

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0176a.1
(440 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
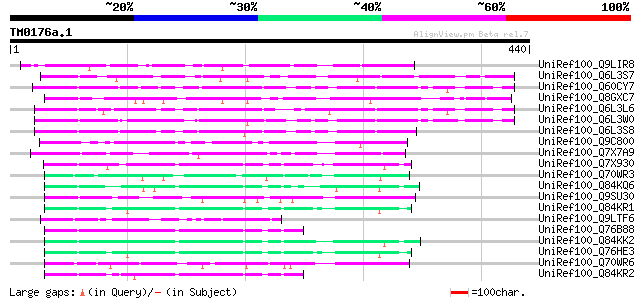
UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana] 104 4e-21
UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum] 98 5e-19
UniRef100_Q60CY7 Putative F-box protein [Solanum demissum] 95 3e-18
UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabid... 91 5e-17
UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum] 90 1e-16
UniRef100_Q6L3W0 Putative F-box protein [Solanum demissum] 88 6e-16
UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum] 82 2e-14
UniRef100_Q9C800 Hypothetical protein F10C21.17 [Arabidopsis tha... 79 3e-13
UniRef100_Q7X7A9 Hypothetical protein At1g11270 [Arabidopsis tha... 77 1e-12
UniRef100_Q7X930 S-locus F-Box protein 3 [Prunus avium] 76 2e-12
UniRef100_Q70WR3 S locus F-box (SLF)-S1E protein [Antirrhinum hi... 75 3e-12
UniRef100_Q84KQ6 F-box [Prunus mume] 74 1e-11
UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis tha... 73 2e-11
UniRef100_Q84KR1 F-box [Prunus mume] 70 2e-10
UniRef100_Q9LTF6 Arabidopsis thaliana genomic DNA, chromosome 5,... 69 2e-10
UniRef100_Q76B88 S haplotype-specific F-box protein 5 [Prunus av... 69 2e-10
UniRef100_Q84KK2 S haplotype-specific F-box protein d [Prunus du... 69 3e-10
UniRef100_Q76HE3 S-locus F-Box protein 7 [Prunus mume] 69 3e-10
UniRef100_Q70WR6 S locus F-box (SLF)-S4D protein [Antirrhinum hi... 68 5e-10
UniRef100_Q84KR2 F-box [Prunus mume] 68 5e-10
>UniRef100_Q9LIR8 Gb|AAF30317.1 [Arabidopsis thaliana]
Length = 364
Score = 104 bits (260), Expect = 4e-21
Identities = 97/341 (28%), Positives = 150/341 (43%), Gaps = 50/341 (14%)
Query: 10 TVLSMNTPHPDNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIK--- 66
T M +PH NLP +++ E+L LPVKSLTR KCVC SW SLI +K
Sbjct: 4 TTREMFSPH--NLPL-----EMMEEILLRLPVKSLTRFKCVCSSWRSLISETLFALKHAL 56
Query: 67 -LHLHRSALHGK-PHLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKK 124
L ++ K P+ ++ S + + C+ L +S V H+ L
Sbjct: 57 ILETSKATTSTKSPYGVI--TTSRYHLKSCCIHSLYNASTVYVSE------HDG--ELLG 106
Query: 125 GDRYHVVGCCNGLICLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFD--VKL 182
D Y VVG C+GL+C H S++ WNP +L ++L ++T + + V
Sbjct: 107 RDYYQVVGTCHGLVCFHVDYDKSLY------LWNPTIKL-QQRLSSSDLETSDDECVVTY 159
Query: 183 SFGYDNSTDTYKVVFFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVN 242
FGYD S D YKVV +++ V + T WR +FP D
Sbjct: 160 GFGYDESEDDYKVVAL----LQQRHQVKIETKIYSTRQKLWRSNTSFPSGVVVADK--SR 213
Query: 243 DGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVV 302
G++++GT+NW + S+ I+S D+ + + L P
Sbjct: 214 SGIYINGTLNWAATSSSSS------------WTIISYDMSRDEFKELPGPVCCGR-GCFT 260
Query: 303 PSVGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKV 343
++G L CL +G + +W MK+FG+ SW++LL +
Sbjct: 261 MTLGDLRGCLSMVCYCKGANADVWVMKEFGEVYSWSKLLSI 301
>UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum]
Length = 372
Score = 97.8 bits (242), Expect = 5e-19
Identities = 113/420 (26%), Positives = 176/420 (41%), Gaps = 75/420 (17%)
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQREN 86
L +++I E+L +P KSL + CV K+W LI S FIK HL A +E
Sbjct: 9 LPHEIIIEILLKVPPKSLLKFMCVSKTWLELI-SSAKFIKTHLELIAN--------DKEY 59
Query: 87 SHH---------DHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGL 137
SHH + + C+ + S S NP +VG NGL
Sbjct: 60 SHHRIIFQESACNFKVCCLPSMLNKERSTELFDIGSPMENPTIYT------WIVGSVNGL 113
Query: 138 ICLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKN---FDVKLSFGYDNSTDTYK 194
