

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0200.8
(213 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
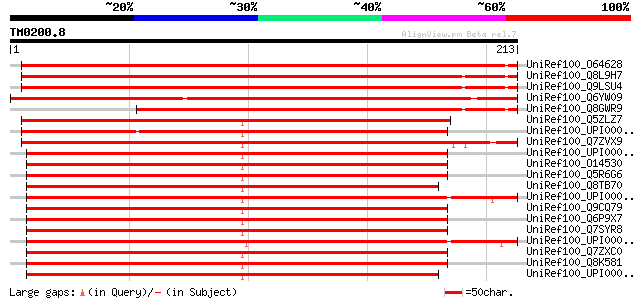
UniRef100_O64628 UPF0071 protein At2g18990 [Arabidopsis thaliana] 326 3e-88
UniRef100_Q8L9H7 Putative ATP binding protein [Arabidopsis thali... 325 5e-88
UniRef100_Q9LSU4 Arabidopsis thaliana genomic DNA, chromosome 3,... 324 8e-88
UniRef100_Q6YW09 Putative ATP binding protein associated with ce... 301 6e-81
UniRef100_Q8GWR9 Hypothetical protein At3g25580/MWL2_20 [Arabido... 252 4e-66
UniRef100_Q5ZLZ7 Hypothetical protein [Gallus gallus] 209 4e-53
UniRef100_UPI00003A9E8C UPI00003A9E8C UniRef100 entry 208 7e-53
UniRef100_Q7ZVX9 ATP binding protein associated with cell differ... 206 3e-52
UniRef100_UPI000036949F UPI000036949F UniRef100 entry 204 1e-51
UniRef100_O14530 Thioredoxin domain containing protein 9 [Homo s... 203 2e-51
UniRef100_Q5R6G6 Hypothetical protein DKFZp468L2419 [Pongo pygma... 203 3e-51
UniRef100_Q8TB70 APACD protein [Homo sapiens] 202 4e-51
UniRef100_UPI000036223A UPI000036223A UniRef100 entry 202 5e-51
UniRef100_Q9CQ79 Thioredoxin domain containing protein 9 [Mus mu... 198 7e-50
UniRef100_Q6P9X7 Txndc9 protein [Rattus norvegicus] 198 9e-50
UniRef100_Q7SYR8 Apacd-prov protein [Xenopus laevis] 197 1e-49
UniRef100_UPI0000318E3B UPI0000318E3B UniRef100 entry 197 2e-49
UniRef100_Q7ZXC0 Apacd-prov protein [Xenopus laevis] 197 2e-49
UniRef100_Q8K581 Thioredoxin domain containing protein 9 [Rattus... 196 3e-49
UniRef100_UPI000036CD43 UPI000036CD43 UniRef100 entry 194 2e-48
>UniRef100_O64628 UPF0071 protein At2g18990 [Arabidopsis thaliana]
Length = 211
Score = 326 bits (835), Expect = 3e-88
Identities = 161/208 (77%), Positives = 187/208 (89%), Gaps = 1/208 (0%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
IQE++EKQVLTV +A+EDKIDDEI++L++LD DDLE LRERRL QMKKMAEK+ RWISLG
Sbjct: 5 IQEILEKQVLTVAKAMEDKIDDEIASLEKLDEDDLEVLRERRLKQMKKMAEKKKRWISLG 64
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQHIETRFVKINAEK 125
HG+YSE+ SEKDFFSVVKAS+RVVCHF+RENWPCKV DKH+ +LAKQHIETRFVKI AEK
Sbjct: 65 HGEYSEIHSEKDFFSVVKASERVVCHFYRENWPCKVMDKHMSILAKQHIETRFVKIQAEK 124
Query: 126 SPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGE 185
SPFLAE+LKI VLPTLALIKN KVDDYVVGF+ELGG DDFSTEDLEER+A+AQVI +GE
Sbjct: 125 SPFLAERLKIVVLPTLALIKNTKVDDYVVGFNELGGKDDFSTEDLEERIARAQVIHYDGE 184
Query: 186 SSIHQARSTAQSKRSVRQATRADSSDSE 213
SS + +ST Q +R+VRQ+ R+D SDSE
Sbjct: 185 SSSLKPKSTTQVRRNVRQSARSD-SDSE 211
>UniRef100_Q8L9H7 Putative ATP binding protein [Arabidopsis thaliana]
Length = 210
Score = 325 bits (833), Expect = 5e-88
