQLPGSRLKTALSARDFDLEMEMLGLGSP 427
LSP + + G+ D + + L LGSP
Sbjct: 252 LSPDAAGALRR--------------GAWAGVGSPVNDVHVSLRQLRLGSP 287
>UniRef100_Q6WEQ1 Zing finger transcription factor PEI1 [Boechera drummondii]
Length = 246
Score = 189 bits (480), Expect = 2e-46
Identities = 80/121 (66%), Positives = 92/121 (75%)
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
Y +D +P INN +YGSDEFRMY++K+K C R SHDWTECP+ H GE A RRDPR+Y Y
Sbjct: 34 YDLDHPIPTINNAIYGSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKATRRDPRRYTY 93
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
V CP FR GAC +GDSCE+AHGVFE WLHPA+YRTR C C RKVCFFAH PE+L
Sbjct: 94 CAVACPAFRNGACHRGDSCEFAHGVFEYWLHPARYRTRACNAGNMCQRKVCFFAHAPEQL 153
Query: 325 R 325
R
Sbjct: 154 R 154
>UniRef100_O65036 Zinc finger transcription factor [Arabidopsis thaliana]
Length = 245
Score = 189 bits (479), Expect = 3e-46
Identities = 79/121 (65%), Positives = 94/121 (77%)
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
Y +D S+P+I++ +YGSDEFRMY++K+K C R SHDWTECP+ H GE A RRDPR+Y Y
Sbjct: 34 YEIDPSIPNIDDAIYGSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKATRRDPRRYTY 93
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
V CP FR GAC +GDSCE+AHGVFE WLHPA+YRTR C C RKVCFFAH PE+L
Sbjct: 94 CAVACPAFRNGACHRGDSCEFAHGVFEYWLHPARYRTRACNAGNLCQRKVCFFAHAPEQL 153
Query: 325 R 325
R
Sbjct: 154 R 154
>UniRef100_O82307 Putative CCCH-type zinc finger protein [Arabidopsis thaliana]
Length = 315
Score = 187 bits (475), Expect = 9e-46
Identities = 99/231 (42%), Positives = 135/231 (57%), Gaps = 31/231 (13%)
Query: 219 YGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPYSCVPCPEFRKGACQ 278
+ SDEFR+Y FK++ C+R SHDWTECPF HPGE ARRRDPRK+ YS CPEFRKG+C+
Sbjct: 86 FSSDEFRIYEFKIRRCARGRSHDWTECPFAHPGEKARRRDPRKFHYSGTACPEFRKGSCR 145
Query: 279 KGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEELRPVYASTGSAMPSP 338
+GDSCE++HGVFE WLHP++YRT+ CKD T C R++CFFAH E+LR + S
Sbjct: 146 RGDSCEFSHGVFECWLHPSRYRTQPCKDGTSCRRRICFFAHTTEQLRVLPCSLDP----- 200
Query: 339 KSYSSNAVDMSVMSPLALSSSSLPMS-----TVSTPPMSPLAGSLSPKSGNLWQNKINLN 393
D+ S LA S +S+ +S +PP+SP G L + N +
Sbjct: 201 --------DLGFFSGLATSPTSILVSPSFSPPSESPPLSPSTGELIASMRKMQLNGGGCS 252
Query: 394 LTPP---SLQLPGSRLKTALSA------RDFDLE----MEMLGLGSPTRQQ 431
+ P +++LP S + A R+F++E ME + G R +
Sbjct: 253 WSSPMRSAVRLPFSSSLRPIQAATWPRIREFEIEEAPAMEFVESGKELRAE 303
>UniRef100_Q6WER2 Zing finger transcription factor PEI1 [Arabidopsis lyrata subsp.