ICL+ +++ WNPA + S+KL K +N + +K FGYD + D YK
Sbjct: 114 ICLYSKIEETVL-------WNPAVK-KSKKLPTLGAKLRNGCSYYLKYGFGYDETRDDYK 165
Query: 195 VVFFEI---EKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVND-GVHLSGT 250
VV + + S+V++++L + WR I F ++ VN G ++G
Sbjct: 166 VVVIQCIYEDSGSCDSVVNIYSLKA----DSWRTINKF------QGNFLVNSPGKFVNGK 215
Query: 251 INWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPK--GLDEVPSVVPSVGVL 308
I W S D T I+SLDL ET+ L P G P + VG
Sbjct: 216 IYWALSADVD---------TFNMCNIISLDLADETWRRLELPDSYGKGSYPLALGVVGSH 266
Query: 309 MNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQLFPLCL 368
++ LC + +EGT+ +W K G SWT++ V D ++ +F+ P
Sbjct: 267 LSVLCL-NCIEGTNSDVWIRKDCGVEVSWTKIFTV---DHPKDLGEFIFFTSIFSVPCYQ 322
Query: 369 HDGDTLILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSSFYYVESLVSPLRV 428
+ D ++L ++ +T + +E ++ ++ YVESLV PL +
Sbjct: 323 SNKDEILLLL--------PPVILTYNGSTRQ---VEVVDRFEECAAAEIYVESLVDPLLI 371
>UniRef100_Q60CY7 Putative F-box protein [Solanum demissum]
Length = 383
Score = 95.1 bits (235), Expect = 3e-18
Identities = 118/415 (28%), Positives = 181/415 (43%), Gaps = 57/415 (13%)
Query: 20 DNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSAL-HGKP 78
D++ L ++LI E+L LP+KSL++ CV KSW L IS F+K H+ +A G
Sbjct: 4 DSIIPLLLPDELITEILLRLPIKSLSKFMCVSKSWLQL-ISSPAFVKKHIKLTANDKGYI 62
Query: 79 HLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLI 138
+ L N+++D + FC ++ L+E I +P R H+VG NGLI
Sbjct: 63 YHRLIFRNTNNDFK-FCPLPPLFTNQQLIEE--ILHIDSPIERTTLST--HIVGSVNGLI 117
Query: 139 CLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFF 198
C+ + +Q WNPA S++L + +K FGYD S D YKVVF
Sbjct: 118 CV------AHVRQREAYIWNPAI-TKSKELPKSTSNLCSDGIKCGFGYDESRDDYKVVFI 170
Query: 199 EIE-KIRRASLVSVFTLDGCNGWNGWRD-IQNFPLLPFSYDDWGVNDGVHLSGTINWMTS 256
+ + ++V++++L N W D +Q LL G ++G + W +S
Sbjct: 171 DYPIRHNHRTVVNIYSL-RTNSWTTLHDQLQGIFLLNL--------HGRFVNGKLYWTSS 221
Query: 257 RDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPK-GLDEVPSVVPSVGVLMNCLCFS 315
Y IT S DL T+ L P G D V VG ++ L ++
Sbjct: 222 --TCINNYKVCNIT-------SFDLADGTWGSLELPSCGKDNSYINVGVVGSDLS-LLYT 271
Query: 316 HDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQLFPLCLH--DGDT 373
L +W MK G SWT+L ++Y Q +H C F +H G+
Sbjct: 272 CQLGAATSDVWIMKHSGVNVSWTKLFTIKYP--QNIKIHR---CVAPAFTFSIHIRHGEI 326
Query: 374 LILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSSFYYVESLVSPLRV 428
L+L + ++YD + T + + I YVESLV+PL++
Sbjct: 327 LLLLD-------SAIMIYDGSTRQLKHTFHVNQCEEI-------YVESLVNPLKI 367
>UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabidopsis thaliana]
Length = 427
Score = 91.3 bits (225), Expect = 5e-17
Identities = 110/441 (24%), Positives = 181/441 (40%), Gaps = 104/441 (23%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
++I E+L LP KS+ R +CV K + +L SD F K+HL L+L+ E+
Sbjct: 39 EIITEILLRLPAKSIGRFRCVSKLFCTLS-SDPGFAKIHLD---------LILRNESVRS 88
Query: 90 DHREFCVTHLSGSSLS-------------------LVENPWI-----------SLAHNPR 119
HR+ V+ + SL L ++P I L + R
Sbjct: 89 LHRKLIVSSHNLYSLDFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGDHLYDDRR 148
Query: 120 YRLKKGDRYH------VVGCCNGLICLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCV 173
LK + + +VG NGL+C+ +G + +NP T F
Sbjct: 149 VMLKLNAKSYRRNWVEIVGSSNGLVCISP-------GEGAVFLYNPTTGDSKRLPENFRP 201
Query: 174 KTKNFD----VKLSFGYDNSTDTYKVVFFEI--EKIRRASLVSVFTLDGCNGWNGWRDIQ 227
K+ ++ FG+D TD YK+V E I AS+ S+ + WR I
Sbjct: 202 KSVEYERDNFQTYGFGFDGLTDDYKLVKLVATSEDILDASVYSLKA-------DSWRRIC 254
Query: 228 NFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYT 287
