

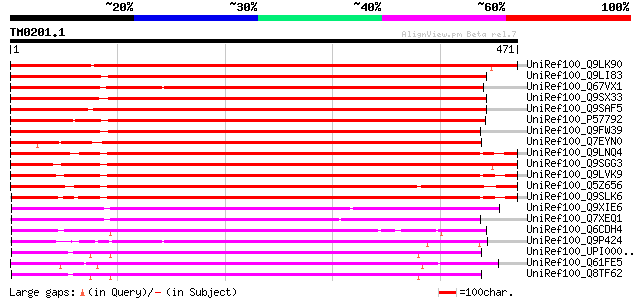
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0201.1
(471 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LK90 Potential phospholipid-transporting ATPase 8 [A... 707 0.0
UniRef100_Q9LI83 Potential phospholipid-transporting ATPase 10 [... 608 e-172
UniRef100_Q67VX1 Putative Potential phospholipid-transporting AT... 605 e-172
UniRef100_Q9SX33 Potential phospholipid-transporting ATPase 9 [A... 601 e-170
UniRef100_Q9SAF5 Potential phospholipid-transporting ATPase 11 [... 600 e-170
UniRef100_P57792 Potential phospholipid-transporting ATPase 12 [... 576 e-163
UniRef100_Q9FW39 Similar to an Arabidopsis putative P-type trans... 568 e-160
UniRef100_Q7EYN0 Putative ATPase [Oryza sativa] 552 e-156
UniRef100_Q9LNQ4 Potential phospholipid-transporting ATPase 4 [A... 534 e-150
UniRef100_Q9SGG3 Potential phospholipid-transporting ATPase 5 [A... 529 e-149
UniRef100_Q9LVK9 Potential phospholipid-transporting ATPase 7 [A... 512 e-143
UniRef100_Q5Z656 Putative ATPase, aminophospholipid transporter ... 510 e-143
UniRef100_Q9SLK6 Potential phospholipid-transporting ATPase 6 [A... 503 e-141
UniRef100_Q9XIE6 Potential phospholipid-transporting ATPase 3 [A... 358 2e-97
UniRef100_Q7XEQ1 Contains similarity to chromaffin granule ATPas... 357 5e-97
UniRef100_Q6CDH4 Yarrowia lipolytica chromosome C of strain CLIB... 324 3e-87
UniRef100_Q9P424 Putative calcium transporting ATPase [Ajellomyc... 322 1e-86
UniRef100_UPI000036A08B UPI000036A08B UniRef100 entry 321 2e-86
UniRef100_Q61FE5 Hypothetical protein CBG11664 [Caenorhabditis b... 321 2e-86
UniRef100_Q8TF62 Potential phospholipid-transporting ATPase IM [... 321 2e-86
>UniRef100_Q9LK90 Potential phospholipid-transporting ATPase 8 [Arabidopsis thaliana]
Length = 1189
Score = 707 bits (1826), Expect = 0.0
Identities = 348/476 (73%), Positives = 401/476 (84%), Gaps = 7/476 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KL+QAG+K+WVLTGDK ETA+NIGYACSLLR+ MK+I++TLDS DI +LEKQGDK+A+ K
Sbjct: 713 KLSQAGVKIWVLTGDKTETAINIGYACSLLREGMKQILVTLDSSDIEALEKQGDKEAVAK 772
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSA-FGLIIDGKSLDYSLNKNLEKSFFELAV 119
AS +SIKKQ+ EG+SQ +A + N+ KE S FGL+IDGKSL Y+L+ LEK F ELA+
Sbjct: 773 ASFQSIKKQLREGMSQ--TAAVTDNSAKENSEMFGLVIDGKSLTYALDSKLEKEFLELAI 830
Query: 120 SCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQA 179
C SVICCRSSPKQKA VTRLVK GTG+T L+IGDGANDVGMLQEA IGVGISGAEGMQA
Sbjct: 831 RCNSVICCRSSPKQKALVTRLVKNGTGRTTLAIGDGANDVGMLQEADIGVGISGAEGMQA 890
Query: 180 VMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQP 239
VMASDFAIAQFRFLERLLLVHGHWCYRRI+LMICYFFYKN+AFGFTLFW+EAYASFSG+P
Sbjct: 891 VMASDFAIAQFRFLERLLLVHGHWCYRRITLMICYFFYKNLAFGFTLFWYEAYASFSGKP 950
Query: 240 AYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWML 299
AYNDWYMS YNVFFTSLPVIALGVFDQDVSA+LCLKYP LY EGV++ LFSW RI+GWML
Sbjct: 951 AYNDWYMSCYNVFFTSLPVIALGVFDQDVSARLCLKYPLLYQEGVQNVLFSWERILGWML 1010
Query: 300 NGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWI 359
NGVISS+ IFFLT N++ QAFR+DGQVVD+ +LGVTMYS VVWTVNCQMA+SINYFTWI
Sbjct: 1011 NGVISSMIIFFLTINTMATQAFRKDGQVVDYSVLGVTMYSSVVWTVNCQMAISINYFTWI 1070
Query: 360 QHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYF 419
QH FIWGSI WY+FL++YG L P S+TA+ VFVE APS +YWL LVV LLPYF
Sbjct: 1071 QHCFIWGSIGVWYLFLVIYGSLPPTFSTTAFQVFVETSAPSPIYWLVLFLVVFSALLPYF 1130
Query: 420 TYRAFQSRFLPMYHDII-QRKRVEGFEV---EISDELPTQVQGKLMHMRERLKQRE 471
TYRAFQ +F PMYHDII +++R E E + ELP QV+ L H+R L +R+
Sbjct: 1131 TYRAFQIKFRPMYHDIIVEQRRTERTETAPNAVLGELPVQVEFTLHHLRANLSRRD 1186
>UniRef100_Q9LI83 Potential phospholipid-transporting ATPase 10 [Arabidopsis thaliana]
Length = 1202
Score = 608 bits (1568), Expect = e-172
Identities = 296/443 (66%), Positives = 352/443 (78%), Gaps = 6/443 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+A SLLRQ+MK+I+I L++P I SLEK G KD +
Sbjct: 718 KLAQAGIKIWVLTGDKMETAINIGFASSLLRQEMKQIIINLETPQIKSLEKSGGKDEIEL 777
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
AS ES+ Q+ EG + + ++ SS AF LIIDGKSL Y+L ++K F +LA S
Sbjct: 778 ASRESVVMQLQEGKALLAASGASSE------AFALIIDGKSLTYALEDEIKKMFLDLATS 831
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK GTGKT L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 832 CASVICCRSSPKQKALVTRLVKSGTGKTTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 891
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
M+SD AIAQFR+LERLLLVHGHWCY RI+ MICYFFYKNI FG T+F +EAY SFSGQPA
Sbjct: 892 MSSDIAIAQFRYLERLLLVHGHWCYSRIASMICYFFYKNITFGVTVFLYEAYTSFSGQPA 951
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDW++S +NVFF+SLPVIALGVFDQDVSA+ C K+P LY EGV++ LFSW RIIGWM N
