

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0193.1
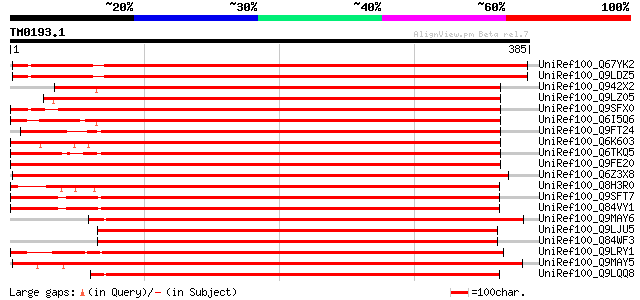
(385 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q67YK2 Hypothetical protein At1g20650 [Arabidopsis tha... 492 e-138
UniRef100_Q9LDZ5 F2D10.13 [Arabidopsis thaliana] 490 e-137
UniRef100_Q942X2 Putative serine/threonine kinase PBS1 protein [... 454 e-126
UniRef100_Q9LZ05 Protein kinase-like [Arabidopsis thaliana] 449 e-125
UniRef100_Q9SFX0 Hypothetical protein F15M4.13 [Arabidopsis thal... 449 e-125
UniRef100_Q6I5Q6 Putative serine/threonine protein kinase [Oryza... 447 e-124
UniRef100_Q9FT24 Protein serine/threonine kinase BNK1 [Brassica ... 445 e-124
UniRef100_Q6K603 Hypothetical protein OJ1789_D08.20 [Oryza sativa] 439 e-122
UniRef100_Q6TKQ5 Protein kinase-like protein [Vitis aestivalis] 438 e-121
UniRef100_Q9FE20 Serine/threonine-protein kinase PBS1 [Arabidops... 438 e-121
UniRef100_Q6Z3X8 Hypothetical protein P0627E10.22 [Oryza sativa] 438 e-121
UniRef100_Q8H3R0 Hypothetical protein P0625E02.115 [Oryza sativa] 434 e-120
UniRef100_Q9SFT7 Hypothetical protein T1B9.28 [Arabidopsis thali... 421 e-116
UniRef100_Q84VY1 At3g07070 [Arabidopsis thaliana] 421 e-116
UniRef100_Q9MAY6 Protein kinase 1 [Populus nigra] 418 e-115
UniRef100_Q9LJU5 Receptor protein kinase-like protein [Arabidops... 418 e-115
UniRef100_Q84WF3 Hypothetical protein At3g20530 [Arabidopsis tha... 416 e-115
UniRef100_Q9LRY1 Receptor protein kinase-like protein [Arabidops... 412 e-114
UniRef100_Q9MAY5 Protein kinase 2 [Populus nigra] 409 e-113
UniRef100_Q9LQQ8 Putative serine/threonine-protein kinase RLCKVI... 406 e-112
>UniRef100_Q67YK2 Hypothetical protein At1g20650 [Arabidopsis thaliana]
Length = 381
Score = 492 bits (1266), Expect = e-138
Identities = 250/383 (65%), Positives = 296/383 (77%), Gaps = 9/383 (2%)
Query: 3 CFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNG 62
CFSCL R KD+ RV+ID+ S + + + T+ GK S G
Sbjct: 4 CFSCLNPRTKDI-RVDIDNARCNSRYQTDSSVHGSDTTGTESISGILVNGKVNSPIPSGG 62
Query: 63 SSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQG 122
A SF F+ELA ATRNF+E NL+GEGGFG+VYKGRL +G+ VA+KQL+ DG QG
Sbjct: 63 -------ARSFTFKELAAATRNFREVNLLGEGGFGRVYKGRLDSGQVVAIKQLNPDGLQG 115
Query: 123 FQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNW 182
+EF++EVLMLSLLHH NLV LIGYCT GDQRLLVYEYMPMGSLEDHLF+L ++EPL+W
Sbjct: 116 NREFIVEVLMLSLLHHPNLVTLIGYCTSGDQRLLVYEYMPMGSLEDHLFDLESNQEPLSW 175
Query: 183 STRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNT 242
+TRMK+AVGAARG+EYLHCTA+PPVIYRDLKSANILLD EF+PKLSDFGLAKLGPVGD T
Sbjct: 176 NTRMKIAVGAARGIEYLHCTANPPVIYRDLKSANILLDKEFSPKLSDFGLAKLGPVGDRT 235
Query: 243 HVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSW 302
HVSTRVMGTYGYCAPEYAMSGKLT+KSDIY FGVVLLEL+TGR+AID ++ GEQNLV+W
Sbjct: 236 HVSTRVMGTYGYCAPEYAMSGKLTVKSDIYCFGVVLLELITGRKAIDLGQKQGEQNLVTW 295
Query: 303 ARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLA 362
+RPY D+++FGH+VDP L+G++P RCL+ AIAI AMCL E+ +RP I DIVVALEYLA
Sbjct: 296 SRPYLKDQKKFGHLVDPSLRGKYPRRCLNYAIAIIAMCLNEEAHYRPFIGDIVVALEYLA 355
Query: 363 SQG-SPEAHYLYGVQQRPSRMER 384
+Q S EA + SR R
Sbjct: 356 AQSRSHEARNVSSPSPEISRTPR 378
>UniRef100_Q9LDZ5 F2D10.13 [Arabidopsis thaliana]
Length = 381
Score = 490 bits (1262), Expect = e-137
Identities = 249/383 (65%), Positives = 295/383 (77%), Gaps = 9/383 (2%)
Query: 3 CFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNG 62
CFSCL R KD+ RV+ID+ S + + + T+ GK G
Sbjct: 4 CFSCLNPRTKDI-RVDIDNARCNSRYQTDSSVHGSDTTGTESISGILVNGKVNSPIPGGG 62
Query: 63 SSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQG 122
A SF F+ELA ATRNF+E NL+GEGGFG+VYKGRL +G+ VA+KQL+ DG QG
Sbjct: 63 -------ARSFTFKELAAATRNFREVNLLGEGGFGRVYKGRLDSGQVVAIKQLNPDGLQG 115
Query: 123 FQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNW 182
+EF++EVLMLSLLHH NLV LIGYCT GDQRLLVYEYMPMGSLEDHLF+L ++EPL+W
Sbjct: 116 NREFIVEVLMLSLLHHPNLVTLIGYCTSGDQRLLVYEYMPMGSLEDHLFDLESNQEPLSW 175
