

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0195a.3
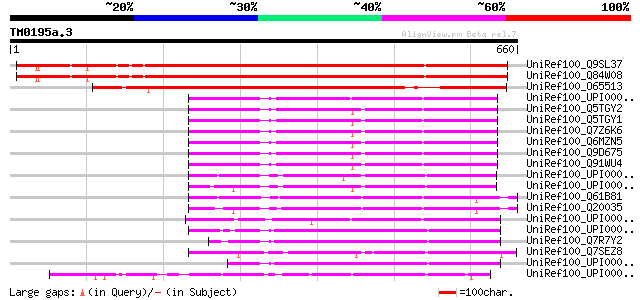
(660 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SL37 Hypothetical protein At2g18100 [Arabidopsis tha... 719 0.0
UniRef100_Q84W08 At2g18100 [Arabidopsis thaliana] 719 0.0
UniRef100_O65513 Hypothetical protein F23E13.100 [Arabidopsis th... 553 e-156
UniRef100_UPI00003AC47A UPI00003AC47A UniRef100 entry 271 3e-71
UniRef100_Q5TGY2 Novel protein [Homo sapiens] 268 3e-70
UniRef100_Q5TGY1 OTTHUMP00000044683 [Homo sapiens] 268 3e-70
UniRef100_Q7Z6K6 Hypothetical protein LOC255104 [Homo sapiens] 268 3e-70
UniRef100_Q6MZN5 Hypothetical protein DKFZp686C23231 [Homo sapiens] 268 3e-70
UniRef100_Q9D675 Mus musculus 0 day neonate skin cDNA, RIKEN ful... 265 4e-69
UniRef100_Q91WU4 RIKEN cDNA 4632413C14 [Mus musculus] 265 4e-69
UniRef100_UPI0000437E80 UPI0000437E80 UniRef100 entry 259 2e-67
UniRef100_UPI00003662F7 UPI00003662F7 UniRef100 entry 259 2e-67
UniRef100_Q61B81 Hypothetical protein CBG13461 [Caenorhabditis b... 237 9e-61
UniRef100_Q20035 Hypothetical protein F35D11.3 [Caenorhabditis e... 236 1e-60
UniRef100_UPI000023484F UPI000023484F UniRef100 entry 234 8e-60
UniRef100_UPI0000466EEF UPI0000466EEF UniRef100 entry 215 3e-54
UniRef100_Q7R7Y2 Hypothetical protein [Plasmodium yoelii yoelii] 215 3e-54
UniRef100_Q7SEZ8 Hypothetical protein [Neurospora crassa] 215 4e-54
UniRef100_UPI000046D1BB UPI000046D1BB UniRef100 entry 211 4e-53
UniRef100_UPI000042EB28 UPI000042EB28 UniRef100 entry 211 4e-53
>UniRef100_Q9SL37 Hypothetical protein At2g18100 [Arabidopsis thaliana]
Length = 655
Score = 719 bits (1855), Expect = 0.0
Identities = 377/654 (57%), Positives = 489/654 (74%), Gaps = 23/654 (3%)
Query: 9 LPPTHRYAAGALFALALHHSQIHQS-----PTT-------AQNAASVSDDPELWIHDNFG 56
L P+ RYAA ALFA+AL+ +QI QS P T + ++ +SD+ +LW+H+ G
Sbjct: 8 LTPSQRYAASALFAIALNQAQISQSKPLGIPATGSTGEPISSDSDPISDEADLWVHEVSG 67
Query: 57 LLSPVLRFLGVDDQSSNGLKETAGSSSQFRHHLGSFLKLLAEE---SDANSSERLDKEAA 113
LL PVLR L +D + +GL+E A +S+Q + H+G+F+KLL+E+ D +SSE ++KE A
Sbjct: 68 LLRPVLRCLQIDSSAWHGLEEIA-ASTQAKDHIGAFIKLLSEDVSDDDDDSSEMVEKETA 126
Query: 114 LTKAVDAMTESMSTASIAESSSGESGGHEQKSQDYLCNTTAPPVVEPHSDSKTKDSESSA 173
L KA DAM +++ ++S++ S E H + + + P E S ++ K++ES
Sbjct: 127 LAKAADAMDQNIRSSSLSVESKMEK--HVEFENECRDKYSVP---EAQSGAEVKETESHG 181
Query: 174 LLPQQKQASTELENVTLEKPLEEASLISYQRKVTVLYALVSACVADTAEVDNKCCRSRQG 233
+ + L V EKP+ E +L+S++RK+ VLY L+SAC+AD E + C R R+G
Sbjct: 182 E-GEIRDGGHNLGIVEHEKPVGEVALLSHERKINVLYELLSACLADKHEENEICTRRRKG 240
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDARH V+LRL+ATW +W +MEA+E+M A S M +G KE+ES+ + W KWKR
Sbjct: 241 YDARHHVALRLLATWFDFQWIKMEAVETMSACSAMALQKSSGNKEEESLSPNSEWAKWKR 300
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
GGI+GAAA+TGGTLMAITGGLAAPAIA G GALAPT G+IVP IGA GFAAAA A G+VA
Sbjct: 301 GGIVGAAALTGGTLMAITGGLAAPAIAAGFGALAPTLGTIVPVIGASGFAAAAEAAGTVA 360
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDF 413
GSVAVAASFGAAGAGLTG+KMA R G +EEFE K IG H + V I ++G +E+DF
Sbjct: 361 GSVAVAASFGAAGAGLTGTKMARRTGDIEEFEFKAIGENHNQGVTVEILVAGFVLKEEDF 420
Query: 414 VKPWEGHYDNSERYVLQYESKNLIALSTAIQDWLTSKIAVELMKGGAMMTVLSTLVAALA 473
VKPWEG N ERY +Q+ES+N+IA+STAIQDWLTS++A+ELM+ GAM TVL++L+AA+
Sbjct: 421 VKPWEGLTSNLERYTVQWESENIIAVSTAIQDWLTSRVAMELMRQGAMRTVLNSLLAAMV 480
Query: 474 WPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIFKCL 533
WPAT++ D IDS+W++AIDRSDKAGK+LAE L KGLQGNRPVTLVGFSLGARV+FKCL
Sbjct: 481 WPATILVAADFIDSRWSIAIDRSDKAGKLLAEALRKGLQGNRPVTLVGFSLGARVVFKCL 540
Query: 534 QFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITFRAS 593
Q LA+++ NA +VE+VV LGAPISI +ENW+ RKMVAGRF+N Y+TNDWTLG+ FRAS
Sbjct: 541 QTLAETE-KNAEIVERVVLLGAPISIKNENWRDVRKMVAGRFINVYATNDWTLGVAFRAS 599
Query: 594 LLSQGLAGIQPVDVPGIENVDVTQLIEGHSSYLWKTQKILEQLELDNYFAVFRS 647
LLSQGLAGIQPV +PGIE+VDVT ++EGHSSYLWKTQ+ILE+LELDN + VFR+
Sbjct: 600 LLSQGLAGIQPVCIPGIEDVDVTDMVEGHSSYLWKTQQILERLELDNSYPVFRN 653
>UniRef100_Q84W08 At2g18100 [Arabidopsis thaliana]
Length = 656
Score = 719 bits (1855), Expect = 0.0
Identities = 379/655 (57%), Positives = 491/655 (74%), Gaps = 24/655 (3%)
Query: 9 LPPTHRYAAGALFALALHHSQIHQS-----PTT-------AQNAASVSDDPELWIHDNFG 56
L P+ RYAA ALFA+AL+ +QI QS P T + ++ +SD+ +LW+H+ G
Sbjct: 8 LTPSQRYAASALFAIALNQAQISQSKPLGIPATGSTGEPISSDSDPISDEADLWVHEVSG 67
Query: 57 LLSPVLRFLGVDDQSSNGLKETAGSSSQFRHHLGSFLKLLAEE---SDANSSERLDKEAA 113
LL PVLR L +D + +GL+E A +S+Q + H+G+F+KLL+E+ D +SSE ++KE A
Sbjct: 68 LLRPVLRCLQIDSSAWHGLEEIA-ASTQAKDHIGAFIKLLSEDVSDDDDDSSEMVEKETA 126
Query: 114 LTKAVDAMTESMSTASIAESSSGESGGHEQKSQDYLCNTTAPPVVEPHSDSKTKDSESSA 173