Sbjct: 952 YNDWFLSLFNVFFSSLPVIALGVFDQDVSARFCYKFPLLYQEGVQNILFSWKRIIGWMFN 1011
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
G IS+LAIFFL S+ +Q F DG+ EILG TMY+ VVW VN QMALSI+YFTW+Q
Sbjct: 1012 GFISALAIFFLCKESLKHQLFDPDGKTAGREILGGTMYTCVVWVVNLQMALSISYFTWVQ 1071
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGSI FWY+FL++YG ++P+ S+ AYMVF+EA AP+ YWL T+ V++ L+PYF
Sbjct: 1072 HIVIWGSIAFWYIFLMIYGAMTPSFSTDAYMVFLEALAPAPSYWLTTLFVMIFALIPYFV 1131
Query: 421 YRAFQSRFLPMYHDIIQRKRVEG 443
Y++ Q RF P YH +IQ R EG
Sbjct: 1132 YKSVQMRFFPKYHQMIQWIRYEG 1154
>UniRef100_Q67VX1 Putative Potential phospholipid-transporting ATPase 8 [Oryza sativa]
Length = 1207
Score = 605 bits (1560), Expect = e-172
Identities = 290/440 (65%), Positives = 352/440 (79%), Gaps = 6/440 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+ACSLLRQ M +I++TL++PDI++LEK GDK+++ +
Sbjct: 724 KLAQAGIKIWVLTGDKMETAINIGFACSLLRQGMTQIIVTLEAPDIIALEKNGDKESIAR 783
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
S + + QI +GI Q+ +S+ T +F LIIDGKSL Y+L +++ F +LA+
Sbjct: 784 ESKQRVMDQIEDGIKQIPPPSQSN-----TESFALIIDGKSLTYALEDDVKFKFLDLALK 838
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK T + L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 839 CASVICCRSSPKQKALVTRLVK-HTNRVTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 897
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
MASDFAIAQFRFLERLLL+HGHWCYRRIS+MICYFFYKN+ FG T+F +EA+ASFSG+PA
Sbjct: 898 MASDFAIAQFRFLERLLLIHGHWCYRRISVMICYFFYKNVTFGVTIFLYEAFASFSGKPA 957
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDW++S YNV FTSLPVIALGVFDQDVS +LCL+YP LY EGV++ LFSW RI+GWM N
Sbjct: 958 YNDWFLSLYNVIFTSLPVIALGVFDQDVSQRLCLQYPGLYQEGVQNILFSWRRILGWMAN 1017
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GVI+++ IF+ T + QAFR+DGQV + LGV MY+ VVW VNCQMALS+NYFT IQ
Sbjct: 1018 GVINAILIFYFCTTAFGIQAFRQDGQVAGLDALGVLMYTCVVWVVNCQMALSVNYFTIIQ 1077
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H FIWGSI WY+FLL YG + P S +AYMVF+E AP+ YWL T+ V+ L+PYF
Sbjct: 1078 HIFIWGSIAVWYLFLLAYGAVDPRFSKSAYMVFIEQVAPALSYWLVTLFAVMATLIPYFC 1137
Query: 421 YRAFQSRFLPMYHDIIQRKR 440
Y A Q RF PM+H+ IQ KR
Sbjct: 1138 YAAIQIRFFPMFHNKIQWKR 1157
>UniRef100_Q9SX33 Potential phospholipid-transporting ATPase 9 [Arabidopsis thaliana]
Length = 1200
Score = 601 bits (1549), Expect = e-170
Identities = 298/443 (67%), Positives = 347/443 (78%), Gaps = 8/443 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+ACSLLRQDMK+I+I L++P+I SLEK G+KD + K
Sbjct: 725 KLAQAGIKIWVLTGDKMETAINIGFACSLLRQDMKQIIINLETPEIQSLEKTGEKDVIAK 784
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
AS E++ QI G +Q+K + + AF LIIDGKSL Y+L+ +++ F ELAVS
Sbjct: 785 ASKENVLSQIINGKTQLKYSGGN--------AFALIIDGKSLAYALDDDIKHIFLELAVS 836
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK G GKT L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 837 CASVICCRSSPKQKALVTRLVKSGNGKTTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 896
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
M+SD AIAQFR+LERLLLVHGHWCYRRIS MICYFFYKNI FGFTLF +E Y +FS PA
Sbjct: 897 MSSDIAIAQFRYLERLLLVHGHWCYRRISTMICYFFYKNITFGFTLFLYETYTTFSSTPA 956
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDW++S YNVFF+SLPVIALGVFDQDVSA+ CLK+P LY EGV++ LFSW RI+GWM N
Sbjct: 957 YNDWFLSLYNVFFSSLPVIALGVFDQDVSARYCLKFPLLYQEGVQNVLFSWRRILGWMFN 1016
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
G S++ IFFL +S+ +QAF DG+ EILG TMY+ +VW VN QMAL+I+YFT IQ
Sbjct: 1017 GFYSAVIIFFLCKSSLQSQAFNHDGKTPGREILGGTMYTCIVWVVNLQMALAISYFTLIQ 1076
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IW SI WY F+ VYG L IS+ AY VFVEA APS YWL T+ VVV L+PYF
Sbjct: 1077 HIVIWSSIVVWYFFITVYGELPSRISTGAYKVFVEALAPSLSYWLITLFVVVATLMPYFI 1136
Query: 421 YRAFQSRFLPMYHDIIQRKRVEG 443
Y A Q F PMYH +IQ R EG
Sbjct: 1137 YSALQMSFFPMYHGMIQWLRYEG 1159
>UniRef100_Q9SAF5 Potential phospholipid-transporting ATPase 11 [Arabidopsis thaliana]
Length = 1203
Score = 600 bits (1547), Expect = e-170
Identities = 288/443 (65%), Positives = 351/443 (79%), Gaps = 4/443 (0%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+ACSLLRQ+MK+I+I L++P I +LEK G+KDA+
Sbjct: 717 KLAQAGIKIWVLTGDKMETAINIGFACSLLRQEMKQIIINLETPHIKALEKAGEKDAIEH 776
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
AS ES+ Q+ EG K+ +S++ AF LIIDGKSL Y+L + +K F +LA
Sbjct: 777 ASRESVVNQMEEG----KALLTASSSASSHEAFALIIDGKSLTYALEDDFKKKFLDLATG 832
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK GTGKT L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 833 CASVICCRSSPKQKALVTRLVKSGTGKTTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 892
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
