

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.11
(131 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
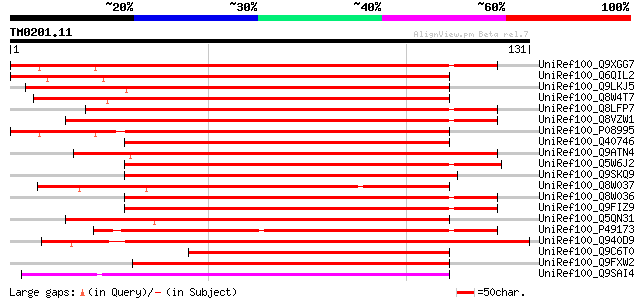
UniRef100_Q9XGG7 Nodulin26-like major intrinsic protein [Pisum s... 154 3e-37
UniRef100_Q6QIL2 NIP2 [Medicago truncatula] 154 6e-37
UniRef100_Q9LKJ5 Multifunctional transport intrinsic membrane pr... 149 2e-35
UniRef100_Q8W4T7 Multifunctional aquaporin [Medicago truncatula] 144 4e-34
UniRef100_Q8LFP7 Aquaporin NIP1.2 [Arabidopsis thaliana] 140 8e-33
UniRef100_Q8VZW1 Aquaporin NIP1.1 [Arabidopsis thaliana] 139 1e-32
UniRef100_P08995 Nodulin-26 [Glycine max] 135 2e-31
UniRef100_Q40746 Major intrinsic protein [Oryza sativa] 121 3e-27
UniRef100_Q9ATN4 NOD26-like membrane integral protein ZmNIP1-1 [... 119 1e-26
UniRef100_Q5W6J2 Hypothetical protein OSJNBb0115F21.2 [Oryza sat... 114 6e-25
UniRef100_Q9SKQ9 Putative major intrinsic (Channel) protein [Ara... 112 2e-24
UniRef100_Q8W037 Aquaporin NIP2.1 [Arabidopsis thaliana] 111 4e-24
UniRef100_Q8W036 Probable aquaporin NIP4.2 [Arabidopsis thaliana] 107 5e-23
UniRef100_Q9FIZ9 Putative aquaporin NIP4.1 [Arabidopsis thaliana] 107 6e-23
UniRef100_Q5QN31 Putative membrane integral protein ZmNIP1-1 [Or... 106 1e-22
UniRef100_P49173 Probable aquaporin NIP-type [Nicotiana alata] 105 3e-22
UniRef100_Q940D9 Early embryogenesis aquaglyceroporin [Pinus taeda] 104 5e-22
UniRef100_Q9C6T0 Putative aquaporin NIP3.1 [Arabidopsis thaliana] 97 8e-20
UniRef100_Q9FXW2 MIP [Adiantum capillus-veneris] 90 1e-17
UniRef100_Q9SAI4 Probable aquaporin NIP6.1 [Arabidopsis thaliana] 87 1e-16
>UniRef100_Q9XGG7 Nodulin26-like major intrinsic protein [Pisum sativum]
Length = 270
Score = 154 bits (390), Expect = 3e-37
Identities = 84/128 (65%), Positives = 98/128 (75%), Gaps = 6/128 (4%)
Query: 1 MADYSG---SNEVILNVNNETSK--KCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVV 55
M D SG +NE+++NVN + S + DS LLQKLVAEV+GTYFLIFAGCA+V
Sbjct: 1 MGDNSGCNETNEIVVNVNKDVSNITQEDSTAHATASLLQKLVAEVVGTYFLIFAGCAAVA 60
Query: 56 VNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLP 115
VN NND VVTLPGI+IVWGLAVMVLVYS+GHISGAHFNPAVT+A ATT+ FP+KQV P
Sbjct: 61 VNKNNDNVVTLPGISIVWGLAVMVLVYSLGHISGAHFNPAVTIAFATTRRFPLKQV-PAY 119
Query: 116 LFKHVGGS 123
+ V GS
Sbjct: 120 IAAQVFGS 127
Score = 32.7 bits (73), Expect = 1.9
Identities = 25/71 (35%), Positives = 39/71 (54%), Gaps = 9/71 (12%)
Query: 33 LQKLVAEVLGTYFLIF--AGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGA 90
LQ V E + T++L+F +G A+ +N + L GIA+ G V++ V G I+GA
Sbjct: 155 LQAFVMEFIITFYLMFIISGVAT-----DNRAIGELAGIAV--GSTVLLNVMFAGPITGA 207
