

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0182.6
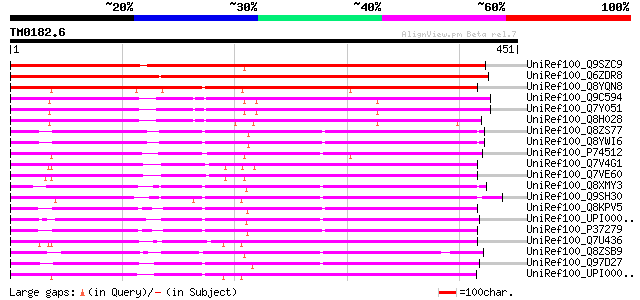
(451 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SZC9 Potential copper-transporting ATPase PAA1 [Arab... 566 e-160
UniRef100_Q6ZDR8 Putative potential copper-transporting ATPase [... 561 e-158
UniRef100_Q8YQN8 Cation-transporting P-type ATPase [Anabaena sp.] 351 2e-95
UniRef100_Q9C594 Metal-transporting ATPase-like protein [Arabido... 323 8e-87
UniRef100_Q7Y051 Paa2 P-type ATPase [Arabidopsis thaliana] 323 8e-87
UniRef100_Q8H028 Hypothetical protein OSJNBa0050H14.2 [Oryza sat... 320 5e-86
UniRef100_Q8ZS77 Cation-transporting ATPase [Anabaena sp.] 298 3e-79
UniRef100_Q8YWI6 Cation-transporting ATPase [Anabaena sp.] 297 4e-79
UniRef100_P74512 Cation-transporting ATPase; E1-E2 ATPase [Synec... 290 8e-77
UniRef100_Q7V4G1 Putative P-type ATPase transporter for copper [... 287 4e-76
UniRef100_Q7VE60 Cation transport ATPase [Prochlorococcus marinus] 283 5e-75
UniRef100_Q8XMY3 Probable copper-transporting ATPase [Clostridiu... 283 7e-75
UniRef100_Q9SH30 Potential copper-transporting ATPase 3 [Arabido... 281 4e-74
UniRef100_Q8KPV5 PacS [Synechococcus sp.] 278 2e-73
UniRef100_UPI00002F1620 UPI00002F1620 UniRef100 entry 278 3e-73
UniRef100_P37279 Cation-transporting ATPase pacS [Synechococcus ... 277 4e-73
UniRef100_Q7U436 Putative P-type ATPase transporter for copper [... 277 5e-73
UniRef100_Q8ZSB9 Cation transporting ATPase [Anabaena sp.] 276 9e-73
UniRef100_Q97D27 Heavy-metal transporting P-type ATPase [Clostri... 275 3e-72
UniRef100_UPI00002E4675 UPI00002E4675 UniRef100 entry 274 3e-72
>UniRef100_Q9SZC9 Potential copper-transporting ATPase PAA1 [Arabidopsis thaliana]
Length = 949
Score = 566 bits (1458), Expect = e-160
Identities = 290/427 (67%), Positives = 350/427 (81%), Gaps = 9/427 (2%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
VAG FTYGVMA+S TFTFW+LFG H+LP+ + GS +SLALQ +CSVLVVACPCALGLA
Sbjct: 502 VAGRFTYGVMALSAATFTFWNLFGAHVLPSALHNGSPMSLALQLSCSVLVVACPCALGLA 561
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TPTA+LVGTSLGA+RGLLLRGG+ILEKF++V+ VVFDKTGTLT G PVVT+V+
Sbjct: 562 TPTAMLVGTSLGARRGLLLRGGDILEKFSLVDTVVFDKTGTLTKGHPVVTEVIIPE---- 617
Query: 121 ANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGS 180
N + + S+VE+L LAAAVESN+ HPVGKAIV AA+A NC K DGTF EEPGS
Sbjct: 618 -NPRHNLNDTWSEVEVLMLAAAVESNTTHPVGKAIVKAARARNCQTMKAEDGTFTEEPGS 676
Query: 181 GAVATIGNRKVYVGTLEWITRHGINNN---ILQEVECKNESFVYVGVNDTLAGLIYFEDE 237
GAVA + N++V VGTLEW+ RHG N L+E E N+S VY+GV++TLA +I FED+
Sbjct: 677 GAVAIVNNKRVTVGTLEWVKRHGATGNSLLALEEHEINNQSVVYIGVDNTLAAVIRFEDK 736
Query: 238 VREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFINEL 297
VREDA VV+ L++Q I VYMLSGDKRNAA +VAS+VGI ++V++GVKP +KK FINEL
Sbjct: 737 VREDAAQVVENLTRQGIDVYMLSGDKRNAANYVASVVGINHERVIAGVKPAEKKNFINEL 796
Query: 298 QKDN-IVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALELSR 356
QK+ IVAMVGDGINDAAALA+S++G+A+GGG GAASEVS ++LM + L+QLLDA+ELSR
Sbjct: 797 QKNKKIVAMVGDGINDAAALASSNVGVAMGGGAGAASEVSPVVLMGNRLTQLLDAMELSR 856
Query: 357 LTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLLLR 416
TM TVKQNLWWAF YNIVGIPIAAGVL P+ GTMLTPS+AGALMG+SS+GVMTNSLLLR
Sbjct: 857 QTMKTVKQNLWWAFGYNIVGIPIAAGVLLPLTGTMLTPSMAGALMGVSSLGVMTNSLLLR 916
Query: 417 FKFSSKQ 423
++F S +
Sbjct: 917 YRFFSNR 923
>UniRef100_Q6ZDR8 Putative potential copper-transporting ATPase [Oryza sativa]
Length = 959
Score = 561 bits (1445), Expect = e-158
Identities = 286/427 (66%), Positives = 354/427 (81%), Gaps = 2/427 (0%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
VAG FTYGVMA+S T+TFWS+FG+ ++PA GSA++LALQ +CSVLV+ACPCALGLA
Sbjct: 507 VAGNFTYGVMALSAATYTFWSIFGSQLVPAAIQHGSAMALALQLSCSVLVIACPCALGLA 566
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TPTAVLVGTSLGA RGLLLRGG+ILEKF+ V+A+VFDKTGTLT+G+PVVTKV+AS +
Sbjct: 567 TPTAVLVGTSLGATRGLLLRGGDILEKFSEVDAIVFDKTGTLTIGKPVVTKVIASHREGD 626
Query: 121 ANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGS 180
N+ + N + EIL LAA VESN+ HP+GKAI++AAQA NCL + DG+F+EEPGS
Sbjct: 627 ENTKDSCNNEWTG-EILSLAAGVESNTTHPLGKAIMEAAQAANCLYLQAKDGSFMEEPGS 685
Query: 181 GAVATIGNRKVYVGTLEWITRHGINNNILQEVECKNESFVYVGVNDTLAGLIYFEDEVRE 240
GAVATIG ++V VGTL+WI RHG+ +N + E +S YV V+ TLAGLI FED++RE
Sbjct: 686 GAVATIGEKQVSVGTLDWIRRHGVLHNPFADGENFGQSVAYVAVDGTLAGLICFEDKLRE 745
Query: 241 DARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFINELQKD 300
D+ ++D LSKQ ISVYMLSGDK++AA +VASLVGI DKV++ VKP +KK FI+ELQK+
Sbjct: 746 DSHQIIDILSKQGISVYMLSGDKKSAAMNVASLVGIQADKVIAEVKPHEKKSFISELQKE 805
Query: 301 N-IVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALELSRLTM 359
+ +VAMVGDGINDAAALA++ +GIA+GGGVGAAS+VSS++LM + LSQL+DALELS+ TM
Sbjct: 806 HKLVAMVGDGINDAAALASADVGIAMGGGVGAASDVSSVVLMGNRLSQLVDALELSKETM 865
Query: 360 TTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLLLRFKF 419
TVKQNLWWAF+YNIVG+PIAAG L PV GT+LTPSIAGALMG SS+GVM NSL LR +
Sbjct: 866 RTVKQNLWWAFLYNIVGLPIAAGALLPVTGTVLTPSIAGALMGFSSVGVMANSLFLRMRL 925
Query: 420 SSKQKQI 426
SS+Q+ I
Sbjct: 926 SSRQQPI 932
>UniRef100_Q8YQN8 Cation-transporting P-type ATPase [Anabaena sp.]