L KA DAM +++ ++S++ S E H + + + P E S ++ K++ES
Sbjct: 127 LAKAADAMDQNIRSSSLSVESKMEK--HVEFENECRDKYSVP---EAQSGAEVKETESHG 181
Query: 174 LLPQQKQASTELENVTLEKPLEEASLISYQRKVTVLYALVSACVADTAEVDNKCCRSRQG 233
+ + L V EKP+ E +L+S++RK+ VLY L+SAC+AD E + C R R+G
Sbjct: 182 E-GEIRDGGHNLGIVEHEKPVGEVALLSHERKINVLYELLSACLADKHEENEICTRRRKG 240
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDARH V+LRL+ATW +W +MEA+E+M A S M +G KE+ES+ + W KWKR
Sbjct: 241 YDARHHVALRLLATWFDFQWIKMEAVETMSACSAMALQKSSGNKEEESLSPNSEWAKWKR 300
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
GGI+GAAA+TGGTLMAITGGLAAPAIA G GALAPT G+IVP IGA GFAAAA A G+VA
Sbjct: 301 GGIVGAAALTGGTLMAITGGLAAPAIAAGFGALAPTLGTIVPVIGASGFAAAAEAAGTVA 360
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVH-QGHLAVSISISGLAFEEKD 412
GSVAVAASFGAAGAGLTG+KMA R G +EEFE K IG H QG LAV I ++G +E+D
Sbjct: 361 GSVAVAASFGAAGAGLTGTKMARRTGDIEEFEFKAIGENHNQGRLAVEILVAGFVLKEED 420
Query: 413 FVKPWEGHYDNSERYVLQYESKNLIALSTAIQDWLTSKIAVELMKGGAMMTVLSTLVAAL 472
FVKPWEG N ERY +Q+ES+N+IA+STAIQDWLTS++A+ELM+ GAM TVL++L+AA+
Sbjct: 421 FVKPWEGLTSNLERYTVQWESENIIAVSTAIQDWLTSRVAMELMRQGAMRTVLNSLLAAM 480
Query: 473 AWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIFKC 532
WPAT++ D +S+W++AIDRSDKAGK+LAE L KGLQGNRPVTLVGFSLGARV+FKC
Sbjct: 481 VWPATILVAADFTNSRWSIAIDRSDKAGKLLAEALRKGLQGNRPVTLVGFSLGARVVFKC 540
Query: 533 LQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITFRA 592
LQ LA+++ NA +VE+VV LGAPISI +ENW+ RKMVAGRF+N Y+TNDWTLG+ FRA
Sbjct: 541 LQTLAETE-KNAEIVERVVLLGAPISIKNENWRDVRKMVAGRFINVYATNDWTLGVAFRA 599
Query: 593 SLLSQGLAGIQPVDVPGIENVDVTQLIEGHSSYLWKTQKILEQLELDNYFAVFRS 647
SLLSQGLAGIQPV +PGIE+VDVT ++EGHSSYLWKTQ+ILE+LELDN + VFR+
Sbjct: 600 SLLSQGLAGIQPVCIPGIEDVDVTDMVEGHSSYLWKTQQILERLELDNSYPVFRN 654
>UniRef100_O65513 Hypothetical protein F23E13.100 [Arabidopsis thaliana]
Length = 516
Score = 553 bits (1426), Expect = e-156
Identities = 303/552 (54%), Positives = 382/552 (68%), Gaps = 53/552 (9%)
Query: 108 LDKEAALTKAVDAMTESMSTASIAESSSGESGGHEQKSQDYLCNTTAPPVVEPHSD-SKT 166
+++E AL KA DAM S+ + ++ + +E + ++ P V D K
Sbjct: 2 VEQEMALAKAADAMVHSIQCSVSIDAKKEKHQEYENECRE---KYAVPEVKSKDVDLDKE 58
Query: 167 KDSESSALLPQQK---------QASTELENVTLEKPLEEASLISYQRKVTVLYALVSACV 217
KD + +A ++ S + E + EK +EE +L+S+QRK+ VLY L+SAC+
Sbjct: 59 KDKKEAAESAAREGLEAGVVIIDGSHKPEVLENEKSVEEVTLLSHQRKINVLYELLSACL 118
Query: 218 ADTAEVDNKCCRSRQGYDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAK 277
+D D KC R R+GYDARHRV+LRL+ATW ++W ++EA+E+MVA S M A K
Sbjct: 119 SDKHHEDKKCKRCRKGYDARHRVALRLLATWFNIEWIKVEAIETMVACSAMAIQKSAEMK 178
Query: 278 EDESIGSENNWDKWKRGGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFI 337
++++ +W KWKRGGIIGAAA+TGGTLMAITGGLAAPAIA G GALAPT G+I+P I
Sbjct: 179 GEDNLTPTTSWAKWKRGGIIGAAAITGGTLMAITGGLAAPAIAAGFGALAPTLGTIIPVI 238
Query: 338 GAGGFAAAATATGSVAGSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVH-QGH 396
GAGGFAAAA+A G+VAGSVAVAASFGAAGAGLTG+KMA RIG L+EFE K IG H QG
Sbjct: 239 GAGGFAAAASAAGTVAGSVAVAASFGAAGAGLTGTKMARRIGDLDEFEFKAIGENHNQGR 298
Query: 397 LAVSISISGLAFEEKDFVKPWEGHYDNSERYVLQYESKNLIALSTAIQDWLTSKIAVELM 456
LAV + ++G+ FEE+DFVKPWEG N ERY LQ+ESKNLI +STAIQDWLTS++A+ELM
Sbjct: 299 LAVEVLVAGVVFEEEDFVKPWEGLTSNLERYTLQWESKNLILVSTAIQDWLTSRLAMELM 358
Query: 457 KGGAMMTVLSTLVAALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRP 516
K GAM TVL++L+ ALAWPAT++ D IDSKW++AIDRSDKAG++LAEVL KGLQGNR
Sbjct: 359 KQGAMHTVLASLLMALAWPATILVAADFIDSKWSIAIDRSDKAGRLLAEVLQKGLQGNR- 417
Query: 517 VTLVGFSLGARVIFKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARK--MVAGR 574
R C N+ N + K MVAGR
Sbjct: 418 ---------YRFHDCC---------------------------NEVNIISIFKSYMVAGR 441
Query: 575 FVNAYSTNDWTLGITFRASLLSQGLAGIQPVDVPGIENVDVTQLIEGHSSYLWKTQKILE 634
F+N Y+TNDWTLGI FRASL+SQGLAGIQP+ +PGIENVDVT ++EGHSSYLWKTQ+ILE
Sbjct: 442 FINVYATNDWTLGIAFRASLISQGLAGIQPICIPGIENVDVTDMVEGHSSYLWKTQQILE 501
Query: 635 QLELDNYFAVFR 646
+LE+D Y+ VFR
Sbjct: 502 RLEIDTYYPVFR 513
>UniRef100_UPI00003AC47A UPI00003AC47A UniRef100 entry
Length = 517
Score = 271 bits (694), Expect = 3e-71
Identities = 161/403 (39%), Positives = 230/403 (56%), Gaps = 18/403 (4%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDAR RV + I L + E+EA+E + SL E + S + K KR
Sbjct: 124 YDARVRVLICHICWLLRIPLEELEALEESLLESLKEQTEEESETAEVSRKKKERRRKLKR 183
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
+IG A V GGT++ +TGGLAAP +A G + IG+ G A A GS A
Sbjct: 184 YLLIGLATVGGGTVIGLTGGLAAPLVAAGAATI----------IGSAG----AAALGSTA 229
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDF 413
G +A+ FGAAGAGLTG KM R+G++EEFE + Q H+ ++I+ A + F