Query: 183 STRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNT 242
+TRMK+AVGAARG+EYLHCTA+PPVIYRDLKSANILLD EF+PKLSDFGLAKLGPVGD T
Sbjct: 176 NTRMKIAVGAARGIEYLHCTANPPVIYRDLKSANILLDKEFSPKLSDFGLAKLGPVGDRT 235
Query: 243 HVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSW 302
HVSTRVMGTYGYCAPEYAMSGKLT+KSDIY FGVVLLEL+TGR+AID ++ GEQNLV+W
Sbjct: 236 HVSTRVMGTYGYCAPEYAMSGKLTVKSDIYCFGVVLLELITGRKAIDLGQKQGEQNLVTW 295
Query: 303 ARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLA 362
+RPY D+++FGH+VDP L+G++P RCL+ AIAI AMCL E+ +RP I DIVVALEYLA
Sbjct: 296 SRPYLKDQKKFGHLVDPSLRGKYPRRCLNYAIAIIAMCLNEEAHYRPFIGDIVVALEYLA 355
Query: 363 SQG-SPEAHYLYGVQQRPSRMER 384
+Q S EA + SR R
Sbjct: 356 AQSRSHEARNVSSPSPEISRTPR 378
>UniRef100_Q942X2 Putative serine/threonine kinase PBS1 protein [Oryza sativa]
Length = 497
Score = 454 bits (1169), Expect = e-126
Identities = 228/344 (66%), Positives = 267/344 (77%), Gaps = 13/344 (3%)
Query: 34 GVNAAAAATDGERKRETKGKGKGVSSSNGS-------------SNGKTAAASFGFRELAD 80
G ++ A DG R G S S+G +N AA +F FRELA
Sbjct: 25 GADSKDARKDGSADRGVSRVGSDKSRSHGGLDSKKDVVIQRDGNNQNIAAQTFTFRELAA 84
Query: 81 ATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTN 140
AT+NF++ L+GEGGFG+VYKGRL TG+AVAVKQL +G QG +EF++EVLMLSLLHHTN
Sbjct: 85 ATKNFRQDCLLGEGGFGRVYKGRLETGQAVAVKQLDRNGLQGNREFLVEVLMLSLLHHTN 144
Query: 141 LVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLH 200
LV LIGYC DGDQRLLVYE+MP+GSLEDHL +L DKEPL+W+TRMK+A GAA+GLEYLH
Sbjct: 145 LVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEPLDWNTRMKIAAGAAKGLEYLH 204
Query: 201 CTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYA 260
A PPVIYRD KS+NILL F+PKLSDFGLAKLGPVGD THVSTRVMGTYGYCAPEYA
Sbjct: 205 DKASPPVIYRDFKSSNILLGEGFHPKLSDFGLAKLGPVGDKTHVSTRVMGTYGYCAPEYA 264
Query: 261 MSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPL 320
M+G+LT+KSD+YSFGVV LEL+TGR+AID ++ GEQNLV+WARP F DRR+F M DP+
Sbjct: 265 MTGQLTVKSDVYSFGVVFLELITGRKAIDNTKPQGEQNLVAWARPLFKDRRKFPKMADPM 324
Query: 321 LQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQ 364
LQGRFP R L+QA+A+ AMCLQEQ RP I D+V AL YLASQ
Sbjct: 325 LQGRFPMRGLYQALAVAAMCLQEQATTRPHIGDVVTALSYLASQ 368
>UniRef100_Q9LZ05 Protein kinase-like [Arabidopsis thaliana]
Length = 395
Score = 449 bits (1155), Expect = e-125
Identities = 224/347 (64%), Positives = 270/347 (77%), Gaps = 8/347 (2%)
Query: 26 SGSSSG------NGGVNAAAAATDGERKRETKGKGKG-VSSSNGSSNGKTAAASFGFREL 78
SG SSG NG ++D K + K +S S + A +F F EL
Sbjct: 7 SGKSSGRNKTRRNGDHKLDRKSSDCSVSTSEKSRAKSSLSESKSKGSDHIVAQTFTFSEL 66
Query: 79 ADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLH 137
A ATRNF++ LIGEGGFG+VYKG L +T + A+KQL H+G QG +EF++EVLMLSLLH
Sbjct: 67 ATATRNFRKECLIGEGGFGRVYKGYLASTSQTAAIKQLDHNGLQGNREFLVEVLMLSLLH 126
Query: 138 HTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLE 197
H NLV LIGYC DGDQRLLVYEYMP+GSLEDHL ++S K+PL+W+TRMK+A GAA+GLE
Sbjct: 127 HPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDISPGKQPLDWNTRMKIAAGAAKGLE 186
Query: 198 YLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAP 257
YLH PPVIYRDLK +NILLD+++ PKLSDFGLAKLGPVGD +HVSTRVMGTYGYCAP
Sbjct: 187 YLHDKTMPPVIYRDLKCSNILLDDDYFPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAP 246
Query: 258 EYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMV 317
EYAM+G+LTLKSD+YSFGVVLLE++TGR+AID+SR GEQNLV+WARP F DRR+F M
Sbjct: 247 EYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDSSRSTGEQNLVAWARPLFKDRRKFSQMA 306
Query: 318 DPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQ 364
DP+LQG++P R L+QA+A+ AMC+QEQP RPLI D+V AL YLASQ
Sbjct: 307 DPMLQGQYPPRGLYQALAVAAMCVQEQPNLRPLIADVVTALSYLASQ 353
>UniRef100_Q9SFX0 Hypothetical protein F15M4.13 [Arabidopsis thaliana]
Length = 381
Score = 449 bits (1154), Expect = e-125
Identities = 227/364 (62%), Positives = 276/364 (75%), Gaps = 10/364 (2%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS 60
M CFSCL ++ D+ R+ ID + + + A+ AT + + G V+