Length = 815
Score = 351 bits (901), Expect = 2e-95
Identities = 201/447 (44%), Positives = 277/447 (61%), Gaps = 33/447 (7%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQG----------------SAVSLALQF 44
VAGYFTYG++ S+ TF FW FGTHI P G S + ++L+
Sbjct: 366 VAGYFTYGILTASLLTFIFWYCFGTHIWPDITVSGGDMEMMMNHAAHITNNSPLLISLKL 425
Query: 45 ACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTV 104
A +V+VVACPCALGLATPTA+LVGT +GA+RGLL++GG++LEK ++ VVFDKTGTLT
Sbjct: 426 AIAVMVVACPCALGLATPTAILVGTGIGAERGLLIKGGDVLEKAHQLDTVVFDKTGTLTT 485
Query: 105 GRPVVTK--VVASTCIENANSSQTIENALSDV------EILRLAAAVESNSVHPVGKAIV 156
G PVVT V A + + + E +L +++LAAAVES + HP+ +AI
Sbjct: 486 GNPVVTDCLVFAEGSPDEISFTAKQERSLFPTPLHPSHSLIQLAAAVESGTHHPLARAIQ 545
Query: 157 DAAQAVNCLDAKVVDGTFLEEPGSGAVATIGNRKVYVGTLEWITRHGIN-----NNILQE 211
AAQ + VD F EPG G A + + V +G +W++ HGI Q+
Sbjct: 546 QAAQQQQLSIPEAVD--FHTEPGMGVSAVVDGQTVLLGNGDWLSWHGITWSETAQQEAQK 603
Query: 212 VECKNESFVYVGVNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVA 271
+ + ++ V V + ++LAGLI +D R DA+ VD L + + V +LSGD+ AA +A
Sbjct: 604 LATQGKTVVGVAIGESLAGLIGVQDTTRPDAQTTVDKLRQMGLRVILLSGDRPEAAHAIA 663
Query: 272 SLVGIPKDKVLSGVKPDQKKKFINELQKDN--IVAMVGDGINDAAALAASHIGIALGGGV 329
+GI + V++GV P +K FI ELQ + VAMVGDGINDA AL+ + +GIAL G
Sbjct: 664 QQLGIDRADVMAGVPPAKKAAFIQELQTKSGAKVAMVGDGINDAPALSQADVGIALHSGT 723
Query: 330 GAASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNG 389
A E + I+LMRD +S +++++ LSR T ++QNL+WAF YN +GIP+AAGVL P G
Sbjct: 724 DVAMETAQIVLMRDRISDVVESIHLSRATFNKIRQNLFWAFAYNTIGIPLAAGVLLPNWG 783
Query: 390 TMLTPSIAGALMGLSSIGVMTNSLLLR 416
+L+PS A ALM SS+ V+TNSLLLR
Sbjct: 784 FVLSPSGAAALMAFSSVSVVTNSLLLR 810
>UniRef100_Q9C594 Metal-transporting ATPase-like protein [Arabidopsis thaliana]
Length = 856
Score = 323 bits (827), Expect = 8e-87
Identities = 198/454 (43%), Positives = 270/454 (58%), Gaps = 43/454 (9%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQ------GSAVSLALQFACSVLVVACP 54
+AG F Y +M++S TF FW G+HI P G A++L+L+ A VLVV+CP
Sbjct: 419 IAGPFVYTIMSLSAMTFAFWYYVGSHIFPDVLLNDIAGPDGDALALSLKLAVDVLVVSCP 478
Query: 55 CALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVA 114
CALGLATPTA+L+GTSLGAKRG L+RGG++LE+ A ++ V DKTGTLT GRPVV+ V +
Sbjct: 479 CALGLATPTAILIGTSLGAKRGYLIRGGDVLERLASIDCVALDKTGTLTEGRPVVSGVAS 538
Query: 115 STCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTF 174
+ E+L++AAAVE + HP+ KAIV+ A+++N L G
Sbjct: 539 L--------------GYEEQEVLKMAAAVEKTATHPIAKAIVNEAESLN-LKTPETRGQ- 582
Query: 175 LEEPGSGAVATIGNRKVYVGTLEWITRHGINNN---------ILQEVECKNES------- 218
L EPG G +A I R V VG+LEW++ + N L + + N S
Sbjct: 583 LTEPGFGTLAEIDGRFVAVGSLEWVSDRFLKKNDSSDMVKLESLLDHKLSNTSSTSRYSK 642
Query: 219 -FVYVG-VNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGI 276
VYVG + + G I D +R+DA V L ++ I +LSGD+ A VA VGI
Sbjct: 643 TVVYVGREGEGIIGAIAISDCLRQDAEFTVARLQEKGIKTVLLSGDREGAVATVAKNVGI 702
Query: 277 PKDKVLSGVKPDQKKKFINELQKD-NIVAMVGDGINDAAALAASHIGIAL--GGGVGAAS 333
+ + P++K +FI+ LQ + VAMVGDGINDA +LA + +GIAL AAS
Sbjct: 703 KSESTNYSLSPEKKFEFISNLQSSGHRVAMVGDGINDAPSLAQADVGIALKIEAQENAAS 762
Query: 334 EVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLT 393
+S+IL+R+ LS ++DAL L++ TM+ V QNL WA YN++ IPIAAGVL P +T
Sbjct: 763 NAASVILVRNKLSHVVDALSLAQATMSKVYQNLAWAIAYNVISIPIAAGVLLPQYDFAMT 822
Query: 394 PSIAGALMGLSSIGVMTNSLLLRFKFSSKQKQIL 427
PS++G LM LSSI V++NSLLL+ S K L
Sbjct: 823 PSLSGGLMALSSIFVVSNSLLLQLHKSETSKNSL 856
>UniRef100_Q7Y051 Paa2 P-type ATPase [Arabidopsis thaliana]
Length = 883
Score = 323 bits (827), Expect = 8e-87
Identities = 198/454 (43%), Positives = 270/454 (58%), Gaps = 43/454 (9%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQ------GSAVSLALQFACSVLVVACP 54
+AG F Y +M++S TF FW G+HI P G A++L+L+ A VLVV+CP
Sbjct: 446 IAGPFVYTIMSLSAMTFAFWYYVGSHIFPDVLLNDIAGPDGDALALSLKLAVDVLVVSCP 505
Query: 55 CALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVA 114
CALGLATPTA+L+GTSLGAKRG L+RGG++LE+ A ++ V DKTGTLT GRPVV+ V +
Sbjct: 506 CALGLATPTAILIGTSLGAKRGYLIRGGDVLERLASIDCVALDKTGTLTEGRPVVSGVAS 565
Query: 115 STCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTF 174
+ E+L++AAAVE + HP+ KAIV+ A+++N L G
Sbjct: 566 L--------------GYEEQEVLKMAAAVEKTATHPIAKAIVNEAESLN-LKTPETRGQ- 609
Query: 175 LEEPGSGAVATIGNRKVYVGTLEWITRHGINNN---------ILQEVECKNES------- 218
