

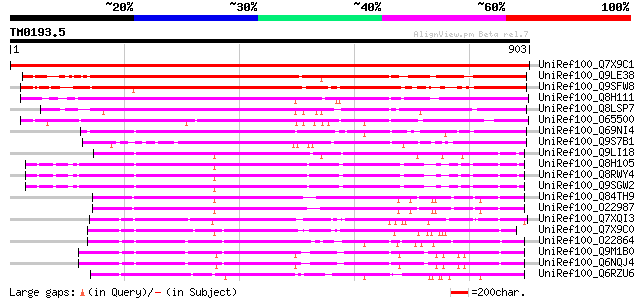
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0193.5
(903 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7X9C1 NIN-like protein 1 [Lotus japonicus] 1818 0.0
UniRef100_Q9LE38 F2D10.12 [Arabidopsis thaliana] 739 0.0
UniRef100_Q9SFW8 Hypothetical protein F15M4.15 [Arabidopsis thal... 702 0.0
UniRef100_Q8H111 Hypothetical protein At2g17150 [Arabidopsis tha... 645 0.0
UniRef100_Q8LSP7 Putataive nodule inception protein [Oryza sativa] 624 e-177
UniRef100_O65500 Hypothetical protein F23E12.170 [Arabidopsis th... 624 e-177
UniRef100_Q69NI4 Putative nodule inception protein [Oryza sativa] 595 e-168
UniRef100_Q9S7B1 Nodule inception protein [Lotus japonicus] 521 e-146
UniRef100_Q9LI18 EST AU057816(S21817) corresponds to a region of... 466 e-129
UniRef100_Q8H105 Hypothetical protein At1g64530 [Arabidopsis tha... 449 e-124
UniRef100_Q8RWY4 Hypothetical protein At1g64530 [Arabidopsis tha... 449 e-124
UniRef100_Q9SGW2 F1N19.10 [Arabidopsis thaliana] 447 e-124
UniRef100_Q84TH9 Hypothetical protein At4g24020 [Arabidopsis tha... 437 e-121
UniRef100_O22987 T19F6.16 protein [Arabidopsis thaliana] 436 e-120
UniRef100_Q7XQI3 OSJNBa0067K08.5 protein [Oryza sativa] 430 e-118
UniRef100_Q7X9C0 NIN-like protein 2 [Lotus japonicus] 416 e-114
UniRef100_O22864 Hypothetical protein At2g43500 [Arabidopsis tha... 410 e-112
UniRef100_Q9M1B0 Hypothetical protein T16L24.130 [Arabidopsis th... 408 e-112
UniRef100_Q6NQJ4 At3g59580 [Arabidopsis thaliana] 405 e-111
UniRef100_Q6RZU6 Hypothetical protein H9-8 [Musa acuminata] 404 e-111
>UniRef100_Q7X9C1 NIN-like protein 1 [Lotus japonicus]
Length = 904
Score = 1818 bits (4710), Expect = 0.0
Identities = 903/904 (99%), Positives = 903/904 (99%), Gaps = 1/904 (0%)
Query: 1 MGDGAVTTTSAAMMEAPTNETSMDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLF 60
MGDGAVTTTSAAMMEAPTNETSMDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLF
Sbjct: 1 MGDGAVTTTSAAMMEAPTNETSMDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLF 60
Query: 61 DPSFSWPALETNEPTHVEDQHESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVV 120
DPSFSWPALETNEPTHVEDQHESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVV
Sbjct: 61 DPSFSWPALETNEPTHVEDQHESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVV 120
Query: 121 EGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPIL 180
EGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPIL
Sbjct: 121 EGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPIL 180
Query: 181 SANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFF 240
SANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFF
Sbjct: 181 SANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFF 240
Query: 241 RSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEV 300
RSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEV
Sbjct: 241 RSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEV 300
Query: 301 VDLTSLKHSSIQNAK-ARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCR 359
VDLTSLKHSSIQNAK ARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCR
Sbjct: 301 VDLTSLKHSSIQNAKQARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCR 360
Query: 360 HSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITM 419
HSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITM
Sbjct: 361 HSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITM 420
Query: 420 LSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSI 479
LSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSI
Sbjct: 421 LSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSI 480
Query: 480 IIQRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSE 539
IIQRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSE
Sbjct: 481 IIQRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSE 540
Query: 540 TVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEV 599
TVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEV
Sbjct: 541 TVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEV 600
Query: 600 LRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGA 659
LRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGA
Sbjct: 601 LRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGA 660
Query: 660 SGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSS 719
SGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSS
Sbjct: 661 SGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSS 720
Query: 720 SCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAK 779
SCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAK
Sbjct: 721 SCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAK 780
Query: 780 LFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEH 839
LFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEH
Sbjct: 781 LFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEH 840
Query: 840 LLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS 899
LLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS
Sbjct: 841 LLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS 900
Query: 900 GMRS 903
GMRS
Sbjct: 901 GMRS 904
>UniRef100_Q9LE38 F2D10.12 [Arabidopsis thaliana]
Length = 844
Score = 739 bits (1907), Expect = 0.0
Identities = 429/887 (48%), Positives = 547/887 (61%), Gaps = 81/887 (9%)
Query: 23 MDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLFDP-SFSWPALETNEPTHVEDQH 81
MD ++M LLLDGCWLE + DGS FL S FDP SF W
Sbjct: 13 MDADFMDGLLLDGCWLETT-DGSEFLNIAPSTSSVSPFDPTSFMW--------------- 56
Query: 82 ESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLG 141
+P +T + S VV Y Q E S+ E + KRWWI P G G
Sbjct: 57 ----SPTQDTSALCTS----GVVSQMYG-QDCVERSSLDEFQWN--KRWWIGPG---GGG 102
Query: 142 PSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYRE 201
S+ E+L++A++ IK + + LIQ+WVPV RG + +L+ PFS D + LA YRE
Sbjct: 103 SSVTERLVQAVEHIKDYTTARGSLIQLWVPVNRGGKRVLTTKEQPFSHDPLCQRLANYRE 162
Query: 202 ISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLA 261
IS + FSAE+D + + GLPGRVF K+PEWTPDVRFF+S+EYPRV HA++ D+ GTLA
Sbjct: 163 ISVNYHFSAEQDDSKALAGLPGRVFLGKLPEWTPDVRFFKSEEYPRVHHAQDCDVRGTLA 222
Query: 262 VPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSY 321
+P+FEQGS+ CLGVIEVVMTT+ + P+LES+C+AL+ VDL S + + K D SY
Sbjct: 223 IPVFEQGSKICLGVIEVVMTTEMVKLRPELESICRALQAVDLRSTELPIPPSLKGCDLSY 282
Query: 322 EAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHSEDNYLHCISPVEQACYVGDP 381
+AALPEI+ +LR AC HKLPLAQTWVSC QQ K GCRH+++NY+HC+S ++ ACYVGDP
Sbjct: 283 KAALPEIRNLLRCACETHKLPLAQTWVSCQQQNKSGCRHNDENYIHCVSTIDDACYVGDP 342
Query: 382 SVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLFGLRAAVA 441
+VR FHEAC EHHLLKGQGVAG+AF+ N P FS+D++ K++YPLSHHA ++GL AVA
Sbjct: 343 TVREFHEACSEHHLLKGQGVAGQAFLTNGPCFSSDVSNYKKSEYPLSHHANMYGLHGAVA 402
Query: 442 IRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDKELERTSS 501
IRLR I++ + D+VLEFFLP +C+D EEQ+ ML +LS I+ RSLR +TDKELE S
Sbjct: 403 IRLRCIHTGSADFVLEFFLPKDCDDLEEQRKMLNALSTIMAHVPRSLRTVTDKELEEESE 462
Query: 502 SVEVMALEDSGFARTVKWSEPQHITSV--ASLEPEEKSSET-----VGGKFSDLREHQED 554
+E + ++ + H S ASLE ++S+ T +G F + +
Sbjct: 463 VIE----REEIVTPKIENASELHGNSPWNASLEEIQRSNNTSNPQNLGLVFDGGDKPNDG 518
Query: 555 SILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKN 614
LK + + + SS G S EK+R KADKTITL+VLRQYF GSLKDAAKN
Sbjct: 519 FGLKRGFDYTMDSNVNESSTFSSGGFSMMAEKKRTKADKTITLDVLRQYFAGSLKDAAKN 578
Query: 615 IGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFP 674
IGVC TTLKR+CRQHGI+RWPSRKIKKVGHSLQK+Q VIDSVQG SG I SFY+NFP
Sbjct: 579 IGVCPTTLKRICRQHGIQRWPSRKIKKVGHSLQKIQRVIDSVQGVSG-PLPIGSFYANFP 637
Query: 675 DLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSM 734
+L S QS+ P S + P P KS SS S S SS CS
Sbjct: 638 NLVS-------------QSQEP-SQQAKTTPPPPPPVQLAKSPVSSYSHSSNSSQCCS-- 681
Query: 735 PEQQPHTSDVACNKDPVVGKDSADV--VLKRIRSEAELKSHSENKAKLFPRSLSQETLGE 792
S+ N S DV LK+ SE EL+S S ++ L SL G
Sbjct: 682 -------SETQLNSGATTDPPSTDVGGALKKTSSEIELQSSSLDETILTLSSLENIPQG- 733
Query: 793 HTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSD 852
T ++ +D R+KV+YG+EK R RM S LL EI +RF++ D