Identities = 162/208 (77%), Positives = 188/208 (89%), Gaps = 2/208 (0%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
IQE+IEKQVLTV +A+EDKIDDEI++L++LD DDLE LRERRL QMKKMAEK+ RW+S+G
Sbjct: 5 IQEIIEKQVLTVARAMEDKIDDEIASLEKLDEDDLEVLRERRLKQMKKMAEKKKRWMSVG 64
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQHIETRFVKINAEK 125
HG+YSE+ SEKDFFSVVKAS+RVVCHF+RENWPCKV DKH+ +LAKQHIETRFVKI AEK
Sbjct: 65 HGEYSEIHSEKDFFSVVKASERVVCHFYRENWPCKVMDKHMSILAKQHIETRFVKIQAEK 124
Query: 126 SPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGE 185
SPFLAE+LKI VLPTLALIKN KVDDYVVGF+ELGG DDFSTEDLEER+A+AQVI EGE
Sbjct: 125 SPFLAERLKIVVLPTLALIKNTKVDDYVVGFNELGGKDDFSTEDLEERIARAQVIHYEGE 184
Query: 186 SSIHQARSTAQSKRSVRQATRADSSDSE 213
SS+ Q +ST Q +R+VRQ+ R+D SDSE
Sbjct: 185 SSLKQ-KSTTQVRRNVRQSARSD-SDSE 210
>UniRef100_Q9LSU4 Arabidopsis thaliana genomic DNA, chromosome 3, P1 clone:MWL2
[Arabidopsis thaliana]
Length = 210
Score = 324 bits (831), Expect = 8e-88
Identities = 161/208 (77%), Positives = 188/208 (89%), Gaps = 2/208 (0%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
IQE+IEKQVLTV +A+EDKIDDEI++L++LD DDLE LRERRL QMKKMAEK+ RW+S+G
Sbjct: 5 IQEIIEKQVLTVAKAMEDKIDDEIASLEKLDEDDLEVLRERRLKQMKKMAEKKKRWMSIG 64
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQHIETRFVKINAEK 125
HG+YSE+ SEKDFFSVVK+S+RVVCHF+RENWPCKV DKH+ +LAKQHIETRFVKI AEK
Sbjct: 65 HGEYSEIHSEKDFFSVVKSSERVVCHFYRENWPCKVMDKHMSILAKQHIETRFVKIQAEK 124
Query: 126 SPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGE 185
SPFLAE+LKI VLPTLALIKN KVDDYVVGF+ELGG DDFSTEDLEER+A+AQVI EGE
Sbjct: 125 SPFLAERLKIVVLPTLALIKNTKVDDYVVGFNELGGKDDFSTEDLEERIARAQVIHYEGE 184
Query: 186 SSIHQARSTAQSKRSVRQATRADSSDSE 213
SS+ Q +ST Q +R+VRQ+ R+D SDSE
Sbjct: 185 SSLKQ-KSTTQVRRNVRQSARSD-SDSE 210
>UniRef100_Q6YW09 Putative ATP binding protein associated with cell differentiation;
Protein 1-4 [Oryza sativa]
Length = 210
Score = 301 bits (772), Expect = 6e-81
Identities = 143/213 (67%), Positives = 183/213 (85%), Gaps = 3/213 (1%)
Query: 1 MDKGKIQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNR 60
MD+ + +++EKQVL+V +AVEDK+D++I+ALDRLD DD+E+LRERR+ QM++ AE+R +
Sbjct: 1 MDQSIVGQILEKQVLSVAKAVEDKLDEQIAALDRLDPDDIEALRERRILQMRRAAERRAK 60
Query: 61 WISLGHGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQHIETRFVK 120
W +LGHG+Y E+P EK+FF+ KASDR+VCHF+R+NWPCKV DKHL +LAKQH+ETRFVK
Sbjct: 61 WRALGHGEYGEVP-EKEFFAAAKASDRLVCHFYRDNWPCKVMDKHLSILAKQHVETRFVK 119
Query: 121 INAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVI 180
++AEK+PFL EKL+I VLPTLAL+KN KV+DYVVGFDELGG DDFSTEDLEERLAKAQVI
Sbjct: 120 VHAEKAPFLTEKLRIVVLPTLALVKNTKVEDYVVGFDELGGKDDFSTEDLEERLAKAQVI 179
