

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0180a.7
(199 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
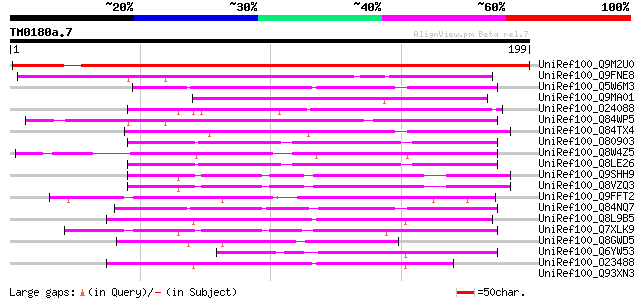
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9M2U0 Hypothetical protein T22E16.50 [Arabidopsis tha... 256 3e-67
UniRef100_Q9FNE8 Arabidopsis thaliana genomic DNA, chromosome 5,... 73 4e-12
UniRef100_Q5W6M3 Hypothetical protein P0015F11.14 [Oryza sativa] 68 1e-10
UniRef100_Q9MA01 F20B17.20 [Arabidopsis thaliana] 66 6e-10
UniRef100_O24088 MtN24 protein [Medicago truncatula] 62 1e-08
UniRef100_Q84WP5 Hypothetical protein At2g36330 [Arabidopsis tha... 61 1e-08
UniRef100_Q84TX4 Hypothetical protein OSJNBa0094J08.19 [Oryza sa... 59 6e-08
UniRef100_O80903 Expressed protein [Arabidopsis thaliana] 57 4e-07
UniRef100_Q8W4Z5 Hypothetical protein f16 [Gossypium hirsutum] 55 8e-07
UniRef100_Q8LE26 Hypothetical protein [Arabidopsis thaliana] 55 8e-07
UniRef100_Q9SHH9 F20D23.10 protein [Arabidopsis thaliana] 53 5e-06
UniRef100_Q8VZQ3 Hypothetical protein At1g17200 [Arabidopsis tha... 53 5e-06
UniRef100_Q9FFT2 Gb|AAD50013.1 [Arabidopsis thaliana] 52 7e-06
UniRef100_Q84NQ7 Hypothetical protein P0034A04.117 [Oryza sativa] 51 2e-05
UniRef100_Q8L9B5 Hypothetical protein At4g16442 [Arabidopsis tha... 51 2e-05
UniRef100_Q7XLK9 OSJNBa0094P09.18 protein [Oryza sativa] 50 3e-05
UniRef100_Q8GWD5 Hypothetical protein At2g39530 [Arabidopsis tha... 48 1e-04
UniRef100_Q6YW53 Integral membrane family protein-like [Oryza sa... 48 2e-04
UniRef100_O23488 Hypothetical protein [Arabidopsis thaliana] 47 4e-04
UniRef100_Q93XN3 Salicylic acid-induced 1 protein [Gossypium hir... 44 0.002
>UniRef100_Q9M2U0 Hypothetical protein T22E16.50 [Arabidopsis thaliana]
Length = 194
Score = 256 bits (653), Expect = 3e-67
Identities = 123/198 (62%), Positives = 149/198 (75%), Gaps = 6/198 (3%)
Query: 2 MRSPQPLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVF 61
MRSP RNG ++ H FHST+ KLRRFNSLIL+ RL SFSFSLAS+VF
Sbjct: 1 MRSPHAFRNGESPTLRDHTH------FHSTVTAQKLRRFNSLILLLRLASFSFSLASAVF 54
Query: 62 MLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFD 121
MLTN+ GS SPHWY +D FRFV ANAIVA+YS+FEM VWE SR T++PE QVWFD
Sbjct: 55 MLTNSRGSASPHWYDFDAFRFVFVANAIVALYSVFEMGTCVWEFSRETTLWPEAFQVWFD 114
Query: 122 FGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSL 181
FGHDQVF+YLLL+A +A A+ RT++ DTCTA+ AFC+QSD+A+ LG+A F+FL F+S
Sbjct: 115 FGHDQVFSYLLLSAGSAAAALARTMRGGDTCTANKAFCLQSDVAIGLGFAAFLFLAFSSC 174
Query: 182 LTGFRVVCFIINGSRFHL 199
+GFRV CF+I GSRFHL
Sbjct: 175 FSGFRVACFLITGSRFHL 192
>UniRef100_Q9FNE8 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MPO12
[Arabidopsis thaliana]
Length = 270
Score = 73.2 bits (178), Expect = 4e-12
Identities = 61/187 (32%), Positives = 95/187 (50%), Gaps = 8/187 (4%)