M+SD AIAQFR+LERLLLVHGHWCY RIS MICYFFYKNI FG T+F +EAY SFS QPA
Sbjct: 893 MSSDIAIAQFRYLERLLLVHGHWCYSRISSMICYFFYKNITFGVTVFLYEAYTSFSAQPA 952
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDW++S +NVFF+SLPVIALGVFDQDVSA+ C K+P LY EGV++ LFSW RIIGWM N
Sbjct: 953 YNDWFLSLFNVFFSSLPVIALGVFDQDVSARYCYKFPLLYQEGVQNLLFSWKRIIGWMFN 1012
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GV ++LAIFFL S+ +Q + +G+ EILG TMY+ VVW VN QMAL+I+YFTW+Q
Sbjct: 1013 GVFTALAIFFLCKESLKHQLYNPNGKTAGREILGGTMYTCVVWVVNLQMALAISYFTWLQ 1072
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGS+ FWY+FL++YG ++P+ S+ AY VF+EA AP+ YWL T+ V+ L+P+F
Sbjct: 1073 HIVIWGSVAFWYIFLMIYGAITPSFSTDAYKVFIEALAPAPSYWLTTLFVMFFALIPFFV 1132
Query: 421 YRAFQSRFLPMYHDIIQRKRVEG 443
+++ Q RF P YH +IQ R EG
Sbjct: 1133 FKSVQMRFFPGYHQMIQWIRYEG 1155
>UniRef100_P57792 Potential phospholipid-transporting ATPase 12 [Arabidopsis thaliana]
Length = 1184
Score = 576 bits (1485), Expect = e-163
Identities = 280/442 (63%), Positives = 346/442 (77%), Gaps = 7/442 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAGIK+WVLTGDKMETA+NIG+ACSLLR+DMK+I+I L++P+I LEK G+KDA+
Sbjct: 720 KLAQAGIKIWVLTGDKMETAINIGFACSLLRRDMKQIIINLETPEIQQLEKSGEKDAIA- 778
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
A E++ QI+ G +Q+K++ ++ AF LIIDGKSL Y+L ++++ F ELA+
Sbjct: 779 ALKENVLHQITSGKAQLKASGGNAK------AFALIIDGKSLAYALEEDMKGIFLELAIG 832
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCRSSPKQKA VTRLVK G+G+T L+IGDGANDVGMLQEA IGVGISG EGMQAV
Sbjct: 833 CASVICCRSSPKQKALVTRLVKTGSGQTTLAIGDGANDVGMLQEADIGVGISGVEGMQAV 892
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
M+SD AIAQFR+LERLLLVHGHWCYRRIS MICYFFYKNI FGFTLF +EAY SFS PA
Sbjct: 893 MSSDIAIAQFRYLERLLLVHGHWCYRRISKMICYFFYKNITFGFTLFLYEAYTSFSATPA 952
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YNDWY+S Y+VFFTSLPVI LG+FDQDVSA CLK+P LY EGV++ LFSW RI+ WM +
Sbjct: 953 YNDWYLSLYSVFFTSLPVICLGIFDQDVSAPFCLKFPVLYQEGVQNLLFSWRRILSWMFH 1012
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
G S++ IFFL S+ +QAF +G+ +ILG TMY+ VVW V+ QM L+I+YFT IQ
Sbjct: 1013 GFCSAIIIFFLCKTSLESQAFNHEGKTAGRDILGGTMYTCVVWVVSLQMVLTISYFTLIQ 1072
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H +WGS+ WY+FL+VYG L +S+ AYMVF+EA AP+ YW+ T+ VV+ ++PYF
Sbjct: 1073 HVVVWGSVVIWYLFLMVYGSLPIRMSTDAYMVFLEALAPAPSYWITTLFVVLSTMMPYFI 1132
Query: 421 YRAFQSRFLPMYHDIIQRKRVE 442
+ A Q RF PM H +Q R E
Sbjct: 1133 FSAIQMRFFPMSHGTVQLLRYE 1154
>UniRef100_Q9FW39 Similar to an Arabidopsis putative P-type transporting ATPase [Oryza
sativa]
Length = 1206
Score = 568 bits (1464), Expect = e-160
Identities = 282/438 (64%), Positives = 336/438 (76%), Gaps = 8/438 (1%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQG-DKDALV 59
KLAQAGIK+WVLTGDKMETA+NIGYACSLLRQ M +I ITL+ PDI++LEK G DK A+
Sbjct: 720 KLAQAGIKIWVLTGDKMETAINIGYACSLLRQGMTQITITLEQPDIIALEKGGGDKAAVA 779
Query: 60 KASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAV 119
KAS E++ KQI+EG ++ + AF LIIDGKSL Y+L ++ + + +LAV
Sbjct: 780 KASKENVVKQINEGKKRIDGSVVGE-------AFALIIDGKSLTYALEEDAKGALMDLAV 832
Query: 120 SCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQA 179
C SVICCRSSPKQKA VTRLVK TGK L+IGDGANDVGM+QEA IGVGISGAEGMQA
Sbjct: 833 GCKSVICCRSSPKQKALVTRLVKESTGKVSLAIGDGANDVGMIQEADIGVGISGAEGMQA 892
Query: 180 VMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQP 239
VMASD +IAQFRFLERLLLVHGHWCY RIS MICYFFYKNI FG TLF +EAY SFSGQ
Sbjct: 893 VMASDVSIAQFRFLERLLLVHGHWCYSRISAMICYFFYKNITFGVTLFLYEAYTSFSGQT 952
Query: 240 AYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWML 299
YNDW +S YNVFFTSLPVIA+GVFDQDVSA+ CL+YP LY EG ++ LF W R++GWM
Sbjct: 953 FYNDWALSTYNVFFTSLPVIAMGVFDQDVSARFCLRYPMLYQEGPQNLLFRWSRLLGWMA 1012
Query: 300 NGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWI 359
GV S + IFFLT+ ++ +QAFRR G+VVD IL T Y+ VVW VN QM ++ NYFT +
Sbjct: 1013 YGVASGVIIFFLTSAALQHQAFRRGGEVVDLAILSGTAYTCVVWAVNAQMTVTANYFTLV 1072
Query: 360 QHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYF 419
QH IWGS+ WYVFLL YG ++PA S+ +M+F + A + YW+ T+LV LLPYF
Sbjct: 1073 QHACIWGSVALWYVFLLAYGAITPAFSTNYFMLFTDGLAAAPSYWVVTLLVPAAALLPYF 1132
Query: 420 TYRAFQSRFLPMYHDIIQ 437
TY A ++RF P YH+ IQ
Sbjct: 1133 TYSAAKTRFFPDYHNKIQ 1150
>UniRef100_Q7EYN0 Putative ATPase [Oryza sativa]
Length = 1171
Score = 552 bits (1423), Expect = e-156
Identities = 274/450 (60%), Positives = 342/450 (75%), Gaps = 21/450 (4%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIG-----------YACSLLRQDMKKIVITLDSPDILSL 49
KLAQAGIK+W+LTGDK+ETAVNIG YAC+LLR+ M+++ ITLD+P ++
Sbjct: 722 KLAQAGIKIWILTGDKLETAVNIGLVPYVAYVPDNYACNLLRKGMEEVYITLDNPGT-NV 780
Query: 50 EKQGDKDALVKASLESIKKQISEGISQVKSAKESSNTDKETSA-FGLIIDGKSLDYSLNK 108