L EPG G +A I R V VG+LEW++ + N L + + N S
Sbjct: 610 LTEPGFGTLAEIDGRFVAVGSLEWVSDRFLKKNDSSDMVKLESLLDHKLSNTSSTSRYSK 669
Query: 219 -FVYVG-VNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGI 276
VYVG + + G I D +R+DA V L ++ I +LSGD+ A VA VGI
Sbjct: 670 TVVYVGREGEGIIGAIAISDCLRQDAEFTVARLQEKGIKTVLLSGDREGAVATVAKNVGI 729
Query: 277 PKDKVLSGVKPDQKKKFINELQKD-NIVAMVGDGINDAAALAASHIGIAL--GGGVGAAS 333
+ + P++K +FI+ LQ + VAMVGDGINDA +LA + +GIAL AAS
Sbjct: 730 KSESTNYSLSPEKKFEFISNLQSSGHRVAMVGDGINDAPSLAQADVGIALKIEAQENAAS 789
Query: 334 EVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLT 393
+S+IL+R+ LS ++DAL L++ TM+ V QNL WA YN++ IPIAAGVL P +T
Sbjct: 790 NAASVILVRNKLSHVVDALSLAQATMSKVYQNLAWAIAYNVISIPIAAGVLLPQYDFAMT 849
Query: 394 PSIAGALMGLSSIGVMTNSLLLRFKFSSKQKQIL 427
PS++G LM LSSI V++NSLLL+ S K L
Sbjct: 850 PSLSGGLMALSSIFVVSNSLLLQLHKSETSKNSL 883
>UniRef100_Q8H028 Hypothetical protein OSJNBa0050H14.2 [Oryza sativa]
Length = 910
Score = 320 bits (820), Expect = 5e-86
Identities = 193/447 (43%), Positives = 264/447 (58%), Gaps = 44/447 (9%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQ------GSAVSLALQFACSVLVVACP 54
+AG F Y VM +S TF+FW GTHI P G ++ L+L+ A VLVV+CP
Sbjct: 471 IAGPFVYTVMTLSAATFSFWYYIGTHIFPEVLLNDISGPDGDSLLLSLKLAVDVLVVSCP 530
Query: 55 CALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVA 114
CALGLATPTA+L+GTSLGAKRGLL+RGG++LE+ A ++A+V DKTGTLT GRPVVT + +
Sbjct: 531 CALGLATPTAILIGTSLGAKRGLLIRGGDVLERLAGIDAIVLDKTGTLTKGRPVVTSIAS 590
Query: 115 STCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTF 174
A + EILRLAAAVE ++HP+ AI++ A+ + LD G
Sbjct: 591 L--------------AYEEAEILRLAAAVEKTALHPIANAIMEEAELLK-LDIPATSGQ- 634
Query: 175 LEEPGSGAVATIGNRKVYVGTLEWI-------------TRHGINNNILQEVECK---NES 218
L EPG G +A + V VGTL+W+ T G + + E ++S
Sbjct: 635 LTEPGFGCLAEVDGCLVAVGTLDWVHNRFETKASSTELTDLGNHLEFVSSSEASSNHSKS 694
Query: 219 FVYVG-VNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIP 277
YVG + + G I D +R+DA+ VD L +++I ++LSGD++ A E + VGI
Sbjct: 695 IAYVGREGEGIIGAIAVSDVLRDDAKATVDRLQQEEILTFLLSGDRKEAVESIGRTVGIR 754
Query: 278 KDKVLSGVKPDQKKKFINELQ-KDNIVAMVGDGINDAAALAASHIGIAL--GGGVGAASE 334
+ + S + P +K I LQ + VAMVGDGINDA +LAA+ +G+A+ AAS+
Sbjct: 755 SENIKSSLTPHEKAGIITALQGEGRRVAMVGDGINDAPSLAAADVGVAMRTNSKESAASD 814
Query: 335 VSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTP 394
+S++L+ + LSQ++DAL LS+ TM V QNL WA YNIV IPIAAGVL P +TP
Sbjct: 815 AASVVLLGNRLSQVMDALSLSKATMAKVHQNLAWAVAYNIVAIPIAAGVLLPQFDFAMTP 874
Query: 395 SIA--GALMGLSSIGVMTNSLLLRFKF 419
S++ G G S + N L R KF
Sbjct: 875 SLSEYGKTTGRSELQAELNLLADRIKF 901
>UniRef100_Q8ZS77 Cation-transporting ATPase [Anabaena sp.]
Length = 753
Score = 298 bits (762), Expect = 3e-79
Identities = 173/427 (40%), Positives = 254/427 (58%), Gaps = 31/427 (7%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
V G+F V+A+++ TF W F + V+LAL VL++ACPCALGLA
Sbjct: 350 VTGWFVPAVIAIALLTFIIWFNFTGN-----------VTLALITTVGVLIIACPCALGLA 398
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TPT+V+VGT GA+ G+L++G LE + +V DKTGT+T G+P VT V N
Sbjct: 399 TPTSVMVGTGKGAENGILIKGAESLELAHKIQTIVLDKTGTITQGKPTVTDFVTVNGTAN 458
Query: 121 ANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGS 180
N ++ +++LAA+VE NS HP+ +A+V AQ+ A V + F GS
Sbjct: 459 GN----------EIRLVQLAASVERNSEHPLAEAVVRYAQSQEVTLADVKE--FEAVAGS 506
Query: 181 GAVATIGNRKVYVGTLEWITRHGINNNILQE----VECKNESFVYVGVNDTLAGLIYFED 236
G + + V +GT W++ GI+ LQ+ +E ++ +++ V+ + GL+ D
Sbjct: 507 GVQGIVSDSLVQIGTQRWMSELGIDTQALQQDKERLEYLGKTAIWIAVDRQIQGLMGISD 566
Query: 237 EVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFINE 296
++ + + L K + V ML+GD R AE +A VGI + VL+ V+PDQK + +
Sbjct: 567 AIKPTSIQAISALQKLGLEVVMLTGDNRRTAETIAREVGIKR--VLAEVRPDQKAATVQK 624
Query: 297 LQKDN-IVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALELS 355
LQ + IVAMVGDGINDA ALA + +G+A+G G A S I L+ L ++ A++LS
Sbjct: 625 LQSEGKIVAMVGDGINDAPALAQADVGMAIGTGTDVAIAASDITLISGDLRSIVTAIQLS 684
Query: 356 RLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLLL 415
R T+ ++QNL++AFIYN+ GIPIAAG+LFP+ G +L P IAGA M SS+ V+TN+L L
Sbjct: 685 RATIRNIRQNLFFAFIYNVAGIPIAAGILFPIFGWLLNPIIAGAAMAFSSVSVVTNALRL 744
Query: 416 RFKFSSK 422
R KF +K
Sbjct: 745 R-KFQAK 750
>UniRef100_Q8YWI6 Cation-transporting ATPase [Anabaena sp.]