Sbjct: 734 ------------TNLLSSQDDDFLRIKVSYGEEKIRLRMRNSRRLRDLLWEIGKRFSIED 781
Query: 853 MSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS 899
MS++D+KYLD+D EWVLLTCD D+EEC+DVC ++ S TIKL +QASS
Sbjct: 782 MSRYDLKYLDEDNEWVLLTCDEDVEECVDVCRTTPSHTIKLLLQASS 828
>UniRef100_Q9SFW8 Hypothetical protein F15M4.15 [Arabidopsis thaliana]
Length = 808
Score = 702 bits (1813), Expect = 0.0
Identities = 412/888 (46%), Positives = 539/888 (60%), Gaps = 110/888 (12%)
Query: 20 ETSMDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLFDPS-FSWPALETNEPTHVE 78
+ +MD +M LLL+GCWLE + D S FL SP S FDPS F W T + ++
Sbjct: 9 DPAMDSSFMDGLLLEGCWLETT-DASEFL-NFSPSTSVAPFDPSSFMWSP--TQDTSNSL 64
Query: 79 DQHESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSP 138
Q Q+ P +S E + S +RWWI P+
Sbjct: 65 SQMYGQDCP----------------------ERSSLEDQNQGRDLSTFNRRWWIGPSGHH 102
Query: 139 GLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLAR 198
G S+ME+L++A+ IK F + LIQ+WVPV RG + +L+ PFS D + LA
Sbjct: 103 GF--SVMERLVQAVTHIKDFTSERGSLIQLWVPVDRGGKRVLTTKEQPFSHDPMCQRLAH 160
Query: 199 YREISEGFQFSAEED-----SKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHARE 253
YREISE +QFS E++ S++LV GLPGRVF KVPEWTPDVRFF+++EYPRV+HA++
Sbjct: 161 YREISENYQFSTEQEDSDSSSRDLV-GLPGRVFLGKVPEWTPDVRFFKNEEYPRVQHAQD 219
Query: 254 FDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQN 313
D+ GTLA+P+FEQGS+ CLGVIEVVMTTQ + P LES+C+AL+ VDL S + +
Sbjct: 220 CDVRGTLAIPVFEQGSQICLGVIEVVMTTQMVKLSPDLESICRALQAVDLRSTEIPIPPS 279
Query: 314 AKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHSEDNYLHCISPVE 373
K D SY+AALPEI+ +LR AC HKLPLAQTWVSC +Q K GCRH+++NY+HC+S ++
Sbjct: 280 LKGPDFSYQAALPEIRNLLRCACETHKLPLAQTWVSCLKQSKTGCRHNDENYIHCVSTID 339
Query: 374 QACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARL 433
ACYVGDP+VR FHEAC EHHLLKGQGV G+AF+ N P FS+D++ K++YPLSHHA +
Sbjct: 340 DACYVGDPTVREFHEACSEHHLLKGQGVVGEAFLTNGPCFSSDVSSYKKSEYPLSHHATM 399
Query: 434 FGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITD 493
FGL VAIRLR I++ + D+VLEFFLP NC D EEQ+ ML +LS I+ RSLR +T
Sbjct: 400 FGLHGTVAIRLRCIHTGSVDFVLEFFLPKNCRDIEEQRKMLNALSTIMAHVPRSLRTVTQ 459
Query: 494 KELERTSSSVEVMALEDSGFARTVKWSEPQHITSV-ASLEPEEKSSETVGGKFSDLREHQ 552
KELE S+ +E + V + ++ T V S+ + G +++ E
Sbjct: 460 KELEEEGDSMVSEVIE-----KGVTLPKIENTTEVHQSISTPQNVGLVFDGGTTEMGELG 514
Query: 553 EDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAA 612
+ G E + F S+ G ++ EK+R KA+K ITL+VLRQYF GSLKDAA
Sbjct: 515 SE---YGKGVSVNENNTF----SSASGFNRVTEKKRTKAEKNITLDVLRQYFAGSLKDAA 567
Query: 613 KNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSN 672
K+IGVC TTLKR+CRQHGI+RWPSRKIKKVGHSLQK+Q VIDSV+G SG I SFY++
Sbjct: 568 KSIGVCPTTLKRICRQHGIQRWPSRKIKKVGHSLQKIQRVIDSVEGVSGHHLPIGSFYAS 627
Query: 673 FPDL-ASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSC 731
FP+L ASP S S QS+ LS S KS SSCS S SC
Sbjct: 628 FPNLAASPEAS-----SLQQQSKITTFLS-------YSHSPPAKSPGSSCSHSS----SC 671
Query: 732 SSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLG 791
SS + V+ +D D K +L RS +
Sbjct: 672 SS--------------ETQVIKEDPTD------------------KTRLVSRSFKE---- 695
Query: 792 EHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVS 851
T+T + S ++ ++D RVKV+Y +EK RF+M S + LL EIA+RF++
Sbjct: 696 --TQTTHLS-------PSSQEDDFLRVKVSYEEEKIRFKMRNSHRLKDLLWEIAKRFSIE 746
Query: 852 DMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS 899
D+S++D+KYLD+D EWVLL CD D+EEC+DVC S TIKL +Q SS
Sbjct: 747 DVSRYDLKYLDEDNEWVLLRCDDDVEECVDVCRSFPGQTIKLLLQLSS 794
>UniRef100_Q8H111 Hypothetical protein At2g17150 [Arabidopsis thaliana]
Length = 909
Score = 645 bits (1664), Expect = 0.0
Identities = 383/924 (41%), Positives = 538/924 (57%), Gaps = 93/924 (10%)
Query: 20 ETSMDFEYMSDLLLDGCWLEASADGSNFLLQQSPPFSSPLFDPSFSWPALETNEPTHVED 79
+T+MD ++M +LL DGCWLE + S +QSP S+ A+ N P
Sbjct: 27 DTAMDLDFMDELLFDGCWLETTDSKSLKQTEQSPSAST----------AMNDNSPFLCFG 76
Query: 80 QHESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPG 139
++ SQ+ N N R Q+E ++E G K WWIAP+ S G
Sbjct: 77 ENPSQD-------------NFSNEETERMFPQAEK---FLLEEAEVG-KSWWIAPSASEG 119
Query: 140 LGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARY 199
S+ E+L++A+ + + +KD L+QIWVP+ + + L+ P + +LA Y
Sbjct: 120 PSSSVKERLLQAISGLNEAVQDKDFLVQIWVPIQQEGKSFLTTWAQPHLFNQEYSSLAEY 179
Query: 200 REISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGT 259
R +SE + F A+E K+ V GLPGRVF K PEWTPDVRFFR DEYPR++ A++ D+ G+
Sbjct: 180 RHVSETYNFPADEGMKDFV-GLPGRVFLQKFPEWTPDVRFFRRDEYPRIKEAQKCDVRGS 238
Query: 260 LAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTS---LKHSSIQNAKA 316
LA+P+FE+GS TCLGV+E+V TTQ++NY +LE +CKALE VDL S L S + +
Sbjct: 239 LALPVFERGSGTCLGVVEIVTTTQKMNYRQELEKMCKALEAVDLRSSSNLNTPSSEFLQV 298
Query: 317 RDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHSEDNYLHCISPVEQAC 376
Y AALPEI++ L + C + PLA +W C +QGK G RHS++N+ C+S ++ AC
Sbjct: 299 YSDFYCAALPEIKDFLATICRSYDFPLALSWAPCARQGKVGSRHSDENFSECVSTIDSAC 358
Query: 377 YVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLFGL 436
V D + F EAC EHHLL+G+G+ GKAF + FF ++ SKT+YPL+HHA++ GL
Sbjct: 359 SVPDEQSKSFWEACSEHHLLQGEGIVGKAFEATKLFFVPEVATFSKTNYPLAHHAKISGL 418
Query: 437 RAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRV-ITDKE 495
AA+A+ L+S S ++VLEFF P C D+E Q+ ML SL + +Q+ RS + I D E
Sbjct: 419 HAALAVPLKS-KSGLVEFVLEFFFPKACLDTEAQQEMLKSLCVTLQQDFRSSNLFIKDLE 477
Query: 496 L--------------------ERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEE 535
L E S E+ E S A +K +E S++ +E
Sbjct: 478 LEVVLPVRETMLFSENLLCGAETVESLTEIQMQESSWIAHMIKANEKGKDVSLSWEYQKE 537
Query: 536 KSSETVGGKFSDLREHQEDSILKGNIECDRE-------------CSPFVE----GNLSSV 578
E G RE+ + + N+ + E P E G + +
Sbjct: 538 DPKELSSG-----RENSQLDPVPNNVPLEAEQLQQASTPGLRVDIGPSTESASTGGGNML 592
Query: 579 GISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRK 638
+ GEK+RAK +KTI LEVLRQYF GSLKDAAK+IGVC TTLKR+CRQHGI RWPSRK
Sbjct: 593 SSRRPGEKKRAKTEKTIGLEVLRQYFAGSLKDAAKSIGVCPTTLKRICRQHGIMRWPSRK 652
Query: 639 IKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNS 698
IKKVGHSL+KLQLV+DSVQGA G S ++DSFY++FP+L SPN+S S +L +E P+
Sbjct: 653 IKKVGHSLKKLQLVMDSVQGAQG-SIQLDSFYTSFPELNSPNMS--SNGPSLKSNEQPSH 709
Query: 699 LSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSAD 758
L+ Q D G ++ E +S SSSCS+ S SS++ N ++ + AD
Sbjct: 710 LNAQTDNGIMAEEN-PRSPSSSCSKSSGSSNNNE--------------NTGNILVAEDAD 754
Query: 759 VVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRV 818
VLKR SEA+L + ++ + K R+ S +T E + S L + + + A +V
Sbjct: 755 AVLKRAHSEAQLHNVNQEETKCLARTQSHKTFKEPLVLDNSSPLTGSSNTSLRARGAIKV 814
Query: 819 KVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEE 878
K T+G+ + RF + SW + L QEIARRFN+ D+S FD+KYLDDD EWVLLTC+ADL E
Sbjct: 815 KATFGEARIRFTLLPSWGFAELKQEIARRFNIDDISWFDLKYLDDDKEWVLLTCEADLVE 874
Query: 879 CIDVCLSSESSTIKLCIQASSGMR 902
CID+ +++ TIK+ + +S ++
Sbjct: 875 CIDIYRLTQTHTIKISLNEASQVK 898
>UniRef100_Q8LSP7 Putataive nodule inception protein [Oryza sativa]
Length = 942
Score = 624 bits (1608), Expect = e-177
Identities = 374/914 (40%), Positives = 522/914 (56%), Gaps = 117/914 (12%)
Query: 54 PFSSPLFDPSFSWPALETNEPTHVEDQHESQEAPLGNTQLVSQSQNMVNVVDGRYNNQSE 113
PFS PLFD S L T P ED + E P S+ + V +
Sbjct: 62 PFS-PLFDIGSSVTTLTTPAPAAGEDDRDEAEMP-------SRGGGGLEVSPAHRGWTFQ 113
Query: 114 TETHSVVEGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNK------DMLIQ 167
T V + P++ E+L RAL+ I + ++ ++L+Q
Sbjct: 114 TAPQEVA-------------------VEPTVKERLRRALERIASQSQSQAQRGDGELLVQ 154
Query: 168 IWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFR 227
+WVP GDR +L+ PF LD ++ LA YR +S +QFSA+E ++ + GLPGRVF
Sbjct: 155 VWVPTRIGDRQVLTTCGQPFWLDRRNQRLANYRTVSMKYQFSADESARADL-GLPGRVFV 213
Query: 228 DKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINY 287
+VPEWTPDVR+F ++EYPRV+HA+ FDI G++A+P+FE SR CLGV+E+VMTTQ++NY
Sbjct: 214 GRVPEWTPDVRYFSTEEYPRVQHAQYFDIRGSVALPVFEPRSRACLGVVELVMTTQKVNY 273
Query: 288 VPQLESVCKALEVVDLTSLKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTW 347
++E++C AL+ VDL S SS +K D SY A +PEI +VLR+ C H LPLAQTW
Sbjct: 274 SAEIENICNALKEVDLRSSDVSSDPRSKVVDASYRAIIPEIMDVLRAVCDTHNLPLAQTW 333