Query: 4 SPQPLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLI----LVFRLISFSFSLAS- 58
+P + NG + + + R P F S+ + + ++ SLI L FR+I+F L S
Sbjct: 79 TPFRVTNGEEEKKVSESRRQLRPSFSSSSSTPRESKWASLIRKALLGFRVIAFVSCLVSF 138
Query: 59 SVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQV 118
SV + G +Y+Y FRF LAAN I VYS F + V+ +S L+
Sbjct: 139 SVMVSDRDKGWAHDSFYNYKEFRFCLAANVIGFVYSGFMICDLVYLLSTSIRRSRHNLRH 198
Query: 119 WFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGF 178
+ +FG DQ+ AYLL ASA+ +A +R + + + ++ F + +VAL Y FV F
Sbjct: 199 FLEFGLDQMLAYLL--ASASTSASIR-VDDWQSNWGADKFPDLARASVALSYVSFVAFAF 255
Query: 179 TSLLTGF 185
SL +G+
Sbjct: 256 CSLASGY 262
>UniRef100_Q5W6M3 Hypothetical protein P0015F11.14 [Oryza sativa]
Length = 204
Score = 68.2 bits (165), Expect = 1e-10
Identities = 41/140 (29%), Positives = 78/140 (55%), Gaps = 6/140 (4%)
Query: 48 RLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISR 107
R+ +++FSL + V M N HG D + HY+ +R+V+A + +Y+ ++V ++
Sbjct: 65 RVAAWAFSLLAFVVMGANDHG-DWRQFEHYEEYRYVVAIGVLAFIYTTLQLVRHGVRLTG 123
Query: 108 GATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVA 167
G + +V V DF DQV AYLL++A +A + ++E A N F S +++
Sbjct: 124 GQDLQGKVA-VLVDFAGDQVTAYLLMSAVSAAIPITNRMRE----GADNVFTDSSAASIS 178
Query: 168 LGYAGFVFLGFTSLLTGFRV 187
+ + F+ L ++L++GF++
Sbjct: 179 MAFFAFLCLALSALVSGFKL 198
>UniRef100_Q9MA01 F20B17.20 [Arabidopsis thaliana]
Length = 393
Score = 65.9 bits (159), Expect = 6e-10
Identities = 33/120 (27%), Positives = 62/120 (51%), Gaps = 7/120 (5%)
Query: 71 SPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAY 130
S +W ++++V+ A A +YSL ++ V+ + G+ + P Q W F DQ+F Y
Sbjct: 267 SSNWSFSPSYQYVVGACAGTVLYSLLQLCLGVYRLVTGSPITPSRFQAWLCFTSDQLFCY 326
Query: 131 LLLAASAAGTAM-------VRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLT 183
L+++A +AG+ + +R D C ++FC +++ L + F+FL +S T
Sbjct: 327 LMMSAGSAGSGVTNLNKTGIRHTPLPDFCKTLSSFCNHVALSLLLVFLSFIFLASSSFFT 386
>UniRef100_O24088 MtN24 protein [Medicago truncatula]
Length = 234
Score = 61.6 bits (148), Expect = 1e-08
Identities = 49/157 (31%), Positives = 82/157 (52%), Gaps = 15/157 (9%)
Query: 46 VFRLISFSFSLASSVFML-----TNTHGS----DSP---HWYHYDTFRFVLAANAIVAVY 93
VF LI+FS AS +L TN + S +P HWY +D FR+ AAN I VY
Sbjct: 77 VFCLIAFSVLGASEQRVLVSENLTNWYSSGFTIQTPYEFHWYKWDEFRYSFAANVIGFVY 136
Query: 94 SLFEMVASV-WEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTC 152
S ++ V + I++ T+ P+ LQ +F+ DQ AY+L++AS++ LK+
Sbjct: 137 SGLQICHLVMYLITKKHTINPK-LQGYFNVAIDQTLAYILMSASSSAATAAHLLKDYWLE 195
Query: 153 TASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRVVC 189
++ F ++ +V++ + F SL++G ++C
Sbjct: 196 HGADTFIEMANASVSMSFLAFGAFALASLVSGI-ILC 231
>UniRef100_Q84WP5 Hypothetical protein At2g36330 [Arabidopsis thaliana]
Length = 283
Score = 61.2 bits (147), Expect = 1e-08