Length = 753
Score = 297 bits (761), Expect = 4e-79
Identities = 174/427 (40%), Positives = 253/427 (58%), Gaps = 31/427 (7%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
V G+F V+A+++ TF W F + V+LAL VL++ACPCALGLA
Sbjct: 350 VTGWFVPAVIAIAILTFIIWYNFMGN-----------VTLALITTVGVLIIACPCALGLA 398
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TPT+V+VGT GA+ G+L++G LE + +V DKTGT+T G+P VT V N
Sbjct: 399 TPTSVMVGTGKGAENGILIKGAESLELAHQIQTIVLDKTGTITQGKPTVTDFVTVDGTAN 458
Query: 121 ANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGS 180
+N ++++++LAA++E NS HP+ +A+V AQ+ A V D F GS
Sbjct: 459 SN----------EIKLIQLAASLERNSEHPLAEAVVRYAQSQEVTLADVTD--FAAVVGS 506
Query: 181 GAVATIGNRKVYVGTLEWITRHGINNNILQE----VECKNESFVYVGVNDTLAGLIYFED 236
G + + V +GT W+ I+ LQ+ +E ++ V++ V+ +AGL+ D
Sbjct: 507 GVQGIVTHHLVQIGTQRWMEELSISTQALQQDKERLEYLGKTAVWLAVDGEIAGLMGIAD 566
Query: 237 EVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFINE 296
++ + + L K + V ML+GD R AE +A VGI + VL+ V+PDQK +
Sbjct: 567 AIKPTSTQAIRALQKLGLEVVMLTGDNRRTAESIAREVGIKR--VLAEVRPDQKAATVQA 624
Query: 297 LQKDN-IVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALELS 355
+Q + IVAMVGDGINDA ALA + +GIA+G G A S I L+ L ++ A++LS
Sbjct: 625 IQAEGKIVAMVGDGINDAPALAQADVGIAIGTGTDVAIAASDITLISGDLQAIVTAIQLS 684
Query: 356 RLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLLL 415
R T+ ++QNL++AFIYN+ GIPIAAG+LFP+ G +L P IAGA M SS+ V+TN+L L
Sbjct: 685 RATIHNIRQNLFFAFIYNVAGIPIAAGILFPIFGWLLNPIIAGAAMAFSSVSVVTNALRL 744
Query: 416 RFKFSSK 422
R KF K
Sbjct: 745 R-KFQPK 750
>UniRef100_P74512 Cation-transporting ATPase; E1-E2 ATPase [Synechocystis sp.]
Length = 780
Score = 290 bits (741), Expect = 8e-77
Identities = 167/430 (38%), Positives = 250/430 (57%), Gaps = 29/430 (6%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQG--SAVSLALQFACSVLVVACPCALG 58
VAG+F YGV+A+++ T FW++ G + P S + LAL+ + SVLVVACPCALG
Sbjct: 362 VAGWFAYGVLAIALVTLGFWAMVGQSLFPEMVADTGLSPLLLALKLSVSVLVVACPCALG 421
Query: 59 LATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCI 118
LATPTA+LVGTSLGA++G+L++GGNILE + FDKTGTLT G +T V I
Sbjct: 422 LATPTAILVGTSLGAEQGILIKGGNILEILQRTTVMAFDKTGTLTQGNLQLTDAVPVADI 481
Query: 119 ENANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEP 178
+ +E+L LAA+VE + HP+ + ++ +AQ + L + ++ E
Sbjct: 482 -------------TGIELLTLAASVEQGTRHPLAQGLISSAQGLELLPVENIE----TEA 524
Query: 179 GSGAVATIGNRKVYVGTLEWITRHGINNN-----ILQEVECKNESFVYVGVNDTLAGLIY 233
G G ++ VG +W+ G+ + ++ + ++ ++V N L G +
Sbjct: 525 GQGVQGWYQGDRLLVGNQQWLMEQGVMGEPQWQTAVDQLLDQGKTVIFVARNQQLQGFLA 584
Query: 234 FEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKF 293
D +R +A+ + L + I+ +L+GD A+ +A VGI ++ + + P K
Sbjct: 585 LRDTLRPEAKATIAQLKQWGIAPLLLTGDHPAIAQAIAMEVGI--EEFQAQMTPQAKVAK 642
Query: 294 INELQKDN---IVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLD 350
I +Q N ++AMVGDGINDA ALA + +GI+L G A E + ++LMR HLS +L
Sbjct: 643 IKAMQGFNPVSVIAMVGDGINDAPALAQADVGISLSGATAVAMETADVVLMRSHLSDVLK 702
Query: 351 ALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMT 410
AL LSR T+ +KQNL WA YN++ IP+AAG P +LTP+IA A+M SSI V+
Sbjct: 703 ALTLSRSTVAKIKQNLLWALGYNLLAIPLAAGAFLPSFAIVLTPAIAAAMMASSSIVVVL 762
Query: 411 NSLLLRFKFS 420
N+L LR++FS
Sbjct: 763 NALALRYQFS 772
>UniRef100_Q7V4G1 Putative P-type ATPase transporter for copper [Prochlorococcus
marinus]
Length = 774
Score = 287 bits (735), Expect = 4e-76
Identities = 177/443 (39%), Positives = 259/443 (57%), Gaps = 45/443 (10%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAY---QG----------------SAVSLA 41
VAG F YGV+++++ TF FW G + P + QG +++ LA
Sbjct: 348 VAGRFCYGVVSLALLTFLFWWQLGARLWPEVLHASGQGLVHGYGHHVPLGGVAETSLGLA 407
Query: 42 LQFACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGT 101
LQ A +VLVVACPCALGLATPT + V + A+RG L RGG+++E A + VVFDKTGT
Sbjct: 408 LQLAIAVLVVACPCALGLATPTVITVASGQAARRGWLFRGGDVIEMAASLRQVVFDKTGT 467
Query: 102 LTVGRPVVTKVVASTCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQA 161
LT+GRP+V VV + + ++L+LAA++E NS HP+ A++ AQ
Sbjct: 468 LTLGRPLVAGVVGTKKPD---------------QLLQLAASLEQNSRHPLAHAVLQEAQR 512
Query: 162 VNC-LDAKVVDGTFLEEPGSGAVATIGNRK--VYVGTLEWITRHGIN--NNILQEVECKN 216
L + + T+ PGSG + + V VGT EW+ G++ + +VE +
Sbjct: 513 HRLSLLSTLATRTY---PGSGLAGELEGVEGTVRVGTPEWLQAEGVHWTAELQADVELSS 569
Query: 217 ---ESFVYVGVNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASL 273
+S V V + + GL+ +D +R D +D L +Q +++ MLSGD+R A E +
Sbjct: 570 LQGQSVVAVALGEEPLGLVAIDDRLRPDVSVALDRLREQGMTLAMLSGDRRQAVERLGQQ 629
Query: 274 VGIPKDKVLSGVKPDQKKKFINELQKDNIVAMVGDGINDAAALAASHIGIALGGGVGAAS 333
+G ++ + PDQK + + L+K ++AMVGDGINDA ALAA+ +GIA+G G A
Sbjct: 630 LGFQPHQLGWQLLPDQKLERLQRLRKAGLLAMVGDGINDAPALAAADLGIAVGTGTQIAQ 689
Query: 334 EVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLT 393
+ + ++L+ D L L +AL L+R TM V+QNL WAF YN++ +PIAAG+L P G +L+
Sbjct: 690 DSADLVLLGDRLEGLPEALLLARRTMAKVRQNLTWAFGYNLIALPIAAGLLLPGFGLLLS 749
Query: 394 PSIAGALMGLSSIGVMTNSLLLR 416
P IA LM LSSI V+ N+L LR
Sbjct: 750 PPIAALLMALSSITVVVNALALR 772
>UniRef100_Q7VE60 Cation transport ATPase [Prochlorococcus marinus]
Length = 774
Score = 283 bits (725), Expect = 5e-75