Query: 348 VSCFQQGKDGCRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFM 407
+ C Q K G RHS+++Y HC+S V++ACYV D SV FH+AC EHHL +G+GV G+AF
Sbjct: 334 IPCICQAKRGSRHSDESYKHCVSTVDEACYVRDCSVLGFHQACSEHHLFRGEGVVGRAFG 393
Query: 408 INQPFFSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDS 467
N+P FS DIT SKT YPLSHHA+LFGLRAAVAI+LRS+ + + D+VLEFFLP+ C ++
Sbjct: 394 TNEPCFSPDITTYSKTQYPLSHHAKLFGLRAAVAIQLRSVKTGSLDFVLEFFLPMKCINT 453
Query: 468 EEQKNMLISLSIIIQRCCRSLRVITDKEL---------ERTSSSVEVMALED------SG 512
EEQ+ ML SLS IQ+ C +LRV+ KEL + T ++ + SG
Sbjct: 454 EEQRAMLNSLSNTIQQVCYTLRVVKPKELVNDGPFEISQPTRPEFYAKSVHEDLDELCSG 513
Query: 513 F---ARTVKWSEPQHITS-VASL----------------------EPEEKSSETV----- 541
RT + ++S +ASL + +E S T
Sbjct: 514 INVPGRTTSLEASEEVSSWIASLVDAQNKGGKGEIDVDLPFGFSKQDDEGFSVTAGWHTS 573
Query: 542 ------GGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTI 595
G FS + H++ + + D S+ K EKRR K +KT+
Sbjct: 574 PVMAPDGSMFSGFKRHEDYDVKENTCSSDP----------SNSNSDKAVEKRRTKTEKTV 623
Query: 596 TLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDS 655
+L+ LR++F GSLK+AAKN+GVC TTLKR+CRQHGI RWPSRKIKKVGHSL+KLQ+VIDS
Sbjct: 624 SLQDLRKHFAGSLKEAAKNLGVCPTTLKRICRQHGINRWPSRKIKKVGHSLKKLQMVIDS 683
Query: 656 VQGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEG--- 712
V G G + ++ S Y NF S L ++ + + ++P + EG
Sbjct: 684 VHGPEG-TVQLSSLYENF---TKTTWSERELQGDVHFPASEQNFQLEPSVPDRPCEGRFT 739
Query: 713 ----ATKSLSSSCSQGSLSSHSCSSMPE-QQPHTS--DVACNKDPVVGKDSADVVLKRI- 764
+ S+S SCSQ S SS CSS+P+ QQ H S +A ++ + ++ ++K
Sbjct: 740 SHTSGSNSISPSCSQSSNSSLGCSSVPKTQQQHGSAPQLAVKEEISMDENQCSTLIKSAS 799
Query: 765 RSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGD 824
+EAEL+ E + + RS SQ L EH E S + K + D+ ++K YG+
Sbjct: 800 HAEAELQMFVEERPTMLFRSQSQVLLSEHKPIENMSNVQKA------RSDSLKIKAIYGE 853
Query: 825 EKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCL 884
E+ FR+ SW ++ L +EI +RF +S + D+KYLDD+ EWVLLTCDADL ECIDV
Sbjct: 854 ERCIFRLQPSWGFQRLKEEIVKRFGISQDTHVDLKYLDDESEWVLLTCDADLLECIDVYK 913
Query: 885 SSESSTIKLCIQAS 898
SS + T+++ + S
Sbjct: 914 SSSNQTVRILVNPS 927
>UniRef100_O65500 Hypothetical protein F23E12.170 [Arabidopsis thaliana]
Length = 1031
Score = 624 bits (1608), Expect = e-177
Identities = 380/967 (39%), Positives = 558/967 (57%), Gaps = 112/967 (11%)
Query: 19 NETSMDFEYMSDLLLDGCWLEASADGSNF--LLQQSPPFSSPLFDPS-----FSWPALET 71
++++MD ++M +LL DGCWLE + DG + + Q S+ + D + + + E
Sbjct: 22 SDSAMDMDFMDELLFDGCWLETT-DGKSLKQTMGQQVSDSTTMNDNNNNSYLYGYQYAEN 80
Query: 72 NEPTHVEDQHESQEAPL--GNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKR 129
H+ ++ ++ P + N V + ++ E + E S+G +R
Sbjct: 81 LSQDHISNEETGRKFPPIPPGFLKIEDLSNQVPFDQSAVMSSAQAEKFLLEE--SEGGRR 138
Query: 130 WWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSL 189
+WIAP S G S+ E+L++A++ + + +KD LIQIW+P+ + + L+ + P
Sbjct: 139 YWIAPRTSQGPSSSVKERLVQAIEGLNEEVQDKDFLIQIWLPIQQEGKNFLTTSEQPHFF 198
Query: 190 DSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVE 249
+ +L RYR++S + F A+EDSKE V GLPGRVF K+PEWTPDVRFFRS+EYPR++
Sbjct: 199 NPKYSSLKRYRDVSVAYNFLADEDSKESV-GLPGRVFLKKLPEWTPDVRFFRSEEYPRIK 257
Query: 250 HAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTS---- 305
A + D+ G+LA+P+FE+GS TCLGV+E+V TTQ++NY P+L+++CKALE V+L S
Sbjct: 258 EAEQCDVRGSLALPVFERGSGTCLGVVEIVTTTQKMNYRPELDNICKALESVNLRSSRSL 317
Query: 306 -----------LKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQG 354
L + ++ ++ R+ + L I S C ++ LPLA TW C +QG
Sbjct: 318 NPPSREVCKNGLLNQTLTSSIHRNNEFWL-LNVICPFCFSFCRVYDLPLALTWAPCARQG 376
Query: 355 KDGCRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFS 414
K G RHS++N+ C+S V+ AC V D R F EAC EHHLL+G+G+ GKAF + FF
Sbjct: 377 KVGSRHSDENFSECVSTVDDACIVPDHQSRHFLEACSEHHLLQGEGIVGKAFNATKLFFV 436
Query: 415 TDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNML 474
++T SKT+YPL+HHA++ GL AA+A+ L++ ++S+ ++VLEFF P C D+E Q++ML
Sbjct: 437 PEVTTFSKTNYPLAHHAKISGLHAALAVPLKNKFNSSVEFVLEFFFPKACLDTEAQQDML 496
Query: 475 ISLSIIIQRCCRSLRVITDKELE---------------------RTSSSVEVMALED--- 510
SLS +Q+ RSL + DKELE T ++ + LE+
Sbjct: 497 KSLSATLQQDFRSLNLFIDKELELEVVFPVREEVVFAENPLINAGTGEDMKPLPLEEISQ 556
Query: 511 ---SGFARTVKWSEPQHITSVA----SLEPEEKSSETVG--------GKFSDLREHQE-- 553
S + +K +E S++ EP+E+ T G G + L E ++
Sbjct: 557 EDSSWISHMIKANEKGKGVSLSWEYQKEEPKEEFMLTSGWDNNQIGSGHNNFLSEAEQFQ 616
Query: 554 ---DSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKD 610
+S L+ +++ E + F G + +G + GEKRR K +KTI LEVLRQYF GSLKD
Sbjct: 617 KVTNSGLRIDMDPSFESASFGVGQ-TLLGSRRPGEKRRTKTEKTIGLEVLRQYFAGSLKD 675
Query: 611 AAKNIG--------------VCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSV 656
AAK+IG VC TTLKR+CRQHGI RWPSRKIKKVGHSL+KLQLVIDSV
Sbjct: 676 AAKSIGESDDSFTQFHFHVAVCPTTLKRICRQHGITRWPSRKIKKVGHSLKKLQLVIDSV 735
Query: 657 QGASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQ-PDLGPLSPEGATK 715
QG G S ++DSFY++FP+L+SP++SG ++ N +S Q P SP
Sbjct: 736 QGVQG-SIQLDSFYTSFPELSSPHMSGTGTSFKNPNAQTENGVSAQGTAAAPKSPP---- 790
Query: 716 SLSSSCSQGSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSE 775
SSSCS S SS CS+ Q +T + N + ++A +LKR RSE L + ++
Sbjct: 791 --SSSCSHSSGSSTCCSTGANQSTNTGTTS-NTVTTLMAENASAILKRARSEVRLHTMNQ 847
Query: 776 NKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSW 835
++ K R+LS +T EH E L + + A +VK T+G+ K
Sbjct: 848 DETKSLSRTLSHKTFSEHPLFENPPRLPENSSRKLKAGGASKVKATFGEAK--------- 898
Query: 836 SYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
EIARRFN+ +++ FD+KYLDDD EWVLLTC+ADLEECID+ SS+S TIK+ +
Sbjct: 899 ------HEIARRFNIDNIAPFDLKYLDDDKEWVLLTCEADLEECIDIYRSSQSRTIKISV 952
Query: 896 QASSGMR 902
+S ++
Sbjct: 953 HEASQVK 959
>UniRef100_Q69NI4 Putative nodule inception protein [Oryza sativa]
Length = 872
Score = 595 bits (1534), Expect = e-168
Identities = 352/825 (42%), Positives = 488/825 (58%), Gaps = 81/825 (9%)
Query: 123 TSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGD-RPILS 181
T+ WWI P+ G S+ E+ +AL +I++ + D+L+Q+WVPV D + +L+
Sbjct: 67 TTTPANSWWIQPS---GASTSVRERFDQALAYIRETQSDADVLVQLWVPVKGNDGQLVLT 123
Query: 182 ANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFR 241
+ PF+LD S +L ++RE+S +QFSA+ S PGLPGRVF ++PEW+PDVR+F
Sbjct: 124 TSGQPFTLDQRSNSLIQFREVSTKYQFSADVASGSS-PGLPGRVFIGRLPEWSPDVRYFT 182
Query: 242 SDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVV 301
S EYPR+ HA+ D+ GT+ +P+FE+G+ +CLGVIE++MT Q++N+ +L ++C AL+ V
Sbjct: 183 SYEYPRINHAQYLDVHGTMGLPVFERGNYSCLGVIELIMTKQKLNFTSELNTICSALQAV 242
Query: 302 DLTSLKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHS 361
+LTS + SSI AK SY+ ALPEI EVLR+AC HKLPLAQTWV+C QQGK G RHS
Sbjct: 243 NLTSTEVSSIPRAKLNSASYKDALPEILEVLRAACITHKLPLAQTWVTCAQQGKRGSRHS 302
Query: 362 EDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLS 421
++NY +CIS ++ ACYV +P ++ FHEAC EHHLL+GQGVAGKAF NQP F DI +
Sbjct: 303 DENYKYCISTIDAACYVNEPRMQSFHEACSEHHLLRGQGVAGKAFTTNQPCFLPDIGSST 362
Query: 422 KTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIII 481
K +YPLSHHA++F L+ AVAIRLR + D+VLEFFLP +C EEQK +L SLS +
Sbjct: 363 KLEYPLSHHAKIFNLKGAVAIRLRCTRTGIADFVLEFFLPTDCEVLEEQKAVLDSLSGTM 422
Query: 482 QRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETV 541
+ C++LRV+TDKE+E ++ M +S R E + EE S T+
Sbjct: 423 RSVCQTLRVVTDKEME--DEAMREMNELNSFSPRGKNKVEELSFGDNTRGDREEASWTTL 480
Query: 542 GG---KFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLE 598
G K SDL E +L + G+ +S SK KRR K +KT++L+
Sbjct: 481 VGTSQKGSDLAELHTHGMLSHGGHGSSQA-----GDQTSKEGSKV--KRRTKTEKTVSLQ 533
Query: 599 VLRQYFPGSLKDAAKNIG------------------------------VCTTTLKRVCRQ 628
VLRQYF GSLKDAAK++G VC TTLKR+CRQ
Sbjct: 534 VLRQYFAGSLKDAAKSLGGQPSHAPYSQNVSIVITLFFRDSCYCMKVQVCPTTLKRICRQ 593
Query: 629 HGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFPDLA---SPNLSGAS 685
HGI RWPSRKIKKV HSL+KLQ +IDSV GA A F++++ Y + + + NLSG+
Sbjct: 594 HGINRWPSRKIKKVDHSLRKLQQIIDSVHGAETA-FQLNTLYKDLTNTSVSSDNNLSGSV 652
Query: 686 LVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHTSDVA 745
V NQ N+L + LSS+ SLS SCS + P S A
Sbjct: 653 TVPLANQ----NNLDFEMH--------QHHRLSSNIPSTSLSHSSCSQSSDSSPSCSGGA 700
Query: 746 CNKDPVVGKDSA-------DVVLKRIRSEA-ELKSHSENKAKLFPRSLSQETLGEHTKTE 797