Sbjct: 230 GIAVMASLFGAAGAGLTGYKMKKRVGAIEEFEFLPLTEGRQLHITIAITGWLCAGKYGSF 289
Query: 414 VKPWEGHYDNSERYVLQYESKNLIALSTAIQDWLTSKIAVELMKGGAMMTVLSTLVAALA 473
PW +SE+Y L +ESK L+ L +A+ D L + + + TVLS +V AL
Sbjct: 290 TAPWSSMLHSSEQYCLAWESKYLMELGSAL-DTLVNGFVNMMAQEALKFTVLSGIVTALT 348
Query: 474 WPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIFKCL 533
WP +L+T ++ID+ W V + RS + GK LA +LL QG RPVTL+GFSLGARVI+ CL
Sbjct: 349 WPTSLLTVANVIDNPWGVCLHRSAEVGKHLAHILLSRQQGKRPVTLIGFSLGARVIYFCL 408
Query: 534 QFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITFRAS 593
Q +A + D+ G++E VV LGAP+ + WKA K+V+GR +N Y DW LG +R S
Sbjct: 409 QEMA-QEKDSQGIIEDVVLLGAPVEGEAKYWKAITKVVSGRIINGYCRGDWLLGFVYRTS 467
Query: 594 LLSQGLAGIQPV--DVPGIENVDVTQLIEGHSSYLWKTQKILE 634
+ +AG+QP+ D + N+D++ ++ GH Y+ + IL+
Sbjct: 468 SVQLNVAGLQPIKLDDRRMVNMDLSSVVSGHLDYMKQMDTILK 510
>UniRef100_Q5TGY2 Novel protein [Homo sapiens]
Length = 580
Score = 268 bits (686), Expect = 3e-70
Identities = 159/406 (39%), Positives = 233/406 (57%), Gaps = 24/406 (5%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDAR RV + + + L V E++ +E M SL E + S + N KWKR
Sbjct: 130 YDARARVLVCHMTSLLQVPLEELDVLEEMFLESLKEIKEEESEMAEASRKKKENRRKWKR 189
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
+IG A V GGT++ +TGGLAAP +A G + IG+ G A A GS A
Sbjct: 190 YLLIGLATVGGGTVIGVTGGLAAPLVAAGAATI----------IGSAG----AAALGSAA 235
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDF 413
G + + FGAAGAGLTG KM R+G++EEF + Q H+ ++++ + + + F
Sbjct: 236 GIAIMTSLFGAAGAGLTGYKMKKRVGAIEEFTFLPLTEGRQLHITIAVTGWLASGKYRTF 295
Query: 414 VKPWEGHYDNSERYVLQYESKNLIALSTAIQ---DWLTSKIAVELMKGGAMMTVLSTLVA 470
PW + E+Y L +E+K L+ L A++ L + +A E +K TVLS +VA
Sbjct: 296 SAPWAALAHSREQYCLAWEAKYLMELGNALETILSGLANMVAQEALK----YTVLSGIVA 351
Query: 471 ALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIF 530
AL WPA+L++ ++ID+ W V + RS + GK LA +LL QG RPVTL+GFSLGARVI+
Sbjct: 352 ALTWPASLLSVANVIDNPWGVCLHRSAEVGKHLAHILLSRQQGRRPVTLIGFSLGARVIY 411
Query: 531 KCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITF 590
CLQ +A + D G++E V+ LGAP+ ++W+ RK+V+GR +N Y DW L +
Sbjct: 412 FCLQEMA-QEKDCQGIIEDVILLGAPVEGEAKHWEPFRKVVSGRIINGYCRGDWLLSFVY 470
Query: 591 RASLLSQGLAGIQPVDVPG--IENVDVTQLIEGHSSYLWKTQKILE 634
R S + +AG+QPV + +ENVD+T ++ GH Y + IL+
Sbjct: 471 RTSSVQLRVAGLQPVLLQDRRVENVDLTSVVSGHLDYAKQMDAILK 516
>UniRef100_Q5TGY1 OTTHUMP00000044683 [Homo sapiens]
Length = 634
Score = 268 bits (686), Expect = 3e-70
Identities = 159/406 (39%), Positives = 233/406 (57%), Gaps = 24/406 (5%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDAR RV + + + L V E++ +E M SL E + S + N KWKR
Sbjct: 130 YDARARVLVCHMTSLLQVPLEELDVLEEMFLESLKEIKEEESEMAEASRKKKENRRKWKR 189
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
+IG A V GGT++ +TGGLAAP +A G + IG+ G A A GS A
Sbjct: 190 YLLIGLATVGGGTVIGVTGGLAAPLVAAGAATI----------IGSAG----AAALGSAA 235
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDF 413
G + + FGAAGAGLTG KM R+G++EEF + Q H+ ++++ + + + F
Sbjct: 236 GIAIMTSLFGAAGAGLTGYKMKKRVGAIEEFTFLPLTEGRQLHITIAVTGWLASGKYRTF 295
Query: 414 VKPWEGHYDNSERYVLQYESKNLIALSTAIQ---DWLTSKIAVELMKGGAMMTVLSTLVA 470
PW + E+Y L +E+K L+ L A++ L + +A E +K TVLS +VA
Sbjct: 296 SAPWAALAHSREQYCLAWEAKYLMELGNALETILSGLANMVAQEALK----YTVLSGIVA 351
Query: 471 ALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIF 530
AL WPA+L++ ++ID+ W V + RS + GK LA +LL QG RPVTL+GFSLGARVI+
Sbjct: 352 ALTWPASLLSVANVIDNPWGVCLHRSAEVGKHLAHILLSRQQGRRPVTLIGFSLGARVIY 411
Query: 531 KCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITF 590
CLQ +A + D G++E V+ LGAP+ ++W+ RK+V+GR +N Y DW L +
Sbjct: 412 FCLQEMA-QEKDCQGIIEDVILLGAPVEGEAKHWEPFRKVVSGRIINGYCRGDWLLSFVY 470
Query: 591 RASLLSQGLAGIQPVDVPG--IENVDVTQLIEGHSSYLWKTQKILE 634
R S + +AG+QPV + +ENVD+T ++ GH Y + IL+
Sbjct: 471 RTSSVQLRVAGLQPVLLQDRRVENVDLTSVVSGHLDYAKQMDAILK 516
>UniRef100_Q7Z6K6 Hypothetical protein LOC255104 [Homo sapiens]
Length = 634
Score = 268 bits (686), Expect = 3e-70
Identities = 159/406 (39%), Positives = 233/406 (57%), Gaps = 24/406 (5%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDAR RV + + + L V E++ +E M SL E + S + N KWKR
Sbjct: 130 YDARARVLVCHMTSLLQVPLEELDVLEEMFLESLKEIKEEESEMAEASRKKKENRRKWKR 189
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
+IG A V GGT++ +TGGLAAP +A G + IG+ G A A GS A
Sbjct: 190 YLLIGLATVGGGTVIGVTGGLAAPLVAAGAATI----------IGSAG----AAALGSAA 235
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDF 413
G + + FGAAGAGLTG KM R+G++EEF + Q H+ ++++ + + + F
Sbjct: 236 GIAIMTSLFGAAGAGLTGYKMKKRVGAIEEFTFLPLTEGRQLHITIAVTGWLASGKYRTF 295