Query: 91 HFNPAVTVAHA 101
NPA ++ A
Sbjct: 208 SMNPARSIGPA 218
>UniRef100_Q6QIL2 NIP2 [Medicago truncatula]
Length = 269
Score = 154 bits (388), Expect = 6e-37
Identities = 80/115 (69%), Positives = 92/115 (79%), Gaps = 4/115 (3%)
Query: 1 MADYSGSN---EVILNVNNETSKKC-DSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVV 56
MA+ S SN E++LNVN + S K DS LQK VAEV+GTYFLIFAGCASV+V
Sbjct: 1 MAENSASNATNEIVLNVNKDVSNKSEDSTSHATASFLQKSVAEVIGTYFLIFAGCASVLV 60
Query: 57 NLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
N NN+ VVTLPGI+IVWGLAVMVLVYS+GHISGAHFNPAVT+A A+TK FP+KQV
Sbjct: 61 NKNNENVVTLPGISIVWGLAVMVLVYSLGHISGAHFNPAVTIAFASTKRFPLKQV 115
Score = 30.4 bits (67), Expect = 9.2
Identities = 25/75 (33%), Positives = 39/75 (51%), Gaps = 9/75 (12%)
Query: 33 LQKLVAEVLGTYFLIF--AGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGA 90
LQ V E + T++ +F +G A+ +N + L GIA+ G V++ V G I+GA
Sbjct: 154 LQAFVIEFIITFYPMFIISGVAT-----DNRAIGELAGIAV--GSTVLLNVMFAGPITGA 206
Query: 91 HFNPAVTVAHATTKS 105
NPA ++ A S
Sbjct: 207 SMNPARSIGPALLHS 221
>UniRef100_Q9LKJ5 Multifunctional transport intrinsic membrane protein 2 [Lotus
japonicus]
Length = 270
Score = 149 bits (375), Expect = 2e-35
Identities = 72/108 (66%), Positives = 90/108 (82%), Gaps = 1/108 (0%)
Query: 5 SGSNEVILNVNNETSKKCDSIEED-CVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV 63
+G++EV++NVN + SK + + + V LQK++AE++GTYF IFAGCAS+VVN NND V
Sbjct: 11 NGAHEVVVNVNKDASKTIEVSDTNFTVSFLQKVIAELVGTYFFIFAGCASIVVNKNNDNV 70
Query: 64 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
VTLPGIA+VWGLAVMVLVYS+GHISGAHFNPA T+A A+TK FP KQV
Sbjct: 71 VTLPGIALVWGLAVMVLVYSLGHISGAHFNPAATIAFASTKRFPWKQV 118
Score = 32.0 bits (71), Expect = 3.2
Identities = 23/69 (33%), Positives = 33/69 (47%), Gaps = 5/69 (7%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
LQ V E + T+FLIF ++ + D IV G V++ V G I+GA
Sbjct: 157 LQAFVIEFIITFFLIF-----ILFGVATDDRAIGEVAGIVVGSTVLLNVLFAGPITGASM 211
Query: 93 NPAVTVAHA 101
NPA ++ A
Sbjct: 212 NPARSIGSA 220
>UniRef100_Q8W4T7 Multifunctional aquaporin [Medicago truncatula]
Length = 276
Score = 144 bits (363), Expect = 4e-34
Identities = 70/106 (66%), Positives = 88/106 (82%), Gaps = 1/106 (0%)
Query: 7 SNEVILNVNNETSKKCD-SIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVT 65
++EV+L+ N ++S C S VP LQKL+AE++GTYFLIFAGCAS+VVN +ND VVT
Sbjct: 11 THEVVLDTNKDSSDTCKGSGSFVSVPFLQKLIAEMVGTYFLIFAGCASIVVNKDNDNVVT 70
Query: 66 LPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
LPGIAIVWGL ++VL+YS+GHISGAHFNPAVT+A ATT+ FP+ QV
Sbjct: 71 LPGIAIVWGLTLLVLIYSLGHISGAHFNPAVTIAFATTRRFPLLQV 116
