

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
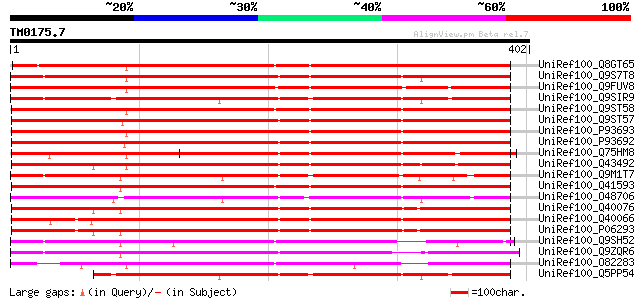
Query= TM0175.7
(402 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8GT65 Serpin-like protein [Citrus paradisi] 394 e-108
UniRef100_Q9S7T8 F16N3.3 protein [Arabidopsis thaliana] 369 e-101
UniRef100_Q9FUV8 Phloem serpin-1 [Cucurbita maxima] 350 5e-95
UniRef100_Q9SIR9 Putative serpin [Arabidopsis thaliana] 320 3e-86
UniRef100_Q9ST58 Serpin [Triticum aestivum] 318 2e-85
UniRef100_Q9ST57 Serpin [Triticum aestivum] 318 2e-85
UniRef100_P93693 Serpin [Triticum aestivum] 317 5e-85
UniRef100_P93692 Serpin [Triticum aestivum] 313 5e-84
UniRef100_Q75HM8 Putative serine protease inhibitor [Oryza sativa] 311 2e-83
UniRef100_Q43492 Serpin [Hordeum vulgare] 309 8e-83
UniRef100_Q9M1T7 Serpin-like protein [Arabidopsis thaliana] 304 3e-81
UniRef100_Q41593 Serpin [Triticum aestivum] 303 4e-81
UniRef100_O48706 Putative serpin [Arabidopsis thaliana] 302 1e-80
UniRef100_Q40076 Protein z-type serpin [Hordeum vulgare] 300 5e-80
UniRef100_Q40066 Protein zx [Hordeum vulgare] 299 8e-80
UniRef100_P06293 Protein Z [Hordeum vulgare] 295 2e-78
UniRef100_Q9SH52 F22C12.22 [Arabidopsis thaliana] 288 2e-76
UniRef100_Q9ZQR6 Putative serpin protein [Arabidopsis thaliana] 283 6e-75
UniRef100_O82283 Putative serpin [Arabidopsis thaliana] 280 7e-74
UniRef100_Q5PP54 At2g25240 [Arabidopsis thaliana] 277 3e-73
>UniRef100_Q8GT65 Serpin-like protein [Citrus paradisi]
Length = 389
Score = 394 bits (1013), Expect = e-108
Identities = 216/389 (55%), Positives = 274/389 (69%), Gaps = 6/389 (1%)
Query: 3 LQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSFL 62
+++SI QT+VALS+TKH+ + E ++ NLVFSP S+H +LS+++ GS G TLD+L SFL
Sbjct: 1 VRESISNQTDVALSLTKHV-ALTEAKDSNLVFSPSSIHVLLSLISAGSKGPTLDQLLSFL 59
Query: 63 RFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYKAT 120
+ S D LNTF S++++ V +D +PS LS N +W DKSL+L ++FKQ++ YKA
Sbjct: 60 KSKSDDQLNTFASELVAVVFADGSPSGGPRLSVANGVWIDKSLSLKNTFKQVVDNVYKAA 119
Query: 121 LALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHTF 180
VD +TK +V REVN W EKETNGL+ E+LP +V T+LIFANALYFKG W TF
Sbjct: 120 SNQVDSQTKAAEVSREVNMWAEKETNGLVKEVLPPGSVDNSTRLIFANALYFKGAWNETF 179
Query: 181 DPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCIF 240
D S T DFHLLNG SI+VPFM+S KK QF+ FDGF VL L YKQG DK+ RFSM F
Sbjct: 180 DSSKTKDYDFHLLNGGSIKVPFMTS-KKNQFVSAFDGFKVLGLPYKQGEDKR-RFSMYFF 237
Query: 241 LPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVVS 300
LP+AKDGLP+L+EK+ SES FL LP Q+V V FRIP+FKI F +E S VLK LG+V