++ + ++ A E I +++ + Q+ K TSA F LIIDG +L ++L
Sbjct: 781 PEEHNGESSGMAPYEQIGRKLEDARRQI--------LQKGTSAPFALIIDGNALTHALMG 832
Query: 109 NLEKSFFELAVSCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIG 168
L+ +F +LAV CASV+CCR SPKQKA +TRLVK KT L+IGDGANDVGMLQEA IG
Sbjct: 833 GLKTAFLDLAVDCASVLCCRISPKQKALITRLVKNRIRKTTLAIGDGANDVGMLQEADIG 892
Query: 169 VGISGAEGMQAVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFW 228
VGISGAEGMQAVMASDFAIAQFRFLERLLLVHGHWCYRRI+ MICYFF+KNI FGFTLFW
Sbjct: 893 VGISGAEGMQAVMASDFAIAQFRFLERLLLVHGHWCYRRIAAMICYFFFKNITFGFTLFW 952
Query: 229 FEAYASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTL 288
FEA+A FS QP YNDW++SFYNV FTSLPVIALGVFD+DVS+++CL+ P L+ +GV +
Sbjct: 953 FEAHAMFSAQPGYNDWFISFYNVAFTSLPVIALGVFDKDVSSRVCLEVPSLHQDGVNNLF 1012
Query: 289 FSWPRIIGWMLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQ 348
FSW RI+ WMLNGV S+ I+F ++++ QA R+DG V F+ILGVTMY+ VVWTVNCQ
Sbjct: 1013 FSWSRILSWMLNGVCCSIIIYFGALHAVLIQAVRQDGHVAGFDILGVTMYTCVVWTVNCQ 1072
Query: 349 MALSINYFTWIQHFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATV 408
+AL I+YFTWIQHF IWGSI WY FL++YG P IS++AY VF EACA S +YWL+T+
Sbjct: 1073 LALYISYFTWIQHFVIWGSILIWYTFLVIYGSFPPTISTSAYHVFWEACASSPLYWLSTL 1132
Query: 409 LVVVCVLLPYFTYRAFQSRFLPMYHDIIQR 438
++VV L+PYF Y+ QS F P + D +QR
Sbjct: 1133 VIVVTALIPYFLYKITQSLFCPQHCDQVQR 1162
>UniRef100_Q9LNQ4 Potential phospholipid-transporting ATPase 4 [Arabidopsis thaliana]
Length = 1216
Score = 534 bits (1375), Expect = e-150
Identities = 267/471 (56%), Positives = 339/471 (71%), Gaps = 24/471 (5%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAG+KLWVLTGDKMETA+NIGY+CSLLRQ MK+I IT+ + + S + + KD
Sbjct: 728 KLAQAGLKLWVLTGDKMETAINIGYSCSLLRQGMKQICITVVNSEGASQDAKAVKD---- 783
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
+I QI++ + VK K+ +AF LIIDGK+L Y+L ++ F LAV
Sbjct: 784 ----NILNQITKAVQMVKLEKDPH------AAFALIIDGKTLTYALEDEMKYQFLALAVD 833
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCR SPKQKA VTRLVK GTGK L+IGDGANDVGM+QEA IGVGISG EGMQAV
Sbjct: 834 CASVICCRVSPKQKALVTRLVKEGTGKITLAIGDGANDVGMIQEADIGVGISGVEGMQAV 893
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
MASDF+IAQFRFLERLL+VHGHWCY+RI+ MICYFFYKNIAFG TLF+FEA+ FSGQ
Sbjct: 894 MASDFSIAQFRFLERLLVVHGHWCYKRIAQMICYFFYKNIAFGLTLFYFEAFTGFSGQSV 953
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YND+Y+ +NV TSLPVIALGVF+QDVS+++CL++P LY +G ++ F W RI+GWM N
Sbjct: 954 YNDYYLLLFNVVLTSLPVIALGVFEQDVSSEICLQFPALYQQGKKNLFFDWYRILGWMGN 1013
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GV SSL IFFL I QAFR GQ D + +G TM++ ++W VN Q+AL++++FTWIQ
Sbjct: 1014 GVYSSLVIFFLNIGIIYEQAFRVSGQTADMDAVGTTMFTCIIWAVNVQIALTVSHFTWIQ 1073
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGSI WY+F+ +YG + P++S Y + VE AP+ +YW+AT LV V +LPYF
Sbjct: 1074 HVLIWGSIGLWYLFVALYGMMPPSLSGNIYRILVEILAPAPIYWIATFLVTVTTVLPYFA 1133
Query: 421 YRAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQGKLMHMRERLKQRE 471
+ +FQ P+ H IIQ ++ ++ ++ D + M RER K RE
Sbjct: 1134 HISFQRFLHPLDHHIIQ--EIKYYKRDVED--------RRMWTRERTKARE 1174
>UniRef100_Q9SGG3 Potential phospholipid-transporting ATPase 5 [Arabidopsis thaliana]
Length = 1228
Score = 529 bits (1363), Expect = e-149
Identities = 260/486 (53%), Positives = 338/486 (69%), Gaps = 28/486 (5%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAG+KLWVLTGDKMETA+NIG+ACSLLRQ M++I IT S+ +G +
Sbjct: 739 KLAQAGLKLWVLTGDKMETAINIGFACSLLRQGMRQICIT-------SMNSEGGSQDSKR 791
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
E+I Q+++ + VK K+ +AF LIIDGK+L Y+L +++ F LAV
Sbjct: 792 VVKENILNQLTKAVQMVKLEKDPH------AAFALIIDGKTLTYALEDDMKYQFLALAVD 845
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCR SPKQKA V RLVK GTGKT L+IGDGANDVGM+QEA IGVGISG EGMQAV
Sbjct: 846 CASVICCRVSPKQKALVVRLVKEGTGKTTLAIGDGANDVGMIQEADIGVGISGVEGMQAV 905
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
MASDF+IAQFRFLERLL+VHGHWCY+RI+ MICYFFYKNIAFG TLF+FEA+ FSGQ
Sbjct: 906 MASDFSIAQFRFLERLLVVHGHWCYKRIAQMICYFFYKNIAFGLTLFYFEAFTGFSGQSV 965
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YND+Y+ +NV TSLPVIALGVF+QDVS+++CL++P LY +G ++ F W RI+GWM N
Sbjct: 966 YNDYYLLLFNVVLTSLPVIALGVFEQDVSSEICLQFPALYQQGTKNLFFDWSRILGWMCN 1025
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GV +SL IFFL I +QAFR +GQ D + +G TM++ ++W N Q+AL++++FTWIQ
Sbjct: 1026 GVYASLVIFFLNIGIIYSQAFRDNGQTADMDAVGTTMFTCIIWAANVQIALTMSHFTWIQ 1085
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGSI WY+F+ +Y + P+ S Y + E AP+ +YW+AT+LV V +LPY
Sbjct: 1086 HVLIWGSIGMWYLFVAIYSMMPPSYSGNIYRILDEILAPAPIYWMATLLVTVAAVLPYVA 1145
Query: 421 YRAFQSRFLPMYHDIIQRKRVEGFEVE---------------ISDELPTQVQGKLMHMRE 465