>UniRef100_Q8LFP7 Aquaporin NIP1.2 [Arabidopsis thaliana]
Length = 294
Score = 140 bits (352), Expect = 8e-33
Identities = 73/104 (70%), Positives = 82/104 (78%), Gaps = 1/104 (0%)
Query: 20 KKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMV 79
KK DS+ VP LQKL+AEVLGTYFLIFAGCA+V VN +DK VTLPGIAIVWGL VMV
Sbjct: 38 KKQDSLLSISVPFLQKLMAEVLGTYFLIFAGCAAVAVNTQHDKAVTLPGIAIVWGLTVMV 97
Query: 80 LVYSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
LVYS+GHISGAHFNPAVT+A A+ FP+KQV P + V GS
Sbjct: 98 LVYSLGHISGAHFNPAVTIAFASCGRFPLKQV-PAYVISQVIGS 140
Score = 31.2 bits (69), Expect = 5.4
Identities = 23/73 (31%), Positives = 36/73 (48%), Gaps = 5/73 (6%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
LQ V E + T++L+F V +N + L G+A+ G V++ V G +SGA
Sbjct: 175 LQSFVIEFIITFYLMFVISG---VATDNRAIGELAGLAV--GSTVLLNVIIAGPVSGASM 229
Query: 93 NPAVTVAHATTKS 105
NP ++ A S
Sbjct: 230 NPGRSLGPAMVYS 242
>UniRef100_Q8VZW1 Aquaporin NIP1.1 [Arabidopsis thaliana]
Length = 296
Score = 139 bits (351), Expect = 1e-32
Identities = 71/109 (65%), Positives = 82/109 (75%), Gaps = 1/109 (0%)
Query: 15 NNETSKKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWG 74
N KK DS+ VP LQKL+AE LGTYFL+F GCASVVVN+ ND VVTLPGIAIVWG
Sbjct: 36 NPRPLKKQDSLLSVSVPFLQKLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAIVWG 95
Query: 75 LAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
L +MVL+YS+GHISGAH NPAVT+A A+ FP+KQV P + V GS
Sbjct: 96 LTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQV-PAYVISQVIGS 143
>UniRef100_P08995 Nodulin-26 [Glycine max]
Length = 271
Score = 135 bits (341), Expect = 2e-31
Identities = 72/117 (61%), Positives = 88/117 (74%), Gaps = 8/117 (6%)
Query: 1 MADYSG---SNEVILNVNNETSK---KCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASV 54
MADYS S EV++NV TS+ + DS+ VP LQKLVAE +GTYFLIFAGCAS+
Sbjct: 1 MADYSAGTESQEVVVNVTKNTSETIQRSDSLVS--VPFLQKLVAEAVGTYFLIFAGCASL 58
Query: 55 VVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
VVN N ++T PGIAIVWGL + VLVY++GHISG HFNPAVT+A A+T+ FP+ QV
Sbjct: 59 VVNENYYNMITFPGIAIVWGLVLTVLVYTVGHISGGHFNPAVTIAFASTRRFPLIQV 115
Score = 33.5 bits (75), Expect = 1.1
Identities = 25/69 (36%), Positives = 36/69 (51%), Gaps = 5/69 (7%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
LQ V E + T+FL+F C V +N V GIAI G +++ V G ++GA
Sbjct: 154 LQAFVFEFIMTFFLMFVICG---VATDNRAVGEFAGIAI--GSTLLLNVIIGGPVTGASM 208
Query: 93 NPAVTVAHA 101
NPA ++ A
Sbjct: 209 NPARSLGPA 217
>UniRef100_Q40746 Major intrinsic protein [Oryza sativa]
Length = 284
Score = 121 bits (304), Expect = 3e-27
Identities = 53/82 (64%), Positives = 69/82 (83%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