Sbjct: 238 LPDAKDGLPTLLEKMGSESKFLDHHLPSQRVEVGDFRIPRFKISFGIEVSKVLKGLGLVL 297
Query: 301 PFSKSNANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTGI 359
PFS +MV SP+ + L V SI K+ IEVNEEGTEA A + V + I
Sbjct: 298 PFSGEGGGLAEMVDSPVGKNLYVSSIFQKSFIEVNEEGTEAAAASAATVVLRSILLLDKI 357
Query: 360 DFVADHPFLFLIREDLTGTILFIGQVLHP 388
DFVADHPF+F+IRED+TG ++FIG VL+P
Sbjct: 358 DFVADHPFVFMIREDMTGLVMFIGHVLNP 386
>UniRef100_Q9S7T8 F16N3.3 protein [Arabidopsis thaliana]
Length = 391
Score = 369 bits (948), Expect = e-101
Identities = 195/393 (49%), Positives = 269/393 (67%), Gaps = 9/393 (2%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MD+++SI Q +V++++ KH+ + Q N++FSP S++ +LS++A GSAG+T D++ S
Sbjct: 1 MDVRESISLQNQVSMNLAKHVITTVS-QNSNVIFSPASINVVLSIIAAGSAGATKDQILS 59
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FL+F S D LN+F S+++S VL+D + + LS N W DKSL+ SFKQL+ YK
Sbjct: 60 FLKFSSTDQLNSFSSEIVSAVLADGSANGGPKLSVANGAWIDKSLSFKPSFKQLLEDSYK 119
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKH 178
A DF++K +V EVNSW EKETNGLITE+LP + +TKLIFANALYFKG W
Sbjct: 120 AASNQADFQSKAVEVIAEVNSWAEKETNGLITEVLPEGSADSMTKLIFANALYFKGTWNE 179
Query: 179 TFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMC 238
FD S+T +FHLL+G + PFM+S+KK Q++ +DGF VL L Y QG+DK+ +FSM
Sbjct: 180 KFDESLTQEGEFHLLDGNKVTAPFMTSKKK-QYVSAYDGFKVLGLPYLQGQDKR-QFSMY 237
Query: 239 IFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGV 298
+LP+A +GL L++K+ S FL +PR++V+VR+F+IPKFK F +ASNVLK LG+
Sbjct: 238 FYLPDANNGLSDLLDKIVSTPGFLDNHIPRRQVKVREFKIPKFKFSFGFDASNVLKGLGL 297
Query: 299 VSPFSKSNANFTKMVVSPL--DELCVESIHHKASIEVNEEGTEAGVA-CSMCDVKLCVSA 355
SPFS T+MV SP LCV +I HKA IEVNEEGTEA A + ++ +
Sbjct: 298 TSPFS-GEEGLTEMVESPEMGKNLCVSNIFHKACIEVNEEGTEAAAASAGVIKLRGLLME 356
Query: 356 RTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
IDFVADHPFL ++ E++TG +LFIGQV+ P
Sbjct: 357 EDEIDFVADHPFLLVVTENITGVVLFIGQVVDP 389
>UniRef100_Q9FUV8 Phloem serpin-1 [Cucurbita maxima]
Length = 389
Score = 350 bits (897), Expect = 5e-95
Identities = 193/391 (49%), Positives = 265/391 (67%), Gaps = 8/391 (2%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
MD++++IR +VA++ITK + +E + N+V SP S++ +LS++A GS G LD+L S
Sbjct: 1 MDIKEAIRNHGDVAMAITKRILQHDEAKGSNVVISPLSIYVLLSLVAAGSKGRPLDQLLS 60
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FL+ +S+D+LN F S ++ V +D + L+FVN +W D+SL+L SF+Q++ +YK
Sbjct: 61 FLKSNSIDNLNAFASHIIDKVFADASSCGGPRLAFVNGVWIDQSLSLKSSFQQVVDKYYK 120
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKH 178
A L VDF TK ++V EVNSWVEK T GLI E+LP +V T+L+ ANALYFK W+