Sbjct: 1 MRCFSCLNTQTNDM-RINIDTLSDLT---------DYASVATKIDPRGTGSKSGILVNGK 50
Query: 61 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQLSHDGR 120
S A SF F+ELA AT+NF+E N+IG+GGFG VYKGRL +G+ VA+KQL+ DG
Sbjct: 51 VNSPKPGGGARSFTFKELAAATKNFREGNIIGKGGFGSVYKGRLDSGQVVAIKQLNPDGH 110
Query: 121 QGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL 180
QG QEF++EV MLS+ HH NLV LIGYCT G QRLLVYEYMPMGSLEDHLF+L D+ PL
Sbjct: 111 QGNQEFIVEVCMLSVFHHPNLVTLIGYCTSGAQRLLVYEYMPMGSLEDHLFDLEPDQTPL 170
Query: 181 NWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGD 240
+W TRMK+AVGAARG+EYLHC P VIYRDLKSANILLD EF+ KLSDFGLAK+GPVG+
Sbjct: 171 SWYTRMKIAVGAARGIEYLHCKISPSVIYRDLKSANILLDKEFSVKLSDFGLAKVGPVGN 230
Query: 241 NTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLV 300
THVSTRVMGTYGYCAPEYAMSG+LT+KSDIYSFGVVLLEL++GR+AID S+ GEQ LV
Sbjct: 231 RTHVSTRVMGTYGYCAPEYAMSGRLTIKSDIYSFGVVLLELISGRKAIDLSKPNGEQYLV 290
Query: 301 SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEY 360
+WARPY D ++FG +VDPLL+G+F RCL+ AI+IT MCL ++ RP I D+VVA EY
Sbjct: 291 AWARPYLKDPKKFGLLVDPLLRGKFSKRCLNYAISITEMCLNDEANHRPKIGDVVVAFEY 350
Query: 361 LASQ 364
+ASQ
Sbjct: 351 IASQ 354
>UniRef100_Q6I5Q6 Putative serine/threonine protein kinase [Oryza sativa]
Length = 491
Score = 447 bits (1150), Expect = e-124
Identities = 230/370 (62%), Positives = 277/370 (74%), Gaps = 17/370 (4%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS 60
M CF C S K A + G +GG A G K +++G G+ S
Sbjct: 1 MGCFPCFGSGGKG--------EAKKGGGGRKDGGSADRRVARVGSDKSKSQG---GLDSR 49
Query: 61 NGS-----SNGKTAAA-SFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGEAVAVKQ 114
+ +NG+ AA +F FRELA AT+NF++ L+GEGGFG+VYKG L G+AVAVKQ
Sbjct: 50 KDAFIPRDANGQPIAAHTFTFRELAAATKNFRQDCLLGEGGFGRVYKGHLENGQAVAVKQ 109
Query: 115 LSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELS 174
L +G QG +EF++EVLMLSLLHH NLV LIGYC DGDQRLLVYE+MP+GSLEDHL ++
Sbjct: 110 LDRNGLQGNREFLVEVLMLSLLHHDNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDIP 169
Query: 175 HDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAK 234
DKEPL+W+TRMK+A GAA+GLE+LH A+PPVIYRD KS+NILL ++PKLSDFGLAK
Sbjct: 170 PDKEPLDWNTRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLGEGYHPKLSDFGLAK 229
Query: 235 LGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRP 294
LGPVGD THVSTRVMGTYGYCAPEYAM+G+LT+KSD+YSFGVV LEL+TGR+AID ++
Sbjct: 230 LGPVGDKTHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDNTKPL 289
Query: 295 GEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDI 354
GEQNLV+WARP F DRR+F M DPLL GRFP R L+QA+A+ AMCLQEQ RP I D+
Sbjct: 290 GEQNLVAWARPLFKDRRKFPKMADPLLAGRFPMRGLYQALAVAAMCLQEQAATRPFIGDV 349
Query: 355 VVALEYLASQ 364
V AL YLASQ
Sbjct: 350 VTALSYLASQ 359
>UniRef100_Q9FT24 Protein serine/threonine kinase BNK1 [Brassica napus]
Length = 376
Score = 445 bits (1145), Expect = e-124
Identities = 224/357 (62%), Positives = 275/357 (76%), Gaps = 17/357 (4%)
Query: 9 SRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNGSSNGKT 68
S K R + D+N +R+ S S + A ++ ++ S S GS N
Sbjct: 10 SSGKTKKRSDSDENLSRNCSVSASERSKAKSSVSE--------------SRSRGSDN--I 53
Query: 69 AAASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHDGRQGFQEFV 127
A +F F ELA ATRNF++ LIGEGGFG+VYKG L +TG+ A+KQL H+G QG +EF+
Sbjct: 54 VAQTFTFSELATATRNFRKECLIGEGGFGRVYKGYLASTGQTAAIKQLDHNGLQGNREFL 113
Query: 128 MEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMK 187
+EVLMLSLLHH NLV LIGYC DGDQRLLVYEYMP+GSLEDHL ++S K+PL+W+TRMK
Sbjct: 114 VEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDISPSKQPLDWNTRMK 173
Query: 188 VAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTR 247
+A GAA+GLEYLH PPVIYRDLK +NILL +++ PKLSDFGLAKLGPVGD +HVSTR
Sbjct: 174 IAAGAAKGLEYLHDKTMPPVIYRDLKCSNILLGDDYFPKLSDFGLAKLGPVGDKSHVSTR 233
Query: 248 VMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYF 307
VMGTYGYCAPEYAM+G+LTLKSD+YSFGVVLLE++TGR+AID SR GEQNLV+WARP F
Sbjct: 234 VMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDNSRCTGEQNLVAWARPLF 293
Query: 308 SDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQ 364
DRR+F M DP++QG++P R L+QA+A+ AMC+QEQP RP+I D+V AL YLASQ
Sbjct: 294 KDRRKFSQMADPMIQGQYPPRGLYQALAVAAMCVQEQPNLRPVIADVVTALTYLASQ 350
>UniRef100_Q6K603 Hypothetical protein OJ1789_D08.20 [Oryza sativa]
Length = 526
Score = 439 bits (1130), Expect = e-122
Identities = 228/402 (56%), Positives = 284/402 (69%), Gaps = 38/402 (9%)
Query: 1 MSCFSCLFSRRKDVSRVEIDD----NATRSGSSSGNGGVNAAAAATDGER---------- 46
M CFSC S ++ ++ ++ S +++ GG ++A GER
Sbjct: 1 MGCFSCFDSPAEEQLNPKVGGPYGGGSSSSAAAAAYGGGGGSSAGRHGERGGGYPDLHHH 60
Query: 47 -------------------KRETKGKGKGV----SSSNGSSNGKTAAASFGFRELADATR 83
+T+ K + S+ ++ +A +F FRELA ATR
Sbjct: 61 HQQQQLPMAAPRVEKLSAGAEKTRVKSNAILREPSAPKDANGNVISAQTFTFRELATATR 120
Query: 84 NFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLV 142
NF+ +GEGGFG+VYKGRL +TG+ VA+KQL+ DG QG +EF++EVLMLSLLHH NLV
Sbjct: 121 NFRPECFLGEGGFGRVYKGRLESTGQVVAIKQLNRDGLQGNREFLVEVLMLSLLHHQNLV 180
Query: 143 RLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCT 202
LIGYC DGDQRLLVYEYM GSLEDHL +L DKE L+W+TRMK+A GAA+GLEYLH
Sbjct: 181 NLIGYCADGDQRLLVYEYMHFGSLEDHLHDLPPDKEALDWNTRMKIAAGAAKGLEYLHDK 240
Query: 203 ADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMS 262
A+PPVIYRD KS+NILLD F+PKLSDFGLAKLGPVGD +HVSTRVMGTYGYCAPEYAM+
Sbjct: 241 ANPPVIYRDFKSSNILLDESFHPKLSDFGLAKLGPVGDKSHVSTRVMGTYGYCAPEYAMT 300
Query: 263 GKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQ 322
G+LT+KSD+YSFGVVLLEL+TGRRAID++R GEQNLVSWARP F+DRR+ M DP L+
Sbjct: 301 GQLTVKSDVYSFGVVLLELITGRRAIDSTRPHGEQNLVSWARPLFNDRRKLPKMADPRLE 360
Query: 323 GRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLASQ 364
GR+P R L+QA+A+ +MC+Q + RPLI D+V AL YLASQ
Sbjct: 361 GRYPMRGLYQALAVASMCIQSEAASRPLIADVVTALSYLASQ 402
>UniRef100_Q6TKQ5 Protein kinase-like protein [Vitis aestivalis]
Length = 376
Score = 438 bits (1127), Expect = e-121
Identities = 225/365 (61%), Positives = 273/365 (74%), Gaps = 15/365 (4%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS 60
M CF C K + + ++ + S+S VN++ DG K S
Sbjct: 1 MGCFPCAGKSNKKGKKNQEKNSKDQIPSTSERLKVNSSV---DGR---------KDASKD 48
Query: 61 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHDG 119
GS + AA +F FRELA AT+NF+ L+GEGGFG+VYKGRL +T + VA+KQL +G
Sbjct: 49 GGSDH--IAAHTFTFRELAAATKNFRAECLLGEGGFGRVYKGRLESTNKIVAIKQLDRNG 106
Query: 120 RQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEP 179
QG +EF++EVLMLSLLHH NLV LIGYC DGDQRLLVYEYM +GSLEDHL +L DK+
Sbjct: 107 LQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEYMALGSLEDHLHDLPPDKKR 166
Query: 180 LNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVG 239
L+W+TRMK+A GAA+GLEYLH A PPVIYRDLK +NILL ++PKLSDFGLAKLGPVG
Sbjct: 167 LDWNTRMKIAAGAAKGLEYLHDKASPPVIYRDLKCSNILLGEGYHPKLSDFGLAKLGPVG 226
Query: 240 DNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNL 299
D THVSTRVMGTYGYCAPEYAM+G+LTLKSD+YSFGVVLLE++TGR+AID S+ GE NL
Sbjct: 227 DKTHVSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRKAIDNSKAAGEHNL 286
Query: 300 VSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
V+WARP F DRR+F M DP+L G++P R L+QA+A+ AMC+QEQP RPLI D+V AL
Sbjct: 287 VAWARPLFKDRRKFSQMADPMLHGQYPLRGLYQALAVAAMCVQEQPNMRPLIADVVTALT 346
Query: 360 YLASQ 364
YLASQ
Sbjct: 347 YLASQ 351
>UniRef100_Q9FE20 Serine/threonine-protein kinase PBS1 [Arabidopsis thaliana]
Length = 456
Score = 438 bits (1127), Expect = e-121
Identities = 222/366 (60%), Positives = 271/366 (73%), Gaps = 2/366 (0%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGER-KRETKGKGKGVSS 59
M CFSC S + + N + S N + + GE+ +T G K
Sbjct: 1 MGCFSCFDSSDDEKLNPVDESNHGQKKQSQPTVSNNISGLPSGGEKLSSKTNGGSKRELL 60
Query: 60 SNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHD 118
G+ AA +F FRELA AT NF +GEGGFG+VYKGRL +TG+ VAVKQL +
Sbjct: 61 LPRDGLGQIAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRN 120
Query: 119 GRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKE 178