VP +QK++AE+ GTYFLIFAGC +V +N + + +T PG+AIVWGLAVMV+VY++GHISG
Sbjct: 44 VPFIQKIIAEIFGTYFLIFAGCGAVTINQSKNGQITFPGVAIVWGLAVMVMVYAVGHISG 103
Query: 90 AHFNPAVTVAHATTKSFPVKQV 111
AHFNPAVT+A AT + FP +QV
Sbjct: 104 AHFNPAVTLAFATCRRFPWRQV 125
Score = 33.1 bits (74), Expect = 1.4
Identities = 23/69 (33%), Positives = 37/69 (53%), Gaps = 5/69 (7%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
+Q LV E + T++L+F V +N + L G+A+ G +++ V G ISGA
Sbjct: 164 VQSLVLEFIITFYLMFVISG---VATDNRAIGELAGLAV--GATILLNVLIAGPISGASM 218
Query: 93 NPAVTVAHA 101
NPA ++ A
Sbjct: 219 NPARSLGPA 227
>UniRef100_Q9ATN4 NOD26-like membrane integral protein ZmNIP1-1 [Zea mays]
Length = 282
Score = 119 bits (299), Expect = 1e-26
Identities = 56/113 (49%), Positives = 81/113 (71%), Gaps = 6/113 (5%)
Query: 17 ETSKKCDSIEEDC------VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIA 70
E +K + ++ C VP +QK++AE+ GTYFL+FAGC +V +N + + +T PG+A
Sbjct: 21 EEGRKEEFADQGCAAMVVSVPFIQKIIAEIFGTYFLMFAGCGAVTINASKNGQITFPGVA 80
Query: 71 IVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
IVWGLAVMV+VY++GHISGAHFNPAVT+A AT+ FP +Q+ L + +G +
Sbjct: 81 IVWGLAVMVMVYAVGHISGAHFNPAVTLAFATSGRFPWRQLPAYVLAQMLGAT 133
Score = 36.2 bits (82), Expect = 0.17
Identities = 23/69 (33%), Positives = 38/69 (54%), Gaps = 5/69 (7%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
+Q LV E++ T++L+F V +N + L G+A+ G +++ V G +SGA
Sbjct: 160 VQSLVIEIITTFYLMFVISG---VATDNRAIGELAGLAV--GATILLNVLIAGPVSGASM 214
Query: 93 NPAVTVAHA 101
NPA +V A
Sbjct: 215 NPARSVGPA 223
>UniRef100_Q5W6J2 Hypothetical protein OSJNBb0115F21.2 [Oryza sativa]
Length = 286
Score = 114 bits (284), Expect = 6e-25
Identities = 58/95 (61%), Positives = 68/95 (71%), Gaps = 1/95 (1%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
V QK++AE+LGT+FLIFAGCA+V VN VT PGI I WGLAVMV+VYS+GHISG
Sbjct: 50 VQFAQKVIAEILGTFFLIFAGCAAVAVNKRTGGTVTFPGICITWGLAVMVMVYSVGHISG 109
Query: 90 AHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGSA 124
AH NPAVT+A AT FP ++V P V GSA
Sbjct: 110 AHLNPAVTLAFATCGRFPWRRV-PAYAAAQVAGSA 143
Score = 35.8 bits (81), Expect = 0.22
Identities = 25/69 (36%), Positives = 36/69 (51%), Gaps = 5/69 (7%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
+Q L E + T++L+F V +N + L G+A+ G V+V V G ISGA
Sbjct: 170 VQSLAMEFIITFYLMFVVSG---VATDNRAIGELAGLAV--GATVLVNVLFAGPISGASM 224
Query: 93 NPAVTVAHA 101
NPA T+ A
Sbjct: 225 NPARTIGPA 233
>UniRef100_Q9SKQ9 Putative major intrinsic (Channel) protein [Arabidopsis thaliana]
Length = 262
Score = 112 bits (279), Expect = 2e-24
Identities = 50/84 (59%), Positives = 66/84 (78%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