N L + ++D GVH +G I+W+ + + Q +++ D++TE +
Sbjct: 255 N---LNYEHNDGSYTSGVHFNGAIHWVFTESRH-----------NQRVVVAFDIQTEEFR 300
Query: 288 LLSPPKGLDEVPSVVPS--VGVLMNCLCFSHDLEGTHFVIWQMKKFGDAKSWTQL-LKVR 344
+ P ++ + VG L LC + H IW M ++G+AKSW+++ + +
Sbjct: 301 EMPVPDEAEDCSHRFSNFVVGSLNGRLCVVNSCYDVHDDIWVMSEYGEAKSWSRIRINLL 360
Query: 345 YQDLQREPLHSMFYCHYQLFPLCLHDGDTLILASYSDRYDGNEAILYDKRANTAERTALE 404
Y+ ++ PLC D +L DG + +LY+ N + +
Sbjct: 361 YRSMK---------------PLCSTKNDEEVLL----ELDG-DLVLYNFETNASSNLGI- 399
Query: 405 TESKYILGFSSFYYVESLVSP 425
K GF + YVESL+SP
Sbjct: 400 CGVKLSDGFEANTYVESLISP 420
>UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum]
Length = 384
Score = 89.7 bits (221), Expect = 1e-16
Identities = 115/416 (27%), Positives = 174/416 (41%), Gaps = 63/416 (15%)
Query: 22 LPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKP--- 78
+P L ++LI E+L LP+KSL++ CV KSW L IS F+K H+ +A +GK
Sbjct: 7 IPLLLLPDELITEILLRLPIKSLSKFMCVSKSWLQL-ISSPAFVKNHIKLTA-NGKGYIY 64
Query: 79 HLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLI 138
H L+ R +D +FC + L+E + ++ R L H+VG NGLI
Sbjct: 65 HRLIFRNT--NDDFKFCPLPSLFTKQQLIEELFDIVSPIERTTLST----HIVGSVNGLI 118
Query: 139 CLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFF 198
C + +Q WNP T S++L + +K FGYD S D YKVVF
Sbjct: 119 CA------AHVRQREAYIWNP-TITKSKELPKSRSNLCSDGIKCGFGYDESHDDYKVVFI 171
Query: 199 EI-EKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSR 257
S+V++++L N W D G+ L +N
Sbjct: 172 NYPSHHNHRSVVNIYSL-RTNSWTTLHD---------------QLQGIFL---LNLHCRF 212
Query: 258 DKSAPYYNPVTI--TVEQLGILSLDLRTETYTLLSPPK-GLDEVPSVVPSVGVLMNCLCF 314
K Y+ T + I S DL T+ L P G D V VG ++ L +
Sbjct: 213 VKEKLYWTSSTCINNYKVCNITSFDLADGTWESLELPSCGKDNSYINVGVVGSDLS-LLY 271
Query: 315 SHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQLFPLCLH--DGD 372
+ + +W MK G SWT+L ++Y Q +H C F +H G+
Sbjct: 272 TCQRGAANSDVWIMKHSGVNVSWTKLFTIKYP--QNIKIHR---CVVPAFTFSIHIRHGE 326
Query: 373 TLILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSSFYYVESLVSPLRV 428
L++ + ++YD + T + + I YVESLV+PL++
Sbjct: 327 ILLVLD-------SAIMIYDGSTRQLKHTFHVNQCEEI-------YVESLVNPLKI 368
>UniRef100_Q6L3W0 Putative F-box protein [Solanum demissum]
Length = 427
Score = 87.8 bits (216), Expect = 6e-16
Identities = 118/416 (28%), Positives = 172/416 (40%), Gaps = 61/416 (14%)
Query: 22 LPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALH-GKPHL 80
+P L ++LI E+L LPVKSL++ CV KSW LI S TF+K H+ +A G H
Sbjct: 48 IPLLLLPDELITEILLKLPVKSLSKFMCVSKSWLQLI-SSPTFVKNHIKLTADDKGYIHH 106
Query: 81 LLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICL 140
L N + + FC S L + L H + H+VG NGLIC+
Sbjct: 107 RLIFRNIDGNFK-FC----SLPPLFTKQQHTEELFHIDSPIERSTLSTHIVGSVNGLICV 161
Query: 141 -HGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFE 199
HG I WNP T S++L F + +K FGYD S D YKVVF
Sbjct: 162 VHGQKEAYI--------WNP-TITKSKELPKFTSNMCSSSIKYGFGYDESRDDYKVVFIH 212
Query: 200 I-----EKIRRASLVSVFTLDGCNGWNGWRD-IQNFPLLPFSYDDWGVNDGVHLSGTINW 253
++V +++L N W +RD +Q F + + G ++G + W
Sbjct: 213 YPYNHSSSSNMTTVVHIYSLRN-NSWTTFRDQLQCFLVNHY---------GRFVNGKLYW 262
Query: 254 MTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLS-PPKGLDEVPSVVPSVGVLMNCL 312
+S Y IT S DL T+ L P G D + VG ++ L
Sbjct: 263 TSS--TCINKYKVCNIT-------SFDLADGTWGSLDLPICGKDNFDINLGVVGSDLS-L 312
Query: 313 CFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVRYQDLQREPLHSMFYCHYQLFPLCLHDGD 372
++ +W MK SWT+L ++Y Q H C +F +H
Sbjct: 313 LYTCQRGAATSDVWIMKHSAVNVSWTKLFTIKYP--QNIKTHR---CFAPVFTFSIHFRH 367