P VG D ++ +++EA + H + P L Q+ + +
Sbjct: 701 TKHSPQVGADQVRSGCLPQHSPVQTLQTEAASINEHFSGQEA--PIDLLQDVAEKANGEQ 758
Query: 798 YQSYLLKTCHKATPKEDAH---RVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMS 854
+ S ++PK+ A+ RVKVT+G EK RFR+ ++ L QEI++R +++DM+
Sbjct: 759 HMSQ-----SPSSPKQTANVGMRVKVTFGSEKVRFRLKPECDFQELKQEISKRLSIADMN 813
Query: 855 KFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQASS 899
VKYLDDD EWVL+TCDADL EC V ++ TIK+ + ++
Sbjct: 814 SLIVKYLDDDSEWVLMTCDADLHECFHVYKLADIQTIKISVHLAA 858
>UniRef100_Q9S7B1 Nodule inception protein [Lotus japonicus]
Length = 878
Score = 521 bits (1342), Expect = e-146
Identities = 343/837 (40%), Positives = 467/837 (54%), Gaps = 116/837 (13%)
Query: 128 KRWWIAPTCSPG--LGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRG-------DRP 178
KRWWI P + S+ E+L+ A+ ++K + N ++LIQIWVP+ RG
Sbjct: 78 KRWWIGPAAAVAGSCNSSVKERLVIAVGYLKDYTRNSNVLIQIWVPLRRGILHDHDYHTN 137
Query: 179 ILSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVR 238
L +NN P E A + +S GF +P P V VR
Sbjct: 138 YLLSNNPP----PQPEAAADHESVSLGFP----------MPAAPNSNLYSNV-----HVR 178
Query: 239 FFRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQ-INYVPQLESVCKA 297
FFRS EYPRV+ A+++ G+LA+P+FE+G+ TCLGV+E+V+T Q INY +V A
Sbjct: 179 FFRSHEYPRVQ-AQQY---GSLALPVFERGTGTCLGVLEIVITNQTTINY-----NVSNA 229
Query: 298 LE-VVDLTSLKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKD 356
L+ VD S + K D+ Y+AA+ EI EV+ S C H LPLA TW C QQGK
Sbjct: 230 LDQAVDFRSSQSFIPPAIKVYDELYQAAVNEIIEVMTSVCKTHNLPLALTWAPCIQQGKC 289
Query: 357 GCRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTD 416
GC S +NY+ C+S V+ AC+VGD + F EAC E+HL +GQG+ G AF ++P F+ D
Sbjct: 290 GCGVSSENYMWCVSTVDSACFVGDLDILGFQEACSEYHLFRGQGIVGTAFTTSKPCFAID 349
Query: 417 ITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYS-SADDYVLEFFLPVNCNDSEEQKNMLI 475
IT SK +YPL+HHA +FGL AAVAI LRS+Y+ SA D+VLEFFLP +C+DSEEQK +L
Sbjct: 350 ITAFSKAEYPLAHHANMFGLHAAVAIPLRSVYTGSAADFVLEFFLPKDCHDSEEQKQLLN 409
Query: 476 SLSIIIQRCCRSLRVI--------------TDKELER----------------TSSSVEV 505
SLS+++Q+ CRSL V+ + +ELE SS
Sbjct: 410 SLSMVVQQACRSLHVVLVEDEYTLPMPSHTSKEELEEEEITITNNHEQKLFVSPSSHESE 469
Query: 506 MALEDSGFARTVK--------------WSEPQH----ITSVASLEPEEKSSETVGGKFSD 547
+ E S A ++ EP+ T+ S ++ ++ F
Sbjct: 470 CSKESSWIAHMMEAQQKGKGVSVSLEYLEEPKEEFKVTTNWDSSTDHDQQAQVFSSDFGQ 529
Query: 548 LREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGS 607
+ + S ++G D+E S SS G K+GEKRR KA+KTI+L+VLRQYF GS
Sbjct: 530 MSSGFKASTVEGG---DQESSYTFGSRRSSSGGRKSGEKRRTKAEKTISLQVLRQYFAGS 586
Query: 608 LKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKID 667
LKDAAK+IGVC TTLKR+CRQHGI RWPSRKIKKVGHSL+KLQLVIDSVQGA GA +I
Sbjct: 587 LKDAAKSIGVCPTTLKRICRQHGITRWPSRKIKKVGHSLKKLQLVIDSVQGAEGA-IQIG 645
Query: 668 SFYSNFPDLASPNLSGA----SLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQ 723
SFY++FP+L+S + S + S N + N+L D G + + KS S+CSQ
Sbjct: 646 SFYASFPELSSSDFSASCRSDSSKKMHNYPDQNNTLYGHGDHGGVVT--SLKSPPSACSQ 703
Query: 724 GSLSSHSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPR 783
+ C+ + + DV + P V + +L R + H E L
Sbjct: 704 TFAGNQPCTII-----NNGDVLMTESPPV----PEALLSR-------RDHCEEAELLNNA 747
Query: 784 SLSQETLG-EHTKTEYQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQ 842
S+ ++T K++ L + + + A RVK T+ DEK RF + W + L
Sbjct: 748 SIQEDTKRFSRPKSQTLPPLSDSSGWNSLETGAFRVKATFADEKIRFSLQPIWGFSDLQL 807
Query: 843 EIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI-QAS 898
EIARRFN++D++ +KYLDDD EWV+L CD DLEEC D+ SS+S TI+L + QAS
Sbjct: 808 EIARRFNLNDVTNILLKYLDDDGEWVVLACDGDLEECKDIHRSSQSRTIRLSLFQAS 864
>UniRef100_Q9LI18 EST AU057816(S21817) corresponds to a region of the predicted gene
[Oryza sativa]
Length = 852
Score = 466 bits (1198), Expect = e-129
Identities = 291/800 (36%), Positives = 448/800 (55%), Gaps = 63/800 (7%)
Query: 146 EKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEG 205
E+L +AL++ K+ + ++ +L+Q+W PV GDR +L+ + PF LD S L +YR +S
Sbjct: 51 ERLTQALRYFKE-STDQHLLVQVWAPVKSGDRYVLTTSGQPFVLDQQSIGLLQYRAVSMM 109
Query: 206 FQFSAE-EDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPI 264
+ FS + E++ EL GLPGRV++ KVPEWTP+V+++ S EYPR+ HA +++ GT+A+P+
Sbjct: 110 YMFSVDGENAGEL--GLPGRVYKQKVPEWTPNVQYYSSTEYPRLNHAISYNVHGTVALPV 167
Query: 265 FEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSYEAA 324
F+ + C+ V+E++MT+++INY +++ VCKALE V+L S + N + ++ ++A
Sbjct: 168 FDPSVQNCIAVVELIMTSKKINYAGEVDKVCKALEAVNLKSTEILDHPNVQICNEGRQSA 227
Query: 325 LPEIQEVLRSACHMHKLPLAQTWVSCFQQG--------KDGCRHSEDNYLH--CISPVEQ 374
L EI E+L C HKLPLAQTWV C + K C + + + C+S +
Sbjct: 228 LVEILEILTVVCEEHKLPLAQTWVPCKYRSVLAHGGGVKKSCLSFDGSCMGEVCMSTSDV 287
Query: 375 ACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLF 434
A +V D + F +AC+EHHL KGQGV+GKAF+ +P FS DI+ K +YPL H+AR+F
Sbjct: 288 AFHVIDAHMWGFRDACVEHHLQKGQGVSGKAFIYRRPCFSKDISQFCKLEYPLVHYARMF 347
Query: 435 GLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDK 494
GL AI L+S+Y+ DDY+LEFFLP NC + ++Q +L S+ +++C R+L+V+ +
Sbjct: 348 GLAGCFAICLQSMYTGDDDYILEFFLPPNCRNEDDQNALLESILARMKKCLRTLKVVGNG 407
Query: 495 ELERTSSSV-EVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETV------GGKFSD 547
+ + V+ +E V + + PE S+ V G K S
Sbjct: 408 DTNEVCLQISNVLIIETEDLKTNVHFENSE---GCFRESPESNGSQRVHEVDNDGNKVSI 464
Query: 548 LREHQ---EDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYF 604
+ E +D+ R +L +K E+RR KA+KTI+L+VL+QYF
Sbjct: 465 MSERHLLADDNSQNNGASVGRPNGSGASDSLHK--SNKPPERRRGKAEKTISLDVLQQYF 522
Query: 605 PGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASF 664
GSLK+AAK++GVC TT+KR+CRQHGI RWPSRKI KV SL KL+ VI+SVQG S A+F
Sbjct: 523 SGSLKNAAKSLGVCPTTMKRICRQHGISRWPSRKINKVNRSLSKLKQVIESVQG-SDAAF 581
Query: 665 KIDSFYSNFPDLASPNLSGASLVSAL-NQSENPNSLSIQPDLGP--------------LS 709
+ S P P+ +L A N+ ++L+++ D +S
Sbjct: 582 NLTSITGPLPIPVGPSSDSQNLEKASPNKVAELSNLAVEGDRDSSLQKPIENDNLAILMS 641
Query: 710 PEGATKSLSSSCSQGSLSSHSCSSMPEQ--QPHTSDVACNKDPV----VGKDSADVVLK- 762
+G + ++ + +SHS SS E TS+ +C+ P V K A +
Sbjct: 642 QQGFIDANNNLQLEADKASHSRSSSGEGSINSRTSEASCHGSPANQTFVCKPIASTFAEP 701
Query: 763 RIRSEAELKSHSENKAKLFPRSL------SQETLGEHTKTEYQSYLLKTCHKATPKEDAH 816
++ EA K + A R L S++ T Q +L ++ + A +
Sbjct: 702 QLIPEAFTKEPFQEPALPLSRMLIEDSGSSKDLKNLFTSAVDQPFLARSSNLALMQNSGT 761
Query: 817 -RVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDAD 875
+K ++ ++ RFR P S S L E+A+R + D+ FD+KYLDDD EWV L C+AD
Sbjct: 762 VTIKASFKEDIVRFRFPCSGSVTALKDEVAKRLRM-DVGMFDIKYLDDDHEWVKLACNAD 820
Query: 876 LEECIDVCLSSESSTIKLCI 895
LEEC+++ S S I+L +
Sbjct: 821 LEECMEI---SGSHVIRLLV 837
>UniRef100_Q8H105 Hypothetical protein At1g64530 [Arabidopsis thaliana]
Length = 841
Score = 449 bits (1155), Expect = e-124
Identities = 307/886 (34%), Positives = 462/886 (51%), Gaps = 86/886 (9%)
Query: 28 MSDLLLDGCW-LEASADGSNFLLQQSPPFSSPLFDP-SFSWPALETNEPTHVEDQHESQE 85
+ DL L G W L+ SNF +SP S P S W ET+ E
Sbjct: 3 LDDLDLSGSWPLDQITFASNF---KSPVIFSSSEQPFSPLWSFSETSGDVG----GELYS 55
Query: 86 APLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLGPSIM 145
A + T+ S + + ++SET T E W I P +P +I
Sbjct: 56 AAVAPTRFTDYSVLLAS-------SESETTTK---ENNQVPSPSWGIMPLENPDSYCAIK 105
Query: 146 EKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEG 205
K+ +AL++ K+ + +L Q+W PV R +L+ + PF L S L +YR +S
Sbjct: 106 AKMTQALRYFKESTGQQHVLAQVWAPVKNRGRYVLTTSGQPFVLGPNSNGLNQYRMVSLT 165
Query: 206 FQFSAE-EDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPI 264
+ FS + E EL GLPGRVFR K+PEWTP+V+++ S E+ R+ HA +++ GTLA+P+
Sbjct: 166 YMFSLDGERDGEL--GLPGRVFRKKLPEWTPNVQYYSSKEFSRLGHALHYNVQGTLALPV 223
Query: 265 FEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSYEAA 324
FE + C+GV+E++MT+ +INY P++E VCKALE V+L + + + + + ++ + A
Sbjct: 224 FEPSRQLCVGVVELIMTSPKINYAPEVEKVCKALEAVNLKTSEILNHETTQICNEGRQNA 283
Query: 325 LPEIQEVLRSACHMHKLPLAQTWVSCFQQG--------KDGCRHSEDNYLH--CISPVEQ 374
L EI E+L C +KLPLAQTWV C + K C + + + C+S +
Sbjct: 284 LAEILEILTVVCETYKLPLAQTWVPCRHRSVLAFGGGFKKSCSSFDGSCMGKVCMSTSDL 343
Query: 375 ACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLF 434
A YV D V F +AC EHHL KGQGVAG+AF F D+T KTDYPL H+AR+F
Sbjct: 344 AVYVVDAHVWGFRDACAEHHLQKGQGVAGRAFQSGNLCFCRDVTRFCKTDYPLVHYARMF 403