Sbjct: 121 AELRQVDFLTKANEVISEVNSWVEKNTYGLIREILPAGSVGSSTQLVLANALYFKAAWQQ 180
Query: 179 TFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMC 238
FD S+T DF+L++G+S++ PFMS E K Q++ FDGF VL L Y QG D + RFSM
Sbjct: 181 AFDASITMKRDFYLIDGSSVKAPFMSGE-KDQYVAVFDGFKVLALPYSQGPDPR-RFSMY 238
Query: 239 IFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGV 298
FLP+ KDGL SLIEKL SE F+ +P +K + F IPKFKI F +E S+VLK+LG+
Sbjct: 239 FFLPDRKDGLASLIEKLDSEPGFIDRHIPCKKQELGGFLIPKFKISFGIEVSDVLKKLGL 298
Query: 299 VSPFSKSNANFTKMVVSPL-DELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSART 357
V PF++ MV SP+ L V +I HKA IEV+EEGT+A A S V +
Sbjct: 299 VLPFTE--GGLLGMVESPVAQNLRVSNIFHKAFIEVDEEGTKA-AASSAVTVGIVSLPIN 355
Query: 358 GIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
IDF+A+ PFL+LIRED +GT+LFIGQVL+P
Sbjct: 356 RIDFIANRPFLYLIREDKSGTLLFIGQVLNP 386
>UniRef100_Q9SIR9 Putative serpin [Arabidopsis thaliana]
Length = 385
Score = 320 bits (821), Expect = 3e-86
Identities = 184/393 (46%), Positives = 249/393 (62%), Gaps = 16/393 (4%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
M+L KSI +V + +TKH+ + NLVFSP S++ +LS++A GS T +++ S
Sbjct: 1 MELGKSIENHNDVVVRLTKHVIATVA-NGSNLVFSPISINVLLSLIAAGSCSVTKEQILS 59
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSFL-LSFVNEMWADKSLALSHSFKQLMTTHYKA 119
FL S DHLN +Q++ T S L LS N +W DK +L SFK L+ YKA
Sbjct: 60 FLMLPSTDHLNLVLAQIID---GGTEKSDLRLSIANGVWIDKFFSLKLSFKDLLENSYKA 116
Query: 120 TLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKL--TKLIFANALYFKGVWK 177
T + VDF +K +V EVN+W E TNGLI ++L +++ + + L+ ANA+YFKG W
Sbjct: 117 TCSQVDFASKPSEVIDEVNTWAEVHTNGLIKQILSRDSIDTIRSSTLVLANAVYFKGAWS 176
Query: 178 HTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSM 237
FD +MT +DFHLL+GTS++VPFM++ + Q++R++DGF VLRL Y + + + FSM
Sbjct: 177 SKFDANMTKKNDFHLLDGTSVKVPFMTNYED-QYLRSYDGFKVLRLPYIEDQRQ---FSM 232
Query: 238 CIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELG 297
I+LPN K+GL L+EK+ SE F +P + V FRIPKFK FE AS VLK++G
Sbjct: 233 YIYLPNDKEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVLKDMG 292
Query: 298 VVSPFSKSNANFTKMVVSPL--DELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSA 355
+ SPF+ + T+MV SP D+L V SI HKA IEV+EEGTEA A V C S
Sbjct: 293 LTSPFN-NGGGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEA--AAVSVGVVSCTSF 349
Query: 356 RTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
R DFVAD PFLF +RED +G ILF+GQVL P
Sbjct: 350 RRNPDFVADRPFLFTVREDKSGVILFMGQVLDP 382
>UniRef100_Q9ST58 Serpin [Triticum aestivum]
Length = 398
Score = 318 bits (814), Expect = 2e-85