+ AFQ P+ H IIQ + G ++E +V K+ H+R
Sbjct: 1146 HIAFQRFLNPLDHHIIQEIKYYGRDIEDARLWTRERTKAREKTKIGFTARVDAKIRHLRS 1205
Query: 466 RLKQRE 471
+L +++
Sbjct: 1206 KLNKKQ 1211
>UniRef100_Q9LVK9 Potential phospholipid-transporting ATPase 7 [Arabidopsis thaliana]
Length = 1247
Score = 512 bits (1318), Expect = e-143
Identities = 257/471 (54%), Positives = 326/471 (68%), Gaps = 23/471 (4%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
KLAQAG+K+WVLTGDKMETA+NIGYACSLLRQ MK+I I L + ++G
Sbjct: 750 KLAQAGLKIWVLTGDKMETAINIGYACSLLRQGMKQIYIALRN-------EEGSSQDPEA 802
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
A+ E+I QI +K K+ +AF LIIDGK+L Y+L +++ F LAV
Sbjct: 803 AARENILMQIINASQMIKLEKDPH------AAFALIIDGKTLTYALEDDIKYQFLALAVD 856
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCR SPKQKA VTRL K GTGKT L+IGDGANDVGM+QEA IGVGISG EGMQAV
Sbjct: 857 CASVICCRVSPKQKALVTRLAKEGTGKTTLAIGDGANDVGMIQEADIGVGISGVEGMQAV 916
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
MASDF+IAQFRFLERLL+VHGHWCY+RI+ MICYFFYKNI FG TLF+FEA+ FSGQ
Sbjct: 917 MASDFSIAQFRFLERLLVVHGHWCYKRIAQMICYFFYKNITFGLTLFYFEAFTGFSGQAI 976
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
YND Y+ +NV TSLPVIALGVF+QDVS+++CL++P LY +G ++ F W RIIGWM N
Sbjct: 977 YNDSYLLLFNVILTSLPVIALGVFEQDVSSEVCLQFPALYQQGPKNLFFDWYRIIGWMAN 1036
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
GV +S+ IF L Q+F GQ D + +G M++ ++W VN Q+AL++++FTWIQ
Sbjct: 1037 GVYASVVIFSLNIGIFHVQSFCSGGQTADMDAMGTAMFTCIIWAVNVQIALTMSHFTWIQ 1096
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H IWGSI WY+FL ++G L P +S + + E AP+ ++WL ++LV+ LPY
Sbjct: 1097 HVLIWGSIVTWYIFLALFGMLPPKVSGNIFHMLSETLAPAPIFWLTSLLVIAATTLPYLA 1156
Query: 421 YRAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQGKLMHMRERLKQRE 471
Y +FQ P+ H IIQ ++ F +++ DE M RER K RE
Sbjct: 1157 YISFQRSLNPLDHHIIQ--EIKHFRIDVQDE--------CMWTRERSKARE 1197
>UniRef100_Q5Z656 Putative ATPase, aminophospholipid transporter (APLT), class I, type
8A, member 1 [Oryza sativa]
Length = 1222
Score = 510 bits (1314), Expect = e-143
Identities = 251/471 (53%), Positives = 332/471 (70%), Gaps = 25/471 (5%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVK 60
+LAQAG+K+WVLTGDKMETA+NIGYACSLLRQ M++I +++ + D ++ + K
Sbjct: 743 RLAQAGLKIWVLTGDKMETAINIGYACSLLRQGMRRICLSIPTDDQVAQDAN-------K 795
Query: 61 ASLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVS 120
A+ ES+ QI+ G VK K+ +AF L+IDGK+L ++L +++ F LA+
Sbjct: 796 AAKESLMSQIANGSQMVKLEKDPD------AAFALVIDGKALTFALEDDMKHMFLNLAIE 849
Query: 121 CASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAV 180
CASVICCR SPKQKA VTRLVK G GKT L+IGDGANDVGM+QEA IGVGISG EGMQAV
Sbjct: 850 CASVICCRVSPKQKALVTRLVKEGIGKTTLAIGDGANDVGMIQEADIGVGISGVEGMQAV 909
Query: 181 MASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPA 240
MASDF+I+QFRFLERLL+VHGHWCY+RI+ MICYFFYKNIAFG T+F+FEA+A FSGQ
Sbjct: 910 MASDFSISQFRFLERLLVVHGHWCYKRIAQMICYFFYKNIAFGLTIFYFEAFAGFSGQSV 969
Query: 241 YNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLN 300
Y+DW+M +NV TSLPVI+LGVF+QDVS+++CL++P LY +G + F W RI+GWM N
Sbjct: 970 YDDWFMLLFNVVLTSLPVISLGVFEQDVSSEICLQFPALYQQGPRNLFFDWYRILGWMAN 1029
Query: 301 GVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
G+ SSLAIFFL +QA R GQ D +G TM++ ++W VN Q+AL++++FTWIQ
Sbjct: 1030 GLYSSLAIFFLNICIFYDQAIRSGGQTADMAAVGTTMFTCIIWAVNMQIALTMSHFTWIQ 1089
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFT 420
H F+WGS+ WY+F++VYG S S Y + +E P+ +YW AT+LV +PY
Sbjct: 1090 HLFVWGSVGTWYLFIIVYG--SALRSRDNYQILLEVLGPAPLYWAATLLVTAACNMPYLI 1147
Query: 421 YRAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQGKLMHMRERLKQRE 471
+ ++Q P+ H +IQ + L V+ + M RER K R+
Sbjct: 1148 HISYQRLCNPLDHHVIQEIKY----------LKKDVEDQTMWKRERSKARQ 1188
>UniRef100_Q9SLK6 Potential phospholipid-transporting ATPase 6 [Arabidopsis thaliana]
Length = 1244
Score = 503 bits (1295), Expect = e-141
Identities = 255/470 (54%), Positives = 326/470 (69%), Gaps = 23/470 (4%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
LAQAG+K+WVLTGDKMETA+NIGYACSLLRQ MK+I I+L + + E + +A K
Sbjct: 753 LAQAGLKIWVLTGDKMETAINIGYACSLLRQGMKQISISLTNVE----ESSQNSEAAAK- 807
Query: 62 SLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSC 121
ESI QI+ +K K+ +AF LIIDGK+L Y+L +++ F LAV C
Sbjct: 808 --ESILMQITNASQMIKIEKDPH------AAFALIIDGKTLTYALKDDVKYQFLALAVDC 859
Query: 122 ASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVM 181
ASVICCR SPKQKA VTRL K GTGKT L+IGDGANDVGM+QEA IGVGISG EGMQAVM
Sbjct: 860 ASVICCRVSPKQKALVTRLAKEGTGKTTLAIGDGANDVGMIQEADIGVGISGVEGMQAVM 919
Query: 182 ASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAY 241
ASDF+IAQFRFLERLL+VHGHWCY+RI+ MICYFFYKNI FG TLF+FE + FSGQ Y