Identities = 164/444 (36%), Positives = 258/444 (57%), Gaps = 45/444 (10%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPA---TAYQG------------------SAVS 39
VAG F YGV+A+S+ TF FW G+++ P ++ QG + +
Sbjct: 346 VAGKFCYGVVALSIFTFIFWWQIGSNLWPEVLNSSGQGLINSHEHMLHSSFGSEAQTPLG 405
Query: 40 LALQFACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKT 99
LA+Q + +VLV+ACPCALGLATPT + V + A++G L +GG++LEK A ++ ++FDKT
Sbjct: 406 LAIQLSIAVLVIACPCALGLATPTVISVASGKAAQKGWLFKGGDVLEKAASIDQIIFDKT 465
Query: 100 GTLTVGRPVVTKVVASTCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAA 159
GTLTVGRPVVT+ + +T + +++++AA++E+NS HP+ AI+ A
Sbjct: 466 GTLTVGRPVVTETLLTTDKD---------------KLIQIAASIENNSRHPIAYAILQKA 510
Query: 160 QAVNCLDAKVVDGTFLEEPGSGAVATIGNRK--VYVGTLEWITRHGINNN-----ILQEV 212
+ N K + + PG G + K V VGT+EW G++ N L+ +
Sbjct: 511 EEFNLPLLKAFNTKSI--PGKGIFGELEGIKGIVRVGTIEWARNEGVSWNKKTDLYLESI 568
Query: 213 ECKNESFVYVGVNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVAS 272
+S V + ++ L G++ +D++R DA+ ++ L KQ + ++SGD+R A + +
Sbjct: 569 TKIGKSVVSISIDKELTGILIIDDQIRNDAKFAINLLRKQGQILRIMSGDRREAVLRIGN 628
Query: 273 LVGIPKDKVLSGVKPDQKKKFINELQKDNIVAMVGDGINDAAALAASHIGIALGGGVGAA 332
+G + P+ K K++ L+ +AMVGDGINDA ALA+S +GIA+G G A
Sbjct: 629 ELGFESQLLNWQQLPEDKLKYLENLKNYGNIAMVGDGINDAPALASSDLGIAIGTGTEIA 688
Query: 333 SEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTML 392
+ + ++L+ D L + A L++ M +KQNL+ AF YNI+ +PIAAG+L P G +L
Sbjct: 689 QDSADLVLLGDSLEGVPQAFSLAKRAMNKIKQNLFLAFGYNIIALPIAAGILLPRFGILL 748
Query: 393 TPSIAGALMGLSSIGVMTNSLLLR 416
+P IA LM LSSI V+ N+L L+
Sbjct: 749 SPPIAAFLMALSSITVVINALYLK 772
>UniRef100_Q8XMY3 Probable copper-transporting ATPase [Clostridium perfringens]
Length = 889
Score = 283 bits (724), Expect = 7e-75
Identities = 169/429 (39%), Positives = 251/429 (58%), Gaps = 35/429 (8%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
++GYF V++++V W + G + + AL SVLV+ACPCALGLA
Sbjct: 491 ISGYFVPIVISLAVIASLAWY-----------FSGESKTFALTIFISVLVIACPCALGLA 539
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TPTA++VGT GA+ G+L++ G LE +N VVFDKTGT+T G+P VT ++
Sbjct: 540 TPTAIMVGTGKGAENGILIKSGEALESTQNLNTVVFDKTGTITEGKPKVTDIIC------ 593
Query: 121 ANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGS 180
EN D E+L LAA+ E S HP+G+AIV A+ N V+D F PG
Sbjct: 594 -------ENISKD-ELLLLAASAEKGSEHPLGEAIVRDAEEKNLELKNVLD--FEAIPGK 643
Query: 181 GAVATIGNRKVYVGTLEWITRHGIN-NNIL---QEVECKNESFVYVGVNDTLAGLIYFED 236
G +I N+ + +G + + IN N+L +E+ K ++ +++ +N+ +AG+I D
Sbjct: 644 GIKCSIENKSILLGNYKLMKDKNINLKNLLATSEELASKGKTPMFIAINEKIAGIIAVAD 703
Query: 237 EVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFINE 296
V+E ++ ++TL K + V ML+GD A+ +A VG+ D+V++ V P +K + I
Sbjct: 704 TVKETSKKAIETLQKMGLEVVMLTGDNLKTAKAIAKEVGV--DRVIAEVLPQEKAEKIKS 761
Query: 297 LQKDNI-VAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALELS 355
LQ + VAMVGDGINDA ALA + IG+A+G G A E + I+LM+ + ++ A++LS
Sbjct: 762 LQDEGKKVAMVGDGINDAPALAIADIGMAIGSGTDIAMESADIVLMKGDILHVVGAIQLS 821
Query: 356 RLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLLL 415
R TM +K+NL+WAF YN +GIP+A GVL G +L P I M SS+ V+ N+L L
Sbjct: 822 RQTMKNIKENLFWAFGYNTLGIPVAMGVLHIFGGPLLNPMIGAFAMSFSSVSVLLNALRL 881
Query: 416 RFKFSSKQK 424
+ KF K
Sbjct: 882 K-KFKPNYK 889
>UniRef100_Q9SH30 Potential copper-transporting ATPase 3 [Arabidopsis thaliana]
Length = 995
Score = 281 bits (718), Expect = 4e-74
Identities = 178/452 (39%), Positives = 261/452 (57%), Gaps = 34/452 (7%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGT-HILPATAYQGSAVS--LALQFACSVLVVACPCAL 57
++ +F V+ +S +T+ W L G H P + S S LALQF SV+V+ACPCAL
Sbjct: 562 ISKFFVPLVIFLSFSTWLAWFLAGKLHWYPESWIPSSMDSFELALQFGISVMVIACPCAL 621
Query: 58 GLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTC 117
GLATPTAV+VGT +GA +G+L++GG LE+ VN +VFDKTGTLT+G+PVV K
Sbjct: 622 GLATPTAVMVGTGVGASQGVLIKGGQALERAHKVNCIVFDKTGTLTMGKPVVVK------ 675
Query: 118 IENANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAV-----NCLDAKVVDG 172
++ ++N + E L AA E NS HP+ KAIV+ A+ N + D
Sbjct: 676 ------TKLLKNMVLR-EFYELVAATEVNSEHPLAKAIVEYAKKFRDDEENPAWPEACD- 727
Query: 173 TFLEEPGSGAVATIGNRKVYVGTLEWITRHGI-----NNNILQEVECKNESFVYVGVNDT 227
F+ G G AT+ R++ VG + H + +L + E ++ + V +N
Sbjct: 728 -FVSITGKGVKATVKGREIMVGNKNLMNDHKVIIPDDAEELLADSEDMAQTGILVSINSE 786
Query: 228 LAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKP 287
L G++ D ++ AR + L +I M++GD A +A VGI D V++ KP
Sbjct: 787 LIGVLSVSDPLKPSAREAISILKSMNIKSIMVTGDNWGTANSIAREVGI--DSVIAEAKP 844
Query: 288 DQKKKFINELQK-DNIVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLS 346
+QK + + ELQ ++VAMVGDGIND+ AL A+ +G+A+G G A E + I+LM+ +L
Sbjct: 845 EQKAEKVKELQAAGHVVAMVGDGINDSPALVAADVGMAIGAGTDIAIEAADIVLMKSNLE 904
Query: 347 QLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSI 406
++ A++LSR T + ++ N WA YN++GIPIAAGVLFP L P IAGA M SS+
Sbjct: 905 DVITAIDLSRKTFSRIRLNYVWALGYNLMGIPIAAGVLFPGTRFRLPPWIAGAAMAASSV 964
Query: 407 GVMTNSLLLRFKFSSKQKQILDMLPKTKIHVD 438
V+ SLLL+ + K+ + LD L +I V+
Sbjct: 965 SVVCCSLLLK---NYKRPKKLDHLEIREIQVE 993
>UniRef100_Q8KPV5 PacS [Synechococcus sp.]