V +QKL+ E +GT+ +IFAGC+++VVN K VTLPGIA+VWGL V V++YSIGH+SG
Sbjct: 50 VSFVQKLIGEFVGTFSMIFAGCSAIVVNETYGKPVTLPGIALVWGLVVTVMIYSIGHVSG 109
Query: 90 AHFNPAVTVAHATTKSFPVKQVFP 113
AHFNPAV++A A++K FP Q P
Sbjct: 110 AHFNPAVSIAFASSKKFPFNQFHP 133
>UniRef100_Q8W037 Aquaporin NIP2.1 [Arabidopsis thaliana]
Length = 288
Score = 111 bits (277), Expect = 4e-24
Identities = 59/113 (52%), Positives = 81/113 (71%), Gaps = 10/113 (8%)
Query: 8 NEVILNVNN----ETSKKCDSIEEDCVPLL-----QKLVAEVLGTYFLIFAGCASVVVNL 58
N V+LN+ +TS + E PLL QKL+AE++GTY+LIFAGCA++ VN
Sbjct: 13 NVVVLNIKASSLADTSLPSNKHESSSPPLLSVHFLQKLLAELVGTYYLIFAGCAAIAVNA 72
Query: 59 NNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
++ VVTL GIA+VWG+ +MVLVY +GH+S AHFNPAVT+A A+++ FP+ QV
Sbjct: 73 QHNHVVTLVGIAVVWGIVIMVLVYCLGHLS-AHFNPAVTLALASSQRFPLNQV 124
Score = 33.5 bits (75), Expect = 1.1
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 8/100 (8%)
Query: 5 SGSNEVILNVNNETSKKCDSIEEDCVPL---LQKLVAEVLGTYFLIFAGCASVVVNLNND 61
S + ++ ++NN+ K + P LQ V E + T FL+ CA +
Sbjct: 139 SATLRLLFDLNNDVCSKKHDVFLGSSPSGSDLQAFVMEFIITGFLMLVVCAVTTTKRTTE 198
Query: 62 KVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHA 101
+ L G+ I G V + V G +SGA NPA ++ A
Sbjct: 199 E---LEGLII--GATVTLNVIFAGEVSGASMNPARSIGPA 233
>UniRef100_Q8W036 Probable aquaporin NIP4.2 [Arabidopsis thaliana]
Length = 283
Score = 107 bits (268), Expect = 5e-23
Identities = 54/94 (57%), Positives = 67/94 (70%), Gaps = 1/94 (1%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
V L QKL+AE++GTYF+IF+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIIFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 90 AHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
AHFNPAVTV A + FP QV PL + + GS
Sbjct: 99 AHFNPAVTVTFAVFRRFPWYQV-PLYIGAQLTGS 131
Score = 36.6 bits (83), Expect = 0.13
Identities = 32/100 (32%), Positives = 51/100 (51%), Gaps = 10/100 (10%)
Query: 34 QKLVAEVLGTYFLIFAGCASVVVNLNNDKVVT--LPGIAIVWGLAVMVLVYSIGHISGAH 91
Q LVAE++ ++ L+F V+ + D T L GIA+ G+ +++ V+ G ISGA
Sbjct: 160 QALVAEIIISFLLMF-----VISGVATDSRATGELAGIAV--GMTIILNVFVAGPISGAS 212
Query: 92 FNPAVTVAHATTKSFPVKQVFPLPLFKHVGGSAKGFHWRF 131
NPA ++ A K ++ + VG A GF + F
Sbjct: 213 MNPARSLGPAIVMG-RYKGIWVYIVGPFVGIFAGGFVYNF 251
>UniRef100_Q9FIZ9 Putative aquaporin NIP4.1 [Arabidopsis thaliana]
Length = 283
Score = 107 bits (267), Expect = 6e-23
Identities = 53/94 (56%), Positives = 66/94 (69%), Gaps = 1/94 (1%)
Query: 30 VPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISG 89