Identities = 178/390 (45%), Positives = 256/390 (65%), Gaps = 6/390 (1%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
D++ SI QT AL + + S + N VFSP SLH LS++A G+ +T D+L +
Sbjct: 8 DVRLSIAHQTRFALRLASTISSNPKSAASNAVFSPVSLHVALSLLAAGAGSATRDQLVAT 67
Query: 62 LRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYKA 119
L V+ L+ QV+ VL+D + + ++F N ++ D SL L SF++L YKA
Sbjct: 68 LGTGEVEGLHALAEQVVQFVLADASSAGGPHVAFANGVFVDASLPLKPSFQELAVCKYKA 127
Query: 120 TLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHT 179
VDF+TK +V +VNSWVEK T+G I ++LP+ +V TKL+ ANALYFKG W
Sbjct: 128 DTQSVDFQTKAAEVATQVNSWVEKVTSGRIKDILPSGSVDNTTKLVLANALYFKGAWTDQ 187
Query: 180 FDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCI 239
FD S T D F+L +G+S++ PFMSS Q++ + DG VL+L YKQG DK+ +FSM I
Sbjct: 188 FDSSGTKNDYFYLPDGSSVQTPFMSS-MDDQYLSSSDGLKVLKLPYKQGGDKR-QFSMYI 245
Query: 240 FLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVV 299
LP A GL +L EKL++E FL+ +PRQ+V +R+F++PKFKI FE EAS++LK LG+
Sbjct: 246 LLPEAPGGLSNLAEKLSAEPDFLERHIPRQRVALRQFKLPKFKISFETEASDLLKCLGLQ 305
Query: 300 SPFSKSNANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTG 358
PFS + A+F++MV SP+ L V S+ H+A +EVNE+GTEA + ++ L +
Sbjct: 306 LPFS-NEADFSEMVDSPMAHGLRVSSVFHQAFVEVNEQGTEAAASTAIKMALLQARPPSV 364
Query: 359 IDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DF+ADHPFLFL+RED++G +LF+G V++P
Sbjct: 365 MDFIADHPFLFLLREDISGVVLFMGHVVNP 394
>UniRef100_Q9ST57 Serpin [Triticum aestivum]
Length = 398
Score = 318 bits (814), Expect = 2e-85
Identities = 177/390 (45%), Positives = 248/390 (63%), Gaps = 6/390 (1%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
D++ SI QT + + S E N+ FSP SLH LS++ G+ G+T D+L +
Sbjct: 8 DVRLSIAHQTRFGFRLASTISSNPESTANNVAFSPVSLHVALSLITAGAGGATRDQLVAT 67
Query: 62 LRFDSVDHLNTFFSQVLSTVLSDTT--PSFLLSFVNEMWADKSLALSHSFKQLMTTHYKA 119
L + L+ QV+ VL+D + S ++F N ++ D SL L SF++L YKA
Sbjct: 68 LGEGEAERLHALAEQVVQFVLADASYADSPRVTFANGVFVDASLPLKPSFQELAVCKYKA 127
Query: 120 TLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHT 179
VDF+TK +V +VNSWVEK T GLI ++LP ++ T+L+ NALYFKG W
Sbjct: 128 EAQSVDFQTKAAEVTAQVNSWVEKVTTGLIKDILPAGSISNTTRLVLGNALYFKGAWTDQ 187
Query: 180 FDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCI 239
FD +T D F+LL+G+SI+ PFM S ++ Q+I + DG VL+L YKQG DK+ +FSM I
Sbjct: 188 FDSRVTKSDYFYLLDGSSIQTPFMYSSEE-QYISSSDGLKVLKLPYKQGGDKR-QFSMYI 245
Query: 240 FLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVV 299
LP A G+ SL EKL++E L+ +PRQKV +R+F++PKFKI F +EAS++LK LG+
Sbjct: 246 LLPEAPSGIWSLAEKLSAEPELLERHIPRQKVALRQFKLPKFKISFGIEASDLLKHLGLQ 305