Length = 747
Score = 278 bits (712), Expect = 2e-73
Identities = 169/422 (40%), Positives = 249/422 (58%), Gaps = 34/422 (8%)
Query: 1 VAGYFTYGVMAVSVTTFTFW-SLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGL 59
V G+F V+A+++ TF W + G V+LAL A VL++ACPCALGL
Sbjct: 349 VTGWFVPAVIAIAILTFVLWFNWIGN------------VTLALITAVGVLIIACPCALGL 396
Query: 60 ATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIE 119
ATPT+++VGT GA+ G+L++ LE + V+ DKTGTLT G+P VT +A I
Sbjct: 397 ATPTSIMVGTGKGAEYGILIKSAESLELAQTIQTVILDKTGTLTQGQPSVTDFLA---IG 453
Query: 120 NANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPG 179
+ + QT+ L AA++E+ S HP+ +AIV +A + V D F PG
Sbjct: 454 DRDQQQTL---------LGWAASLENYSEHPLAEAIVRYGEAQGITLSTVTD--FEAIPG 502
Query: 180 SGAVATIGNRKVYVGTLEWITRHGINNNILQ----EVECKNESFVYVGVNDTLAGLIYFE 235
SG + + +GT W+ GI + LQ + E ++ V V + L ++
Sbjct: 503 SGVQGQVEGIWLQIGTQRWLGELGIETSALQNQWEDWEAAGKTVVGVAADGHLQAILSIA 562
Query: 236 DEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFIN 295
D+++ + VV +L + + V ML+GD R A+ +A VGI + VL+ V+PDQK +
Sbjct: 563 DQLKPSSVAVVRSLQRLGLQVVMLTGDNRRTADAIAQAVGITQ--VLAEVRPDQKAAQVA 620
Query: 296 ELQ-KDNIVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALEL 354
+LQ + +VAMVGDGINDA ALA + +GIA+G G A S I L+ L ++ A++L
Sbjct: 621 QLQSRGQVVAMVGDGINDAPALAQADVGIAIGTGTDVAIAASDITLISGDLQGIVTAIQL 680
Query: 355 SRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLL 414
SR TMT ++QNL++AFIYN+ GIPIAAG+L+P+ G +L+P +AGA M SS+ V+TN+L
Sbjct: 681 SRATMTNIRQNLFFAFIYNVAGIPIAAGILYPLLGWLLSPMLAGAAMAFSSVSVVTNALR 740
Query: 415 LR 416
LR
Sbjct: 741 LR 742
>UniRef100_UPI00002F1620 UPI00002F1620 UniRef100 entry
Length = 584
Score = 278 bits (710), Expect = 3e-73
Identities = 164/423 (38%), Positives = 246/423 (57%), Gaps = 31/423 (7%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
V+G+F V+AV+V F W ++G P AY L A +VL++ACPCALGLA
Sbjct: 186 VSGWFVPVVIAVAVVAFIAWGIWGPE--PRFAY-------GLVAAVAVLIIACPCALGLA 236
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TP +++VG GA G+L++ LE V+ +V DKTGTLT GRP VT+VV + +
Sbjct: 237 TPMSIMVGVGRGAGLGVLIKNAEALEHMEKVDTLVVDKTGTLTEGRPAVTQVVPAPGFDE 296
Query: 121 ANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGS 180
A E+LRLAA+VE S HP+ AIV+AA+ + V D F G
Sbjct: 297 A-------------ELLRLAASVERASEHPLALAIVEAAKDRAIATSDVTD--FDSPTGR 341
Query: 181 GAVATIGNRKVYVGTLEWITRHGINNNIL----QEVECKNESFVYVGVNDTLAGLIYFED 236
GA+ T+ R++ +G ++ G+ L + + +++G++ T+ G D
Sbjct: 342 GALGTVEGRRIVLGNARFLADEGVATEALAGQADALRRDGATAIFIGIDGTVGGAFAIAD 401
Query: 237 EVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFINE 296
V+ + L + I V ML+GD R AE VA +GI D+V + V PDQK + +
Sbjct: 402 PVKATTPEALAALKAEGIRVVMLTGDNRTTAEAVARRLGI--DEVEAEVLPDQKSAVVAK 459
Query: 297 LQKDN-IVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALELS 355
+++ +VAM GDG+NDA ALAA+ +GIA+G G A E + + L++ L+ ++ A +LS
Sbjct: 460 FKREGRVVAMAGDGVNDAPALAAADVGIAMGSGTDVAIESAGVTLLKGDLTGIVRARKLS 519
Query: 356 RLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLLL 415
+ TM+ ++QNL +AFIYN+ G+P+AAG L+P+ G +L+P IA A M LSS+ V+TN+L L
Sbjct: 520 QATMSNIRQNLVFAFIYNVAGVPVAAGALYPLFGILLSPIIAAAAMALSSVSVVTNALRL 579
Query: 416 RFK 418
K
Sbjct: 580 NRK 582
>UniRef100_P37279 Cation-transporting ATPase pacS [Synechococcus sp.]