Query: 435 GLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDK 494
L + A+ L+S Y+ D+YVLEFFLP D EQ +L SL +++ SL+V+++
Sbjct: 404 KLTSCFAVCLKSTYTGDDEYVLEFFLPPAITDKSEQDCLLGSLLQTMKQHYSSLKVVSET 463
Query: 495 ELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLREHQED 554
EL + S+EV+ + G + EP I A + + K S + E+
Sbjct: 464 ELCENNMSLEVVEASEDGMVYSK--LEPIRIHHPAQISKDYLELNAPEQKVSLNSDFMEN 521
Query: 555 SILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKN 614
+ + +E + P E K E++R K +KTI+LEVL+QYF GSLKDAAK+
Sbjct: 522 NEVDDGVERFQTLDPIPEAK-----TVKKSERKRGKTEKTISLEVLQQYFAGSLKDAAKS 576
Query: 615 IGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFP 674
+GVC TT+KR+CRQHGI RWPSRKI KV SL +L+ VIDSVQGA G+ +P
Sbjct: 577 LGVCPTTMKRICRQHGISRWPSRKINKVNRSLTRLKHVIDSVQGADGSLNLTSLSPRPWP 636
Query: 675 DLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSM 734
P + L ++ P S S +L + E S+ S
Sbjct: 637 HQIPP------IDIQLAKNCPPTSTSPLSNLQDVKIENRDAEDSAGSS------------ 678
Query: 735 PEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHT 794
TS +C +P+ + L +H++ ++ +L
Sbjct: 679 ------TSRASCKVNPI------------CETRFRLPTHNQEPSRQV--ALDDSDSSSKN 718
Query: 795 KTEYQSYLLKTCH-KATPKEDAHR---VKVTYGDEKTRFRM-PKSWSYEHLLQEIARRFN 849
T + ++L TC A+P H+ +K TY ++ RF++ P+S S L Q++A+R
Sbjct: 719 MTNFWAHL--TCQDTASPTILQHKLVSIKATYREDIIRFKISPESVSITELKQQVAKRLK 776
Query: 850 VSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
+ + + F++KYLDDD EWV ++CDADL EC+D +++++T++L +
Sbjct: 777 L-ETAAFELKYLDDDREWVSVSCDADLSECLDTS-AAKANTLRLSV 820
>UniRef100_Q8RWY4 Hypothetical protein At1g64530 [Arabidopsis thaliana]
Length = 841
Score = 449 bits (1154), Expect = e-124
Identities = 307/886 (34%), Positives = 462/886 (51%), Gaps = 86/886 (9%)
Query: 28 MSDLLLDGCW-LEASADGSNFLLQQSPPFSSPLFDP-SFSWPALETNEPTHVEDQHESQE 85
+ DL L G W L+ SNF +SP S P S W ET+ E
Sbjct: 3 LDDLDLSGSWPLDQITFASNF---KSPVIFSSSEQPFSPLWSFSETSGDVG----GELYS 55
Query: 86 APLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLGPSIM 145
A + T+ S + + ++SET T E W I P +P +I
Sbjct: 56 AAVAPTRFTDYSVLLAS-------SESETTTK---ENNQVPSPSWGIMPLENPDSYCAIK 105
Query: 146 EKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEG 205
K+ +AL++ K+ + +L Q+W PV R +L+ + PF L S L +YR +S
Sbjct: 106 AKMTQALRYFKESTGQQHVLAQVWAPVKNRGRYVLTTSGQPFVLGPNSNGLNQYRMVSLT 165
Query: 206 FQFSAE-EDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPI 264
+ FS + E EL GLPGRVFR K+PEWTP+V+++ S E+ R+ HA +++ GTLA+P+
Sbjct: 166 YMFSLDGERDGEL--GLPGRVFRKKLPEWTPNVQYYSSKEFSRLGHALHYNVQGTLALPV 223
Query: 265 FEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSYEAA 324
FE + C+GV+E++MT+ +INY P++E VCKALE V+L + + + + + ++ + A
Sbjct: 224 FEPSRQLCVGVVELIMTSPKINYAPEVEKVCKALEAVNLKTSEILNHETTQICNEGRQNA 283
Query: 325 LPEIQEVLRSACHMHKLPLAQTWVSCFQQG--------KDGCRHSEDNYLH--CISPVEQ 374
L EI E+L C +KLPLAQTWV C + K C + + + C+S +
Sbjct: 284 LAEILEILTVVCETYKLPLAQTWVPCRYRSVLAFGGGFKKSCSSFDGSCMGKVCMSTSDL 343
Query: 375 ACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHARLF 434
A YV D V F +AC EHHL KGQGVAG+AF F D+T KTDYPL H+AR+F
Sbjct: 344 AVYVVDAHVWGFRDACAEHHLQKGQGVAGRAFQSGNLCFCRDVTRFCKTDYPLVHYARMF 403
Query: 435 GLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVITDK 494
L + A+ L+S Y+ D+YVLEFFLP D EQ +L SL +++ SL+V+++
Sbjct: 404 KLTSCFAVCLKSTYTGDDEYVLEFFLPPAITDKSEQDCLLGSLLQTMKQHYSSLKVVSET 463
Query: 495 ELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLREHQED 554
EL + S+EV+ + G + EP I A + + K S + E+
Sbjct: 464 ELCENNMSLEVVEASEDGMVYSK--LEPIRIHHPAQISKDYLELNAPEQKVSLNSDFMEN 521
Query: 555 SILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAAKN 614
+ + +E + P E K E++R K +KTI+LEVL+QYF GSLKDAAK+
Sbjct: 522 NEVDDGVERFQTLDPIPEAK-----TVKKSERKRGKTEKTISLEVLQQYFAGSLKDAAKS 576
Query: 615 IGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDSFYSNFP 674
+GVC TT+KR+CRQHGI RWPSRKI KV SL +L+ VIDSVQGA G+ +P
Sbjct: 577 LGVCPTTMKRICRQHGISRWPSRKINKVNRSLTRLKHVIDSVQGADGSLNLTSLSPRPWP 636
Query: 675 DLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSM 734
P + L ++ P S S +L + E S+ S
Sbjct: 637 HQIPP------IDIQLAKNCPPTSTSPLSNLQDVKIENRDAEDSAGSS------------ 678
Query: 735 PEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHT 794
TS +C +P+ + L +H++ ++ +L
Sbjct: 679 ------TSRASCKVNPI------------CETRFRLPTHNQEPSRQV--ALDDSDSSSKN 718
Query: 795 KTEYQSYLLKTCH-KATPKEDAHR---VKVTYGDEKTRFRM-PKSWSYEHLLQEIARRFN 849
T + ++L TC A+P H+ +K TY ++ RF++ P+S S L Q++A+R
Sbjct: 719 MTNFWAHL--TCQDTASPTILQHKLVSIKATYREDIIRFKISPESVSITELKQQVAKRLK 776
Query: 850 VSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
+ + + F++KYLDDD EWV ++CDADL EC+D +++++T++L +
Sbjct: 777 L-ETAAFELKYLDDDREWVSVSCDADLSECLDTS-AAKANTLRLSV 820
>UniRef100_Q9SGW2 F1N19.10 [Arabidopsis thaliana]
Length = 847
Score = 447 bits (1150), Expect = e-124
Identities = 310/892 (34%), Positives = 463/892 (51%), Gaps = 92/892 (10%)
Query: 28 MSDLLLDGCW-LEASADGSNFLLQQSPPFSSPLFDP-SFSWPALETNEPTHVEDQHESQE 85
+ DL L G W L+ SNF +SP S P S W ET+ E
Sbjct: 3 LDDLDLSGSWPLDQITFASNF---KSPVIFSSSEQPFSPLWSFSETSGDVG----GELYS 55
Query: 86 APLGNTQLVSQSQNMVNVVDGRYNNQSETETHSVVEGTSDGVKRWWIAPTCSPGLGPSIM 145
A + T+ S + + ++SET T E W I P +P +I
Sbjct: 56 AAVAPTRFTDYSVLLAS-------SESETTTK---ENNQVPSPSWGIMPLENPDSYCAIK 105
Query: 146 EKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREISEG 205
K+ +AL++ K+ + +L Q+W PV R +L+ + PF L S L +YR +S
Sbjct: 106 AKMTQALRYFKESTGQQHVLAQVWAPVKNRGRYVLTTSGQPFVLGPNSNGLNQYRMVSLT 165
Query: 206 FQFSAE-EDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVPI 264
+ FS + E EL GLPGRVFR K+PEWTP+V+++ S E+ R+ HA +++ GTLA+P+
Sbjct: 166 YMFSLDGERDGEL--GLPGRVFRKKLPEWTPNVQYYSSKEFSRLGHALHYNVQGTLALPV 223
Query: 265 FEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTS---LKHSSIQ---NAKARD 318
FE + C+GV+E++MT+ +INY P++E VCKALE V+L + L H + Q + +
Sbjct: 224 FEPSRQLCVGVVELIMTSPKINYAPEVEKVCKALEAVNLKTSEILNHETTQVICESNICN 283
Query: 319 KSYEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQG--------KDGCRHSEDNYLH--C 368
+ + AL EI E+L C +KLPLAQTWV C + K C + + + C
Sbjct: 284 EGRQNALAEILEILTVVCETYKLPLAQTWVPCRHRSVLAFGGGFKKSCSSFDGSCMGKVC 343
Query: 369 ISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLS 428
+S + A YV D V F +AC EHHL KGQGVAG+AF F D+T KTDYPL
Sbjct: 344 MSTSDLAVYVVDAHVWGFRDACAEHHLQKGQGVAGRAFQSGNLCFCRDVTRFCKTDYPLV 403
Query: 429 HHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSL 488
H+AR+F L + A+ L+S Y+ D+YVLEFFLP D EQ +L SL +++ SL
Sbjct: 404 HYARMFKLTSCFAVCLKSTYTGDDEYVLEFFLPPAITDKSEQDCLLGSLLQTMKQHYSSL 463
Query: 489 RVITDKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDL 548
+V+++ EL + S+EV+ + G + EP I A + + K S
Sbjct: 464 KVVSETELCENNMSLEVVEASEDGMVYSK--LEPIRIHHPAQISKDYLELNAPEQKVSLN 521
Query: 549 REHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSL 608
+ E++ + +E + P E K E++R K +KTI+LEVL+QYF GSL
Sbjct: 522 SDFMENNEVDDGVERFQTLDPIPEAK-----TVKKSERKRGKTEKTISLEVLQQYFAGSL 576
Query: 609 KDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFKIDS 668
KDAAK++GVC TT+KR+CRQHGI RWPSRKI KV SL +L+ VIDSVQGA G+
Sbjct: 577 KDAAKSLGVCPTTMKRICRQHGISRWPSRKINKVNRSLTRLKHVIDSVQGADGSLNLTSL 636
Query: 669 FYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSLSS 728
+P P + L ++ P S S +L + E S+ S
Sbjct: 637 SPRPWPHQIPP------IDIQLAKNCPPTSTSPLSNLQDVKIENRDAEDSAGSS------ 684
Query: 729 HSCSSMPEQQPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQE 788
TS +C +P+ + L +H++ ++ +L
Sbjct: 685 ------------TSRASCKVNPI------------CETRFRLPTHNQEPSRQV--ALDDS 718
Query: 789 TLGEHTKTEYQSYLLKTCH-KATPKEDAHR---VKVTYGDEKTRFRM-PKSWSYEHLLQE 843
T + ++L TC A+P H+ +K TY ++ RF++ P+S S L Q+
Sbjct: 719 DSSSKNMTNFWAHL--TCQDTASPTILQHKLVSIKATYREDIIRFKISPESVSITELKQQ 776
Query: 844 IARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
+A+R + + + F++KYLDDD EWV ++CDADL EC+D +++++T++L +
Sbjct: 777 VAKRLKL-ETAAFELKYLDDDREWVSVSCDADLSECLDTS-AAKANTLRLSV 826
>UniRef100_Q84TH9 Hypothetical protein At4g24020 [Arabidopsis thaliana]
Length = 959
Score = 437 bits (1125), Expect = e-121
Identities = 290/827 (35%), Positives = 428/827 (51%), Gaps = 106/827 (12%)