Query: 373 TLILASYSDRYDGNEAILYDKRANTAERTALETESKYILGFSSFYYVESLVSPLRV 428
+ IL + ++YD + T+ + + I YVESLV+PL +
Sbjct: 368 SEILLLLR-----SAIMIYDGSTRQLKHTSDVMQCEEI-------YVESLVNPLTI 411
>UniRef100_Q6L3S8 Putative F-Box protein [Solanum demissum]
Length = 327
Score = 82.4 bits (202), Expect = 2e-14
Identities = 96/329 (29%), Positives = 143/329 (43%), Gaps = 37/329 (11%)
Query: 22 LPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSA-LHGKPHL 80
+P L ++LI E+L LP+KSL + CV KSW L IS F+K H+ +A G +
Sbjct: 7 IPPLLLPDELITEILLKLPIKSLLKFMCVSKSWLQL-ISSPAFVKNHIKLTADDKGYIYH 65
Query: 81 LLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICL 140
L N++ D + FC + L++ + + R L H+VG NGLIC
Sbjct: 66 RLIFRNTNDDFK-FCPLPPLFTQQQLIKELYHIDSPIERTTLST----HIVGSVNGLICA 120
Query: 141 HGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVF--F 198
+ +Q WNP T S++L + +K FGYD S D YKVVF +
Sbjct: 121 ------AHVRQREAYIWNP-TITKSKELPKSRSNLCSDGIKCGFGYDESRDDYKVVFIDY 173
Query: 199 EIEKIRRASLVSVFTLDGCNGWNGWRD-IQNFPLLPFSYDDWGVNDGVHLSGTINWMTSR 257
I + ++V++++L W D +Q F LL G ++G + W +S
Sbjct: 174 PIHRHNHRTVVNIYSL-RTKSWTTLHDQLQGFFLLNL--------HGRFVNGKLYWTSS- 223
Query: 258 DKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPK-GLDEVPSVVPSVGVLMNCLCFSH 316
Y IT S DL T+ L P G D V VG ++ L ++
Sbjct: 224 -SCINNYKVCNIT-------SFDLADGTWERLELPSCGKDNSYINVGVVGSDLS-LLYTC 274
Query: 317 DLEGTHFVIWQMKKFGDAKSWTQLLKVRY 345
+W MK G SWT+L ++Y
Sbjct: 275 QRGAATSDVWIMKHSGVNVSWTKLFTIKY 303
>UniRef100_Q9C800 Hypothetical protein F10C21.17 [Arabidopsis thaliana]
Length = 441
Score = 79.0 bits (193), Expect = 3e-13
Identities = 85/316 (26%), Positives = 136/316 (42%), Gaps = 51/316 (16%)
Query: 26 QLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRE 85
+L + L+ E+L LPVK L RLK + K W SLI SD HL L LL+++
Sbjct: 96 ELPDVLVEEILQRLPVKYLVRLKSISKGWKSLIESD------HLAEKHLR-----LLEKK 144
Query: 86 NSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSV 145
+ +E +T +S S+ + + D V G CNGL+C+ Y +
Sbjct: 145 ---YGLKEIKITVERSTSKSICIKFFSRRSGMNAINSDSDDLLRVPGSCNGLVCV--YEL 199
Query: 146 DSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKV-VFFEIEKIR 204
DS++ + NP T + + + + FG D T TYKV V + +++
Sbjct: 200 DSVY----IYLLNPMTGVTRT-----LTPPRGTKLSVGFGIDVVTGTYKVMVLYGFDRVG 250
Query: 205 RASLVSVFTLDGCNGWNGWRD-IQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPY 263
VF LD N WR + +P S + V ++G++ W+ + D S
Sbjct: 251 TV----VFDLD----TNKWRQRYKTAGPMPLSCIPTPERNPVFVNGSLFWLLASDFSE-- 300
Query: 264 YNPVTITVEQLGILSLDLRTETYTLLSPPKGLD--EVPSVVPSVGVLMNCLCFSHDLEGT 321
IL +DL TE + LS P +D +V S + L + LC S+ +G
Sbjct: 301 ------------ILVMDLHTEKFRTLSQPNDMDDVDVSSGYIYMWSLEDRLCVSNVRQGL 348
Query: 322 HFVIWQMKKFGDAKSW 337
H +W + + ++ W
Sbjct: 349 HSYVWVLVQDELSEKW 364
>UniRef100_Q7X7A9 Hypothetical protein At1g11270 [Arabidopsis thaliana]
Length = 312
Score = 76.6 bits (187), Expect = 1e-12
Identities = 88/324 (27%), Positives = 137/324 (42%), Gaps = 44/324 (13%)
Query: 18 HPDNLPRTQLHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGK 77
H ++ + L +D++ +L LPV+SL R KCV W S I S Q F + L R
Sbjct: 26 HCSSVVKLLLPHDVVGLILERLPVESLLRFKCVSNQWKSTIES-QCFQERQLIRRMESRG 84
Query: 78 PHLLLQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGL 137
P +L+ + D ++ + GSS+ +R G C GL
Sbjct: 85 PDVLV--VSFADDEDKYGRKAVFGSSIV------------STFRFPTLHTLICYGSCEGL 130
Query: 138 ICLH-GYSVDSIFQQGWLSFWN---PATRLMSEKLGCFCVKTKNFDV-KLSFGYDNSTDT 192
IC++ YS + + + W+ P + L F K +F KL+FG D T
Sbjct: 131 ICIYCVYSPNIVVNPA--TKWHRSCPLSNLQQFLDDKFEKKEYDFPTPKLAFGKDKLNGT 188
Query: 193 YKVVF-FEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTI 251