Sbjct: 920 ASDFSIAQFRFLERLLVVHGHWCYKRIAQMICYFFYKNITFGLTLFYFECFTGFSGQSIY 979
Query: 242 NDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNG 301
ND Y+ +NV TSLPVI+LGVF+QDV + +CL++P LY +G ++ F W RI+GWM NG
Sbjct: 980 NDSYLLLFNVVLTSLPVISLGVFEQDVPSDVCLQFPALYQQGPKNLFFDWYRILGWMGNG 1039
Query: 302 VISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQH 361
V +S+ IF L Q+FR DGQ D +G M++ ++W VN Q+AL++++FTWIQH
Sbjct: 1040 VYASIVIFTLNLGIFHVQSFRSDGQTADMNAMGTAMFTCIIWAVNVQIALTMSHFTWIQH 1099
Query: 362 FFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCVLLPYFTY 421
IWGSI WYVFL +YG L +S + + VE AP+ ++WL ++LV+ LPY +
Sbjct: 1100 VMIWGSIGAWYVFLALYGMLPVKLSGNIFHMLVEILAPAPIFWLTSLLVIAATTLPYLFH 1159
Query: 422 RAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQVQGKLMHMRERLKQRE 471
++Q P+ H IIQ ++ F +++ DE M RE+ K RE
Sbjct: 1160 ISYQRSVNPLDHHIIQ--EIKHFRIDVEDE--------RMWKREKSKARE 1199
>UniRef100_Q9XIE6 Potential phospholipid-transporting ATPase 3 [Arabidopsis thaliana]
Length = 1213
Score = 358 bits (919), Expect = 2e-97
Identities = 193/456 (42%), Positives = 275/456 (59%), Gaps = 8/456 (1%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
L++AGIK+WVLTGDKMETA+NI YAC+L+ +MK+ VI+ ++ I E++GD+ + +
Sbjct: 697 LSRAGIKIWVLTGDKMETAINIAYACNLINNEMKQFVISSETDAIREAEERGDQVEIARV 756
Query: 62 SLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSC 121
E +K+++ + + + + + + K L+IDGK L Y+L+ +L L+++C
Sbjct: 757 IKEEVKRELKKSLEEAQHSLHTVAGPK----LSLVIDGKCLMYALDPSLRVMLLSLSLNC 812
Query: 122 ASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVM 181
SV+CCR SP QKA+VT LV+ G K LSIGDGANDV M+Q AH+G+GISG EGMQAVM
Sbjct: 813 TSVVCCRVSPLQKAQVTSLVRKGAQKITLSIGDGANDVSMIQAAHVGIGISGMEGMQAVM 872
Query: 182 ASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAY 241
ASDFAIAQFRFL LLLVHG W Y RI ++ YFFYKN+ F T FWF FSGQ Y
Sbjct: 873 ASDFAIAQFRFLTDLLLVHGRWSYLRICKVVMYFFYKNLTFTLTQFWFTFRTGFSGQRFY 932
Query: 242 NDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNG 301
+DW+ S +NV FT+LPVI LG+F++DVSA L +YP LY EG+ ++ F W + W +
Sbjct: 933 DDWFQSLFNVVFTALPVIVLGLFEKDVSASLSKRYPELYREGIRNSFFKWRVVAVWATSA 992
Query: 302 VISSLAIF-FLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQ 360
V SL + F+TT+S A G+V + +++ +V VN ++ L N T
Sbjct: 993 VYQSLVCYLFVTTSSF--GAVNSSGKVFGLWDVSTMVFTCLVIAVNVRILLMSNSITRWH 1050
Query: 361 HFFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCV-LLPYF 419
+ + GSI W VF VY + + FV S Y+ T+L+V V LL F
Sbjct: 1051 YITVGGSILAWLVFAFVYCGIMTPHDRNENVYFVIYVLMSTFYFYFTLLLVPIVSLLGDF 1110
Query: 420 TYRAFQSRFLPMYHDIIQRKRVEGFEVEISDELPTQ 455
++ + F P + I+Q + +D+L +
Sbjct: 1111 IFQGVERWFFPYDYQIVQEIHRHESDASKADQLEVE 1146
>UniRef100_Q7XEQ1 Contains similarity to chromaffin granule ATPase II homolog [Oryza
sativa]
Length = 1142
Score = 357 bits (915), Expect = 5e-97
Identities = 190/437 (43%), Positives = 261/437 (59%), Gaps = 6/437 (1%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
L+ AGIK+WVLTGDKMETA+NI YACSL+ DMK+ +I+ ++ I E +GD + +
Sbjct: 626 LSAAGIKIWVLTGDKMETAINIAYACSLVNNDMKQFIISSETDVIREAEDRGDPVEIARV 685
Query: 62 SLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSC 121
ES+K+ + + + + S+ K LIIDG+ L Y+L+ L L++ C
Sbjct: 686 IKESVKQSLKSYHEEARGSLISTPGQK----LALIIDGRCLMYALDPTLRVDLLGLSLIC 741
Query: 122 ASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVM 181
SV+CCR SP QKA+V LVK G K LSIGDGANDV M+Q AH+G+GISG EGMQAVM
Sbjct: 742 HSVVCCRVSPLQKAQVASLVKKGAHKITLSIGDGANDVSMIQAAHVGIGISGQEGMQAVM 801
Query: 182 ASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAY 241
ASDFAIAQFR+L LLLVHG W Y R+ +I YFFYKN+ F T FWF FSGQ Y
Sbjct: 802 ASDFAIAQFRYLTDLLLVHGRWSYLRLCKVITYFFYKNLTFTLTQFWFTFQTGFSGQRFY 861
Query: 242 NDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNG 301
+DW+ S YNV FT+LPVI +G+FD+DVSA L KYP LY EG+ +T F W I W
Sbjct: 862 DDWFQSLYNVIFTALPVIMVGLFDKDVSASLSKKYPKLYQEGIRNTFFKWRVIAVWAFFA 921
Query: 302 VISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQH 361
S+ +F+ T + G+++ + ++ VV TVN ++ +S N T +
Sbjct: 922 FYQSI-VFYYFTAAASRYGHGSSGKILGLWDVSTMAFTCVVVTVNLRLLMSCNSITRWHY 980
Query: 362 FFIWGSIFFWYVFLLVYGYLSPAISSTAYMVFVEACAPSAVYWLATVLVV-VCVLLPYFT 420
+ GSI W++F+ +Y + + + FV S ++ T+L+V + L F
Sbjct: 981 ISVAGSITAWFMFIFIYSAIMTSFDRQENVYFVIYVLMSTFFFYLTLLLVPIIALFGDFL 1040
Query: 421 YRAFQSRFLPMYHDIIQ 437
Y + Q F P + +IQ
Sbjct: 1041 YLSIQRWFFPYDYQVIQ 1057
>UniRef100_Q6CDH4 Yarrowia lipolytica chromosome C of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 1768
Score = 324 bits (831), Expect = 3e-87
Identities = 196/454 (43%), Positives = 264/454 (57%), Gaps = 25/454 (5%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