G QG +EF++EVLMLSLLHH NLV LIGYC DGDQRLLVYE+MP+GSLEDHL +L DKE
Sbjct: 121 GLQGNREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKE 180
Query: 179 PLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPV 238
L+W+ RMK+A GAA+GLE+LH A+PPVIYRD KS+NILLD F+PKLSDFGLAKLGP
Sbjct: 181 ALDWNMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPT 240
Query: 239 GDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQN 298
GD +HVSTRVMGTYGYCAPEYAM+G+LT+KSD+YSFGVV LEL+TGR+AID+ GEQN
Sbjct: 241 GDKSHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQN 300
Query: 299 LVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVAL 358
LV+WARP F+DRR+F + DP L+GRFP+R L+QA+A+ +MC+QEQ RPLI D+V AL
Sbjct: 301 LVAWARPLFNDRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTAL 360
Query: 359 EYLASQ 364
YLA+Q
Sbjct: 361 SYLANQ 366
>UniRef100_Q6Z3X8 Hypothetical protein P0627E10.22 [Oryza sativa]
Length = 390
Score = 438 bits (1126), Expect = e-121
Identities = 225/371 (60%), Positives = 275/371 (73%), Gaps = 3/371 (0%)
Query: 3 CFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSSNG 62
CF+CL + + + + S N A++ ++ + + SNG
Sbjct: 4 CFACLGAGGGKMKKKKKSPPQIPPASERDNPPNLASSTVMKQDQDSFQLAANEDILVSNG 63
Query: 63 SS-NGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRL-TTGEAVAVKQLSHDGR 120
SS N + AA +F FRELA AT NF+ L+GEGGFG+VYKG L T + VA+KQL +G
Sbjct: 64 SSENRRIAARTFTFRELAAATSNFRVDCLLGEGGFGRVYKGYLETVDQVVAIKQLDRNGL 123
Query: 121 QGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEPL 180
QG +EF++EVLMLS+LHH NLV LIGYC DGDQRLLVYEYMP+GSLEDHL + K L
Sbjct: 124 QGNREFLVEVLMLSMLHHPNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLHDPPPGKSRL 183
Query: 181 NWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGD 240
+W+TRMK+A GAA+GLEYLH A+PPVIYRDLK +NILL ++PKLSDFGLAKLGP+GD
Sbjct: 184 DWNTRMKIAAGAAKGLEYLHDKANPPVIYRDLKCSNILLGEGYHPKLSDFGLAKLGPIGD 243
Query: 241 NTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLV 300
+HVSTRVMGTYGYCAPEYAM+G+LTLKSD+YSFGVVLLE++TGRRAID +R GEQNLV
Sbjct: 244 KSHVSTRVMGTYGYCAPEYAMTGQLTLKSDVYSFGVVLLEIITGRRAIDNTRAAGEQNLV 303
Query: 301 SWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEY 360
+WARP F DRR+F M DP L G++PSR L+QA+A+ AMC+QEQP RPLI D+V AL Y
Sbjct: 304 AWARPLFKDRRKFPQMADPALHGQYPSRGLYQALAVAAMCVQEQPTMRPLIGDVVTALAY 363
Query: 361 LASQG-SPEAH 370
LASQ PEAH
Sbjct: 364 LASQTYDPEAH 374
>UniRef100_Q8H3R0 Hypothetical protein P0625E02.115 [Oryza sativa]
Length = 479
Score = 434 bits (1115), Expect = e-120
Identities = 226/377 (59%), Positives = 271/377 (70%), Gaps = 34/377 (9%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNA--AAAATDGERK-------RETK 51
MSCF C SSG GGV+A AA + G R R
Sbjct: 1 MSCFPC--------------------SGSSGKGGVDAKSVAALSPGPRPAASAAPDRSNS 40
Query: 52 GKGKGVSSSN-----GSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTT 106
+G G+ + GSS A F FRELA AT+NF++ L+GEGGFG+VYKG++
Sbjct: 41 SRGSGIKKDDSVRRGGSSANDGPAKIFTFRELAVATKNFRKDCLLGEGGFGRVYKGQMEN 100
Query: 107 GEAVAVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSL 166
G+ +AVKQL +G QG +EF++EVLMLSLLHH NLVRLIGYC DGDQRLLVYEYM +GSL
Sbjct: 101 GQVIAVKQLDRNGLQGNREFLVEVLMLSLLHHPNLVRLIGYCADGDQRLLVYEYMLLGSL 160
Query: 167 EDHLFELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPK 226
E+HL + K+PL+W+ RMK+AVGAA+GLEYLH A+PPVIYRD KS+NILL ++ PK
Sbjct: 161 ENHLHDRPPGKKPLDWNARMKIAVGAAKGLEYLHDKANPPVIYRDFKSSNILLGEDYYPK 220
Query: 227 LSDFGLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRR 286
LSDFGLAKLGPVGD THVSTRVMGTYGYCAPEYAM+G+LT+KSD+YSFGVV LEL+TGR+
Sbjct: 221 LSDFGLAKLGPVGDKTHVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRK 280
Query: 287 AIDTSRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPK 346
AID ++ GEQNLV+WARP F DRR+F M DP LQG +P R L+QA+A+ +MCLQE
Sbjct: 281 AIDHTQPAGEQNLVAWARPLFRDRRKFCQMADPSLQGCYPKRGLYQALAVASMCLQENAT 340
Query: 347 FRPLITDIVVALEYLAS 363
RPLI DIV AL YLAS
Sbjct: 341 SRPLIADIVTALSYLAS 357
>UniRef100_Q9SFT7 Hypothetical protein T1B9.28 [Arabidopsis thaliana]