Query: 181 FLEGESSIHQARSTAQSKRSVRQATRADSSDSE 213
FL+GE H ++ +KRSVRQ+ +SSDSE
Sbjct: 180 FLDGEGPAHASKQA--TKRSVRQSDTGNSSDSE 210
>UniRef100_Q8GWR9 Hypothetical protein At3g25580/MWL2_20 [Arabidopsis thaliana]
Length = 158
Score = 252 bits (644), Expect = 4e-66
Identities = 123/160 (76%), Positives = 144/160 (89%), Gaps = 2/160 (1%)
Query: 54 MAEKRNRWISLGHGDYSELPSEKDFFSVVKASDRVVCHFFRENWPCKVADKHLGVLAKQH 113
MAEK+ RW+S+GHG+YSE+ SEKDFFSVVK+S+RVVCHF+RENWPCKV DKH+ +LAKQH
Sbjct: 1 MAEKKKRWMSIGHGEYSEIHSEKDFFSVVKSSERVVCHFYRENWPCKVMDKHMSILAKQH 60
Query: 114 IETRFVKINAEKSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEER 173
IETRFVKI AEKSPFLAE+LKI VLPTLALIKN KVDDYVVGF+ELGG DDFSTEDLEER
Sbjct: 61 IETRFVKIQAEKSPFLAERLKIVVLPTLALIKNTKVDDYVVGFNELGGKDDFSTEDLEER 120
Query: 174 LAKAQVIFLEGESSIHQARSTAQSKRSVRQATRADSSDSE 213
+A+AQV+ EGESS+ Q +ST Q +R+VRQ+ R+D SDSE
Sbjct: 121 IARAQVVHYEGESSLKQ-KSTTQVRRNVRQSARSD-SDSE 158
>UniRef100_Q5ZLZ7 Hypothetical protein [Gallus gallus]
Length = 209
Score = 209 bits (532), Expect = 4e-53
Identities = 95/181 (52%), Positives = 137/181 (75%), Gaps = 1/181 (0%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
+Q+V+E ++L + VE+ +D E+ LD++D D+LE L++RRL +KK +++ W+S G
Sbjct: 10 LQKVLENEILQTAKVVEEHLDAEMQKLDQMDEDELEHLKQRRLEALKKAQQQKQEWLSKG 69
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAE 124
HG+Y E+PSE+DFF VK S VVCHF+R+ + C++ DKHL VLAK+H+ET+F+K+NAE
Sbjct: 70 HGEYREIPSERDFFQEVKESKNVVCHFYRDTTFRCQIMDKHLTVLAKKHVETKFLKLNAE 129
Query: 125 KSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
KSPFL E+L+I V+PT+ALIK+ K DY+VGF +LG TDDF+TE LE RL A +I G
Sbjct: 130 KSPFLCERLRIKVIPTVALIKDGKTQDYIVGFTDLGNTDDFTTETLEWRLGCADIINYSG 189
Query: 185 E 185
+
Sbjct: 190 K 190
>UniRef100_UPI00003A9E8C UPI00003A9E8C UniRef100 entry
Length = 226
Score = 208 bits (530), Expect = 7e-53
Identities = 97/180 (53%), Positives = 138/180 (75%), Gaps = 2/180 (1%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
+Q+V+E ++L + VE+ +D E+ LD++D D+LE L++RRL +KK A+++N W+S G
Sbjct: 11 LQKVLENEILQTAKVVEEHLDAEMQKLDQMDEDELEHLKQRRLEALKK-AQQQNEWLSKG 69
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAE 124
HG+Y E+PSE+DFF VK S VVCHF+R+ + C++ DKHL VLAK+H+ET+F+K+NAE
Sbjct: 70 HGEYREIPSERDFFQEVKESKNVVCHFYRDTTFRCQIMDKHLTVLAKKHVETKFLKLNAE 129
Query: 125 KSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
KSPFL E+L+I V+PT+ALIK+ K DY+VGF +LG TDDF+TE LE RL A +I G
Sbjct: 130 KSPFLCERLRIKVIPTVALIKDGKTQDYIVGFTDLGNTDDFTTETLEWRLGCADIINYSG 189
>UniRef100_Q7ZVX9 ATP binding protein associated with cell differentiation
[Brachydanio rerio]
Length = 226