V L QKL+AE++GTYF++F+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIVFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 90 AHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
AHFNPAVTV A + FP QV PL + GS
Sbjct: 99 AHFNPAVTVTFAIFRRFPWHQV-PLYIGAQFAGS 131
Score = 37.7 bits (86), Expect = 0.058
Identities = 26/66 (39%), Positives = 38/66 (57%), Gaps = 5/66 (7%)
Query: 36 LVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPA 95
LVAE++ ++ L+F V +N V L GIA+ G+ +MV V+ G ISGA NPA
Sbjct: 162 LVAEIIISFLLMFVISG---VATDNRAVGELAGIAV--GMTIMVNVFVAGPISGASMNPA 216
Query: 96 VTVAHA 101
++ A
Sbjct: 217 RSLGPA 222
>UniRef100_Q5QN31 Putative membrane integral protein ZmNIP1-1 [Oryza sativa]
Length = 303
Score = 106 bits (265), Expect = 1e-22
Identities = 56/110 (50%), Positives = 70/110 (62%), Gaps = 13/110 (11%)
Query: 15 NNETSKKCDSIEEDCVPLLQK-------------LVAEVLGTYFLIFAGCASVVVNLNND 61
++ +S +C + V +QK ++AE+LGTYF+IFAGC +VVVN +
Sbjct: 32 SSSSSSRCHGNDVISVQFMQKVHPWCMCMNKNLLILAEILGTYFMIFAGCGAVVVNQSTG 91
Query: 62 KVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
VT PGI VWGL VMVLVY++ HISGAHFNPAVTVA AT F KQV
Sbjct: 92 GAVTFPGICAVWGLVVMVLVYTVSHISGAHFNPAVTVAFATCGRFRWKQV 141
>UniRef100_P49173 Probable aquaporin NIP-type [Nicotiana alata]
Length = 270
Score = 105 bits (261), Expect = 3e-22
Identities = 56/102 (54%), Positives = 70/102 (67%), Gaps = 4/102 (3%)
Query: 22 CDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLV 81
C S+ V +LQKL+AE +GTYF+IFAGC SV VN V T PGI + WGL VMV+V
Sbjct: 33 CSSVS--VVVILQKLIAEAIGTYFVIFAGCGSVAVNKIYGSV-TFPGICVTWGLIVMVMV 89
Query: 82 YSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGS 123
Y++G+ISGAHFNPAVT+ + FP KQV PL + + GS
Sbjct: 90 YTVGYISGAHFNPAVTITFSIFGRFPWKQV-PLYIIAQLMGS 130
Score = 31.6 bits (70), Expect = 4.1
Identities = 21/71 (29%), Positives = 37/71 (51%), Gaps = 5/71 (7%)
Query: 34 QKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFN 93
Q L E++ ++ L+F V ++ + + GIA+ G+ + + V+ G ISGA N
Sbjct: 159 QSLAIEIIISFLLMFVISG---VATDDRAIGQVAGIAV--GMTITLNVFVAGPISGASMN 213
Query: 94 PAVTVAHATTK 104
PA ++ A K
Sbjct: 214 PARSIGPAIVK 224
>UniRef100_Q940D9 Early embryogenesis aquaglyceroporin [Pinus taeda]
Length = 264
Score = 104 bits (259), Expect = 5e-22
Identities = 56/127 (44%), Positives = 81/127 (63%), Gaps = 8/127 (6%)
Query: 9 EVILNV----NNETSKKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVV 64
E++L V + C S+ +P ++K+VAE +GT+FLIF GC SVVV+ ++ +
Sbjct: 4 EILLGVAPMEEGRRGRSCGSL----LPSVRKVVAEFIGTFFLIFVGCGSVVVDKISNGSI 59
Query: 65 TLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQVFPLPLFKHVGGSA 124
T G+++VWG+A M+++YSIGHISGAH NPAVT+A A K FP QV + + G +