YK V+ + + R + + D N N WR + P P+ +D+ D V+ G++
Sbjct: 189 YKQVWLYNSSEFRLDDVTTCEVFDFSN--NAWRYVH--PASPYRINDY--QDPVYSDGSV 242
Query: 252 NWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNC 311
+W+T +S ILS L TET+ +L L E V S+ +L N
Sbjct: 243 HWLTEGKESK--------------ILSFHLHTETFQVLCEAPFLRERDPVGDSMCILDNR 288
Query: 312 LCFSHDLEGTHFVIWQMKKFGDAK 335
LC S ++ G +IW + G K
Sbjct: 289 LCVS-EINGPAQLIWSLDSSGGNK 311
>UniRef100_Q7X930 S-locus F-Box protein 3 [Prunus avium]
Length = 376
Score = 75.9 bits (185), Expect = 2e-12
Identities = 79/323 (24%), Positives = 132/323 (40%), Gaps = 44/323 (13%)
Query: 29 NDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSAL-HGKPHLL------ 81
N+++ ++L LP KSL R C CKSW+ L I +F++ HLHR+ H +LL
Sbjct: 8 NEILIDILVRLPAKSLIRFLCTCKSWSDL-IGSSSFVRTHLHRNVTKHAHVYLLCLHHPQ 66
Query: 82 LQRENSHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLH 141
+R+N + D + + SL N N + L+ + + + G NGL+C+
Sbjct: 67 FERQNDNDDPYD-----IEELQWSLFSNEKFEQFSNLSHPLENTEHFRIYGSSNGLVCM- 120
Query: 142 GYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIE 201
S + + + WNP+ R + K V L FG+ + YK V +
Sbjct: 121 --SDEILNFDSPIQIWNPSVRKFRTLPMSTNINMKFSHVSLQFGFHPGVNDYKAVRM-MH 177
Query: 202 KIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSA 261
+ A V V++L + W+ I+ P P+ W + G +G + +
Sbjct: 178 TNKGALAVEVYSLK----TDCWKMIEVIP--PWLKCTWKHHKGTFFNGVAYHIIEK---- 227
Query: 262 PYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCLCFSH---DL 318
P+ I+S D +E + P + + V + CL F D
Sbjct: 228 ---GPI------CSIMSFDSGSEKFEEFITPDAICSPRELCIDVYKELICLIFGFYGCDE 278
Query: 319 EGTHFV-IWQMKKFGDAKSWTQL 340
EG V +W +++ K W QL
Sbjct: 279 EGMDKVDLWVLQE----KRWKQL 297
>UniRef100_Q70WR3 S locus F-box (SLF)-S1E protein [Antirrhinum hispanicum]
Length = 384
Score = 75.5 bits (184), Expect = 3e-12
Identities = 86/330 (26%), Positives = 132/330 (39%), Gaps = 46/330 (13%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
D++ E++ LPVKSL R +CV KS+ +II FI H R + LLL R
Sbjct: 9 DMVIEIMVQLPVKSLVRFRCVSKSF-CVIIKSSNFINNHFLRR--QTRDTLLLIRRYFPS 65
Query: 90 DHREFCVTHLSGSSLSLVENPW--ISLAHNPRYRLKKGDRYH-----VVGCCNGLICLHG 142
+ ++ S L E W +S+ RL+ Y ++G CNGLIC
Sbjct: 66 PQEDDALSFHKPDSPGLEEEVWAKLSIPFLSDLRLRYDQPYFPQSVIILGPCNGLIC--- 122
Query: 143 YSVDSIFQQGWLSFWNPATRLMSEKLGC-FCVKTKNFDVKLSFGYDNSTDTYKVVFFEIE 201
IF ++ NPA R + C FC + + + G+ N FF+I
Sbjct: 123 -----IFYDDFIISCNPALREFKKLPPCPFCCPKRFYSNIIGQGFGNCDSN----FFKIV 173
Query: 202 KIRRASLVSVFTLDG--------CNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINW 253
+R VS + D + WR I+ +L Y V +G +W
Sbjct: 174 LVRTIKSVSDYNRDKPYIMVHLYNSNTQSWRLIEGEAVL-VQYIFSSPCTDVFFNGACHW 232
Query: 254 MTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCLC 313
+ PY IL+ D+ TE ++ P G E+ + L CL
Sbjct: 233 -NAGVFGIPYPG---------SILTFDISTEIFSEFEYPDGFRELYGGCLCLTALSECLS 282
Query: 314 ---FSHDLEGTHFV-IWQMKKFGDAKSWTQ 339
++ + F+ IW MK +G++ SWT+
Sbjct: 283 VIRYNDSTKDPQFIEIWVMKVYGNSDSWTK 312
>UniRef100_Q84KQ6 F-box [Prunus mume]
Length = 374
Score = 73.6 bits (179), Expect = 1e-11
Identities = 83/341 (24%), Positives = 137/341 (39%), Gaps = 68/341 (19%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSAL-HGKPHLLLQRENSH 88
+++ ++L LPVKSL R C CKSW+ L I +F+ HLHR+ H +LL
Sbjct: 9 EIVIDILVRLPVKSLVRFLCTCKSWSDL-IGSSSFVSTHLHRNVTKHAHVYLL------- 60
Query: 89 HDHREFCVTHLSGSSLSLVENPWI------SLAHNPRYR--------LKKGDRYHVVGCC 134
C+ H + L ++P++ S+ N + L+ + Y + G
Sbjct: 61 ------CLNHPNVEYLDDRDDPYVKQEFQWSIFPNEIFEECSKLTHPLRSTEDYMIYGSS 114