Score = 206 bits (525), Expect = 3e-52
Identities = 103/216 (47%), Positives = 152/216 (69%), Gaps = 10/216 (4%)
Query: 6 IQEVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLG 65
+ + +E+QVL + VE+++D E+ L+R+D D+LE L+ERRL +KK +++ WIS G
Sbjct: 9 VAKALEQQVLQSARMVEEQLDAELEKLERMDEDELELLKERRLEALKKAQKQKQEWISKG 68
Query: 66 HGDYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAE 124
HG+Y E+PSEKDFF VK S VVCHF+R++ + CK+ DKHL +LAK+H+ET+F+K+N E
Sbjct: 69 HGEYREIPSEKDFFPEVKESKSVVCHFYRDSTFRCKILDKHLAILAKKHLETKFIKLNVE 128
Query: 125 KSPFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
K+PFL E+L+I V+PTLAL+K+ K DY+VGF +LG TD+F TE LE RL + +I G
Sbjct: 129 KAPFLTERLRIKVIPTLALVKDGKTKDYIVGFSDLGNTDEFPTEMLEWRLGCSDIINYSG 188
Query: 185 E----SSIHQ---ARSTAQSKRSVRQATRADSSDSE 213
++ Q ++ T K+++R +A SDSE
Sbjct: 189 NLLEPPTVGQKTGSKFTKVEKKTIR--GKAYDSDSE 222
>UniRef100_UPI000036949F UPI000036949F UniRef100 entry
Length = 226
Score = 204 bits (520), Expect = 1e-51
Identities = 90/178 (50%), Positives = 135/178 (75%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+V+E Q+L + VE+ +D EI LD++D D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 KVLEHQLLQTTKLVEEHLDSEIQKLDQMDEDELERLKEKRLQALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSE+DFF VK S+ VVCHF+R++ + CK+ D+HL +L+K+H+ET+F+K+N EK+
Sbjct: 72 EYREIPSERDFFQEVKESENVVCHFYRDSTFRCKILDRHLAILSKKHLETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L+I V+PTLAL+K+ K DYVVGF +LG TDDF+TE LE RL + ++ G
Sbjct: 132 PFLCERLRIKVIPTLALLKDGKTQDYVVGFTDLGNTDDFTTETLEWRLGSSDILNYSG 189
>UniRef100_O14530 Thioredoxin domain containing protein 9 [Homo sapiens]
Length = 226
Score = 203 bits (517), Expect = 2e-51
Identities = 90/178 (50%), Positives = 134/178 (74%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+V+E Q+L + VE+ +D EI LD++D D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 KVLEHQLLQTTKLVEEHLDSEIQKLDQMDEDELERLKEKRLQALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSE+DFF VK S+ VVCHF+R++ + CK+ D+HL +L+K+H+ET+F+K+N EK+
Sbjct: 72 EYREIPSERDFFQEVKESENVVCHFYRDSTFRCKILDRHLAILSKKHLETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L I V+PTLAL+K+ K DYVVGF +LG TDDF+TE LE RL + ++ G
Sbjct: 132 PFLCERLHIKVIPTLALLKDGKTQDYVVGFTDLGNTDDFTTETLEWRLGSSDILNYSG 189
>UniRef100_Q5R6G6 Hypothetical protein DKFZp468L2419 [Pongo pygmaeus]
Length = 226
Score = 203 bits (516), Expect = 3e-51
Identities = 89/178 (50%), Positives = 135/178 (75%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+V+E Q+L + VE+ +D EI LD++D D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 KVLEHQLLQTTKLVEEHLDSEIQKLDQMDEDELERLKEKRLQALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSE++FF VK S+ VVCHF+R++ + CK+ D+HL +L+K+H+ET+F+K+N EK+