Query: 414 VKPWEGHYDNSERYVLQYESKNLIALSTAIQ---DWLTSKIAVELMKGGAMMTVLSTLVA 470
PW + E+Y L +E+K L+ L A++ L + +A E +K TVLS +VA
Sbjct: 296 SAPWAALAHSREQYCLAWEAKYLMELGNALETILSGLANMVAQEALK----YTVLSGIVA 351
Query: 471 ALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIF 530
AL WPA+L++ ++ID+ W V + RS + GK LA +LL QG RPVTL+GFSLGARVI+
Sbjct: 352 ALTWPASLLSVANVIDNPWGVCLHRSAEVGKHLAHILLSRQQGRRPVTLIGFSLGARVIY 411
Query: 531 KCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITF 590
CLQ +A + D G++E V+ LGAP+ ++W+ RK+V+GR +N Y DW L +
Sbjct: 412 FCLQEMA-QEKDCQGIIEDVILLGAPVEGEAKHWEPFRKVVSGRIINGYCRGDWLLSFVY 470
Query: 591 RASLLSQGLAGIQPVDVPG--IENVDVTQLIEGHSSYLWKTQKILE 634
R S + +AG+QPV + +ENVD+T ++ GH Y + IL+
Sbjct: 471 RTSSVQLHVAGLQPVLLQDRRVENVDLTSVVSGHLDYAKQMDAILK 516
>UniRef100_Q6MZN5 Hypothetical protein DKFZp686C23231 [Homo sapiens]
Length = 634
Score = 268 bits (686), Expect = 3e-70
Identities = 159/406 (39%), Positives = 233/406 (57%), Gaps = 24/406 (5%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDAR RV + + + L V E++ +E M SL E + S + N KWKR
Sbjct: 130 YDARARVLVCHMTSLLQVPLEELDVLEEMFLESLKEIKEEESEMAEASRKKKENRRKWKR 189
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
+IG A V GGT++ +TGGLAAP +A G + IG+ G A A GS A
Sbjct: 190 YLLIGLATVGGGTVIGVTGGLAAPLVAAGAATI----------IGSAG----AAALGSAA 235
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDF 413
G + + FGAAGAGLTG KM R+G++EEF + Q H+ ++++ + + + F
Sbjct: 236 GIAIMTSLFGAAGAGLTGYKMKKRVGAIEEFTFLPLTEGRQLHITIAVTGWLASGKYRTF 295
Query: 414 VKPWEGHYDNSERYVLQYESKNLIALSTAIQ---DWLTSKIAVELMKGGAMMTVLSTLVA 470
PW + E+Y L +E+K L+ L A++ L + +A E +K TVLS +VA
Sbjct: 296 SAPWAALAHSREQYCLAWEAKYLMELGNALETILSGLANMVAQEALK----YTVLSGIVA 351
Query: 471 ALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIF 530
AL WPA+L++ ++ID+ W V + RS + GK LA +LL QG RPVTL+GFSLGARVI+
Sbjct: 352 ALTWPASLLSVANVIDNPWGVCLHRSAEVGKHLAHILLSRQQGRRPVTLIGFSLGARVIY 411
Query: 531 KCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITF 590
CLQ +A + D G++E V+ LGAP+ ++W+ RK+V+GR +N Y DW L +
Sbjct: 412 FCLQEMA-QEKDCQGIIEDVILLGAPVEGEAKHWEPFRKVVSGRIINGYCRGDWLLSFVY 470
Query: 591 RASLLSQGLAGIQPVDVPG--IENVDVTQLIEGHSSYLWKTQKILE 634
R S + +AG+QPV + +ENVD+T ++ GH Y + IL+
Sbjct: 471 RTSSVQLHVAGLQPVLLQDRRVENVDLTSVVSGHLDYAKQMDAILK 516
>UniRef100_Q9D675 Mus musculus 0 day neonate skin cDNA, RIKEN full-length enriched
library, clone:4632413C14 product:DJ301K23.1 (NOVEL
PROTEIN SIMILAR TO PREDICTED YEAST AND WORM PROTEINS)
homolog (Mus musculus 10 days neonate skin cDNA, RIKEN
full-length enriched library, clone:4732490M16
product:DJ301K23.1 (NOVEL PROTEIN SIMILAR TO PREDICTED
YEAST AND WORM PROTEINS) homolog) [Mus musculus]
Length = 631
Score = 265 bits (676), Expect = 4e-69
Identities = 159/406 (39%), Positives = 234/406 (57%), Gaps = 24/406 (5%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDAR RV + + + L V E++ +E + SL ++ E + S + KWKR
Sbjct: 127 YDARARVLVCHVTSLLQVPMEELDILEEVFLESLKDAKEEESETAEASRKRKEKRRKWKR 186
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
+IG A V GGT++ +TGGLAAP +A G + IG+ G A A GSVA
Sbjct: 187 YLLIGLATVGGGTVIGVTGGLAAPLVAAGAATI----------IGSAG----AAALGSVA 232
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDF 413
G + + FGAAGAGLTG KM R+G++EEF + Q H+ ++I+ + + F
Sbjct: 233 GIAVMTSLFGAAGAGLTGYKMKKRVGAIEEFMFLPLTEGRQLHITIAITGWLGSGRYRTF 292
Query: 414 VKPWEGHYDNSERYVLQYESKNLIALSTAIQ---DWLTSKIAVELMKGGAMMTVLSTLVA 470
PW + E+Y L +E+K L+ L A++ L + +A E +K TVLS +VA
Sbjct: 293 NAPWMALARSQEQYCLAWEAKYLMELGNALETILSGLANMVAQEALK----YTVLSGIVA 348
Query: 471 ALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIF 530
AL WPA+L++ ++ID+ W V + RS + GK LA +LL QG RPVTL+GFSLGARVI+
Sbjct: 349 ALTWPASLLSVANVIDNPWGVCLHRSAEVGKHLAHILLSRQQGRRPVTLIGFSLGARVIY 408
Query: 531 KCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITF 590
CLQ +A + D G++E VV LGAP+ + ++W+ R +V+GR +N Y DW L +
Sbjct: 409 FCLQEMA-QEQDCQGIIEDVVLLGAPVEGDPKHWEPFRNVVSGRIINGYCRGDWLLSFVY 467
Query: 591 RASLLSQGLAGIQPVDVPG--IENVDVTQLIEGHSSYLWKTQKILE 634
R S + +AG+QPV + +ENVD+T ++ GH Y + IL+
Sbjct: 468 RTSSVQLRVAGLQPVLLQDRRMENVDLTSVVNGHLDYAKQMDAILK 513
>UniRef100_Q91WU4 RIKEN cDNA 4632413C14 [Mus musculus]
Length = 631
Score = 265 bits (676), Expect = 4e-69
Identities = 159/406 (39%), Positives = 234/406 (57%), Gaps = 24/406 (5%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDAR RV + + + L V E++ +E + SL ++ E + S + KWKR
Sbjct: 127 YDARARVLVCHVTSLLQVPMEELDILEEVFLESLKDTKEEESETAEASRKRKEKRRKWKR 186
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
+IG A V GGT++ +TGGLAAP +A G + IG+ G A A GSVA