Identities = 50/186 (26%), Positives = 84/186 (44%), Gaps = 12/186 (6%)
Query: 7 PLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLI----LVFRLISFSFSLAS-SVF 61
P R A G RSS R + A + R ++ L FRL +L S S+
Sbjct: 99 PTRKSARVGSG----RSSGQRSGAVSAILRRSRREEVVKFSALGFRLSEVVLALISFSIM 154
Query: 62 MLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFD 121
T G + Y +RF L+ N + VYS F+ + + + + L+ F+
Sbjct: 155 AADKTKGWSGDSFDRYKEYRFCLSVNVVAFVYSSFQACDLAYHLVKEKHLISHHLRPLFE 214
Query: 122 FGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSL 181
F DQV AYLL++AS TA V + + + + F + ++A+ + F+ F+SL
Sbjct: 215 FIIDQVLAYLLMSAS---TAAVTRVDDWVSNWGKDEFTEMASASIAMSFLAFLAFAFSSL 271
Query: 182 LTGFRV 187
++G+ +
Sbjct: 272 ISGYNL 277
>UniRef100_Q84TX4 Hypothetical protein OSJNBa0094J08.19 [Oryza sativa]
Length = 156
Score = 59.3 bits (142), Expect = 6e-08
Identities = 37/150 (24%), Positives = 74/150 (48%), Gaps = 6/150 (4%)
Query: 45 LVFRLISFSFSLASSVFMLTNTHGSDSPHWY-HYDTFRFVLAANAIVAVYSLFEMVASVW 103
L+ R +++ FSL + V M +N HG + +Y + + L + I +Y+ ++ V
Sbjct: 10 LLLRAVAWLFSLLALVVMASNKHGHGGAQDFDNYPEYTYCLGISIIAVLYTTAQVTRDVH 69
Query: 104 EISRGATVFP-EVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQS 162
+S G V DF DQV AYLL++A +A + +++ A N F +
Sbjct: 70 RLSWGRDVIAGRKAAAVVDFAGDQVVAYLLMSALSAAAPVTDYMRQ----AADNLFTDSA 125
Query: 163 DIAVALGYAGFVFLGFTSLLTGFRVVCFII 192
A+++ + F+ G ++L++G+ + ++
Sbjct: 126 AAAISMAFLAFLAAGLSALVSGYNLAMEVL 155
>UniRef100_O80903 Expressed protein [Arabidopsis thaliana]
Length = 188
Score = 56.6 bits (135), Expect = 4e-07
Identities = 35/142 (24%), Positives = 72/142 (50%), Gaps = 9/142 (6%)
Query: 46 VFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEI 105
+ R I FSL + + M++N HG ++ Y+ +R+VLA + I +Y+ ++ A +
Sbjct: 50 ITRGICLLFSLIAFLIMVSNKHGYGR-NFNDYEEYRYVLAISIISTLYTAWQTFAHFSK- 107
Query: 106 SRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIA 165
+F + DF DQ+ AYLL++A+++ + +E N F + A
Sbjct: 108 ---REIFDRRTSILVDFSGDQIVAYLLISAASSAIPLTNIFREGQ----DNIFTDSAASA 160
Query: 166 VALGYAGFVFLGFTSLLTGFRV 187
+++ F+ L ++L +G+++
Sbjct: 161 ISMAIFAFIALALSALFSGYKL 182
>UniRef100_Q8W4Z5 Hypothetical protein f16 [Gossypium hirsutum]
Length = 207
Score = 55.5 bits (132), Expect = 8e-07
Identities = 49/195 (25%), Positives = 83/195 (42%), Gaps = 34/195 (17%)
Query: 3 RSPQPLRNGADGGDNNNNHRSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVFM 62
RSP L + N N ++S R TM RL+ + +A+ V M
Sbjct: 15 RSPMALMGSSR---NENQEVNTSMRTAETM--------------LRLVPMALGVAALVVM 57
Query: 63 LTNTHGSD--SPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVL-QVW 119
L N+ +D S + FR+++ AN I A YSL + + A P + + W
Sbjct: 58 LKNSQSNDFGSVSYSDLGAFRYLVHANGICAGYSLLSAIIA-------AVPSPSTMPRAW 110
Query: 120 FDFGHDQVFAYLLLAASAAGTAMVRTLKETDT-------CTASNAFCVQSDIAVALGYAG 172
F DQ+ Y++L A+A T ++ + D+ C FC ++ IAV + +