Sbjct: 72 EYREIPSEREFFQEVKESENVVCHFYRDSTFRCKILDRHLAILSKKHLETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L+I V+PTLAL+K+ K DYVVGF +LG TDDF+TE LE RL + ++ G
Sbjct: 132 PFLCERLRIKVIPTLALLKDGKTQDYVVGFTDLGNTDDFTTETLEWRLGSSDILNYSG 189
>UniRef100_Q8TB70 APACD protein [Homo sapiens]
Length = 188
Score = 202 bits (515), Expect = 4e-51
Identities = 89/174 (51%), Positives = 133/174 (76%), Gaps = 1/174 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+V+E Q+L + VE+ +D EI LD++D D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 KVLEHQLLQTTKLVEEHLDSEIQKLDQMDEDELERLKEKRLQALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSE+DFF VK S+ VVCHF+R++ + CK+ D+HL +L+K+H+ET+F+K+N EK+
Sbjct: 72 EYREIPSERDFFQEVKESENVVCHFYRDSTFRCKILDRHLAILSKKHLETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVI 180
PFL E+L I V+PTLAL+K+ K DYVVGF +LG TDDF+TE LE RL + ++
Sbjct: 132 PFLCERLHIKVIPTLALLKDGKTQDYVVGFTDLGNTDDFTTETLEWRLGSSDIL 185
>UniRef100_UPI000036223A UPI000036223A UniRef100 entry
Length = 220
Score = 202 bits (514), Expect = 5e-51
Identities = 103/211 (48%), Positives = 147/211 (68%), Gaps = 6/211 (2%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+ I K + T + VED++D E++ L +LD DDLE LRERRL ++K +++ +++ GHG
Sbjct: 7 DAITKILETSAKIVEDELDAELNQLKQLDEDDLERLRERRLEALRKAQKRKQEYLNKGHG 66
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSEKDFFS VK SD+VVCHF+R + + CK+ DKHL LAK+H+ET+F+K+N EK+
Sbjct: 67 EYREIPSEKDFFSEVKESDKVVCHFYRSSTFRCKILDKHLATLAKKHVETKFIKLNVEKA 126
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGES 186
PFL E+L+I ++PTLAL+ + K DYVVGF +LG TD+FSTE LE RL A VI G +
Sbjct: 127 PFLTERLRIKIIPTLALLLDGKSKDYVVGFTDLGNTDEFSTEMLEWRLGCADVINYSG-N 185
Query: 187 SIHQARSTAQSKRSV----RQATRADSSDSE 213
++ T +S R V ++ R DS+
Sbjct: 186 TMEPPSMTQKSGRKVTKLEKKTIRGGGLDSD 216
>UniRef100_Q9CQ79 Thioredoxin domain containing protein 9 [Mus musculus]
Length = 226
Score = 198 bits (504), Expect = 7e-50
Identities = 91/178 (51%), Positives = 132/178 (74%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
EV+E Q L + VE+ +D EI LD++ D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 EVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+ SE+DFF VK S++VVCHF+R+ + CK+ D+HL +LAK+H+ET+F+K+N EK+
Sbjct: 72 EYREIGSERDFFQEVKESEKVVCHFYRDTTFRCKILDRHLAILAKKHLETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L+I V+PTLAL+++ K DYVVGF +LG TDDF+TE LE RL + VI G
Sbjct: 132 PFLCERLRIKVIPTLALLRDGKTQDYVVGFTDLGNTDDFTTETLEWRLGCSDVINYSG 189
>UniRef100_Q6P9X7 Txndc9 protein [Rattus norvegicus]
Length = 259
Score = 198 bits (503), Expect = 9e-50