Sbjct: 187 YLLIGLATVGGGTVIGVTGGLAAPLVAAGAATI----------IGSAG----AAALGSVA 232
Query: 354 GSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDF 413
G + + FGAAGAGLTG KM R+G++EEF + Q H+ ++I+ + + F
Sbjct: 233 GIAVMTSLFGAAGAGLTGYKMKKRVGAIEEFMFLPLTEGRQLHITIAITGWLGSGRYRTF 292
Query: 414 VKPWEGHYDNSERYVLQYESKNLIALSTAIQ---DWLTSKIAVELMKGGAMMTVLSTLVA 470
PW + E+Y L +E+K L+ L A++ L + +A E +K TVLS +VA
Sbjct: 293 NAPWMALARSQEQYCLAWEAKYLMELGNALETILSGLANMVAQEALK----YTVLSGIVA 348
Query: 471 ALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVIF 530
AL WPA+L++ ++ID+ W V + RS + GK LA +LL QG RPVTL+GFSLGARVI+
Sbjct: 349 ALTWPASLLSVANVIDNPWGVCLHRSAEVGKHLAHILLSRQQGRRPVTLIGFSLGARVIY 408
Query: 531 KCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGITF 590
CLQ +A + D G++E VV LGAP+ + ++W+ R +V+GR +N Y DW L +
Sbjct: 409 FCLQEMA-QEQDCQGIIEDVVLLGAPVEGDPKHWEPFRNVVSGRIINGYCRGDWLLSFVY 467
Query: 591 RASLLSQGLAGIQPVDVPG--IENVDVTQLIEGHSSYLWKTQKILE 634
R S + +AG+QPV + +ENVD+T ++ GH Y + IL+
Sbjct: 468 RTSSVQLRVAGLQPVLLQDRRMENVDLTSVVNGHLDYAKQMDAILK 513
>UniRef100_UPI0000437E80 UPI0000437E80 UniRef100 entry
Length = 497
Score = 259 bits (661), Expect = 2e-67
Identities = 160/406 (39%), Positives = 235/406 (57%), Gaps = 27/406 (6%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
YDAR RV +R ++ L V ++E E + L + E A+E + K +R
Sbjct: 105 YDARARVLIRHVSCLLRVTQQQLEDFEETLGEKLREA-GEESAEESSRRQKKERGRKLRR 163
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
+IG A V GGT++ +TGGLAAP +A G GA+ +GAGG A +ATG
Sbjct: 164 YLLIGLATVGGGTVIGVTGGLAAPLVAAGAGAV----------LGAGGAAVLGSATG--- 210
Query: 354 GSVAVAAS-FGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKD 412
+A+ AS FGAAGAGLTG KM R+G++EEFE + HL ++++ + +
Sbjct: 211 --IAIMASLFGAAGAGLTGYKMNKRVGAIEEFEFLPMSSGKHLHLTIAVTGWLCSGKYSS 268
Query: 413 FVKPWEGHYDNSERYVLQYES---KNLIALSTAIQDWLTSKIAVELMKGGAMMTVLSTLV 469
F PW E+Y L++ES K+L ++ ++ D L S +A E +K TVLS +V
Sbjct: 269 FQAPWSSLGACGEQYCLKWESRYLKDLGSILDSLWDGLVSVVAQEALK----YTVLSGIV 324
Query: 470 AALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVI 529
AL WPA+L+ +ID+ W V + RS + GK LA+VL QG RPV L+GFSLGARVI
Sbjct: 325 TALTWPASLLAVASVIDNPWGVCLSRSAEVGKHLAQVLRSRQQGKRPVNLIGFSLGARVI 384
Query: 530 FKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGIT 589
+ CL+ LA+ G + G+VE VV LGAP+ +++ W+ ++VAG+ VN Y DW LG
Sbjct: 385 YFCLEELANDQG-SEGVVEDVVLLGAPVDGSEKAWQRLSRVVAGKIVNGYCRGDWLLGYL 443
Query: 590 FRASLLSQGLAGIQPVDVPG--IENVDVTQLIEGHSSYLWKTQKIL 633
+R+S + +AG+QP+ I NVD++ +++GH Y+ + IL
Sbjct: 444 YRSSSVQLCVAGLQPISSKDRKIVNVDLSSVVKGHLDYMRQMDTIL 489
>UniRef100_UPI00003662F7 UPI00003662F7 UniRef100 entry
Length = 496
Score = 259 bits (661), Expect = 2e-67
Identities = 164/409 (40%), Positives = 237/409 (57%), Gaps = 33/409 (8%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWD---K 290
YDAR RV +R ++ L V ++E E +L+ L E G + +E + K
Sbjct: 104 YDARARVLIRHVSCLLRVCSQQLEEFEQ----TLVEKLGEGGEESEEESSRRRRKERGRK 159
Query: 291 WKRGGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATG 350
+R +IG A V GGT++ +TGGLAAP +A G A+ +GAGG AA +ATG
Sbjct: 160 LRRYLLIGLATVGGGTVIGVTGGLAAPLVAAGATAV----------LGAGGAAALGSATG 209
Query: 351 SVAGSVAVAAS-FGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFE 409
+A+ AS FGAAGAGLTG KM +G++EEFE + HL ++++ + +
Sbjct: 210 -----IAIMASLFGAAGAGLTGYKMNKCVGAIEEFEFLPLSPGKHLHLTIAVTGWLCSGK 264
Query: 410 EKDFVKPWEGHYDNSERYVLQYESKNLIALSTAIQ---DWLTSKIAVELMKGGAMMTVLS 466
F PW + E+Y L +ES+ L L +A+ D L S +A E +K TVLS
Sbjct: 265 YSSFQGPWVSLGEYGEQYCLVWESRFLRDLGSAMAFLLDGLVSIVAQEALK----YTVLS 320
Query: 467 TLVAALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGA 526
+VAAL WPA+L+ +ID+ W V ++RS + GK LA+VL QG RPV+L+GFSLGA
Sbjct: 321 GIVAALTWPASLLAAASVIDNPWCVCLNRSAEVGKHLAQVLRSRQQGKRPVSLIGFSLGA 380
Query: 527 RVIFKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTL 586
RVI+ CLQ LA+ G + G+VE VV LGAP+ +++ W+ ++VAG+ VN Y DW L
Sbjct: 381 RVIYYCLQELANDQG-SEGVVEDVVLLGAPVDGSEKAWEKMTRVVAGKIVNGYCRGDWLL 439
Query: 587 GITFRASLLSQGLAGIQPVDVPG--IENVDVTQLIEGHSSYLWKTQKIL 633
G +R+S +AG+QPV + I NVD++ ++ GH Y+ + IL
Sbjct: 440 GFLYRSSAAQLSVAGLQPVAIQNRRIINVDLSSVVNGHLDYMRQMDTIL 488
>UniRef100_Q61B81 Hypothetical protein CBG13461 [Caenorhabditis briggsae]
Length = 625
Score = 237 bits (604), Expect = 9e-61
Identities = 152/434 (35%), Positives = 231/434 (53%), Gaps = 34/434 (7%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNW--DKW 291
YD+R+RV LR + T LGV W E E +E +A +L++ + E + E K
Sbjct: 157 YDSRYRVLLRHLTTLLGVVWTEFEDVEDSLASTLLDE--QFVETEQSRVVREKTARNKKI 214
Query: 292 KRGGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGS 351