Query: 300 SPFSKSNANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTG 358
PFS A+ ++MV SP+ + L + S+ HK +EVNE GTEA A V L S +
Sbjct: 306 LPFS-DEADLSEMVDSPMPQGLRISSVFHKTFVEVNETGTEAAAATIAKAVLLSASPPSD 364
Query: 359 IDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DF+ADHPFLFLIRED +G +LFIG V++P
Sbjct: 365 MDFIADHPFLFLIREDTSGVVLFIGHVVNP 394
>UniRef100_P93693 Serpin [Triticum aestivum]
Length = 399
Score = 317 bits (811), Expect = 5e-85
Identities = 176/390 (45%), Positives = 253/390 (64%), Gaps = 5/390 (1%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
D++ SI QT AL + + S + N FSP SLH+ LS++A G+ +T D+L +
Sbjct: 8 DVRLSIAHQTRFALRLASTISSNPKSAASNAAFSPVSLHSALSLLAAGAGSATRDQLVAT 67
Query: 62 LRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTHYKA 119
L V+ + QV+ VL+D + + ++F N ++ D SL L SF++L YKA
Sbjct: 68 LGTGEVEGGHALAEQVVQFVLADASSAGGPRVAFANGVFVDASLLLKPSFQELAVCKYKA 127
Query: 120 TLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHT 179
VDF+TK +V +VNSWVEK T+G I +LP+ +V TKL+ ANALYFKG W
Sbjct: 128 ETQSVDFQTKAAEVTTQVNSWVEKVTSGRIKNILPSGSVDNTTKLVLANALYFKGAWTDQ 187
Query: 180 FDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCI 239
FD T D F+LL+G+S++ PFMSS Q+I + DG VL+L YKQG D + +FSM I
Sbjct: 188 FDSYGTKNDYFYLLDGSSVQTPFMSSMDDDQYISSSDGLKVLKLPYKQGGDNR-QFSMYI 246
Query: 240 FLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVV 299
LP A GL SL EKL++E FL+ +PRQ+V +R+F++PKFKI F +EAS++LK LG+
Sbjct: 247 LLPEAPGGLSSLAEKLSAEPDFLERHIPRQRVAIRQFKLPKFKISFGMEASDLLKCLGLQ 306
Query: 300 SPFSKSNANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTG 358
PFS A+F++MV SP+ + L V S+ H+A +EVNE+GTEA + ++ V +
Sbjct: 307 LPFS-DEADFSEMVDSPMPQGLRVSSVFHQAFVEVNEQGTEAAASTAIKMVPQQARPPSV 365
Query: 359 IDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DF+ADHPFLFL+RED++G +LF+G V++P
Sbjct: 366 MDFIADHPFLFLLREDISGVVLFMGHVVNP 395
>UniRef100_P93692 Serpin [Triticum aestivum]
Length = 398
Score = 313 bits (802), Expect = 5e-84
Identities = 177/390 (45%), Positives = 246/390 (62%), Gaps = 6/390 (1%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
D++ SI QT A + + S E N FSP SLH LS++ G+ G+T ++L +
Sbjct: 8 DVRLSIAHQTRFAFRLASAISSNPESTVNNAAFSPVSLHVALSLITAGAGGATRNQLAAT 67
Query: 62 LRFDSVDHLNTFFSQVLSTVLSDTTP--SFLLSFVNEMWADKSLALSHSFKQLMTTHYKA 119
L V+ L+ QV+ VL+D + ++F N ++ D SL L SF++L YKA
Sbjct: 68 LGEGEVEGLHALAEQVVQFVLADASNIGGPRVAFANGVFVDASLQLKPSFQELAVCKYKA 127
Query: 120 TLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHT 179
VDF+TK +V +VNSWVEK T GLI ++LP ++ T+L+ NALYFKG W
Sbjct: 128 EAQSVDFQTKAAEVTAQVNSWVEKVTTGLIKDILPAGSIDNTTRLVLGNALYFKGAWTDQ 187