Query: 135 NGLICLHGYSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYK 194
NGL+C+ S + + + WNP+ + + K V L FG+ + YK
Sbjct: 115 NGLVCV---SDEILNFDSPILIWNPSVKKFRTPPMSININIKFSYVALQFGFHPGVNDYK 171
Query: 195 VVFFEIEKIRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWM 254
V + + A + V++L G + W+ I+ P P+ W HL GT
Sbjct: 172 AVRM-MRTNKNALAIEVYSL----GTDSWKMIEAIP--PWLKCTW-----QHLKGT---- 215
Query: 255 TSRDKSAPYYNPVTITVEQLG----ILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMN 310
++N V V Q G I+S D +E + P + + V
Sbjct: 216 --------FFNGVAYHVIQKGPIFSIISFDSGSEEFEEFIAPDAISTPWGLCIDVYREQI 267
Query: 311 CL---CFSHDLEGTHFV-IWQMKKFGDAKSWTQLLKVRYQD 347
CL C+ EG + +W +++ K W QL Y +
Sbjct: 268 CLLFDCYPFGEEGMEKIDLWVLQE----KMWKQLRPFIYPE 304
>UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis thaliana]
Length = 408
Score = 72.8 bits (177), Expect = 2e-11
Identities = 87/334 (26%), Positives = 142/334 (42%), Gaps = 55/334 (16%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQRENSHH 89
D++ ++ LP K+L R + + K LI +D FI+ HLHR G ++L R
Sbjct: 7 DIVNDIFLRLPAKTLVRCRALSKPCYHLI-NDPDFIESHLHRVLQTGDHLMILLRGAL-- 63
Query: 90 DHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDSIF 149
R + V S S+S VE+P +K+G V G NGLI L D
Sbjct: 64 --RLYSVDLDSLDSVSDVEHP-----------MKRGGPTEVFGSSNGLIGLSNSPTD--- 107
Query: 150 QQGWLSFWNPATR----LMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVF---FEIEK 202
L+ +NP+TR L + + V GYD+ +D YKVV F+I+
Sbjct: 108 ----LAVFNPSTRQIHRLPPSSIDLPDGSSTRGYVFYGLGYDSVSDDYKVVRMVQFKIDS 163
Query: 203 IRRASL-----VSVFTLDGCNGWNGWRDIQN----FPLLPFSYDD--WGVNDGVHLSGTI 251
V VF+L N W+ I++ LL + Y + GV ++
Sbjct: 164 EDELGCSFPYEVKVFSLKK----NSWKRIESVASSIQLLFYFYYHLLYRRGYGVLAGNSL 219
Query: 252 NWMTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGL-DEVPSVVPSVGVLMN 310
+W+ R +N I+ DL E + ++ P+ + + + +GVL
Sbjct: 220 HWVLPRRPGLIAFNL---------IVRFDLALEEFEIVRFPEAVANGNVDIQMDIGVLDG 270
Query: 311 CLCFSHDLEGTHFVIWQMKKFGDAKSWTQLLKVR 344
CLC + + ++ +W MK++ SWT++ V+
Sbjct: 271 CLCLMCNYDQSYVDVWMMKEYNVRDSWTKVFTVQ 304
>UniRef100_Q84KR1 F-box [Prunus mume]
Length = 377
Score = 69.7 bits (169), Expect = 2e-10
Identities = 78/323 (24%), Positives = 127/323 (39%), Gaps = 46/323 (14%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSAL-HGKPHLLLQRENSH 88
+++ ++L LP KSL R C CKSW L I +F+ +HL+R+ H +LL H
Sbjct: 9 EILIDILVRLPAKSLVRFLCTCKSWGDL-IGSSSFVSIHLNRNVTKHAHVYLLC----LH 63
Query: 89 HDHREFCVTH-----LSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGY 143
H + E V S+ N + L +RY + G NGL+C+
Sbjct: 64 HPNFERQVDPDDPYVEQKFHWSIFSNETFEECSKLSHPLGSTERYGIYGSSNGLVCI--- 120
Query: 144 SVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKI 203
S + + + WNP+ R + + K V L FG+ + YKVV +
Sbjct: 121 SDEILNFDSPIHIWNPSVRKLRTPPMSANINVKFSHVALQFGFHPGLNDYKVVRM-MRTN 179
Query: 204 RRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPY 263
+ A V V++L + W+ I+ P P+ W + G +G + + P
Sbjct: 180 KNALAVEVYSL----RTDSWKMIETIP--PWLKCTWQHHKGTFFNGVAYHIIEK---GPL 230
Query: 264 YNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCL------CFSHD 317
++ I+S D +E + P + + V CL C D
Sbjct: 231 FS----------IMSFDSGSEEFEEFLAPDAICNSWGLCIDVYKEQICLLFTFYDCEEED 280
Query: 318 LEGTHFVIWQMKKFGDAKSWTQL 340
+E + F + Q K+ W QL
Sbjct: 281 MEKSDFWVLQEKR------WKQL 297
>UniRef100_Q9LTF6 Arabidopsis thaliana genomic DNA, chromosome 5, BAC clone:F6N7
[Arabidopsis thaliana]
Length = 351
Score = 69.3 bits (168), Expect = 2e-10
Identities = 67/205 (32%), Positives = 92/205 (44%), Gaps = 29/205 (14%)
Query: 27 LHNDLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQREN 86