Length = 747
Score = 277 bits (709), Expect = 4e-73
Identities = 168/422 (39%), Positives = 249/422 (58%), Gaps = 34/422 (8%)
Query: 1 VAGYFTYGVMAVSVTTFTFW-SLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGL 59
V G+F V+A+++ TF W + G V+LAL A V+++ACPCALGL
Sbjct: 349 VTGWFVPAVIAIAILTFLLWFNWIGN------------VTLALITAVGVMIIACPCALGL 396
Query: 60 ATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIE 119
ATPT+++VGT GA+ G+L++ LE + V+ DKTGTLT G+P VT +A I
Sbjct: 397 ATPTSIMVGTGKGAEYGILIKSAESLELAQTIQTVILDKTGTLTQGQPSVTDFLA---IG 453
Query: 120 NANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPG 179
+ + QT+ L AA++E+ S HP+ +AIV +A + V D F PG
Sbjct: 454 DRDQQQTL---------LGWAASLENYSEHPLAEAIVRYGEAQGITLSTVTD--FEAIPG 502
Query: 180 SGAVATIGNRKVYVGTLEWITRHGINNNILQ----EVECKNESFVYVGVNDTLAGLIYFE 235
SG + + +GT W+ GI + LQ + E ++ V V + L ++
Sbjct: 503 SGVQGQVEGIWLQIGTQRWLGELGIETSALQNQWEDWEAAGKTVVGVAADGHLQAILSIA 562
Query: 236 DEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFIN 295
D+++ + VV +L + + V ML+GD R A+ +A VGI + VL+ V+PDQK +
Sbjct: 563 DQLKPSSVAVVRSLQRLGLQVVMLTGDNRRTADAIAQAVGITQ--VLAEVRPDQKAAQVA 620
Query: 296 ELQ-KDNIVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALEL 354
+LQ + +VAMVGDGINDA ALA + +GIA+G G A S I L+ L ++ A++L
Sbjct: 621 QLQSRGQVVAMVGDGINDAPALAQADVGIAIGTGTDVAIAASDITLISGDLQGIVTAIQL 680
Query: 355 SRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLL 414
SR TMT ++QNL++AFIYN+ GIPIAAG+L+P+ G +L+P +AGA M SS+ V+TN+L
Sbjct: 681 SRATMTNIRQNLFFAFIYNVAGIPIAAGILYPLLGWLLSPMLAGAAMAFSSVSVVTNALR 740
Query: 415 LR 416
LR
Sbjct: 741 LR 742
>UniRef100_Q7U436 Putative P-type ATPase transporter for copper [Synechococcus sp.]
Length = 771
Score = 277 bits (708), Expect = 5e-73
Identities = 175/446 (39%), Positives = 257/446 (57%), Gaps = 48/446 (10%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGT----HILPATAY---QG------------------ 35
VAG F YGV+A+++ TF FW LFG ++ A+A QG
Sbjct: 342 VAGRFCYGVIALALATFLFWWLFGASQWPQVMQASAPGMPQGHLMTSGHAMHHGGLGSGA 401
Query: 36 -SAVSLALQFACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAV 94
S V LALQ + +VLVVACPCALGLATPT + V T L A+RG L RGG+++E A ++ V
Sbjct: 402 TSPVGLALQLSIAVLVVACPCALGLATPTVITVATGLAAQRGWLFRGGDVIETAAGLDHV 461
Query: 95 VFDKTGTLTVGRPVVTKVVASTCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGKA 154
VFDKTGTLT+GRP+VT V A + AL +L+LAA++E +S HP+ A
Sbjct: 462 VFDKTGTLTLGRPLVTSVWAK------------DAAL----LLQLAASLEQSSRHPLAHA 505
Query: 155 IVDAAQAVNCLDAKVVDGTFLEEPGSGAVATIGN--RKVYVGTLEWITRHGI--NNNILQ 210
++ AQ + + V T + G G V + + + VG +W+ G+ ++
Sbjct: 506 LLQEAQRRDLTLLEPVQVTTVS--GQGLVGEVEGWPQPIRVGRPDWLGSFGVALSDEART 563
Query: 211 EVECKNESFVYVGVNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHV 270
+ + S V V L GL+ ED++R D ++ L +Q +++ + SGD+ A +
Sbjct: 564 WLAQADGSVVAVAHGSALLGLVQIEDQLRADVVPALERLRQQGLALAIFSGDREPAVRAL 623
Query: 271 ASLVGIPKDKVLSGVKPDQKKKFINELQKDNIVAMVGDGINDAAALAASHIGIALGGGVG 330
+G + + P+QK + + EL++ VAMVGDGINDA ALAA+ +GIA+G G
Sbjct: 624 GQQLGFEAADLGWQMLPEQKLQRLEELRQRGRVAMVGDGINDAPALAAADLGIAIGTGTQ 683
Query: 331 AASEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGT 390
A + + ++LM D L L +AL L+R T+ V+QNL+WAF YN++ +P+AAG L P G
Sbjct: 684 IAQDTAGMVLMGDRLDNLPEALTLARRTLAKVRQNLFWAFGYNLIALPLAAGALLPSQGV 743
Query: 391 MLTPSIAGALMGLSSIGVMTNSLLLR 416
+L+P +A LM +SSI V+ N+LLLR
Sbjct: 744 LLSPPLAALLMAISSITVVLNALLLR 769
>UniRef100_Q8ZSB9 Cation transporting ATPase [Anabaena sp.]