Length = 414
Score = 421 bits (1083), Expect = e-116
Identities = 214/364 (58%), Positives = 266/364 (72%), Gaps = 7/364 (1%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS 60
M+CFSC + K + D++ R+G +G T E + + K
Sbjct: 1 MNCFSCFYFHEKKKVPRDSDNSYRRNGEVTGRDNNK-----THPENPKTVNEQNKNNDED 55
Query: 61 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLT-TGEAVAVKQLSHDG 119
+N AA +F FRELA AT+NF++ LIGEGGFG+VYKG+L TG VAVKQL +G
Sbjct: 56 KEVTNN-IAAQTFSFRELATATKNFRQECLIGEGGFGRVYKGKLEKTGMIVAVKQLDRNG 114
Query: 120 RQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEP 179
QG +EF++EVLMLSLLHH +LV LIGYC DGDQRLLVYEYM GSLEDHL +L+ D+ P
Sbjct: 115 LQGNKEFIVEVLMLSLLHHKHLVNLIGYCADGDQRLLVYEYMSRGSLEDHLLDLTPDQIP 174
Query: 180 LNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVG 239
L+W TR+++A+GAA GLEYLH A+PPVIYRDLK+ANILLD EFN KLSDFGLAKLGPVG
Sbjct: 175 LDWDTRIRIALGAAMGLEYLHDKANPPVIYRDLKAANILLDGEFNAKLSDFGLAKLGPVG 234
Query: 240 DNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNL 299
D HVS+RVMGTYGYCAPEY +G+LT KSD+YSFGVVLLEL+TGRR IDT+R EQNL
Sbjct: 235 DKQHVSSRVMGTYGYCAPEYQRTGQLTTKSDVYSFGVVLLELITGRRVIDTTRPKDEQNL 294
Query: 300 VSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
V+WA+P F + RF + DP L+G FP + L+QA+A+ AMCLQE+ RPL++D+V AL
Sbjct: 295 VTWAQPVFKEPSRFPELADPSLEGVFPEKALNQAVAVAAMCLQEEATVRPLMSDVVTALG 354
Query: 360 YLAS 363
+L +
Sbjct: 355 FLGT 358
>UniRef100_Q84VY1 At3g07070 [Arabidopsis thaliana]
Length = 414
Score = 421 bits (1083), Expect = e-116
Identities = 214/364 (58%), Positives = 266/364 (72%), Gaps = 7/364 (1%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS 60
M+CFSC + K + D++ R+G +G T E + + K
Sbjct: 1 MNCFSCFYFHEKKKVPRDSDNSYRRNGEVTGRDNNK-----THPENPKTVNEQNKNNDED 55
Query: 61 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLT-TGEAVAVKQLSHDG 119
+N AA +F FRELA AT+NF++ LIGEGGFG+VYKG+L TG VAVKQL +G
Sbjct: 56 KEVTNN-IAAQTFSFRELATATKNFRQECLIGEGGFGRVYKGKLEKTGMIVAVKQLDRNG 114
Query: 120 RQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEP 179
QG +EF++EVLMLSLLHH +LV LIGYC DGDQRLLVYEYM GSLEDHL +L+ D+ P
Sbjct: 115 LQGNKEFIVEVLMLSLLHHKHLVNLIGYCADGDQRLLVYEYMSRGSLEDHLLDLTPDQIP 174
Query: 180 LNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVG 239
L+W TR+++A+GAA GLEYLH A+PPVIYRDLK+ANILLD EFN KLSDFGLAKLGPVG
Sbjct: 175 LDWDTRIRIALGAAMGLEYLHDRANPPVIYRDLKAANILLDGEFNAKLSDFGLAKLGPVG 234
Query: 240 DNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNL 299
D HVS+RVMGTYGYCAPEY +G+LT KSD+YSFGVVLLEL+TGRR IDT+R EQNL
Sbjct: 235 DKQHVSSRVMGTYGYCAPEYQRTGQLTTKSDVYSFGVVLLELITGRRVIDTTRPKDEQNL 294
Query: 300 VSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
V+WA+P F + RF + DP L+G FP + L+QA+A+ AMCLQE+ RPL++D+V AL
Sbjct: 295 VTWAQPVFKEPSRFPELADPSLEGVFPEKALNQAVAVAAMCLQEEATVRPLMSDVVTALG 354
Query: 360 YLAS 363
+L +
Sbjct: 355 FLGT 358
>UniRef100_Q9MAY6 Protein kinase 1 [Populus nigra]
Length = 405
Score = 418 bits (1074), Expect = e-115
Identities = 208/325 (64%), Positives = 257/325 (79%), Gaps = 3/325 (0%)
Query: 59 SSNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTT-GEAVAVKQLSH 117
S + SNGK A +F F ELA AT NF+ +GEGGFGKVYKG L +AVA+KQL
Sbjct: 73 SKDRRSNGKQAQ-TFTFEELAAATSNFRSDCFLGEGGFGKVYKGYLDKINQAVAIKQLDR 131
Query: 118 DGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDK 177
+G QG +EFV+EV+ LSL H NLV+LIG+C +GDQRLLVYEYMP+GSLE+HL ++ ++
Sbjct: 132 NGVQGIREFVVEVVTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPLGSLENHLHDIPPNR 191
Query: 178 EPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGP 237
+PL+W+TRMK+A GAA+GLEYLH PPVIYRDLK +NILL ++PKLSDFGLAK+GP
Sbjct: 192 QPLDWNTRMKIAAGAAKGLEYLHNEMKPPVIYRDLKCSNILLGEGYHPKLSDFGLAKVGP 251
Query: 238 VGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQ 297
GD THVSTRVMGTYGYCAP+YAM+G+LT KSDIYSFGVVLLEL+TGR+AID + GEQ
Sbjct: 252 SGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDQRKERGEQ 311
Query: 298 NLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVA 357