+ DL+ E+L LPVK L R CVCK W + II + FI L+ RS+ +P+++
Sbjct: 2 ISEDLLVEILLRLPVKPLARCLCVCKLW-ATIIRSRYFINLYQSRSSTR-QPYVMF---- 55
Query: 87 SHHDHREFCVTH-LSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSV 145
+ D C H S S SLV S ++ H C NGLIC+ S
Sbjct: 56 ALRDIFTSCRWHFFSSSQPSLVTKATCSANNSS----------HTPDCVNGLICVEYMS- 104
Query: 146 DSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKIRR 205
Q W+S NPATR L + F K GYD YKV+FF + +
Sbjct: 105 -----QLWIS--NPATR--KGVLVPQSAPHQKFR-KWYMGYDPINYQYKVLFFSKQYLLS 154
Query: 206 ASLVSVFTLDGCNGWNGWRDIQNFP 230
+ VFTL+G W +++N P
Sbjct: 155 PYKLEVFTLEGQGSWK-MIEVENIP 178
>UniRef100_Q76B88 S haplotype-specific F-box protein 5 [Prunus avium]
Length = 377
Score = 69.3 bits (168), Expect = 2e-10
Identities = 59/222 (26%), Positives = 93/222 (41%), Gaps = 13/222 (5%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRS-ALHGKPHLL-LQRENS 87
+++ ++L LP KSL R C CKSW+ L + +F+ HLHR+ H HLL L N
Sbjct: 9 EILIDILVRLPAKSLVRFLCTCKSWSDL-VGSSSFVSTHLHRNITKHAHVHLLCLHHPNV 67
Query: 88 HHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDS 147
SL N + L + Y + G NGL+C+ S +
Sbjct: 68 RRQVNPDDPYVTQEFQWSLFPNETFEECSKLSHPLGSTEHYGIYGSSNGLVCI---SDEI 124
Query: 148 IFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKIRRAS 207
+ + WNP+ R + K V L FG+ ++ + YKVV + + A
Sbjct: 125 LNFDSPILMWNPSVRKFRTAPTSTNINLKFAYVALQFGFHHAVNDYKVVRM-MRTNKDAL 183
Query: 208 LVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSG 249
V V++L + W+ I+ P P+ W + G +G
Sbjct: 184 AVEVYSL----RTDSWKMIEAIP--PWLKCTWQHHRGTFFNG 219
>UniRef100_Q84KK2 S haplotype-specific F-box protein d [Prunus dulcis]
Length = 376
Score = 68.6 bits (166), Expect = 3e-10
Identities = 71/325 (21%), Positives = 129/325 (38%), Gaps = 34/325 (10%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSAL-HGKPHLLLQRENSH 88
+++ ++L LP KSL R C CKSW+ L I +F+ HLHR+ H + +LL +
Sbjct: 9 EILIDILVRLPAKSLVRFLCTCKSWSDL-IGSSSFVGTHLHRNVTGHAQAYLLCLHHPNF 67
Query: 89 HDHREFCVTHLSGS-SLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVDS 147
R+ + SL N + + L + Y + G NGL+C+ S +
Sbjct: 68 ECQRDDDDPYFKEELQWSLFSNVTFEESSKLSHPLGSTEHYVIYGSSNGLVCI---SDEI 124
Query: 148 IFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKIRRAS 207
+ + WNP+ + + + K V L FG+ + + Y+ V + + A
Sbjct: 125 MNFDSPIHIWNPSVKKLRTTPISTSINIKFSHVALQFGFHSGVNDYRAVRM-LRTNQNAL 183
Query: 208 LVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPYYNPV 267
V +++L + W I+ P P+ W + G +G + + +
Sbjct: 184 AVEIYSL----RTDSWTMIEAIP--PWLKCTWQHHQGTFFNGVAYHIIEKGPT------- 230
Query: 268 TITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCLCFSH---DLEGTHFV 324
++S D +E + P + + + V CL F + + EG +
Sbjct: 231 ------FSVISFDSGSEEFEEFIAPDAICSLWRLCIHVYKEQICLLFGYYSCEEEGMENI 284
Query: 325 -IWQMKKFGDAKSWTQLLKVRYQDL 348
+W +++ K W QL Y L
Sbjct: 285 DLWVLQE----KRWKQLCPFIYDPL 305
>UniRef100_Q76HE3 S-locus F-Box protein 7 [Prunus mume]
Length = 377
Score = 68.6 bits (166), Expect = 3e-10
Identities = 78/323 (24%), Positives = 127/323 (39%), Gaps = 46/323 (14%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSAL-HGKPHLLLQRENSH 88
+++ ++L LP KSL R C CKSW L I +F+ +HL+R+ H +LL H
Sbjct: 9 EILIDILVRLPAKSLVRFLCTCKSWGDL-IGSSSFVSIHLNRNVTKHAHVYLLC----LH 63
Query: 89 HDHREFCVTH-----LSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGY 143
H + E V S+ N + L +RY + G NGL+C+
Sbjct: 64 HPNFERQVDPDDPYVEQKFHWSIFSNETFEECSKLSHPLGSTERYGIYGSSNGLVCI--- 120
Query: 144 SVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEKI 203
S + + + WNP+ R + + K V L FG+ + YKVV +