Query: 144 IMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREIS 203
I E++ +AL++ K+ + + +L Q+W PV + R +L+ PF L+ L +YR IS
Sbjct: 147 IKERMTQALRYFKE-STEQHVLAQVWAPVRKNGRDLLTTLGQPFVLNPNGNGLNQYRMIS 205
Query: 204 EGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVP 263
+ FS + +S ++ GLPGRVFR K+PEWTP+V+++ S E+ R++HA +++ GTLA+P
Sbjct: 206 LTYMFSVDSES-DVELGLPGRVFRQKLPEWTPNVQYYSSKEFSRLDHALHYNVRGTLALP 264
Query: 264 IFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSYEA 323
+F ++C+GV+E++MT+++I+Y P+++ VCKALE V+L S + Q + ++S +
Sbjct: 265 VFNPSGQSCIGVVELIMTSEKIHYAPEVDKVCKALEAVNLKSSEILDHQTTQICNESRQN 324
Query: 324 ALPEIQEVLRSACHMHKLPLAQTWVSCFQQG---------KDGCRHSEDNYLH--CISPV 372
AL EI EVL C H LPLAQTWV C Q G K C + + + C+S
Sbjct: 325 ALAEILEVLTVVCETHNLPLAQTWVPC-QHGSVLANGGGLKKNCTSFDGSCMGQICMSTT 383
Query: 373 EQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHAR 432
+ ACYV D V F +AC+EHHL KGQGVAG+AF+ F DIT KT YPL H+A
Sbjct: 384 DMACYVVDAHVWGFRDACLEHHLQKGQGVAGRAFLNGGSCFCRDITKFCKTQYPLVHYAL 443
Query: 433 LFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVIT 492
+F L AI L+S Y+ D Y+LEFFLP + D +EQ +L S+ + ++ +SLRV +
Sbjct: 444 MFKLTTCFAISLQSSYTGDDSYILEFFLPSSITDDQEQDLLLGSILVTMKEHFQSLRVAS 503
Query: 493 DKELERTSSSVE---VMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLR 549
+ + + AL D E E++ FS +
Sbjct: 504 GVDFGEDDDKLSFEIIQALPD---------------------EKVHSKIESIRVPFSGFK 542
Query: 550 EHQEDSILKGNIECDRECSPFVEGNLSSV-GI---SKTGEKRRAKADKTITLEVLRQYFP 605
+ +++L + N+++V G+ K EK+R K +KTI+L+VL+QYF
Sbjct: 543 SNATETMLIPQPVVQSSDPVNEKINVATVNGVVKEKKKTEKKRGKTEKTISLDVLQQYFT 602
Query: 606 GSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFK 665
GSLKDAAK++GVC TT+KR+CRQHGI RWPSRKIKKV S+ KL+ VI+SVQG G
Sbjct: 603 GSLKDAAKSLGVCPTTMKRICRQHGISRWPSRKIKKVNRSITKLKRVIESVQGTDGGLDL 662
Query: 666 IDSFYSNFP---------DLASPNLSGASLVSALNQSEN-------PNSLSIQPDLGPLS 709
S+ P L SPN S + N S N PN + P+L P +
Sbjct: 663 TSMAVSSIPWTHGQTSAQPLNSPNGSKPPELPNTNNSPNHWSSDHSPNEPNGSPELPPSN 722
Query: 710 PEGATKSLSSSCSQGSLSSH-SCSS-------MPEQQP------------------HTSD 743
G +S + S G+ +SH SC +P Q P H
Sbjct: 723 --GHKRSRTVDESAGTPTSHGSCDGNQLDEPKVPNQDPLFTVGGSPGLLFPPYSRDHDVS 780
Query: 744 VACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLL 803
A P S D + +A S++ L P + + + +
Sbjct: 781 AASFAMPNRLLGSIDHFRGMLIEDA---GSSKDLRNLCPTAAFDDKFQDTNWMNNDNNSN 837
Query: 804 KTCHKATPKEDAHR---------------VKVTYGDEKTRFRMPKSWSYEHLLQEIARRF 848
+ A PKE+A +K +Y D+ RFR+ L E+A+R
Sbjct: 838 NNLY-APPKEEAIANVACEPSGSEMRTVTIKASYKDDIIRFRISSGSGIMELKDEVAKRL 896
Query: 849 NVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
V D FD+KYLDDD EWVL+ CDADL+EC+++ SS + ++L +
Sbjct: 897 KV-DAGTFDIKYLDDDNEWVLIACDADLQECLEIPRSSRTKIVRLLV 942
>UniRef100_O22987 T19F6.16 protein [Arabidopsis thaliana]
Length = 959
Score = 436 bits (1121), Expect = e-120
Identities = 289/827 (34%), Positives = 428/827 (50%), Gaps = 106/827 (12%)
Query: 144 IMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYREIS 203
I E++ +AL++ K+ + + +L Q+W PV + R +L+ PF L+ L +YR IS
Sbjct: 147 IKERMTQALRYFKE-STEQHVLAQVWAPVRKNGRDLLTTLGQPFVLNPNGNGLNQYRMIS 205
Query: 204 EGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTLAVP 263
+ FS + +S ++ GLPGRVFR K+PEWTP+V+++ S E+ R++HA +++ GTLA+P
Sbjct: 206 LTYMFSVDSES-DVELGLPGRVFRQKLPEWTPNVQYYSSKEFSRLDHALHYNVRGTLALP 264
Query: 264 IFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKSYEA 323
+F ++C+GV+E++MT+++I+Y P+++ VCKALE V+L S + Q + ++S +
Sbjct: 265 VFNPSGQSCIGVVELIMTSEKIHYAPEVDKVCKALEAVNLKSSEILDHQTTQICNESRQN 324
Query: 324 ALPEIQEVLRSACHMHKLPLAQTWVSCFQQG---------KDGCRHSEDNYLH--CISPV 372
AL EI EVL C H LPLAQTWV C Q G K C + + + C+S
Sbjct: 325 ALAEILEVLTVVCETHNLPLAQTWVPC-QHGSVLANGGGLKKNCTSFDGSCMGQICMSTT 383
Query: 373 EQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHAR 432
+ ACYV D V F +AC+EHHL KGQGVAG+AF+ F DIT KT YPL H+A
Sbjct: 384 DMACYVVDAHVWGFRDACLEHHLQKGQGVAGRAFLNGGSCFCRDITKFCKTQYPLVHYAL 443
Query: 433 LFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVIT 492
+F L AI L+S Y+ D Y+LEFFLP + D +EQ +L S+ + ++ +SLRV +
Sbjct: 444 MFKLTTCFAISLQSSYTGDDSYILEFFLPSSITDDQEQDLLLGSILVTMKEHFQSLRVAS 503
Query: 493 DKELERTSSSVE---VMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLR 549
+ + + AL D + E++ FS +
Sbjct: 504 GVDFGEDDDKLSFEIIQALPDKKVHSKI---------------------ESIRVPFSGFK 542
Query: 550 EHQEDSILKGNIECDRECSPFVEGNLSSV-GI---SKTGEKRRAKADKTITLEVLRQYFP 605
+ +++L + N+++V G+ K EK+R K +KTI+L+VL+QYF
Sbjct: 543 SNATETMLIPQPVVQSSDPVNEKINVATVNGVVKEKKKTEKKRGKTEKTISLDVLQQYFT 602
Query: 606 GSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASGASFK 665
GSLKDAAK++GVC TT+KR+CRQHGI RWPSRKIKKV S+ KL+ VI+SVQG G
Sbjct: 603 GSLKDAAKSLGVCPTTMKRICRQHGISRWPSRKIKKVNRSITKLKRVIESVQGTDGGLDL 662
Query: 666 IDSFYSNFP---------DLASPNLSGASLVSALNQSEN-------PNSLSIQPDLGPLS 709
S+ P L SPN S + N S N PN + P+L P +
Sbjct: 663 TSMAVSSIPWTHGQTSAQPLNSPNGSKPPELPNTNNSPNHWSSDHSPNEPNGSPELPPSN 722
Query: 710 PEGATKSLSSSCSQGSLSSH-SCSS-------MPEQQP------------------HTSD 743
G +S + S G+ +SH SC +P Q P H
Sbjct: 723 --GHKRSRTVDESAGTPTSHGSCDGNQLDEPKVPNQDPLFTVGGSPGLLFPPYSRDHDVS 780
Query: 744 VACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLL 803
A P S D + +A S++ L P + + + +
Sbjct: 781 AASFAMPNRLLGSIDHFRGMLIEDA---GSSKDLRNLCPTAAFDDKFQDTNWMNNDNNSN 837
Query: 804 KTCHKATPKEDAHR---------------VKVTYGDEKTRFRMPKSWSYEHLLQEIARRF 848
+ A PKE+A +K +Y D+ RFR+ L E+A+R
Sbjct: 838 NNLY-APPKEEAIANVACEPSGSEMRTVTIKASYKDDIIRFRISSGSGIMELKDEVAKRL 896
Query: 849 NVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCI 895
V D FD+KYLDDD EWVL+ CDADL+EC+++ SS + ++L +
Sbjct: 897 KV-DAGTFDIKYLDDDNEWVLIACDADLQECLEIPRSSRTKIVRLLV 942
>UniRef100_Q7XQI3 OSJNBa0067K08.5 protein [Oryza sativa]
Length = 936
Score = 430 bits (1105), Expect = e-118
Identities = 296/829 (35%), Positives = 425/829 (50%), Gaps = 107/829 (12%)
Query: 140 LGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARY 199
+G S+ ++++ AL ++ + L Q+W+PV + +LS PF LD + LA Y
Sbjct: 135 VGSSLADRMLMALSLFRE-SLGSGALAQVWMPVEQEGHVVLSTCEQPFLLD---QVLAGY 190
Query: 200 REISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGT 259
RE+S F FSA+E+ L PGLPGRVF VPEWT V ++ EY R+EHA +I G+
Sbjct: 191 REVSRHFVFSAKEEPG-LQPGLPGRVFISGVPEWTSSVLYYNRPEYLRMEHALHHEIRGS 249
Query: 260 LAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDK 319
LA+PI++ +C V E+V ++ ++ ++++VC AL+ V+L + K SS N K +
Sbjct: 250 LAMPIYDPSKDSCCAVFELVTRKEKPDFSAEMDNVCNALQAVNLKATKGSS--NQKFYTE 307
Query: 320 SYEAALPEIQEVLRSACHMHKLPLAQTWVSC-------FQQGKDGCRHSEDNYLHCISPV 372
+ + A EI +VLR+ CH H LPLA TWV + GKDG S+ I
Sbjct: 308 NQKFAFTEILDVLRAICHAHMLPLALTWVPTSNGIDGGYVVGKDGASFSQSGKT-IIRIH 366
Query: 373 EQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHAR 432
E ACYV D ++ F +AC HL KGQG+AG+A N PFFS DI S DYPL+HHAR
Sbjct: 367 ESACYVNDGKMQGFLQACARRHLEKGQGIAGRALKSNLPFFSPDIREYSIEDYPLAHHAR 426
Query: 433 LFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVIT 492
F L AAVAIRLRS Y+ DDY+LEFFLPV+C S EQ+ +L +LS +QR C+SLR +
Sbjct: 427 KFSLHAAVAIRLRSTYTGNDDYILEFFLPVSCKGSGEQQMLLNNLSSTMQRICKSLRTVY 486
Query: 493 DKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLREHQ 552
+ E++ ++G A + + + + + G F D
Sbjct: 487 EAEVDNV----------NAGTAAVFRKNNESCLPTGHTESSSHGDQSITGASFEDTSLAN 536
Query: 553 EDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSLKDAA 612
+ +++ E + P SS+G EK+R+ A+K I+L+VLR+YF GSLKDAA
Sbjct: 537 KPGVMEP--ELAEQVQP------SSIG---HAEKKRSTAEKNISLDVLRKYFSGSLKDAA 585
Query: 613 KNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQG---------ASGAS 663
K++GVC TTLKR+CR HGI RWPSRKI KV SL+K+Q VI+SV G A+G+
Sbjct: 586 KSLGVCPTTLKRICRHHGISRWPSRKINKVNRSLKKIQTVINSVHGVDRSLQYDPATGSL 645
Query: 664 FKIDSF-----YSNFPDLASPNL-------------SGASL------------VSALNQS 693
+ S + + L +P++ G SL +S + +S
Sbjct: 646 VPVVSLPEKLTFPSCDGLPTPSVGKTVEENSDLKSEEGCSLPDGSQRQSCQLQISDVKKS 705
Query: 694 ENPNSLSIQPDLGPLSPEGATKSLSSSCSQGSL----------------SSHSCSSMPEQ 737
N + I AT +S +QG L ++ S S P
Sbjct: 706 -NEDEFHIGSGNSDFYGANATAKSNSEVTQGPLCPTGAFSALHLKGTDCTNPSSSLRPSS 764