KR +IGAA GG L+ +TGGLAAP +A G L GG A A AT
Sbjct: 215 KRYLMIGAAGGVGGVLIGLTGGLAAPLVAASAGMLI-----------GGGTAVAGLAT-- 261
Query: 352 VAGSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIG-GVHQG-HLAVSISISGLAFE 409
AG+ + + G AGAG TG KM R+G++EEF ++ + GV L VS I
Sbjct: 262 TAGAAVLGTTMGVAGAGFTGYKMKKRVGAIEEFTVETLAEGVSLSCTLVVSGWIDSDTSP 321
Query: 410 EKDFVKPWEGHYDNSERYVLQYESKNLIALSTAIQDWLTSKIAVELMKGGAMMTVLSTLV 469
E+ FV W E+Y L+YES L+ L AI+ ++ ++V + + + T L+ LV
Sbjct: 322 EQAFVHQWRHLRHTKEQYTLRYESNYLMELGNAIEYLMSFAVSVAIQQT-LLETALAGLV 380
Query: 470 AALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGFSLGARVI 529
+A+AWP L++ ++D+ W V + R+ + G+ LAEVLL G RP+TL+GFSLGARVI
Sbjct: 381 SAVAWPVALISVSSVLDNPWNVCVSRAAEVGEHLAEVLLSRSHGKRPITLIGFSLGARVI 440
Query: 530 FKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGIT 589
F CL ++ ++ G++E V+ LGAP++ + + W +V+GR +N Y DW L
Sbjct: 441 FHCLLTMS-KRSESLGIIEDVILLGAPVTASPKEWSKVCSVVSGRVINGYCETDWLLRFL 499
Query: 590 FRASLLSQGLAGIQPVD---VPGIENVDVTQLIEGHSSYLWKTQKILEQLELDNYFAVFR 646
+R +AG PVD I N +++ +++GH Y + ++L + +
Sbjct: 500 YRTMSAQFRIAGTGPVDNRNSKKIYNYNLSHIVKGHMDYSKRLTEVLHAVGV-------- 551
Query: 647 SEQERPHEEKSIVN 660
+ PH E S+V+
Sbjct: 552 --KVGPHSEDSVVD 563
>UniRef100_Q20035 Hypothetical protein F35D11.3 [Caenorhabditis elegans]
Length = 617
Score = 236 bits (603), Expect = 1e-60
Identities = 151/441 (34%), Positives = 234/441 (52%), Gaps = 49/441 (11%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWD---- 289
YD+R+RV LR + T LGV W E E +E +A +L+ E++ + SE++
Sbjct: 158 YDSRYRVFLRHLTTLLGVVWTEFEDVEDSLASTLL---------EEQFVESEHSRTVREK 208
Query: 290 -----KWKRGGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAA 344
K KR +IGAA GG L+ +TGGLAAP +A G L IG G A
Sbjct: 209 TARNKKIKRYLMIGAAGGVGGVLIGLTGGLAAPLVAASAGML----------IGGGAVAG 258
Query: 345 AATATGSVAGSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIG-GVHQG-HLAVSIS 402
AT AG+ + + G AGAG TG KM R+G++EEF ++ + GV L VS
Sbjct: 259 LATT----AGAAVLGTTMGVAGAGFTGYKMKKRVGAIEEFSVETLSEGVSLSCSLVVSGW 314
Query: 403 ISGLAFEEKDFVKPWEGHYDNSERYVLQYESKNLIALSTAIQDWLTSKIAVELMKGGAMM 462
I ++ FV W E+Y L+YES L+ L AI+ ++ ++V + + +
Sbjct: 315 IESDTSPDQAFVHQWRHLRHTKEQYTLRYESNYLMELGNAIEYLMSFAVSVAIQQT-LLE 373
Query: 463 TVLSTLVAALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVGF 522
T L+ LV+A+AWP L++ ++D+ W V + R+ + G+ LAEVLL G RP+TL+GF
Sbjct: 374 TALAGLVSAVAWPVALMSVSSVLDNPWNVCVSRAAEVGEQLAEVLLSRSHGKRPITLIGF 433
Query: 523 SLGARVIFKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTN 582
SLGARVIF CL ++ ++ G++E V+ LGAP++ + + W +V+GR +N Y
Sbjct: 434 SLGARVIFHCLLTMS-KRSESVGIIEDVILLGAPVTASPKEWSKVCTVVSGRVINGYCET 492
Query: 583 DWTLGITFRASLLSQGLAGIQPVD---VPGIENVDVTQLIEGHSSYLWKTQKILEQLELD 639
DW L +R +AG P+D I N +++ +++GH Y + ++L + +
Sbjct: 493 DWLLRFLYRTMSAQFRIAGTGPIDNRNSKKIYNYNLSHIVKGHMDYSKRLTEVLNAVGV- 551
Query: 640 NYFAVFRSEQERPHEEKSIVN 660
+ PH E S+V+
Sbjct: 552 ---------KVGPHSEDSVVD 563
>UniRef100_UPI000023484F UPI000023484F UniRef100 entry
Length = 873
Score = 234 bits (596), Expect = 8e-60
Identities = 159/426 (37%), Positives = 235/426 (54%), Gaps = 31/426 (7%)
Query: 230 SRQGYDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKED-ESIGSENNW 288
S + Y+A RV + A+ LG+ + E V+ L+++ + A D ++ G E++
Sbjct: 376 SLEHYNANSRVLMLYAASSLGLGIKTLNQDEVKVSRGLLDAALQLPANSDAQAQGRESDP 435
Query: 289 D-KWKRGGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAAT 347
KWK +G A+V G L+ +TGGLAAP IA GLG + G G AAA
Sbjct: 436 SRKWK----VGIASVAGAVLIGVTGGLAAPFIAAGLGTVMGGLGL--------GATAAAG 483
Query: 348 ATGSVAGS-VAVAASFGAAGAGLTGSKMATRIGSLEEFELKEIGG-----------VHQG 395
G+VAGS V V FGA G +TG M +++F I G +
Sbjct: 484 YLGAVAGSGVIVGGIFGAYGGRMTGRMMDKYAREVDDFAFLPIHGSRKHFNDEKEAARED 543
Query: 396 H-LAVSISISGLAFEEKDFVKPWEGHYDNSERYVLQYESKNLIALSTAIQDWLTSKIAVE 454
H L V+I I+G EE +FV PW +SE + L++E+K L+ L AI D L + A
Sbjct: 544 HRLRVTIGITGWLTEEDNFVVPWRVIGKDSEVFGLRWETKPLLDLGHAI-DRLVTSAAWS 602
Query: 455 LMKGGAMMTVLSTLVAALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGN 514
+ T+ + L++AL P L+ + D+ ++VA R+DKAG+VLA+ L+ +QG
Sbjct: 603 AGRHVLTKTIFAGLISALTLPVGLMKVAGIADNPFSVAKSRADKAGEVLADALISKVQGE 662
Query: 515 RPVTLVGFSLGARVIFKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGR 574
RPVTL+G+SLG+RVIF CLQ L S GL+E +F+GAP + +W+ R++V+GR
Sbjct: 663 RPVTLIGYSLGSRVIFSCLQTL--SKRHAYGLIESAIFMGAPTPSDTPDWRRMRRVVSGR 720
Query: 575 FVNAYSTNDWTLGITFRASLLSQGLAGIQPVD-VPGIENVDVTQLIEGHSSYLWKTQKIL 633
VN YS ND L +R S L G+AG+Q ++ VPG+EN+DV+ +I GH Y + +IL
Sbjct: 721 LVNVYSGNDSVLAFLYRTSSLQLGVAGLQAIENVPGVENLDVSDIINGHLRYQYLLGRIL 780
Query: 634 EQLELD 639
+ L+
Sbjct: 781 RTVGLE 786