lyrata]
Length = 246
Score = 187 bits (474), Expect = 1e-45
Identities = 78/121 (64%), Positives = 93/121 (76%)
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
Y +D +P+I++ +YGSDEFRMY++K+K C R SHDWTECP+ H GE A RRDPR+Y Y
Sbjct: 35 YEIDPPIPNIDDAIYGSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKATRRDPRRYTY 94
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
V CP FR GAC +GDSCE+AHGVFE WLHPA+YRTR C C RKVCFFAH PE+L
Sbjct: 95 CAVACPAFRNGACHRGDSCEFAHGVFEYWLHPARYRTRACNAGNLCQRKVCFFAHAPEQL 154
Query: 325 R 325
R
Sbjct: 155 R 155
>UniRef100_Q6V5H6 Cu2+ plastocyanin-like [Arabidopsis arenosa]
Length = 247
Score = 187 bits (474), Expect = 1e-45
Identities = 78/121 (64%), Positives = 93/121 (76%)
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
Y +D +P+I++ +YGSDEFRMY++K+K C R SHDWTECP+ H GE A RRDPR+Y Y
Sbjct: 34 YEIDPPIPNIDDAIYGSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKATRRDPRRYTY 93
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
V CP FR GAC +GDSCE+AHGVFE WLHPA+YRTR C C RKVCFFAH PE+L
Sbjct: 94 CAVACPAFRNGACHRGDSCEFAHGVFEYWLHPARYRTRACNAGNLCQRKVCFFAHAPEQL 153
Query: 325 R 325
R
Sbjct: 154 R 154
>UniRef100_Q6V5C7 Zn-finger transcription factor [Arabidopsis arenosa]
Length = 247
Score = 187 bits (474), Expect = 1e-45
Identities = 78/121 (64%), Positives = 93/121 (76%)
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
Y +D +P+I++ +YGSDEFRMY++K+K C R SHDWTECP+ H GE A RRDPR+Y Y
Sbjct: 34 YEIDPPIPNIDDAIYGSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKATRRDPRRYTY 93
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
V CP FR GAC +GDSCE+AHGVFE WLHPA+YRTR C C RKVCFFAH PE+L
Sbjct: 94 CAVACPAFRNGACHRGDSCEFAHGVFEYWLHPARYRTRACNAGNLCQRKVCFFAHAPEQL 153
Query: 325 R 325
R
Sbjct: 154 R 154
>UniRef100_Q6V5E1 Zn-finger transcription factor [Olimarabidopsis pumila]
Length = 246
Score = 186 bits (472), Expect = 2e-45
Identities = 78/121 (64%), Positives = 92/121 (75%)
Query: 205 YPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENARRRDPRKYPY 264
Y +D +P I++ +YGSDEFRMY++K+K C R SHDWTECP+ H GE A RRDPR+Y Y
Sbjct: 34 YDIDPPIPTIDDAIYGSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKATRRDPRRYTY 93
Query: 265 SCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVCFFAHRPEEL 324
V CP FR GAC +GDSCE+AHGVFE WLHPA+YRTR C C RKVCFFAH PE+L
Sbjct: 94 CAVACPAFRNGACHRGDSCEFAHGVFEYWLHPARYRTRACNAGNLCQRKVCFFAHAPEQL 153
Query: 325 R 325
R
Sbjct: 154 R 154
>UniRef100_Q6V5K4 Zinc finger transcription factor [Brassica oleracea]
Length = 246
Score = 185 bits (470), Expect = 3e-45
Identities = 80/130 (61%), Positives = 96/130 (73%), Gaps = 1/130 (0%)
Query: 196 SKEGSDKKEYPVDVSLPDINNGLYGSDEFRMYSFKVKPCSRAYSHDWTECPFVHPGENAR 255
SK G+ + Y +D LP +N+ +Y SDEFRMY++K+K C R SHDWTECP+ H GE A
Sbjct: 26 SKPGN-ARAYEIDPPLPTVNDVIYSSDEFRMYAYKIKRCPRTRSHDWTECPYAHRGEKAT 84
Query: 256 RRDPRKYPYSCVPCPEFRKGACQKGDSCEYAHGVFESWLHPAQYRTRLCKDETGCGRKVC 315
RRDPR+Y Y V CP FR GAC +GD+CE+AHGVFE WLHPA+YRTR C C RKVC
Sbjct: 85 RRDPRRYSYCAVACPAFRNGACHRGDTCEFAHGVFEYWLHPARYRTRACNAGNMCQRKVC 144
Query: 316 FFAHRPEELR 325
FFAH PE+LR
Sbjct: 145 FFAHAPEQLR 154
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.129 0.379
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,129,021,659
Number of Sequences: 2790947
Number of extensions: 49368766
Number of successful extensions: 199653
Number of sequences better than 10.0: 1860
Number of HSP's better than 10.0 without gapping: 146
Number of HSP's successfully gapped in prelim test: 1768
Number of HSP's that attempted gapping in prelim test: 190446
Number of HSP's gapped (non-prelim): 6628
length of query: 664
length of database: 848,049,833
effective HSP length: 134
effective length of query: 530
effective length of database: 474,062,935
effective search space: 251253355550
effective search space used: 251253355550
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0204.2