Sbjct: 60 THLGVSLVWGMAAMIVIYSIGHISGAHLNPAVTLALAAVKRFPWVQVPGYIVAQVFGSIS 119
Query: 125 KGFHWRF 131
GF RF
Sbjct: 120 AGFLLRF 126
Score = 31.6 bits (70), Expect = 4.1
Identities = 24/69 (34%), Positives = 33/69 (47%), Gaps = 5/69 (7%)
Query: 33 LQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHF 92
+Q E++ T L+F A V + V L G+AI G + + V G ISGA
Sbjct: 144 MQSFALEIITTSLLVFVVSA---VATDTKAVGELGGLAI--GATIAMNVAISGPISGASM 198
Query: 93 NPAVTVAHA 101
NPA T+ A
Sbjct: 199 NPARTIGSA 207
>UniRef100_Q9C6T0 Putative aquaporin NIP3.1 [Arabidopsis thaliana]
Length = 269
Score = 97.1 bits (240), Expect = 8e-20
Identities = 43/66 (65%), Positives = 55/66 (83%)
Query: 46 LIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKS 105
+IFAGC+++VVN K VTLPGIA+VWGL V V++YSIGH+SGAHFNPAV++A A++K
Sbjct: 1 MIFAGCSAIVVNETYGKPVTLPGIALVWGLVVTVMIYSIGHVSGAHFNPAVSIAFASSKK 60
Query: 106 FPVKQV 111
FP QV
Sbjct: 61 FPFNQV 66
>UniRef100_Q9FXW2 MIP [Adiantum capillus-veneris]
Length = 282
Score = 90.1 bits (222), Expect = 1e-17
Identities = 40/80 (50%), Positives = 59/80 (73%)
Query: 32 LLQKLVAEVLGTYFLIFAGCASVVVNLNNDKVVTLPGIAIVWGLAVMVLVYSIGHISGAH 91
+LQK+ AE++ TY L+FAGC + +V+ + +T G++ +GL VM+++YS+GHISGAH
Sbjct: 48 ILQKVGAELISTYILVFAGCGAAMVDEKSGGAITHFGVSAAFGLVVMIMIYSVGHISGAH 107
Query: 92 FNPAVTVAHATTKSFPVKQV 111
NPAVT+A AT + FP QV
Sbjct: 108 MNPAVTLAFATVRHFPWAQV 127
>UniRef100_Q9SAI4 Probable aquaporin NIP6.1 [Arabidopsis thaliana]
Length = 305
Score = 86.7 bits (213), Expect = 1e-16
Identities = 52/108 (48%), Positives = 62/108 (57%), Gaps = 1/108 (0%)
Query: 4 YSGSNEVILNVNNETSKKCDSIEEDCVPLLQKLVAEVLGTYFLIFAGCASVVVNLNNDKV 63
+S NE L C S+ V L +KL AE +GT LIFAG A+ +VN D
Sbjct: 51 FSVDNEWALEDGRLPPVTC-SLPPPNVSLYRKLGAEFVGTLILIFAGTATAIVNQKTDGA 109
Query: 64 VTLPGIAIVWGLAVMVLVYSIGHISGAHFNPAVTVAHATTKSFPVKQV 111
TL G A GLAVM+++ S GHISGAH NPAVT+A A K FP K V
Sbjct: 110 ETLIGCAASAGLAVMIVILSTGHISGAHLNPAVTIAFAALKHFPWKHV 157
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 216,642,384
Number of Sequences: 2790947
Number of extensions: 7812789
Number of successful extensions: 22339
Number of sequences better than 10.0: 1045
Number of HSP's better than 10.0 without gapping: 915
Number of HSP's successfully gapped in prelim test: 145
Number of HSP's that attempted gapping in prelim test: 20885
Number of HSP's gapped (non-prelim): 1333
length of query: 131
length of database: 848,049,833
effective HSP length: 107
effective length of query: 24
effective length of database: 549,418,504
effective search space: 13186044096
effective search space used: 13186044096
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0201.11