Sbjct: 111 TFFLLDQILTYVILGAAAVSTEVLYLANKGDSAITWSAACGTFAGFCHKATIAVVITFVA 170
Query: 173 FVFLGFTSLLTGFRV 187
+ SL++ +R+
Sbjct: 171 VICYAVLSLVSSYRL 185
>UniRef100_Q8LE26 Hypothetical protein [Arabidopsis thaliana]
Length = 188
Score = 55.5 bits (132), Expect = 8e-07
Identities = 34/142 (23%), Positives = 72/142 (49%), Gaps = 9/142 (6%)
Query: 46 VFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVASVWEI 105
+ R I FSL + + M++N HG ++ Y+ +R+VL+ + I +Y+ ++ A +
Sbjct: 50 ITRGICLLFSLIAFLIMVSNKHGYGR-NFNDYEEYRYVLSISIISTLYTAWQTFAHFSK- 107
Query: 106 SRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQSDIA 165
+F + DF DQ+ AYLL++A+++ + +E N F + A
Sbjct: 108 ---REIFDRRTSILVDFSGDQIVAYLLISAASSAIPLTNIFREGQ----DNIFTDSAASA 160
Query: 166 VALGYAGFVFLGFTSLLTGFRV 187
+++ F+ L ++L +G+++
Sbjct: 161 ISMAIFAFIALALSALFSGYKL 182
>UniRef100_Q9SHH9 F20D23.10 protein [Arabidopsis thaliana]
Length = 204
Score = 52.8 bits (125), Expect = 5e-06
Identities = 46/151 (30%), Positives = 68/151 (44%), Gaps = 19/151 (12%)
Query: 46 VFRLISFSFSLASSVFML----TNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVAS 101
+ RL +A+ V ML TN GS S + + FR+++ AN I A YSL A+
Sbjct: 36 MLRLAPVGLCVAALVVMLKDSETNEFGSIS--YSNLTAFRYLVHANGICAGYSLLS--AA 91
Query: 102 VWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQ 161
+ + R ++ P +VW F DQ+ YL+LAA A ++ D+ + C
Sbjct: 92 IAAMPRSSSTMP---RVWTFFCLDQLLTYLVLAAGAVSAEVLYLAYNGDSAITWSDAC-- 146
Query: 162 SDIAVALGYAGFVFLGFTSLLTGFRVVCFII 192
Y GF S++ F VVCF I
Sbjct: 147 ------SSYGGFCHRATASVIITFFVVCFYI 171
>UniRef100_Q8VZQ3 Hypothetical protein At1g17200 [Arabidopsis thaliana]
Length = 204
Score = 52.8 bits (125), Expect = 5e-06
Identities = 46/151 (30%), Positives = 68/151 (44%), Gaps = 19/151 (12%)
Query: 46 VFRLISFSFSLASSVFML----TNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVAS 101
+ RL +A+ V ML TN GS S + + FR+++ AN I A YSL A+
Sbjct: 36 MLRLAPVGLCVAALVVMLKDSETNEFGSIS--YSNLTAFRYLVHANGICAGYSLLS--AA 91
Query: 102 VWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQ 161
+ + R ++ P +VW F DQ+ YL+LAA A ++ D+ + C
Sbjct: 92 IAAMPRSSSTMP---RVWTFFCLDQLLTYLVLAAGAVSAEVLYLAYNGDSAITWSDAC-- 146
Query: 162 SDIAVALGYAGFVFLGFTSLLTGFRVVCFII 192
Y GF S++ F VVCF I
Sbjct: 147 ------SSYGGFCHRATASVIITFFVVCFYI 171
>UniRef100_Q9FFT2 Gb|AAD50013.1 [Arabidopsis thaliana]
Length = 194
Score = 52.4 bits (124), Expect = 7e-06
Identities = 52/186 (27%), Positives = 84/186 (44%), Gaps = 26/186 (13%)
Query: 16 DNNNNH-----RSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVFMLTNTHGSD 70
DNNNN+ RSSS + A L+ +S + RL S+A+ +TN +
Sbjct: 3 DNNNNNTREEERSSSSKQQQPQAPMSLKIIDSCL---RLSVVPLSVATIWLTVTNHESNP 59
Query: 71 SPHWYHYDTF---RFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQV 127
Y++ ++++ +AI A+Y+L V+S W V V + W F DQV
Sbjct: 60 DYGNLEYNSIMGLKYMVGVSAISAIYALLSTVSS-W-------VTCLVSKAWLFFIPDQV 111