LA+AGIKLWVLTGDK+ETA+NIG++C+LL DM+ +VI D+ D K A KA
Sbjct: 1041 LAKAGIKLWVLTGDKVETAINIGFSCNLLNNDMELLVIRADTDD-----NDSTKGATPKA 1095
Query: 62 SLE-SIKKQISEGISQVKSAKESSNTDKETSA----FGLIIDGKSLDYSLNKNLEKSFFE 116
++ SI+K +S+ S S +E + S F +IIDG++L Y+L + F
Sbjct: 1096 AVRRSIEKYLSQYFSMSGSYEELEAAKNDHSPPKGNFAVIIDGEALTYALQSEISTQFLL 1155
Query: 117 LAVSCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEG 176
L C SV+CCR SP QKA V RLVK LSIGDGANDV M+QEA +GVGI+G EG
Sbjct: 1156 LCKQCRSVLCCRVSPAQKAAVVRLVKNTLTVMTLSIGDGANDVAMIQEADVGVGIAGEEG 1215
Query: 177 MQAVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFS 236
QAVM SD+AI QFRFL+RLLLVHG W Y+R++ MI FFYKN+ F FTLFW+ + +F
Sbjct: 1216 RQAVMCSDYAIGQFRFLDRLLLVHGRWDYKRLAEMIPNFFYKNLVFTFTLFWYGCFNTFD 1275
Query: 237 GQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIG 296
Y+ + FYN+ FTSLP+I LGV DQDV +C+ P LY G+ + R +
Sbjct: 1276 AAYLYDYTIVMFYNLAFTSLPIIFLGVLDQDVPDYICIAVPQLYRSGILGIEWGMRRFVE 1335
Query: 297 WMLNGVISSLAIFFLTTNSIINQA-FRRDGQVVDFE-ILGVTMYSIVVWTVNCQMALSIN 354
+ ++G+ SL FF N A R DG +D +G+ + SI V + C M + +N
Sbjct: 1336 YTVDGLYQSLVCFFFPFLMFYNTASVRSDGLAMDHRFFMGIPVASICV--IACNMYVIMN 1393
Query: 355 YFTWIQHFFIWGSIFFWYV-FLLVYGYLSPAISSTAYMVFVEACAP----SAVYWLATVL 409
+ W W SI + + LLVY ++ ST + F +A AP S YW +L
Sbjct: 1394 QYRW-----DWVSILIFSISILLVYFWIGVYTCSTFSIEFYKA-APMVFGSTTYWAVLLL 1447
Query: 410 VVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEG 443
VV LLP+F +F F P DI++ + +G
Sbjct: 1448 GVVAALLPHFAVLSFNKIFRPRDIDIVREEWHKG 1481
>UniRef100_Q9P424 Putative calcium transporting ATPase [Ajellomyces capsulata]
Length = 1305
Score = 322 bits (825), Expect = 1e-86
Identities = 190/449 (42%), Positives = 263/449 (58%), Gaps = 30/449 (6%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
L AGIK+WVLTGD+ ETA+NIG +C L+ +DM +++ +S AL
Sbjct: 818 LQTAGIKVWVLTGDRQETAINIGMSCKLISEDMALLIVNEES-------------AL--- 861
Query: 62 SLESIKKQISEGISQVKSAKESSNTDKETSAFGLIIDGKSLDYSLNKNLEKSFFELAVSC 121
+ K +S+ + QV+S ++ + D ET A LIIDGKSL Y+L K++EK F +LAV C
Sbjct: 862 ---ATKDNLSKKLQQVQS--QAGSPDSETLA--LIIDGKSLTYALEKDMEKIFLDLAVMC 914
Query: 122 ASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGMQAVM 181
+VICCR SP QKA V +L + +L+IGDGANDV M+Q AH+GVGISG EG+QA
Sbjct: 915 KAVICCRVSPLQKALVVKLQR-HLKALLLAIGDGANDVSMIQAAHVGVGISGVEGLQAAR 973
Query: 182 ASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSGQPAY 241
++D AIAQFRFL +LLLVHG W Y+RIS +I Y FYKNIA T FW+ SFSGQ Y
Sbjct: 974 SADVAIAQFRFLRKLLLVHGAWSYQRISKVILYSFYKNIALYMTQFWYSFQNSFSGQVIY 1033
Query: 242 NDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGWMLNG 301
W +SFYNVFFT +P A+G+FDQ +SA+L +YP LY G + F W+ NG
Sbjct: 1034 ESWTLSFYNVFFTVMPPFAMGIFDQFISARLLDRYPQLYQLGQKGVFFKMHSFWSWIGNG 1093
Query: 302 VISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFTWIQH 361
SL +FL+ + +G++ G +Y+ V+ TV + AL N +T
Sbjct: 1094 FYHSLIAYFLSQAIFLWDLPLANGKLAGHWFWGTALYTAVLATVLGKAALVTNIWTKYTF 1153
Query: 362 FFIWGSIFFWYVFLLVYGYLSPAIS---STAYMVFVEACAPSAVYWLATVLVVVCVLLPY 418
I GS+ FL VYG+ +P I ST Y + S V+WL +++ V L+
Sbjct: 1154 IAIPGSMIIRMGFLPVYGFSAPRIGAGFSTEYEGIIPNLFQSLVFWLMAIVLPVVCLVRD 1213
Query: 419 FTYRAFQSRFLPM-YHDI--IQRKRVEGF 444
F ++ + + P YH + IQ+ V+ +
Sbjct: 1214 FAWKYIKRMYFPQAYHHVQEIQKYNVQDY 1242
>UniRef100_UPI000036A08B UPI000036A08B UniRef100 entry
Length = 848
Score = 321 bits (823), Expect = 2e-86
Identities = 173/444 (38%), Positives = 255/444 (56%), Gaps = 11/444 (2%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
L+ A IK+WVLTGDK ETA+NIGYAC++L DM + + + + E+ K
Sbjct: 319 LSLANIKIWVLTGDKQETAINIGYACNMLTDDMNDVFVIAGNNAVEVREELRK----AKQ 374
Query: 62 SLESIKKQISEG--ISQVKSAKESSNTDKETSA--FGLIIDGKSLDYSLNKNLEKSFFEL 117
+L + S G + + K E + +ET + LII+G SL ++L +++ EL
Sbjct: 375 NLFGQNRNFSNGHVVCEKKQQLELDSIVEETLTGDYALIINGHSLAHALESDVKNDLLEL 434
Query: 118 AVSCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGM 177
A C +VICCR +P QKA+V LVK L+IGDGANDV M++ AHIG+GISG EG+
Sbjct: 435 ACMCKTVICCRVTPLQKAQVVELVKKYRNAVTLAIGDGANDVSMIKSAHIGIGISGQEGL 494
Query: 178 QAVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSG 237
QAV+ASD++ AQFR+L+RLLLVHG W Y R+ +CYFFYKN AF FWF + FS
Sbjct: 495 QAVLASDYSFAQFRYLQRLLLVHGRWSYFRMCKFLCYFFYKNFAFTLVHFWFGFFCGFSA 554
Query: 238 QPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGW 297
Q Y+ W+++ +N+ +TSLPV+A+G+FDQDVS + + P LY G + LF+ +
Sbjct: 555 QTVYDQWFITLFNIVYTSLPVLAMGIFDQDVSDQNSMDCPQLYKPGQLNLLFNKRKFFIC 614
Query: 298 MLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFT 357
+L+G+ +SL +FF+ + N A + D++ VTM + +V V+ Q+AL +Y+T
Sbjct: 615 VLHGIYTSLVLFFIPYGAFYNVAGEDGQHIADYQSFAVTMATSLVIVVSVQIALDTSYWT 674
Query: 358 WIQHFFIWGSIFFWYVFLLVY---GYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCV 414