Identities = 90/178 (50%), Positives = 132/178 (73%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
EV+E Q L + VE+ +D EI LD++ D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 45 EVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQKQEWLSKGHG 104
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+ SE+DFF VK S++VVCHF+R+ + CK+ D+HL +LAK+H+ET+F+K+N EK+
Sbjct: 105 EYREIGSERDFFQEVKESEKVVCHFYRDTTFRCKILDRHLAILAKKHLETKFLKLNVEKA 164
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L+I V+PTLAL+++ K DY+VGF +LG TDDF+TE LE RL + VI G
Sbjct: 165 PFLCERLRIKVIPTLALLRDGKTQDYIVGFTDLGNTDDFTTETLEWRLGCSDVINYSG 222
>UniRef100_Q7SYR8 Apacd-prov protein [Xenopus laevis]
Length = 226
Score = 197 bits (502), Expect = 1e-49
Identities = 89/178 (50%), Positives = 134/178 (75%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+V+E Q+L + +E+++D E+ LD+ D D++E L+ERRL +KK+ +++ W+S GHG
Sbjct: 12 KVMENQLLQTAKIMEEQLDAELEKLDKTDEDEMELLKERRLEALKKVQKQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSE++FF VK S VVCHF++++ + CK+ DKHL +LAK+H+ET+F+K+N EK+
Sbjct: 72 EYREIPSEREFFQEVKESKNVVCHFYKDSTFRCKILDKHLPMLAKKHVETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L I V+PTLAL+K+ K DY+VGF +LG TD+F+TE LE RL A +I G
Sbjct: 132 PFLCERLHIKVIPTLALVKDGKTKDYIVGFTDLGNTDEFTTETLEWRLGCAGIINYSG 189
>UniRef100_UPI0000318E3B UPI0000318E3B UniRef100 entry
Length = 221
Score = 197 bits (501), Expect = 2e-49
Identities = 102/213 (47%), Positives = 146/213 (67%), Gaps = 8/213 (3%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+ I K + + VE+++D E++ L ++D DDLE LRERRL +KK +++ ++ GHG
Sbjct: 7 DAIAKVLEMSAKIVEEELDAELNQLKQMDEDDLERLRERRLEALKKAHKRKQELLTKGHG 66
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFRENWP-CKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSEKDFFS VK S++VVCHF+R + CK+ DKHL +LAK+H+ET+F+K+N EK+
Sbjct: 67 EYREIPSEKDFFSEVKESEKVVCHFYRNSTSRCKILDKHLAILAKKHVETKFIKLNVEKA 126
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEGES 186
PFL E+L+I ++PTLAL+ + K DYVVGF +LG TD+FSTE LE RL A VI G +
Sbjct: 127 PFLTERLRIKIIPTLALLVDGKSKDYVVGFTDLGNTDEFSTEMLEWRLGCADVINYSG-N 185
Query: 187 SIHQARSTAQSKRSVRQAT------RADSSDSE 213
++ T +S R V + RA SDS+
Sbjct: 186 TLEPPLMTQKSGRKVTKVEKKAIRGRAYESDSD 218
>UniRef100_Q7ZXC0 Apacd-prov protein [Xenopus laevis]
Length = 263
Score = 197 bits (500), Expect = 2e-49
Identities = 89/178 (50%), Positives = 133/178 (74%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+V+E Q+L + +E+++D E+ LD+ D D++E L+ERRL +KK +++ W+S GHG
Sbjct: 49 KVMENQLLQTAKIMEEQLDAELEKLDKTDEDEMELLKERRLEALKKAQKQKQEWLSKGHG 108