Query: 738 QPHTSDVACNKDPVVGKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTE 797
+ + + P + ++ D++ EAE K H S S T +
Sbjct: 765 ESTRNQIVGRNSPSIQQEDLDML---DNHEAEDKDHMHPSTSGMTDSSSGSASSHPTFKQ 821
Query: 798 YQSYLLKTCHKATPKEDAHRVKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFD 857
LK A+P A VK TY + RF+ S + HLL+EIA+RF + +
Sbjct: 822 NTRSALKDA--ASP---ALTVKATYNGDTVRFKFLPSMGWYHLLEEIAKRFKL-PTGAYQ 875
Query: 858 VKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKL------CIQASSG 900
+KY DD+ EWV+L D+DL+EC+DV S S +KL CI +SSG
Sbjct: 876 LKYKDDEDEWVILANDSDLQECVDVLDSIGSRIVKLQVRDLPCIVSSSG 924
>UniRef100_Q7X9C0 NIN-like protein 2 [Lotus japonicus]
Length = 972
Score = 416 bits (1069), Expect = e-114
Identities = 296/813 (36%), Positives = 428/813 (52%), Gaps = 97/813 (11%)
Query: 135 TCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSE 194
T S GPS+ E+++RAL + K+ + +L Q+WVP+ G + ILS + P+ LD +
Sbjct: 160 TISRPPGPSLDERMLRALSFFKE-SAGGGILAQVWVPLEHGGQVILSTSEQPYLLD---Q 215
Query: 195 NLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREF 254
LA YRE+S F+F AE GLPGRVF KVPEWT +V ++ +EY RVEHAR +
Sbjct: 216 MLAGYREVSRTFKFPAEGKPGGF-SGLPGRVFVSKVPEWTSNVGYYSKNEYLRVEHARNY 274
Query: 255 DICGTLAVPIFEQGSRT-CLGVIEVVMTTQQINYVPQLESVCKALEVVDL-TSLKHSSIQ 312
+ GT+A PIF+ S C V+E+V T + ++ +LE VC AL++V+L T++
Sbjct: 275 KVRGTIAFPIFDTHSELPCCAVLELVTTKEMSDFDRELEVVCHALQLVNLRTTMPLRIFP 334
Query: 313 NAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSC-FQQG----------KDGCRHS 361
+ +K AAL EI +VL+S CH H+LPLA TW+ C + +G K+G S
Sbjct: 335 ECYSNNK--RAALAEIVDVLKSVCHAHRLPLALTWIPCCYTEGPKGEAMRIQIKEGHSSS 392
Query: 362 EDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLS 421
+ L CI E ACYV D + F AC+EH L +G+G+AGKA N PFF D+
Sbjct: 393 GEKVLLCIE--ESACYVTDRLMEGFVRACIEHPLEEGKGIAGKALQSNHPFFYPDVKEYD 450
Query: 422 KTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIII 481
++YPL HHAR L AAVAIRLRS +++ DDY+LEFFLPVN S EQ+ +L +LS +
Sbjct: 451 ISEYPLVHHARKCNLSAAVAIRLRSTHTNDDDYILEFFLPVNMRGSSEQQLLLDNLSGTM 510
Query: 482 QRCCRSLRVITDKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETV 541
QR CRSLR ++D E R S+ GF E +++ S + L E +
Sbjct: 511 QRICRSLRTVSDVESSRIEST-------HMGF-------ENKNLPSFSPLSRENSQIPLI 556
Query: 542 GGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLR 601
+++ + S L+ P+ N S G + EK R A+K ++L VL+
Sbjct: 557 NANQDSVQKSLKASRLRNK----GSKPPY---NQVSNGSRRQVEKNRGTAEKNVSLSVLQ 609
Query: 602 QYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQGASG 661
Q+F GSLKDAAK+IGVC TTLKR+CR HGI RWPSRKI KV SL+K+Q V+DSVQG
Sbjct: 610 QHFSGSLKDAAKSIGVCPTTLKRICRHHGISRWPSRKINKVNSSLKKIQTVLDSVQGVE- 668
Query: 662 ASFKID----SFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSP------- 710
+S K D +F + + + + L + +P ++ P P
Sbjct: 669 SSLKFDPSVGAFVAGGSTIQGIDTHKSLLFPEKSTIRDPRPITQDAVSVPAVPCGESEKS 728
Query: 711 ----EGATKSLSSS---CSQGS-------LSSHSCS------SMPEQQPHTSDVA----- 745
EG K ++S C + S SSHS S EQ SD+A
Sbjct: 729 AIKLEGKLKKTNASLVDCCEDSKSMAMDDRSSHSSSLWTKAQGSSEQASLGSDLAKRDKW 788
Query: 746 -CNKDPV---------VGKDSA----DVVLKRIRSEAELKSHSENKAKLFPRSLSQETLG 791
N D + VG+ S D + + ++ ++ + + L S +
Sbjct: 789 VLNNDGLRVENLKCNTVGQSSGSFTDDEMGIDVDNDEVVELNHPTCSSLTGSSNGSSPMI 848
Query: 792 EHTKTEYQSYLLKTCHKATPKEDAHR--VKVTYGDEKTRFRMPKSWSYEHLLQEIARRFN 849
+ + QS+ ++ K+T + + VK TY + RF+ S L +E+A RF
Sbjct: 849 HGSSSSSQSFENQSKGKSTTVDRGSKIIVKATYRKDIIRFKFDPSAGCFKLYEEVAARFK 908
Query: 850 VSDMSKFDVKYLDDDLEWVLLTCDADLEECIDV 882
+ + F +KYLDD+ EWV+L D+DL+EC+D+
Sbjct: 909 LQN-GTFQLKYLDDEEEWVMLVSDSDLQECVDI 940
>UniRef100_O22864 Hypothetical protein At2g43500 [Arabidopsis thaliana]
Length = 947
Score = 410 bits (1053), Expect = e-112
Identities = 277/807 (34%), Positives = 428/807 (52%), Gaps = 78/807 (9%)
Query: 135 TCSPGLGPSIMEKLIRALK-WIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGS 193
T L S+ EK+++AL +++ + +L Q+W P+ GD+ +LS + + LD
Sbjct: 154 TIPRSLSHSLDEKMLKALSLFMESSGSGEGILAQVWTPIKTGDQYLLSTCDQAYLLDP-- 211
Query: 194 ENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHARE 253
++YRE+S F F+AE + PGLPGRVF VPEWT +V ++++DEY R++HA +
Sbjct: 212 -RFSQYREVSRRFTFAAEANQCSF-PGLPGRVFISGVPEWTSNVMYYKTDEYLRMKHAID 269
Query: 254 FDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQN 313
++ G++A+PI E +C V+E+V + ++ N+ +++SVC+AL+ V+L + ++I
Sbjct: 270 NEVRGSIAIPILEASGTSCCAVMELVTSKEKPNFDMEMDSVCRALQAVNLRT---AAIPR 326
Query: 314 AKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSCF--QQGKDGCRHSEDNYLHCISP 371
+ S AL EIQ+VLR+ CH HKLPLA W+ C Q + + S +N + CI
Sbjct: 327 PQYLSSSQRDALAEIQDVLRTVCHAHKLPLALAWIPCRKDQSIRVSGQKSGENCILCIE- 385
Query: 372 VEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLSHHA 431
E ACYV D + F AC+EH L + +G+ GKAF+ NQPFFS+D+ ++YP+ HA
Sbjct: 386 -ETACYVNDMEMEGFVHACLEHCLREKEGIVGKAFISNQPFFSSDVKAYDISEYPIVQHA 444
Query: 432 RLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSLRVI 491
R +GL AAVAI+LRS Y+ DDY+LE FLPV+ S EQ+ +L SLS +QR CR+LR +
Sbjct: 445 RKYGLNAAVAIKLRSTYTGEDDYILELFLPVSMKGSLEQQLLLDSLSGTMQRICRTLRTV 504
Query: 492 TD-KELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDLRE 550
++ ++ + + + S F +T Q I SL+ E S+ ++ FS +
Sbjct: 505 SEVGSTKKEGTKPGFRSSDMSNFPQTTSSENFQTI----SLDSEFNSTRSM---FSGMSS 557
Query: 551 HQEDSIL--KGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSL 608
+E+SI +G +E D V ++T EK+++ +K ++L L+Q+F GSL
Sbjct: 558 DKENSITVSQGTLEQD-------------VSKARTPEKKKSTTEKNVSLSALQQHFSGSL 604
Query: 609 KDAAKNIG-------------VCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDS 655
KDAAK++G C TTLKR+CRQHGI RWPSRKI KV SL+K+Q V+DS
Sbjct: 605 KDAAKSLGGETSAYFQAWVYFFCPTTLKRICRQHGIMRWPSRKINKVNRSLRKIQTVLDS 664
Query: 656 VQGASGASFKIDSFYSNF----PDLASPNLSGASLVSALNQSENPNSLSIQPD------L 705
VQG G K DS F P + + + L S N + S PD
Sbjct: 665 VQGVEG-GLKFDSATGEFIAVRPFIQEID-TQKGLSSLDNDAHARRSQEDMPDDTSFKLQ 722
Query: 706 GPLSPEGATK----SLSSSCSQGSLSSHSCSSMP------EQQPHTSD---VACNKDPVV 752
S + A K + + GS + S P E + S+ CN V
Sbjct: 723 EAKSVDNAIKLEEDTTMNQARPGSFMEVNASGQPWAWMAKESGLNGSEGIKSVCNLSSVE 782
Query: 753 GKDSADVVLKRIRSEAELKSHSENKAKLFPRSLSQETLGEHTKTEYQSYLLKTCHKATPK 812
D D ++ S E + S + + + S L + T + + H +
Sbjct: 783 ISDGMDPTIRCSGSIVE-PNQSMSCSISDSSNGSGAVLRGSSSTSMEDWNQMRTHNSNSS 841
Query: 813 EDAHR---VKVTYGDEKTRFRMPKSWSYEHLLQEIARRFNVSDMSKFDVKYLDDDLEWVL 869
E VK +Y ++ RF+ S L +E+ +RF + D S F +KYLDD+ EWV+
Sbjct: 842 ESGSTTLIVKASYREDTVRFKFEPSVGCPQLYKEVGKRFKLQDGS-FQLKYLDDEEEWVM 900
Query: 870 LTCDADLEECIDVCLSSESSTIKLCIQ 896
L D+DL+EC+++ ++K ++
Sbjct: 901 LVTDSDLQECLEILHGMGKHSVKFLVR 927
>UniRef100_Q9M1B0 Hypothetical protein T16L24.130 [Arabidopsis thaliana]
Length = 894
Score = 408 bits (1048), Expect = e-112
Identities = 278/833 (33%), Positives = 433/833 (51%), Gaps = 97/833 (11%)
Query: 120 VEGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPI 179
+EG+ KR + L S+ EK+++AL +F+ + +L Q W P+ GD+ +
Sbjct: 82 LEGSYACEKRPLDCTSVPRSLSHSLDEKMLKALSLFMEFS-GEGILAQFWTPIKTGDQYM 140
Query: 180 LSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRF 239
LS + + LDS L+ YRE S F FSAE + PGLPGRVF VPEWT +V +
Sbjct: 141 LSTCDQAYLLDS---RLSGYREASRRFTFSAEANQCSY-PGLPGRVFISGVPEWTSNVMY 196
Query: 240 FRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALE 299
+++ EY R++HA + ++ G++A+P+ E +C V+E+V ++ N+ ++ SVC+AL+
Sbjct: 197 YKTAEYLRMKHALDNEVRGSIAIPVLEASGSSCCAVLELVTCREKPNFDVEMNSVCRALQ 256
Query: 300 VVDLTSLKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSC-FQQGKDG- 357
V+L + S+I + + + AL EI++VLR+ C+ H+LPLA W+ C + +G +
Sbjct: 257 AVNLQT---STIPRRQYLSSNQKEALAEIRDVLRAVCYAHRLPLALAWIPCSYSKGANDE 313
Query: 358 -----CRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPF 412
++S++ L CI E +CYV D + F AC+EH+L +GQG+ GKA + N+P
Sbjct: 314 LVKVYGKNSKECSLLCIE--ETSCYVNDMEMEGFVNACLEHYLREGQGIVGKALISNKPS 371
Query: 413 FSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKN 472
FS+D+ +YPL HAR FGL AAVA +LRS ++ +DY+LEFFLPV+ S EQ+
Sbjct: 372 FSSDVKTFDICEYPLVQHARKFGLNAAVATKLRSTFTGDNDYILEFFLPVSMKGSSEQQL 431
Query: 473 MLISLSIIIQRCCRSLRVITDKE----LERTSSSVEVMALEDSGFARTVKWSEPQHITSV 528
+L SLS +QR CR+L+ ++D E E S SVE+ L PQ SV
Sbjct: 432 LLDSLSGTMQRLCRTLKTVSDAESIDGTEFGSRSVEMTNL-------------PQATVSV 478