NLV+WARP F DRR F MVDPLLQG++P R L+QA+AI AMC+QEQP RP ++D+V+A
Sbjct: 312 NLVAWARPMFKDRRNFSCMVDPLLQGQYPIRGLYQALAIAAMCVQEQPNMRPAVSDLVMA 371
Query: 358 LEYLAS-QGSPEAHYLYGVQQRPSR 381
L YLAS + P+ H + ++ PSR
Sbjct: 372 LNYLASHKYDPQVHSVQDSRRSPSR 396
>UniRef100_Q9LJU5 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 377
Score = 418 bits (1074), Expect = e-115
Identities = 203/299 (67%), Positives = 246/299 (81%), Gaps = 2/299 (0%)
Query: 66 GKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGE-AVAVKQLSHDGRQGFQ 124
G +A F FREL AT+NF N +GEGGFG+VYKG++ T E VAVKQL +G QG +
Sbjct: 54 GNISAHIFTFRELCVATKNFNPDNQLGEGGFGRVYKGQIETPEQVVAVKQLDRNGYQGNR 113
Query: 125 EFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKE-PLNWS 183
EF++EV+MLSLLHH NLV L+GYC DGDQR+LVYEYM GSLEDHL EL+ +K+ PL+W
Sbjct: 114 EFLVEVMMLSLLHHQNLVNLVGYCADGDQRILVYEYMQNGSLEDHLLELARNKKKPLDWD 173
Query: 184 TRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTH 243
TRMKVA GAARGLEYLH TADPPVIYRD K++NILLD EFNPKLSDFGLAK+GP G TH
Sbjct: 174 TRMKVAAGAARGLEYLHETADPPVIYRDFKASNILLDEEFNPKLSDFGLAKVGPTGGETH 233
Query: 244 VSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWA 303
VSTRVMGTYGYCAPEYA++G+LT+KSD+YSFGVV LE++TGRR IDT++ EQNLV+WA
Sbjct: 234 VSTRVMGTYGYCAPEYALTGQLTVKSDVYSFGVVFLEMITGRRVIDTTKPTEEQNLVTWA 293
Query: 304 RPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLA 362
P F DRR+F M DPLL+G++P + L+QA+A+ AMCLQE+ RP+++D+V ALEYLA
Sbjct: 294 SPLFKDRRKFTLMADPLLEGKYPIKGLYQALAVAAMCLQEEAATRPMMSDVVTALEYLA 352
>UniRef100_Q84WF3 Hypothetical protein At3g20530 [Arabidopsis thaliana]
Length = 386
Score = 416 bits (1068), Expect = e-115
Identities = 202/299 (67%), Positives = 245/299 (81%), Gaps = 2/299 (0%)
Query: 66 GKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTTGE-AVAVKQLSHDGRQGFQ 124
G +A F FREL AT+NF N +GEGGFG+VYKG++ T E VAVKQL +G QG +
Sbjct: 63 GNISAHIFTFRELCVATKNFNPDNQLGEGGFGRVYKGQIETPEQVVAVKQLDRNGYQGNR 122
Query: 125 EFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKE-PLNWS 183
EF++EV+MLSLLHH NLV L+GYC DGDQR+LVYEYM GSLEDHL EL+ +K+ PL+W
Sbjct: 123 EFLVEVMMLSLLHHQNLVNLVGYCADGDQRILVYEYMQNGSLEDHLLELARNKKKPLDWD 182
Query: 184 TRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVGDNTH 243
TRMKVA GAARGLEYLH TADPPVIYRD K++NILLD EFNPKLSDFGLAK+GP G H
Sbjct: 183 TRMKVAAGAARGLEYLHETADPPVIYRDFKASNILLDEEFNPKLSDFGLAKVGPTGGEIH 242
Query: 244 VSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNLVSWA 303
VSTRVMGTYGYCAPEYA++G+LT+KSD+YSFGVV LE++TGRR IDT++ EQNLV+WA
Sbjct: 243 VSTRVMGTYGYCAPEYALTGQLTVKSDVYSFGVVFLEMITGRRVIDTTKPTEEQNLVTWA 302
Query: 304 RPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALEYLA 362
P F DRR+F M DPLL+G++P + L+QA+A+ AMCLQE+ RP+++D+V ALEYLA
Sbjct: 303 SPLFKDRRKFTLMADPLLEGKYPIKGLYQALAVAAMCLQEEAATRPMMSDVVTALEYLA 361
>UniRef100_Q9LRY1 Receptor protein kinase-like protein [Arabidopsis thaliana]
Length = 381
Score = 412 bits (1059), Expect = e-114
Identities = 213/367 (58%), Positives = 263/367 (71%), Gaps = 21/367 (5%)
Query: 1 MSCFSCLFSRRKDVSRVEIDDNATRSGSSSGNGGVNAAAAATDGERKRETKGKGKGVSSS 60
MSCFSC S+ D N G + A + T G+ ++
Sbjct: 1 MSCFSCFSSKVLD------------------NEGSSMPAPYKQPNSPKRTTGEVVA-KNA 41
Query: 61 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTT-GEAVAVKQLSHDG 119
NG SN A F FRELA AT+NF++ LIGEGGFG+VYKG+L + VAVKQL +G
Sbjct: 42 NGPSNNM-GARIFTFRELATATKNFRQECLIGEGGFGRVYKGKLENPAQVVAVKQLDRNG 100
Query: 120 RQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEP 179
QG +EF++EVLMLSLLHH NLV LIGYC DGDQRLLVYEYMP+GSLEDHL +L ++P
Sbjct: 101 LQGQREFLVEVLMLSLLHHRNLVNLIGYCADGDQRLLVYEYMPLGSLEDHLLDLEPGQKP 160
Query: 180 LNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVG 239
L+W+TR+K+A+GAA+G+EYLH ADPPVIYRDLKS+NILLD E+ KLSDFGLAKLGPVG
Sbjct: 161 LDWNTRIKIALGAAKGIEYLHDEADPPVIYRDLKSSNILLDPEYVAKLSDFGLAKLGPVG 220
Query: 240 DNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNL 299
D HVS+RVMGTYGYCAPEY +G LT KSD+YSFGVVLLEL++GRR IDT R EQNL