Query: 128 FAYLLLAASAAGTAMVRTLKETDTCTASNAFCVQ-----SDIAVALGYAGFV--FLGFTS 180
AY++ + A T +V L + D + C S + +ALG FV F F S
Sbjct: 112 LAYVMTTSVAGATEIVYLLNKGDKIVTWSEMCSSYPHYCSKLTIALGLHVFVLFFFLFLS 171
Query: 181 LLTGFR 186
+++ +R
Sbjct: 172 VISAYR 177
>UniRef100_Q84NQ7 Hypothetical protein P0034A04.117 [Oryza sativa]
Length = 199
Score = 51.2 bits (121), Expect = 2e-05
Identities = 34/147 (23%), Positives = 66/147 (44%), Gaps = 9/147 (6%)
Query: 41 NSLILVFRLISFSFSLASSVFMLTNTHGSDSPHWYHYDTFRFVLAANAIVAVYSLFEMVA 100
+ ++LV R + +L + + + HG D + Y +R++L + ++YS + A
Sbjct: 52 DGIVLVLRAAAALLALVAMALVASCRHG-DWMEFTRYQEYRYLLGVAVVASLYSALQ-AA 109
Query: 101 SVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETDTCTASNAFCV 160
+ R T + + DF DQ YLL+ AS+A + ++ N F
Sbjct: 110 RTFRRMRAGTAYAAT---FLDFAGDQAVGYLLITASSAALPITIRMRS----AVVNTFTD 162
Query: 161 QSDIAVALGYAGFVFLGFTSLLTGFRV 187
+++ + F L F++L+ GFR+
Sbjct: 163 VVAASISFAFLAFAALAFSALIAGFRL 189
>UniRef100_Q8L9B5 Hypothetical protein At4g16442 [Arabidopsis thaliana]
Length = 182
Score = 50.8 bits (120), Expect = 2e-05
Identities = 44/161 (27%), Positives = 67/161 (41%), Gaps = 14/161 (8%)
Query: 38 RRFNSLILVFRLISFSFSLASSVFMLTNTHGS------DSPHWYHYDTFRFVLAANAIVA 91
RR L+ R F+L + + ++T+T + + F++ AN I A
Sbjct: 6 RRMRLTELLLRCSISVFALLALILVVTDTEVKLIFTIKKTAKYTDMKAVVFLVVANGIAA 65
Query: 92 VYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETD- 150
VYSL + V V +G +F + L W F DQ AYL +AA AA +E +
Sbjct: 66 VYSLLQSVRCVVGTMKGKVLFSKPL-AWAFFSGDQAMAYLNVAAIAATAESGVIAREGEE 124
Query: 151 ------TCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGF 185
CT FC Q I V+ + + F S ++ F
Sbjct: 125 DLQWMRVCTMYGKFCNQMAIGVSSALLASIAMVFVSCISAF 165
>UniRef100_Q7XLK9 OSJNBa0094P09.18 protein [Oryza sativa]
Length = 206
Score = 50.1 bits (118), Expect = 3e-05
Identities = 48/177 (27%), Positives = 82/177 (46%), Gaps = 22/177 (12%)
Query: 22 RSSSPRFHSTMAEHKLRRFNSLILVFRLISFSFSLASSVFML----TNTHGSDSPHWYHY 77
+SS S+ A ++R +L+ R +A+ ML TN +G+ S +
Sbjct: 18 QSSGDLQASSAAAARVRPVETLL---RAAPLGLCVAAMAIMLRNSVTNEYGTVS--YSDL 72
Query: 78 DTFRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASA 137
F++++ AN + A YSL A + R AT+ + W F DQVF YL+LAA A
Sbjct: 73 GGFKYLVYANGLCAAYSLAS--AFYIAVPRPATLS----RSWVVFLLDQVFTYLILAAGA 126
Query: 138 AGTAMV-------RTLKETDTCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRV 187
A ++ + + ++ C FC Q+ +VA+ +A SL++ +R+
Sbjct: 127 ASAELLYLAYNGDKEVTWSEACGVFGGFCRQARTSVAITFASVACYILLSLISSYRL 183
>UniRef100_Q8GWD5 Hypothetical protein At2g39530 [Arabidopsis thaliana]
Length = 178
Score = 48.1 bits (113), Expect = 1e-04
Identities = 35/114 (30%), Positives = 62/114 (53%), Gaps = 9/114 (7%)
Query: 42 SLILVFRLISFSFSLASSVFMLTNTH----GSDSPHWYHYDTF--RFVLAANAIVAVYSL 95
+++L+ R+++ +F L + V + TNT S S D + R++L+A I VY++