>UniRef100_UPI0000466EEF UPI0000466EEF UniRef100 entry
Length = 889
Score = 215 bits (548), Expect = 3e-54
Identities = 144/410 (35%), Positives = 213/410 (51%), Gaps = 27/410 (6%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
++AR ++ A L + + +E +A L+N+L + DES ++N R
Sbjct: 502 HNARIQLYFHRFAKCLKINELLILRIEENLASDLINALQPS---TDESTKNKNI-----R 553
Query: 294 GGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVA 353
I AAA+ GG L+A+T GLAAP I GL AL GAGG + T S A
Sbjct: 554 RIKIAAAALGGGALVALTAGLAAPGIVAGLIAL-----------GAGG--SGVTTFLSTA 600
Query: 354 GSVAVAAS-FGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKD 412
G +A S FGA GAGLTG K + RI +++ FE + G L+V++ +SG + D
Sbjct: 601 GGIAFIVSLFGAGGAGLTGYKYSRRIANIKVFEFLMLNGSISKSLSVAVCVSGEIQTDDD 660
Query: 413 FVKPWEGHYDNS--ERYVLQYESKNLIALSTAIQDWLTSKIAVELMKGGAMMTVLSTLVA 470
PW + NS + Y L++E+ L L + I+ L+ + A+ K T+ S L A
Sbjct: 661 ITLPWINAFPNSYCDLYALKWENHLLKTLGSLIETMLSQEFAITASKIWLQYTIASALTA 720
Query: 471 ALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLL-KGLQGNRPVTLVGFSLGARVI 529
ALAWP L+ +D+ + V +R+ +AG++LAE L + G RPV L+G+S+GARVI
Sbjct: 721 ALAWPLALIKYASNLDNVYLVIRERAQQAGRILAEALSDRNTVGQRPVVLIGYSVGARVI 780
Query: 530 FKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYSTNDWTLGIT 589
F CL++L N +V +F+G P + + W R V R +N YS NDW LG
Sbjct: 781 FYCLKYLYAKKLYN--IVSNAIFIGLPATTSTRVWNQIRMTVTNRVINVYSKNDWLLGFL 838
Query: 590 FRASLLSQGLAGIQPVDVPGIENVDVTQLIEGHSSYLWKTQKILEQLELD 639
+R +AG+ P++ PGIEN D + +I H Y K + I + D
Sbjct: 839 YRYMEWKLSVAGLMPINDPGIENYDASGIIHSHLDYKRKLKDIFRLINFD 888
>UniRef100_Q7R7Y2 Hypothetical protein [Plasmodium yoelii yoelii]
Length = 1196
Score = 215 bits (548), Expect = 3e-54
Identities = 140/385 (36%), Positives = 202/385 (52%), Gaps = 27/385 (7%)
Query: 259 MESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKRGGIIGAAAVTGGTLMAITGGLAAPA 318
+E +A L+N+L + +E+ +K R I AAA+ GG L+A T GLAAP
Sbjct: 834 IEENLASDLINALQPST--------NESTKNKHIRRIKIAAAALGGGALVAFTAGLAAPG 885
Query: 319 IAHGLGALAPTFGSIVPFIGAGGFAAAATATGSVAGSVAVAAS-FGAAGAGLTGSKMATR 377
I GL AL GAGG + T S AG +A S FGA GAGLTG K + R
Sbjct: 886 IVAGLIAL-----------GAGG--SGVTTFLSTAGGLAFIVSLFGAGGAGLTGYKYSRR 932
Query: 378 IGSLEEFELKEIGGVHQGHLAVSISISGLAFEEKDFVKPWEGHYDNS--ERYVLQYESKN 435
I +++ FE + G L+VS+ +SG + D PW + N + Y L++E+
Sbjct: 933 IANIKIFEFLMLNGSISKSLSVSVCVSGEIQTDDDITLPWMNAFPNCYCDLYALKWENNL 992
Query: 436 LIALSTAIQDWLTSKIAVELMKGGAMMTVLSTLVAALAWPATLVTTFDLIDSKWAVAIDR 495
L L + I+ L+ + A+ K T+ STL AALAWP L+ +D+ + V +R
Sbjct: 993 LKTLGSLIETMLSQEFAITASKIWLQYTIASTLSAALAWPLALIKYASNLDNVYLVIRER 1052
Query: 496 SDKAGKVLAEVLL-KGLQGNRPVTLVGFSLGARVIFKCLQFLADSDGDNAGLVEKVVFLG 554
+ +AG++LAE L K G RPV L+G+S+GARVIF CL++L N +V +F+G
Sbjct: 1053 AQQAGRILAEALSDKNTVGQRPVILIGYSVGARVIFYCLKYLYSKKLYN--IVSNAIFIG 1110
Query: 555 APISINDENWKAARKMVAGRFVNAYSTNDWTLGITFRASLLSQGLAGIQPVDVPGIENVD 614
P + + + W R V R +N YS NDW LG +R +AG+ P++ PGIEN D
Sbjct: 1111 LPATTSTKIWSQIRMTVTNRVINVYSKNDWLLGFLYRYMEWKLNVAGLMPINDPGIENYD 1170
Query: 615 VTQLIEGHSSYLWKTQKILEQLELD 639
+ +I H Y K + I + D
Sbjct: 1171 ASGIIHSHLDYKRKLKDIFSLINFD 1195
>UniRef100_Q7SEZ8 Hypothetical protein [Neurospora crassa]
Length = 949
Score = 215 bits (547), Expect = 4e-54
Identities = 145/442 (32%), Positives = 243/442 (54%), Gaps = 32/442 (7%)
Query: 234 YDARHRVSLRLIATWLGVKWNEMEAMESMVAFSLMNSLSEAGAKEDESIGSENNWDKWKR 293
Y A R L + + L + + + + E+ VA SL+ S + A+E + ++ ++ K
Sbjct: 225 YTAESRTFLVYLCSALELPLSFLNSQEAEVATSLVESSTSPEAREQQMSAAKEAEERKKA 284
Query: 294 GGI-----IGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFAAAATA 348
+ +G A+V G ++ +TGGLAAPA+A +G L + G G F
Sbjct: 285 NQVGRFWKVGLASVAGAAIIGVTGGLAAPAVAGVIGGLMGSVG----LGGVASFLGVFWM 340
Query: 349 TGSVAGSVAVAASFGAAGAGLTGSKMATRIGSLEEFELKEI-GGVHQGHLAVSISISGLA 407
G++ G++ FGA GA +TG + +E+F + GG +Q L ++I I+G
Sbjct: 341 NGALVGTL-----FGALGAQMTGEIVDQYAREVEDFAFLPLSGGTNQERLRLTIGINGWL 395
Query: 408 FEE-KDFVKPWEGHYDNSER-YVLQYESKNLIALSTAIQDWLTSK----IAVELMKGGAM 461
E PW S + L+YE ++ L A++ +TS + VEL+K
Sbjct: 396 TESLSSLTAPWLALDPRSSNLFALRYEVAAMLNLGNAMKAMVTSAAWTYVKVELLK---- 451
Query: 462 MTVLSTLVAALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLLKGLQGNRPVTLVG 521
TVL+TL +AL WP +L++ +D+ +++A +RS+KAGK+LA+ L+ +QG RPVTL+G
Sbjct: 452 RTVLATLWSAL-WPISLLSMASALDNPFSLAKNRSEKAGKILADALINRVQGERPVTLIG 510
Query: 522 FSLGARVIFKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAYST 581
+SLGARVI+ CL+ LA+ GLV++V+ +GAP+ + + W+ R +V+G+ N S
Sbjct: 511 YSLGARVIYSCLRTLAERRA--FGLVDEVILIGAPVPSDRDAWEMMRSVVSGKLWNVCSG 568
Query: 582 NDWTLGITFRASLLSQGLAGIQPV-DVPGIENVDVTQLIEGHSSYLWKTQKILEQLEL-- 638