Sbjct: 221 DTLHVSSRVMGTYGYCAPEYQRTGYLTNKSDVYSFGVVLLELISGRRVIDTMRPSHEQNL 280
Query: 300 VSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
V+WA P F D R+ + DPLL+G +P + L+QAIA+ AMCL E+P RPL++D++ AL
Sbjct: 281 VTWALPIFRDPTRYWQLADPLLRGDYPEKSLNQAIAVAAMCLHEEPTVRPLMSDVITALS 340
Query: 360 YLASQGS 366
+L + +
Sbjct: 341 FLGASSN 347
>UniRef100_Q9MAY5 Protein kinase 2 [Populus nigra]
Length = 406
Score = 409 bits (1051), Expect = e-113
Identities = 213/391 (54%), Positives = 272/391 (69%), Gaps = 13/391 (3%)
Query: 3 CFSCLFSRRKDVSRVEI--------DDNATRSGSSSGNGGVNAA---AAATDGERKRETK 51
CFS S+R + S ++ D SGS VN + A+ D + + K
Sbjct: 5 CFSGKSSKRSENSSIDENNSNIKRKDQTQLTSGSMKVKPYVNDSREEGASKDDQLSLDVK 64
Query: 52 GKGKGVSSSNGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTT-GEAV 110
S N A +F F EL AT NF+ +GEGGFGKVYKG L + V
Sbjct: 65 SLNLKDEISKDIRNNGNPAQTFTFEELVAATDNFRSDCFLGEGGFGKVYKGYLEKINQVV 124
Query: 111 AVKQLSHDGRQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHL 170
A+KQL +G QG +EFV+EVL LSL + NLV+LIG+C +GDQRLLVYEYMP+GSLE+HL
Sbjct: 125 AIKQLDQNGLQGIREFVVEVLTLSLADNPNLVKLIGFCAEGDQRLLVYEYMPLGSLENHL 184
Query: 171 FELSHDKEPLNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDF 230
++ +++PL+W+ RMK+A GAA+GLEYLH PPVIYRDLK +NILL ++PKLSDF
Sbjct: 185 HDIPPNRQPLDWNARMKIAAGAAKGLEYLHNEMAPPVIYRDLKCSNILLGEGYHPKLSDF 244
Query: 231 GLAKLGPVGDNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDT 290
GLAK+GP GD+THVSTRVMGTYGYCAP+YAM+G+LT KSD+YSFGVVLLEL+TGR+AID
Sbjct: 245 GLAKVGPSGDHTHVSTRVMGTYGYCAPDYAMTGQLTFKSDVYSFGVVLLELITGRKAIDQ 304
Query: 291 SRRPGEQNLVSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPL 350
++ EQNLV+WARP F DRR F MVDP LQG++P + L+QA+AI AMC+QEQP RP
Sbjct: 305 TKERSEQNLVAWARPMFKDRRNFSGMVDPFLQGQYPIKGLYQALAIAAMCVQEQPNMRPA 364
Query: 351 ITDIVVALEYLAS-QGSPEAHYLYGVQQRPS 380
++D+V+AL YLAS + P+ H ++RPS
Sbjct: 365 VSDVVLALNYLASHKYDPQIHPFKDPRRRPS 395
>UniRef100_Q9LQQ8 Putative serine/threonine-protein kinase RLCKVII [Arabidopsis
thaliana]
Length = 423
Score = 406 bits (1044), Expect = e-112
Identities = 200/304 (65%), Positives = 243/304 (79%), Gaps = 2/304 (0%)
Query: 61 NGSSNGKTAAASFGFRELADATRNFKEANLIGEGGFGKVYKGRLTT-GEAVAVKQLSHDG 119
N GK A +F F+ELA+AT NF+ +GEGGFGKV+KG + + VA+KQL +G
Sbjct: 80 NDQVTGKKAQ-TFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNG 138
Query: 120 RQGFQEFVMEVLMLSLLHHTNLVRLIGYCTDGDQRLLVYEYMPMGSLEDHLFELSHDKEP 179
QG +EFV+EVL LSL H NLV+LIG+C +GDQRLLVYEYMP GSLEDHL L K+P
Sbjct: 139 VQGIREFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKP 198
Query: 180 LNWSTRMKVAVGAARGLEYLHCTADPPVIYRDLKSANILLDNEFNPKLSDFGLAKLGPVG 239
L+W+TRMK+A GAARGLEYLH PPVIYRDLK +NILL ++ PKLSDFGLAK+GP G
Sbjct: 199 LDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSG 258
Query: 240 DNTHVSTRVMGTYGYCAPEYAMSGKLTLKSDIYSFGVVLLELLTGRRAIDTSRRPGEQNL 299
D THVSTRVMGTYGYCAP+YAM+G+LT KSDIYSFGVVLLEL+TGR+AID ++ +QNL
Sbjct: 259 DKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNL 318
Query: 300 VSWARPYFSDRRRFGHMVDPLLQGRFPSRCLHQAIAITAMCLQEQPKFRPLITDIVVALE 359
V WARP F DRR F MVDPLLQG++P R L+QA+AI+AMC+QEQP RP+++D+V+AL
Sbjct: 319 VGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLALN 378
Query: 360 YLAS 363
+LAS
Sbjct: 379 FLAS 382
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 649,234,119
Number of Sequences: 2790947
Number of extensions: 27587379
Number of successful extensions: 145595
Number of sequences better than 10.0: 18030
Number of HSP's better than 10.0 without gapping: 7827
Number of HSP's successfully gapped in prelim test: 10207
Number of HSP's that attempted gapping in prelim test: 105507
Number of HSP's gapped (non-prelim): 21850
length of query: 385
length of database: 848,049,833
effective HSP length: 129
effective length of query: 256
effective length of database: 488,017,670
effective search space: 124932523520
effective search space used: 124932523520
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0193.1