Sbjct: 15 TVLLLLRVLTAAFLLITVVLISTNTVTLEISSTSIKLPFNDVYAYRYMLSAAVIGLVYAV 74
Query: 96 FEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKET 149
++ ++ + + G T L FDF D+V +YLL SAAG + + LK+T
Sbjct: 75 VQLFLTISQFATGKT---HPLTYQFDFYGDKVISYLLATGSAAGFGVSKDLKDT 125
>UniRef100_Q6YW53 Integral membrane family protein-like [Oryza sativa]
Length = 201
Score = 47.8 bits (112), Expect = 2e-04
Identities = 37/115 (32%), Positives = 55/115 (47%), Gaps = 15/115 (13%)
Query: 80 FRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAG 139
FR+++ NAI A YS+ ++ S R T F W F DQ AYLLL +++A
Sbjct: 68 FRYLVWINAITAAYSVASILLSS---CRFITRFD-----WLIFLLDQASAYLLLTSASAA 119
Query: 140 TAMVRTLKETD-------TCTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRV 187
+V +E D C+ FC + ++VAL A + SL++ FRV
Sbjct: 120 AEVVYLAREGDREVSWGEVCSYFGRFCGAATVSVALNAAALLCFMALSLISAFRV 174
>UniRef100_O23488 Hypothetical protein [Arabidopsis thaliana]
Length = 578
Score = 46.6 bits (109), Expect = 4e-04
Identities = 41/146 (28%), Positives = 61/146 (41%), Gaps = 14/146 (9%)
Query: 38 RRFNSLILVFRLISFSFSLASSVFMLTNTHGS------DSPHWYHYDTFRFVLAANAIVA 91
RR L+ R F+L + + ++T+T + + F++ AN I A
Sbjct: 129 RRMRLTELLLRCSISVFALLALILVVTDTEVKLIFTIKKTAKYTDMKAVVFLVVANGIAA 188
Query: 92 VYSLFEMVASVWEISRGATVFPEVLQVWFDFGHDQVFAYLLLAASAAGTAMVRTLKETD- 150
VYSL + V V +G +F + L W F DQ AYL +AA AA +E +
Sbjct: 189 VYSLLQSVRCVVGTMKGKVLFSKPL-AWAFFSGDQAMAYLNVAAIAATAESGVIAREGEE 247
Query: 151 ------TCTASNAFCVQSDIAVALGY 170
CT FC Q I V+ +
Sbjct: 248 DLQWMRVCTMYGKFCNQMAIGVSSAF 273
>UniRef100_Q93XN3 Salicylic acid-induced 1 protein [Gossypium hirsutum]
Length = 169
Score = 43.9 bits (102), Expect = 0.002
Identities = 30/116 (25%), Positives = 53/116 (44%), Gaps = 15/116 (12%)
Query: 80 FRFVLAANAIVAVYSLFEMVASVWEISRGATVFPEVL-QVWFDFGHDQVFAYLLLAASAA 138
FR++ AN I A YSL + + A P + + W F DQ+ Y++L A+A
Sbjct: 12 FRYLAHANGICAGYSLLSAIIA-------AVPSPSTMPRAWTFFLLDQILTYVILGAAAV 64
Query: 139 GTAMVRTLKETDT-------CTASNAFCVQSDIAVALGYAGFVFLGFTSLLTGFRV 187
T ++ + D+ C FC ++ IAV + + + SL++ +R+
Sbjct: 65 STEVLYLANKGDSAITWSAACGTFAGFCHKATIAVVITFVAVICYAVLSLVSSYRL 120
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.328 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 308,333,418
Number of Sequences: 2790947
Number of extensions: 10790316
Number of successful extensions: 38269
Number of sequences better than 10.0: 40
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 38220
Number of HSP's gapped (non-prelim): 49
length of query: 199
length of database: 848,049,833
effective HSP length: 121
effective length of query: 78
effective length of database: 510,345,246
effective search space: 39806929188
effective search space used: 39806929188
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 71 (32.0 bits)
Lotus: description of TM0180a.7