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSE++FF VK S VVCHF++++ + CK+ DKHL +LAK+H+ET+F+K+N EK+
Sbjct: 109 EYREIPSEREFFQEVKESKNVVCHFYKDSTFRCKILDKHLPMLAKKHVETKFLKLNVEKA 168
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L I V+PTLAL+K+ K DY+VGF +LG TD+F+TE LE RL A +I G
Sbjct: 169 PFLCERLHIKVIPTLALVKDGKTKDYIVGFTDLGNTDEFTTETLEWRLGCAGIINYSG 226
>UniRef100_Q8K581 Thioredoxin domain containing protein 9 [Rattus norvegicus]
Length = 226
Score = 196 bits (498), Expect = 3e-49
Identities = 90/178 (50%), Positives = 131/178 (73%), Gaps = 1/178 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
EV+E Q L + VE+ +D EI LD++ D+LE L+E+RL ++K +++ W+S GHG
Sbjct: 12 EVLENQFLQAAKLVENHLDSEIQKLDQIGEDELELLKEKRLAALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+ SE+DFF VK S++VVCHF+R+ + CK+ D+HL +LAK+H+ET+F+K N EK+
Sbjct: 72 EYREIGSERDFFQEVKESEKVVCHFYRDTTFRCKILDRHLAILAKKHLETKFLKPNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVIFLEG 184
PFL E+L+I V+PTLAL+++ K DY+VGF +LG TDDF+TE LE RL + VI G
Sbjct: 132 PFLCERLRIKVIPTLALLRDGKTQDYIVGFTDLGNTDDFTTETLEWRLGCSDVINYSG 189
>UniRef100_UPI000036CD43 UPI000036CD43 UniRef100 entry
Length = 188
Score = 194 bits (492), Expect = 2e-48
Identities = 85/174 (48%), Positives = 132/174 (75%), Gaps = 1/174 (0%)
Query: 8 EVIEKQVLTVVQAVEDKIDDEISALDRLDSDDLESLRERRLHQMKKMAEKRNRWISLGHG 67
+V+E Q+L + VE+ +D EI LD++D D+LE ++E+RL ++K +++ W+S GHG
Sbjct: 12 KVLENQLLQTTKLVEEHLDSEIQKLDQMDEDELEHIKEKRLEALRKAQQQKQEWLSKGHG 71
Query: 68 DYSELPSEKDFFSVVKASDRVVCHFFREN-WPCKVADKHLGVLAKQHIETRFVKINAEKS 126
+Y E+PSE+DFF VK S +VVCHF+R++ + CK+ D+ L +L+++H+ET+F+K+N EK+
Sbjct: 72 EYREIPSERDFFQEVKESKKVVCHFYRDSTFRCKILDRFLAILSEKHLETKFLKLNVEKA 131
Query: 127 PFLAEKLKITVLPTLALIKNAKVDDYVVGFDELGGTDDFSTEDLEERLAKAQVI 180
PFL E+L I V+PTLAL+K+ K DYVVGF +LG TDDF+TE LE L+ + ++
Sbjct: 132 PFLCERLHIKVIPTLALVKDGKAQDYVVGFTDLGNTDDFTTETLEWGLSCSDIL 185
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.132 0.362
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 318,734,029
Number of Sequences: 2790947
Number of extensions: 12629681
Number of successful extensions: 44401
Number of sequences better than 10.0: 552
Number of HSP's better than 10.0 without gapping: 246
Number of HSP's successfully gapped in prelim test: 306
Number of HSP's that attempted gapping in prelim test: 44058
Number of HSP's gapped (non-prelim): 564
length of query: 213
length of database: 848,049,833
effective HSP length: 122
effective length of query: 91
effective length of database: 507,554,299
effective search space: 46187441209
effective search space used: 46187441209
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0200.8