ND+ LG +R S + G+AG+Q + + G+EN+D+T ++GH Y KILE+ +
Sbjct: 569 NDYLLGFIYRTSSIQLGVAGLQEIKGIEGVENLDLTGTVKGHMRYPDILGKILEKCGVEI 628
Query: 639 -DNYFAVFRSEQERPHEEKSIV 659
+ + +EQ+ EE+ I+
Sbjct: 629 KEGFGGDIEAEQDLTREEEIIL 650
>UniRef100_UPI000046D1BB UPI000046D1BB UniRef100 entry
Length = 1086
Score = 211 bits (538), Expect = 4e-53
Identities = 134/360 (37%), Positives = 192/360 (53%), Gaps = 19/360 (5%)
Query: 284 SENNWDKWKRGGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFGSIVPFIGAGGFA 343
+E+ +K R I AAA+ GG L+A T GLAAP I GL AL GAGG
Sbjct: 741 NESTKNKHIRRIKIAAAALGGGALVAFTAGLAAPGIVAGLIAL-----------GAGG-- 787
Query: 344 AAATATGSVAGSVAVAAS-FGAAGAGLTGSKMATRIGSLEEFELKEIGGVHQGHLAVSIS 402
+ T S AG +A S FGA GAGLTG K + RI +++ FE + G L+VS+
Sbjct: 788 SGVTTFLSTAGGLAFIVSLFGAGGAGLTGYKYSRRIANIKIFEFLMLNGNISKSLSVSVC 847
Query: 403 ISGLAFEEKDFVKPWEGHYDNS--ERYVLQYESKNLIALSTAIQDWLTSKIAVELMKGGA 460
+SG + D PW + + + Y L++E+ L L + I+ L+ + A+ K
Sbjct: 848 VSGEIQTDDDITLPWMNAFPSCYCDLYALKWENNLLKTLGSLIETMLSQEFAITASKIWL 907
Query: 461 MMTVLSTLVAALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLL-KGLQGNRPVTL 519
T+ STL AALAWP L+ +D+ + V +R+ +AG++LAE L K G RPV L
Sbjct: 908 QYTLASTLSAALAWPLALIKYASNLDNVYLVIRERAQQAGRILAEALSDKNTVGQRPVIL 967
Query: 520 VGFSLGARVIFKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAARKMVAGRFVNAY 579
+G+S+GARVIF CL++L N +V +F+G P + + + W R V R +N Y
Sbjct: 968 IGYSVGARVIFYCLKYLYSKKLYN--IVSNAIFIGLPATTSTKIWSQIRMTVTNRVINVY 1025
Query: 580 STNDWTLGITFRASLLSQGLAGIQPVDVPGIENVDVTQLIEGHSSYLWKTQKILEQLELD 639
S NDW LG +R +AG+ P++ PGIEN D + +I H Y K + I + D
Sbjct: 1026 SKNDWLLGFLYRYMEWKLNVAGLMPINDPGIENYDASGIIHSHLDYKRKLKDIFSLINFD 1085
>UniRef100_UPI000042EB28 UPI000042EB28 UniRef100 entry
Length = 1121
Score = 211 bits (538), Expect = 4e-53
Identities = 194/601 (32%), Positives = 290/601 (47%), Gaps = 78/601 (12%)
Query: 52 HDNFGLLSPVLRF--LGVDDQSSNGL--KETAGSSSQFRHH-LGSFLKLLAEESDANSSE 106
+D+ ++SP G DD++ L ET +Q + L SF K + A+SS
Sbjct: 356 NDSSTIISPQANVNPFGSDDENEGSLAVSETRAPGAQSKAQTLSSFEKDEGDIGGAHSSP 415
Query: 107 RLDK-----EAALTKAVDAMT----ESMSTASIAESSSGESGGHEQKSQDYLCNTTAPPV 157
+ +AA D T +S+ T ++S+S + E+ S D T P
Sbjct: 416 FSEPSNSPLDAAFASTADRPTPCRPQSLQTEDASQSTSDAT---EKPSVD----TKQTPK 468
Query: 158 VEPHSDSKTKDSESSALLPQQKQASTEL----ENVTLEKPLEEASLISYQRKVTVLYALV 213
EP + D ++ ALLP ST L E+VTL+ + TVL L
Sbjct: 469 GEPVTLFDAADPQAPALLPALPGVSTSLSTTDEHVTLDI------------RWTVLCDLF 516
Query: 214 SACVADTAEVDNKCCRSRQGYDARHRVSLRLIATWLGVKWNEMEAMESMV--AFSLMNSL 271
+AD+ +DAR R L +A LG W ++ E+ V A + +
Sbjct: 517 LVLIADSV------------FDARSRAFLVRVAAALGFDWLDVVRFENRVTEALEIQEGM 564
Query: 272 SEAGAKEDESIGSENNWDKWKRGGIIGAAAVTGGTLMAITGGLAAPAIAHGLGALAPTFG 331
+ G + E I + KR ++G AAV GG ++ ++ GL AP I GLG T G
Sbjct: 565 EKKG--QHEIIEGRRKAARNKRYAMMGLAAVGGGLVIGLSAGLMAPFIGAGLGTAFATVG 622
Query: 332 SIVPFIGAGGFAAAATATGSVAGSVAVAASFGA-AGAGLTGSKMATRIGSLEEFELKEIG 390
G GF A A G AV + G GA + G MA R + FELK +
Sbjct: 623 ----ITGTTGFLAGA-------GGAAVITTGGVLTGANIAGKGMARRTREVRIFELKPLH 671
Query: 391 GVHQGHLAVSISISGLAFEEKDFVKPWEGHYDN--SERYVLQYESKNLIALSTAIQDWLT 448
+ + I++ G + D V+ D + + + +E + + + A++ LT
Sbjct: 672 --NDKRVNCYITMGGFMASKVDDVRLPFSVLDPVVGDVFSVLWEPEMMAEMGNALKI-LT 728
Query: 449 SKIAVELMKGGAMMTVLSTLVAALAWPATLVTTFDLIDSKWAVAIDRSDKAGKVLAEVLL 508
S+I ++ + TV++ L++AL WP L LID+ W+ A+DR+ AG VLA+V++
Sbjct: 729 SEILTQVGQQVLQATVMTALMSALQWPLILTKLGYLIDNPWSNALDRARAAGLVLADVII 788
Query: 509 KGLQGNRPVTLVGFSLGARVIFKCLQFLADSDGDNAGLVEKVVFLGAPISINDENWKAAR 568
+ G RP++L+GFSLGAR IF L LA GLV++V G ++ + + W R
Sbjct: 789 QRHAGVRPISLIGFSLGARAIFYALIELARQKA--YGLVQEVFIFGTTVTASRDTWLDVR 846
Query: 569 KMVAGRFVNAYSTNDWTLGITFRASLLSQGL---AGIQPVD-VPGIENVDVTQLIEGHSS 624
+VAGRFVN Y+TNDW LG FRA+ S GL AG++PV+ V G+ENVDVT++I GH S
Sbjct: 847 SVVAGRFVNGYATNDWMLGYLFRAT--SGGLNTVAGLRPVETVWGLENVDVTEIITGHMS 904
Query: 625 Y 625
Y
Sbjct: 905 Y 905
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,039,940,477
Number of Sequences: 2790947
Number of extensions: 42801794
Number of successful extensions: 137915
Number of sequences better than 10.0: 630
Number of HSP's better than 10.0 without gapping: 129
Number of HSP's successfully gapped in prelim test: 529
Number of HSP's that attempted gapping in prelim test: 130597
Number of HSP's gapped (non-prelim): 4329
length of query: 660
length of database: 848,049,833
effective HSP length: 134
effective length of query: 526
effective length of database: 474,062,935
effective search space: 249357103810
effective search space used: 249357103810
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0195a.3