Length = 724
Score = 276 bits (706), Expect = 9e-73
Identities = 171/428 (39%), Positives = 241/428 (55%), Gaps = 43/428 (10%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
V GYF V+ ++V F W L G A+ +VL+++CPCALG+A
Sbjct: 332 VTGYFVPAVVGIAVLAFLGWLLVGNFPQALLAF------------IAVLIISCPCALGIA 379
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TP A++VG GA+ G+L+RGG ILE+ ++ V+FDKTGTLT G P VT +V E
Sbjct: 380 TPAALMVGVGKGAEGGILIRGGEILERAEKLSTVIFDKTGTLTRGEPSVTDIVPLA--ER 437
Query: 121 ANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGS 180
N EILRLAAAVE+ S HP+G+AIV AA+ KV F PG
Sbjct: 438 PND-----------EILRLAAAVEAGSEHPLGEAIVRAARHQRLDIPKV--SNFEAIPGH 484
Query: 181 GAVATIGNRKVYVGTLEWITRHGINNN-----ILQEVECKNESFVYVGVNDTLAGLIYFE 235
G I ++ +G G N IL +E ++ + VG N L G++
Sbjct: 485 GIRGAINRDRILLGNRRLFREQGYQINPATEEILTRLESDGKTAMLVGCNGLLMGIVAVA 544
Query: 236 DEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFIN 295
D ++ +A+ + L ++ + V ML+GD + AE +A +GI DKV++ V P K + +
Sbjct: 545 DTIKPEAKEAIAALRREKVKVVMLTGDNQRTAEAIARQLGI--DKVIAEVLPGDKAQVVK 602
Query: 296 ELQK-DNIVAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALEL 354
++Q+ +VAMVGDG+NDA ALA + IGIA+G G A E IIL+++ + ++ ++ L
Sbjct: 603 DIQRRGEVVAMVGDGVNDAPALATADIGIAIGSGADVAKETGGIILVKNDVRDVVTSIRL 662
Query: 355 SRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSLL 414
SR TM +KQNL+WAFIYN +GIPIAA L L P IA A M LSS+ V+ NS L
Sbjct: 663 SRATMLKIKQNLFWAFIYNSIGIPIAAFGL-------LNPMIAAAAMALSSLSVIVNSSL 715
Query: 415 LR-FKFSS 421
L+ FK S+
Sbjct: 716 LKGFKLST 723
>UniRef100_Q97D27 Heavy-metal transporting P-type ATPase [Clostridium acetobutylicum]
Length = 818
Score = 275 bits (702), Expect = 3e-72
Identities = 164/424 (38%), Positives = 251/424 (58%), Gaps = 34/424 (8%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQGSAVSLALQFACSVLVVACPCALGLA 60
V+GYF V+ +++ + W L G ++ + L SVLV+ACPCALGLA
Sbjct: 418 VSGYFVPVVITLAIISSLAWYLSGENL-----------TFTLTIFISVLVIACPCALGLA 466
Query: 61 TPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKTGTLTVGRPVVTKVVASTCIEN 120
TPTA++VGT GA+ G+L++ G LE + +VFDKTGT+T G+P VT + A
Sbjct: 467 TPTAIMVGTGKGAEYGVLIKNGTALENTHKIKTIVFDKTGTITEGKPKVTDIKAI----- 521
Query: 121 ANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAAQAVNCLDAKVVDGTFLEEPGS 180
+ +S+ E+L++AA+VE S HP+G+AIV A+ KV D F G
Sbjct: 522 --------DEVSEEELLKIAASVEKASEHPLGEAIVKEAEFKGMEFLKVSD--FKSVTGH 571
Query: 181 GAVATIGNRKVYVGTLEWITRHGIN-NNILQEVEC---KNESFVYVGVNDTLAGLIYFED 236
G A I +++V +G + + + I ++L V+ + ++ +Y+ ++ + G+I D
Sbjct: 572 GIEALIDSKRVLLGNKKLMDNNNIEVKSVLDYVDDLAKQGKTPMYIAIDKQVKGIIAVAD 631
Query: 237 EVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVASLVGIPKDKVLSGVKPDQKKKFINE 296
V+E + + L I V M++GD + +A+ +A VGI D+VL+ V P+ K + +
Sbjct: 632 SVKESSAKAIKKLHDMGIEVAMITGDNKRSADAIAKKVGI--DRVLAEVLPEDKASEVKK 689
Query: 297 LQKDNI-VAMVGDGINDAAALAASHIGIALGGGVGAASEVSSIILMRDHLSQLLDALELS 355
LQ VAMVGDGINDA ALA + IG+A+G G A E + I+LM+ L ++ A+ELS
Sbjct: 690 LQAGGKKVAMVGDGINDAPALAQADIGMAIGKGTDIAMESADIVLMKSDLMDVITAIELS 749
Query: 356 RLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTMLTPSIAGALMGLSSIGVMTNSL-L 414
+ T+ +K+NL+WAF YN++GIP+A G+L+ G +L P IA A M LSS+ V+TN+L L
Sbjct: 750 KKTIKNIKENLFWAFGYNVIGIPVAMGILYIFGGPLLNPMIAAAAMSLSSVSVLTNALRL 809
Query: 415 LRFK 418
RFK
Sbjct: 810 KRFK 813
>UniRef100_UPI00002E4675 UPI00002E4675 UniRef100 entry
Length = 439
Score = 274 bits (701), Expect = 3e-72
Identities = 160/443 (36%), Positives = 250/443 (56%), Gaps = 45/443 (10%)
Query: 1 VAGYFTYGVMAVSVTTFTFWSLFGTHILPATAYQG---------------------SAVS 39
+AG FTY V+ ++ ++F FW I P + ++
Sbjct: 13 IAGKFTYFVLIIATSSFFFWWKGAKQIWPDLLINNHHDLITTSNHTLHNSLGSNAENFLT 72
Query: 40 LALQFACSVLVVACPCALGLATPTAVLVGTSLGAKRGLLLRGGNILEKFAMVNAVVFDKT 99
LA+Q + +VLV+ACPCALGLATPT + V + AK+G+L +GG+ +E + +N ++FDKT
Sbjct: 73 LAIQLSIAVLVIACPCALGLATPTVITVASGKAAKKGVLFKGGDKIEMASKINHIIFDKT 132
Query: 100 GTLTVGRPVVTKVVASTCIENANSSQTIENALSDVEILRLAAAVESNSVHPVGKAIVDAA 159
GTLT G+P + + N + +L+++A++ES S HP+ A+V+ A
Sbjct: 133 GTLTRGKPFIIDYL---------------NTADKLFLLKISASLESQSRHPIASALVNEA 177
Query: 160 QAVNCLDAKVVDGTFLEEPGSGAVATIGN--RKVYVGTLEWITRHGI-----NNNILQEV 212
+ N K+ + E G G + + ++ +G++EW+ G+ + IL+
Sbjct: 178 KKQNLGLLKIKN--IHTESGRGISGELDSIDGEINIGSVEWLNSKGVIIDRKSKEILETE 235
Query: 213 ECKNESFVYVGVNDTLAGLIYFEDEVREDARHVVDTLSKQDISVYMLSGDKRNAAEHVAS 272
E K+ S + V +N L G I D +RED+ + V L + + + +LSGD++ +A
Sbjct: 236 ENKSHSVIGVCINKKLLGFILLGDLLREDSINSVQKLRENNYQIKILSGDRKETVVELAK 295
Query: 273 LVGIPKDKVLSGVKPDQKKKFINELQKDNIVAMVGDGINDAAALAASHIGIALGGGVGAA 332
+ IP+ ++ + P+ K K I L+K N V M+GDGINDA ALAAS++GIA+G G A
Sbjct: 296 KLDIPEAEIKWDLLPEMKLKIIENLKKSNKVVMIGDGINDAPALAASNLGIAVGSGTQIA 355
Query: 333 SEVSSIILMRDHLSQLLDALELSRLTMTTVKQNLWWAFIYNIVGIPIAAGVLFPVNGTML 392
+ ++LM D LS L +L L++ T+ +KQNL+WAF YN++ IPIA G+LFP G +L
Sbjct: 356 KANADVVLMGDQLSGLPYSLSLAKRTIGKIKQNLFWAFGYNLIAIPIAGGILFPKYGILL 415
Query: 393 TPSIAGALMGLSSIGVMTNSLLL 415
TPSIA LM SSI V+ N+L L
Sbjct: 416 TPSIAALLMATSSITVVINALSL 438
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 690,651,345
Number of Sequences: 2790947
Number of extensions: 27772309
Number of successful extensions: 99571
Number of sequences better than 10.0: 2765
Number of HSP's better than 10.0 without gapping: 2325
Number of HSP's successfully gapped in prelim test: 440
Number of HSP's that attempted gapping in prelim test: 90750
Number of HSP's gapped (non-prelim): 5614
length of query: 451
length of database: 848,049,833
effective HSP length: 131
effective length of query: 320
effective length of database: 482,435,776
effective search space: 154379448320
effective search space used: 154379448320
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0182.6