Query: 529 ASLEPE--EKSSETVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEK 586
S + + FS++ ++ + + +E S ++ EK
Sbjct: 479 GSFHTTFLDTDVNSTRSTFSNISSNKRNEMAGSQGTLQQEISG-----------ARRLEK 527
Query: 587 RRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSL 646
+++ +K ++L VL+QYF GSLKDAAK++GVC TTLKR+CRQHGI RWPSRKI KV SL
Sbjct: 528 KKSSTEKNVSLNVLQQYFSGSLKDAAKSLGVCPTTLKRICRQHGIMRWPSRKINKVNRSL 587
Query: 647 QKLQLVIDSVQGASGASFKIDSFYSNFPDL--------ASPNLSGASL-VSALNQSENPN 697
+K+Q V+DSVQG G K DS F + +LS A +Q +
Sbjct: 588 RKIQTVLDSVQGVEG-GLKFDSVTGEFVAVGPFIQEFGTQKSLSSHDEDALARSQGDMDE 646
Query: 698 SLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHT-----SDVACNKDPVV 752
+S++P L S +G L ++H ++P T S + + D +
Sbjct: 647 DVSVEP-LEVKSHDGGGVKLEEDVE----TNHQAGPGSLKKPWTWISKQSGLIYSDDTDI 701
Query: 753 G-------KDSADVVLKRIRSEAELKSHSENKA-----------KLFPRSLSQET----- 789
G KD D+ ++R S L N S+S +
Sbjct: 702 GKRSEEVNKDKEDLCVRRCLSSVALAGDGMNTRIERGNGTVEPNHSISSSMSDSSNSSGA 761
Query: 790 --LGEHTKTEYQSYLLKTCHKATPKEDAH---RVKVTYGDEKTRFRM-PKSWSYEHLLQE 843
LG + + Q++ H + + + VK TY ++ RF++ P L +E
Sbjct: 762 VLLGSSSASLEQNWNQIRTHNNSGESGSSSTLTVKATYREDTVRFKLDPYVVGCSQLYRE 821
Query: 844 IARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQ 896
+A+RF + + F +KYLDD+ EWV+L D+DL EC ++ T+K ++
Sbjct: 822 VAKRFKLQE-GAFQLKYLDDEEEWVMLVTDSDLHECFEILNGMRKHTVKFLVR 873
>UniRef100_Q6NQJ4 At3g59580 [Arabidopsis thaliana]
Length = 894
Score = 405 bits (1042), Expect = e-111
Identities = 277/833 (33%), Positives = 432/833 (51%), Gaps = 97/833 (11%)
Query: 120 VEGTSDGVKRWWIAPTCSPGLGPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPI 179
+EG+ KR + L S+ EK+++AL +F+ + +L Q W P+ GD+ +
Sbjct: 82 LEGSYACEKRPLDCTSVPRSLSHSLDEKMLKALSLFMEFS-GEGILAQFWTPIKTGDQYM 140
Query: 180 LSANNLPFSLDSGSENLARYREISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRF 239
LS + + LDS L+ YRE S F FSAE + PGLPGRVF VPEWT +V +
Sbjct: 141 LSTCDQAYLLDS---RLSGYREASRRFTFSAEANQCSY-PGLPGRVFISGVPEWTSNVMY 196
Query: 240 FRSDEYPRVEHAREFDICGTLAVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALE 299
+++ EY R++HA + ++ G++A+P+ E +C V+E+V ++ N+ ++ SVC+AL+
Sbjct: 197 YKTAEYLRMKHALDNEVRGSIAIPVLEASGSSCCAVLELVTCREKPNFDVEMNSVCRALQ 256
Query: 300 VVDLTSLKHSSIQNAKARDKSYEAALPEIQEVLRSACHMHKLPLAQTWVSC-FQQGKDG- 357
V+L + S+I + + + AL EI++VLR+ C+ H+LPLA W+ C + +G +
Sbjct: 257 AVNLQT---STIPRRQYLSSNQKEALAEIRDVLRAVCYAHRLPLALAWIPCSYSKGANDE 313
Query: 358 -----CRHSEDNYLHCISPVEQACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPF 412
++S++ L CI E +CYV D + AC+EH+L +GQG+ GKA + N+P
Sbjct: 314 LVKVYGKNSKECSLLCIE--ETSCYVNDMEMEGLVNACLEHYLREGQGIVGKALISNKPS 371
Query: 413 FSTDITMLSKTDYPLSHHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKN 472
FS+D+ +YPL HAR FGL AAVA +LRS ++ +DY+LEFFLPV+ S EQ+
Sbjct: 372 FSSDVKTFDICEYPLVQHARKFGLNAAVATKLRSTFTGDNDYILEFFLPVSMKGSSEQQL 431
Query: 473 MLISLSIIIQRCCRSLRVITDKE----LERTSSSVEVMALEDSGFARTVKWSEPQHITSV 528
+L SLS +QR CR+L+ ++D E E S SVE+ L PQ SV
Sbjct: 432 LLDSLSGTMQRLCRTLKTVSDAESIDGTEFGSRSVEMTNL-------------PQATVSV 478
Query: 529 ASLEPE--EKSSETVGGKFSDLREHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEK 586
S + + FS++ ++ + + +E S ++ EK
Sbjct: 479 GSFHTTFLDTDVNSTRSTFSNISSNKRNEMAGSQGTLQQEISG-----------ARRLEK 527
Query: 587 RRAKADKTITLEVLRQYFPGSLKDAAKNIGVCTTTLKRVCRQHGIKRWPSRKIKKVGHSL 646
+++ +K ++L VL+QYF GSLKDAAK++GVC TTLKR+CRQHGI RWPSRKI KV SL
Sbjct: 528 KKSSTEKNVSLNVLQQYFSGSLKDAAKSLGVCPTTLKRICRQHGIMRWPSRKINKVNRSL 587
Query: 647 QKLQLVIDSVQGASGASFKIDSFYSNFPDL--------ASPNLSGASL-VSALNQSENPN 697
+K+Q V+DSVQG G K DS F + +LS A +Q +
Sbjct: 588 RKIQTVLDSVQGVEG-GLKFDSVTGEFVAVGPFIQEFGTQKSLSSHDEDALARSQGDMDE 646
Query: 698 SLSIQPDLGPLSPEGATKSLSSSCSQGSLSSHSCSSMPEQQPHT-----SDVACNKDPVV 752
+S++P L S +G L ++H ++P T S + + D +
Sbjct: 647 DVSVEP-LEVKSHDGGGVKLEEDVE----TNHQAGPGSLKKPWTWISKQSGLIYSDDTDI 701
Query: 753 G-------KDSADVVLKRIRSEAELKSHSENKA-----------KLFPRSLSQET----- 789
G KD D+ ++R S L N S+S +
Sbjct: 702 GKRSEEVNKDKEDLCVRRCLSSVALAGDGMNTRIERGNGTVEPNHSISSSMSDSSNSSGA 761
Query: 790 --LGEHTKTEYQSYLLKTCHKATPKEDAH---RVKVTYGDEKTRFRM-PKSWSYEHLLQE 843
LG + + Q++ H + + + VK TY ++ RF++ P L +E
Sbjct: 762 VLLGSSSASLEQNWNQIRTHNNSGESGSSSTLTVKATYREDTVRFKLDPYVVGCSQLYRE 821
Query: 844 IARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQ 896
+A+RF + + F +KYLDD+ EWV+L D+DL EC ++ T+K ++
Sbjct: 822 VAKRFKLQE-GAFQLKYLDDEEEWVMLVTDSDLHECFEILNGMRKHTVKFLVR 873
>UniRef100_Q6RZU6 Hypothetical protein H9-8 [Musa acuminata]
Length = 1382
Score = 404 bits (1039), Expect = e-111
Identities = 290/837 (34%), Positives = 424/837 (50%), Gaps = 122/837 (14%)
Query: 141 GPSIMEKLIRALKWIKQFNWNKDMLIQIWVPVPRGDRPILSANNLPFSLDSGSENLARYR 200
G S+ ++++RAL +K+ + +L Q+W+P+ +G+ +LS + PF D +NLA YR
Sbjct: 546 GVSLPDRMLRALSLLKESSCGGAILAQVWMPIRQGNHYVLSTSEQPFLCD---QNLAGYR 602
Query: 201 EISEGFQFSAEEDSKELVPGLPGRVFRDKVPEWTPDVRFFRSDEYPRVEHAREFDICGTL 260
E+S F FS + D+ L LPGRVF PEWT +V ++ EY RV++A D+ G+L
Sbjct: 603 EVSRHFTFSTK-DAPGLFLELPGRVFISGRPEWTSNVIYYNRFEYLRVDYAVIHDVRGSL 661
Query: 261 AVPIFEQGSRTCLGVIEVVMTTQQINYVPQLESVCKALEVVDLTSLKHSSIQNAKARDKS 320
AVPIF+ +C V+E+V T ++ N+ ++ESV KAL+ V+L S+K + Q + KS
Sbjct: 662 AVPIFDPDGCSCHAVLELVTTIEKPNFDTEMESVSKALQAVNLRSIKAQAHQQSFT--KS 719
Query: 321 YEAALPEIQEVLRSACHMHKLPLAQTWVSCFQQGKDGCRHS---EDNYLHCISPVEQ--- 374
+ EI +V R CH H LPLA TW+ + DG + E + + + P +
Sbjct: 720 QISIFSEIHDVSRVICHAHMLPLAITWIPIWCD--DGAIYEAKFEKDDIGVMKPTSRRTI 777
Query: 375 ------ACYVGDPSVRFFHEACMEHHLLKGQGVAGKAFMINQPFFSTDITMLSKTDYPLS 428
ACYV D ++ F AC EHHL KGQGVAGKA N PFFS D+ + +YPL+
Sbjct: 778 LCIQKLACYVNDRQMKDFLHACAEHHLEKGQGVAGKALRSNYPFFSPDVRVYDIREYPLA 837
Query: 429 HHARLFGLRAAVAIRLRSIYSSADDYVLEFFLPVNCNDSEEQKNMLISLSIIIQRCCRSL 488
HHAR F LRAAVA RL+S Y+ DDY++EFFLP+NC SEEQ+ +L LS ++R SL
Sbjct: 838 HHARRFDLRAAVAFRLKSTYTGNDDYIVEFFLPINCRGSEEQQLLLSYLSSTMRRIHGSL 897
Query: 489 RVITDKELERTSSSVEVMALEDSGFARTVKWSEPQHITSVASLEPEEKSSETVGGKFSDL 548
R + D E+ E+M + G S+ S + + +SET +
Sbjct: 898 RTVVDAEI----GGSEIMRV---GNHNEASLGSSSTAFSMKSSQLMDGNSETTAEMHFGV 950
Query: 549 REHQEDSILKGNIECDRECSPFVEGNLSSVGISKTGEKRRAKADKTITLEVLRQYFPGSL 608
+ N+E + + + L S+ + T EK+R+ A+K I+ VL+ YF GSL
Sbjct: 951 Q----------NMESNEQSAGAHHEQLKSISMKHT-EKKRSTAEKNISFSVLQHYFSGSL 999
Query: 609 KDAAKNIG-----------VCTTTLKRVCRQHGIKRWPSRKIKKVGHSLQKLQLVIDSVQ 657
KDAA +IG VC TTLKR CRQ+GI RWPSRKIKKV SLQK+Q VI SVQ
Sbjct: 1000 KDAANSIGDHFFEHVITVAVCPTTLKRACRQYGILRWPSRKIKKVNRSLQKIQKVIRSVQ 1059
Query: 658 GASGASFKIDSFYSNFPDLASPNLSGASLVSALNQSENPNSLSIQPDLGPLSPEGATKSL 717
G GA K D S LV++++ ENP +S +P L P ++
Sbjct: 1060 GVDGA-LKYDP-------------STRCLVASVSPPENPPLISSEPKGQDLMPASSSHHS 1105
Query: 718 SSSCSQGSLSSH-------------SCSS----MPEQQPH---TSDVA----CNKD---- 749
++ S G + C + +P H TSD A N
Sbjct: 1106 ETNHSIGKVEQDYFFHGRNLRGTMLKCETNKLGIPSNDCHRDFTSDGALLPYANMQGALS 1165
Query: 750 -PVVGKDSADVVLKR----IRSEAELKSHSENKAKLFPRSLSQETLGEHT-KTEYQSYLL 803
P KD++D + + S N+ ++ R+ S L + + E ++
Sbjct: 1166 WPSYSKDASDSSYNSKEAVCQGSKDGLSFMTNECQIMSRNFSFVALHQMAMEVECNDGII 1225
Query: 804 KTCHKATPKEDAHR------------------------VKVTYGDEKTRFRMPKSWSYEH 839
+ H ++ D+ VK TY + RF+ S H
Sbjct: 1226 EHSHPSSGMTDSSNGRALNHPSFEKSKALISQIGPLITVKATYNGDTIRFKFLLSMGSHH 1285
Query: 840 LLQEIARRFNVSDMSKFDVKYLDDDLEWVLLTCDADLEECIDVCLSSESSTIKLCIQ 896
L +EI RRF + F ++++D+D EWVLL D+DL+ECI+V + S T+KL ++
Sbjct: 1286 LFEEIERRFKLL-AGTFQLEHMDNDEEWVLLVNDSDLQECINVPNNIGSKTVKLQVR 1341
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.131 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,498,335,805
Number of Sequences: 2790947
Number of extensions: 63310328
Number of successful extensions: 164618
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 59
Number of HSP's that attempted gapping in prelim test: 164191
Number of HSP's gapped (non-prelim): 222
length of query: 903
length of database: 848,049,833
effective HSP length: 137
effective length of query: 766
effective length of database: 465,690,094
effective search space: 356718612004
effective search space used: 356718612004
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0193.5