Sbjct: 121 SDEILNFDSPIHIWNPSVRKLRTPPMSANINVKFSHVALQFGFHPGPNDYKVVRM-MRTN 179
Query: 204 RRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSGTINWMTSRDKSAPY 263
+ A V V++L + W+ I+ P P+ W + G +G + + P
Sbjct: 180 KNALAVEVYSL----RTDFWKMIETIP--PWLKCTWQHHKGTFFNGVAYHIIEK---GPL 230
Query: 264 YNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCL------CFSHD 317
++ I+S D +E + P + + V CL C D
Sbjct: 231 FS----------IMSFDSGSEEFEEFLAPDAICNSWGLCIDVYKEQICLLFTFYDCEEED 280
Query: 318 LEGTHFVIWQMKKFGDAKSWTQL 340
+E + F + Q K+ W QL
Sbjct: 281 MEKSDFWVLQEKR------WKQL 297
>UniRef100_Q70WR6 S locus F-box (SLF)-S4D protein [Antirrhinum hispanicum]
Length = 374
Score = 68.2 bits (165), Expect = 5e-10
Identities = 79/327 (24%), Positives = 137/327 (41%), Gaps = 48/327 (14%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSALHGKPHLLLQR---EN 86
D++ E+L WLPVKSL RLKC K + ++I Q FI H+ + LL++R +
Sbjct: 10 DILKEILVWLPVKSLIRLKCASKHLD-MLIKSQAFITSHMIKQR-RNDGMLLVRRILPPS 67
Query: 87 SHHDHREFCVTHLSGSSLSLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHGYSVD 146
+++D F + L + P I+L NP + V+G CNG++C+ G
Sbjct: 68 TYNDVFSFHDVNSPELEEVLPKLP-ITLLSNPDEASFNPNIVDVLGPCNGIVCITG---- 122
Query: 147 SIFQQGWLSFWNPATR----LMSEKLGCFCVKTKNFDVKLSFGYDNS-TDTYKVVFFE-- 199
Q + NPA R L S + C + + ++ G+ ++ T+ +KV+
Sbjct: 123 ----QEDIILCNPALREFRKLPSAPISC---RPPCYSIRTGGGFGSTCTNNFKVILMNTL 175
Query: 200 -IEKIRRASLVSVFTLDGCNGWNGWRDIQNFPL---LPFSY--DDWGVNDGVHLSGTINW 253
++ L N + WR+I +F + + FSY + H +G +
Sbjct: 176 YTARVDGRDAQHRIHLYNSNN-DSWREINDFAIVMPVVFSYQCSELFFKGACHWNGRTSG 234
Query: 254 MTSRDKSAPYYNPVTITVEQLGILSLDLRTETYTLLSPPKGLDEVPSVVPSVGVLMNCLC 313
T+ D IL+ D+ TE + P G + + +L C
Sbjct: 235 ETTPDV----------------ILTFDVSTEVFGQFEHPSGFKLCTGLQHNFMILNECFA 278
Query: 314 -FSHDLEGTHFVIWQMKKFGDAKSWTQ 339
++ +W MK++G +SWT+
Sbjct: 279 SVRSEVVRCLIEVWVMKEYGIKQSWTK 305
>UniRef100_Q84KR2 F-box [Prunus mume]
Length = 376
Score = 68.2 bits (165), Expect = 5e-10
Identities = 60/227 (26%), Positives = 97/227 (42%), Gaps = 23/227 (10%)
Query: 30 DLIAEVLSWLPVKSLTRLKCVCKSWNSLIISDQTFIKLHLHRSAL-HGKPHLLLQRENSH 88
+++ ++L LP KSL R C CKSW+ L I +F++ HL R+ H +LL H
Sbjct: 9 EILIDILVRLPAKSLVRFLCTCKSWSDL-IGSSSFVRTHLDRNVTKHAHVYLLC----LH 63
Query: 89 HDHREFCVTHLSGSSL------SLVENPWISLAHNPRYRLKKGDRYHVVGCCNGLICLHG 142
H + E C + + SL N + L + Y + G NGL+C+
Sbjct: 64 HPNFE-CAIDPNDPYIEEEVQWSLFSNETFEQCSKLSHPLGSTEHYVIYGSSNGLVCI-- 120
Query: 143 YSVDSIFQQGWLSFWNPATRLMSEKLGCFCVKTKNFDVKLSFGYDNSTDTYKVVFFEIEK 202
S + + + WNP+ R + + K V L FG+ + YK V +
Sbjct: 121 -SDEILNFDSPIHIWNPSARKLKTPPISTNINIKFSCVVLQFGFHPGVNDYKAVRM-MRT 178
Query: 203 IRRASLVSVFTLDGCNGWNGWRDIQNFPLLPFSYDDWGVNDGVHLSG 249
+ A V V++L N W+ I+ P P+ W + G+ +G
Sbjct: 179 NKNALAVEVYSL----RTNSWKMIEAIP--PWLKCTWQHHKGIFFNG 219
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.137 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 789,650,727
Number of Sequences: 2790947
Number of extensions: 33724794
Number of successful extensions: 73475
Number of sequences better than 10.0: 390
Number of HSP's better than 10.0 without gapping: 235
Number of HSP's successfully gapped in prelim test: 155
Number of HSP's that attempted gapping in prelim test: 72990
Number of HSP's gapped (non-prelim): 489
length of query: 440
length of database: 848,049,833
effective HSP length: 130
effective length of query: 310
effective length of database: 485,226,723
effective search space: 150420284130
effective search space used: 150420284130
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0176a.1