+I H FIWGSI ++ L G + ++ WL +L V
Sbjct: 675 FINHVFIWGSIAIYFSILFTMHSNGIFGIFPNQFPFVGNARHSLTQKCIWLVILLTTVAS 734
Query: 415 LLPYFTYRAFQSRFLPMYHDIIQR 438
++P +R + P D I+R
Sbjct: 735 VMPVVAFRFLKVDLYPTLSDQIRR 758
>UniRef100_Q61FE5 Hypothetical protein CBG11664 [Caenorhabditis briggsae]
Length = 1213
Score = 321 bits (823), Expect = 2e-86
Identities = 186/485 (38%), Positives = 274/485 (56%), Gaps = 33/485 (6%)
Query: 1 KLAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPD--------------- 45
+L++A IK+WVLTGDK ETA+NI Y+C LL + K+IV+ +
Sbjct: 588 RLSEANIKIWVLTGDKTETAINIAYSCRLLTDETKEIVVVDGQTESEVEVQLKDTRNTFE 647
Query: 46 -ILSLEKQGDKDALVKASLESIKKQISEGISQVKSA---------KESSNTDKETSAFGL 95
IL+L G + + +E+I + S+ +S +S K + + ET L
Sbjct: 648 QILALPSPGGPGSKPRIEIETIHED-SDIVSSARSMDRNIVTPDLKSAELAENETGGVAL 706
Query: 96 IIDGKSLDYSLNKNLEKSFFELAVSCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDG 155
+I+G SL ++L LE++F E+A C +VICCR +P QKA+V LVK LSIGDG
Sbjct: 707 VINGDSLAFALGPRLERTFLEVACMCNAVICCRVTPLQKAQVVDLVKRNKKAVTLSIGDG 766
Query: 156 ANDVGMLQEAHIGVGISGAEGMQAVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYF 215
ANDV M++ AHIGVGISG EGMQAV+ASD+++ QF++LERLLLVHG W Y R++ + YF
Sbjct: 767 ANDVSMIKTAHIGVGISGQEGMQAVLASDYSVGQFKYLERLLLVHGRWSYIRMAKFLRYF 826
Query: 216 FYKNIAFGFTLFWFEAYASFSGQPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLK 275
FYKN AF T FW+ + +S Q ++ ++ YN+FFT+LPV+A+G DQDV L+
Sbjct: 827 FYKNFAFTLTNFWYSFFCGYSAQTVFDAVLIACYNLFFTALPVLAMGSLDQDVDDHYSLR 886
Query: 276 YPFLYLEGVEDTLFSWPRIIGWMLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGV 335
YP LYL G + F+ I +L+G+ SSL IFF+ + N A + D+ L
Sbjct: 887 YPKLYLPGQFNLFFNMRIFIYSVLHGMFSSLVIFFIPYGAFYNAAASSGKDLDDYSALAF 946
Query: 336 TMYSIVVWTVNCQMALSINYFTWIQHFFIWGS-IFFWYVFLLVYGYLS-----PAISSTA 389
T ++ ++ V Q+A +Y+T I HF IWGS + ++ V L+Y +L SS +
Sbjct: 947 TTFTALIVVVTGQIAFDTSYWTAISHFTIWGSLVLYFLVCFLLYEWLPVSWIVKTSSSIS 1006
Query: 390 YMVFVEACAPSAVYWLATVLVVVCVLLPYFTYRAFQSRFLPMYHDIIQRKRVEGFEVEIS 449
Y V + +W + ++V V +LLP R F P + D ++ +R G +
Sbjct: 1007 YGVAFRTMV-TPHFWFSILMVSVVLLLPVMLNRFFWFDTHPSFADRLRIRRKMGKKPSSK 1065
Query: 450 DELPT 454
D+ T
Sbjct: 1066 DDKKT 1070
>UniRef100_Q8TF62 Potential phospholipid-transporting ATPase IM [Homo sapiens]
Length = 1192
Score = 321 bits (823), Expect = 2e-86
Identities = 174/444 (39%), Positives = 255/444 (57%), Gaps = 11/444 (2%)
Query: 2 LAQAGIKLWVLTGDKMETAVNIGYACSLLRQDMKKIVITLDSPDILSLEKQGDKDALVKA 61
L+ A IK+WVLTGDK ETA+NIGYAC++L DM + + + + E+ K
Sbjct: 663 LSLANIKIWVLTGDKQETAINIGYACNMLTDDMNDVFVIAGNNAVEVREELRK----AKQ 718
Query: 62 SLESIKKQISEG--ISQVKSAKESSNTDKETSA--FGLIIDGKSLDYSLNKNLEKSFFEL 117
+L + S G + + K E + +ET + LII+G SL ++L +++ EL
Sbjct: 719 NLFGQNRNFSNGHVVCEKKQQLELDSIVEETITGDYALIINGHSLAHALESDVKNDLLEL 778
Query: 118 AVSCASVICCRSSPKQKARVTRLVKLGTGKTILSIGDGANDVGMLQEAHIGVGISGAEGM 177
A C +VICCR +P QKA+V LVK L+IGDGANDV M++ AHIGVGISG EG+
Sbjct: 779 ACMCKTVICCRVTPLQKAQVVELVKKYRNAVTLAIGDGANDVSMIKSAHIGVGISGQEGL 838
Query: 178 QAVMASDFAIAQFRFLERLLLVHGHWCYRRISLMICYFFYKNIAFGFTLFWFEAYASFSG 237
QAV+ASD++ AQFR+L+RLLLVHG W Y R+ +CYFFYKN AF FWF + FS
Sbjct: 839 QAVLASDYSFAQFRYLQRLLLVHGRWSYFRMCKFLCYFFYKNFAFTLVHFWFGFFCGFSA 898
Query: 238 QPAYNDWYMSFYNVFFTSLPVIALGVFDQDVSAKLCLKYPFLYLEGVEDTLFSWPRIIGW 297
Q Y+ W+++ +N+ +TSLPV+A+G+FDQDVS + + P LY G + LF+ +
Sbjct: 899 QTVYDQWFITLFNIVYTSLPVLAMGIFDQDVSDQNSVDCPQLYKPGQLNLLFNKRKFFIC 958
Query: 298 MLNGVISSLAIFFLTTNSIINQAFRRDGQVVDFEILGVTMYSIVVWTVNCQMALSINYFT 357
+L+G+ +SL +FF+ + N A + D++ VTM + +V V+ Q+AL +Y+T
Sbjct: 959 VLHGIYTSLVLFFIPYGAFYNVAGEDGQHIADYQSFAVTMATSLVIVVSVQIALDTSYWT 1018
Query: 358 WIQHFFIWGSIFFWYVFLLVY---GYLSPAISSTAYMVFVEACAPSAVYWLATVLVVVCV 414
+I H FIWGSI ++ L G + ++ WL +L V
Sbjct: 1019 FINHVFIWGSIAIYFSILFTMHSNGIFGIFPNQFPFVGNARHSLTQKCIWLVILLTTVAS 1078
Query: 415 LLPYFTYRAFQSRFLPMYHDIIQR 438
++P +R + P D I+R
Sbjct: 1079 VMPVVAFRFLKVDLYPTLSDQIRR 1102
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 745,830,999
Number of Sequences: 2790947
Number of extensions: 30840667
Number of successful extensions: 136298
Number of sequences better than 10.0: 1518
Number of HSP's better than 10.0 without gapping: 690
Number of HSP's successfully gapped in prelim test: 828
Number of HSP's that attempted gapping in prelim test: 133504
Number of HSP's gapped (non-prelim): 2484
length of query: 471
length of database: 848,049,833
effective HSP length: 131
effective length of query: 340
effective length of database: 482,435,776
effective search space: 164028163840
effective search space used: 164028163840
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0201.1