Query: 180 FDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCI 239
FDP T DDF+LL+G+SI+ PFM S ++ Q+I + DG VL+L YKQG DK+ +FSM I
Sbjct: 188 FDPRATQSDDFYLLDGSSIQTPFMYSSEE-QYISSSDGLKVLKLPYKQGGDKR-QFSMYI 245
Query: 240 FLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVV 299
LP A GL SL EKL++E FL+ +PRQKV +R+F++PKFKI +EAS++LK LG++
Sbjct: 246 LLPEALSGLWSLAEKLSAEPEFLEQHIPRQKVALRQFKLPKFKISLGIEASDLLKGLGLL 305
Query: 300 SPFSKSNANFTKMVVSPL-DELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTG 358
PF + A+ ++MV SP+ L + SI HKA +EVNE GTEA V +
Sbjct: 306 LPFG-AEADLSEMVDSPMAQNLYISSIFHKAFVEVNETGTEAAATTIAKVVLRQAPPPSV 364
Query: 359 IDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DF+ DHPFLFLIRED +G +LFIG V++P
Sbjct: 365 LDFIVDHPFLFLIREDTSGVVLFIGHVVNP 394
>UniRef100_Q75HM8 Putative serine protease inhibitor [Oryza sativa]
Length = 653
Score = 311 bits (798), Expect = 2e-83
Identities = 188/398 (47%), Positives = 256/398 (64%), Gaps = 12/398 (3%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQE---KNLVFSPFSLHAILSVMAVGSAGSTLDEL 58
DL+ SI QT AL + L S +N+ FSP SLH LS++A G+ G+T D+L
Sbjct: 4 DLRVSIAHQTSFALRLAAALSSPAHPAGGAGRNVAFSPLSLHVALSLVAAGAGGATRDQL 63
Query: 59 FSFLRFD-SVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTT 115
S L S + L+ F Q++ VL+D + + ++F + ++ D SL+L +F +
Sbjct: 64 ASALGGPGSAEGLHAFAEQLVQLVLADASGAGGPRVAFADGVFVDASLSLKKTFGDVAVG 123
Query: 116 HYKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGV 175
YKA VDF+TK +V +VNSWVEK T+GLI E+LP +V T+L+ NALYFKG
Sbjct: 124 KYKAETHSVDFQTKAAEVASQVNSWVEKVTSGLIKEILPPGSVDHTTRLVLGNALYFKGA 183
Query: 176 WKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRF 235
W FD S T +FHLL+G S++ PFMS+ KK Q+I ++D VL+L Y+QG DK+ +F
Sbjct: 184 WTEKFDASKTKDGEFHLLDGKSVQAPFMSTSKK-QYILSYDNLKVLKLPYQQGGDKR-QF 241
Query: 236 SMCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKE 295
SM I LP A+DGL SL EKL SE FL+ +P ++V V +F++PKFKI F EAS++LK
Sbjct: 242 SMYILLPEAQDGLWSLAEKLNSEPEFLEKHIPTRQVTVGQFKLPKFKISFGFEASDLLKS 301
Query: 296 LGVVSPFSKSNANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVS 354
LG+ PFS S A+ T+MV SP + L V S+ HK+ +EVNEEGTEA A + V S
Sbjct: 302 LGLHLPFS-SEADLTEMVDSPEGKNLFVSSVFHKSFVEVNEEGTEAAAATAA--VITLRS 358
Query: 355 ARTGIDFVADHPFLFLIREDLTGTILFIGQVLHPEGAA 392
A DFVADHPFLFLI+ED+TG +LF+G V++P AA
Sbjct: 359 APIAEDFVADHPFLFLIQEDMTGVVLFVGHVVNPLLAA 396
Score = 226 bits (575), Expect = 1e-57
Identities = 128/258 (49%), Positives = 171/258 (65%), Gaps = 7/258 (2%)
Query: 132 QVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHTFDPSMTFVDDFH 191
+V +VNSWV++ T+GLI + +++ TKL+ ANALYFKG W FD S T +FH
Sbjct: 398 EVLGQVNSWVDRVTSGLIKNIATPRSINHNTKLVLANALYFKGAWAEKFDVSKTEDGEFH 457
Query: 192 LLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCIFLPNAKDGLPSL 251
LL+G S++ PFMS+ KK Q++ ++D VL+L Y QG DK+ +FSM I LP A+DGL SL
Sbjct: 458 LLDGESVQAPFMSTRKK-QYLSSYDSLKVLKLPYLQGGDKR-QFSMYILLPEAQDGLWSL 515
Query: 252 IEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVVSPFSKSNANFTK 311
EKL SE F++ +P + V V +F++PKFKI F AS +LK LG+ F S + +
Sbjct: 516 AEKLNSEPEFMENHIPMRPVHVGQFKLPKFKISFGFGASGLLKGLGLPLLFG-SEVDLIE 574
Query: 312 MVVSP-LDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTGIDFVADHPFLFL 370
MV SP L V S+ HK+ IEVNEEGTEA A V + S ++FVADHPF+FL
Sbjct: 575 MVDSPGAQNLFVSSVFHKSFIEVNEEGTEATAAVM---VSMEHSRPRRLNFVADHPFMFL 631
Query: 371 IREDLTGTILFIGQVLHP 388
IRED+TG ILFIG V++P
Sbjct: 632 IREDVTGVILFIGHVVNP 649
>UniRef100_Q43492 Serpin [Hordeum vulgare]
Length = 397
Score = 309 bits (792), Expect = 8e-83
Identities = 174/392 (44%), Positives = 248/392 (62%), Gaps = 11/392 (2%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
DL+ SI QT L + + S E N+ FSP SLH LS++A G+ G+T D+L +
Sbjct: 8 DLRLSIAHQTRFGLRLASAISSDPESAATNVAFSPVSLHVALSLVAAGARGATRDQLVAV 67
Query: 62 LR---FDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTH 116
L + L + QV+ VL+D + + ++F N ++ D SL+L SF++L +
Sbjct: 68 LGGGGAGEAEALQSLAEQVVQFVLADASINSGPRIAFANGVFVDASLSLKPSFQELAVCN 127
Query: 117 YKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVW 176
YK+ + VDF+TK + +VNSWV+ T GLI E+LP ++ T+L+ NALYFKG+W
Sbjct: 128 YKSEVQSVDFKTKAPEAASQVNSWVKNVTAGLIEEILPAGSIDNTTRLVLGNALYFKGLW 187
Query: 177 KHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFS 236
FD S T DDFHLLNG++++ PFMSS K Q++ + DG VL+L Y+ G D + +FS
Sbjct: 188 TKKFDESKTKYDDFHLLNGSTVQTPFMSSTNK-QYLSSSDGLKVLKLPYQHGGDNR-QFS 245
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M I LP A DGL L +KL++E FL+ ++P ++V V +F +PKFKI F EA+ +LK L
Sbjct: 246 MYILLPEAHDGLSRLAQKLSTEPDFLENRIPTEEVEVGQFMLPKFKISFGFEANKLLKTL 305
Query: 297 GVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSAR 356
G+ PFS AN ++MV SP+ L + S+ HK +EV+EEGT+AG A DV + S
Sbjct: 306 GLQLPFS-LEANLSEMVNSPMG-LYISSVFHKTFVEVDEEGTKAGAATG--DVIVDRSLP 361
Query: 357 TGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DFVA+HPFLFLIRED+ G +LFIG V +P
Sbjct: 362 IRMDFVANHPFLFLIREDIAGVVLFIGHVANP 393
>UniRef100_Q9M1T7 Serpin-like protein [Arabidopsis thaliana]
Length = 393
Score = 304 bits (779), Expect = 3e-81
Identities = 175/402 (43%), Positives = 249/402 (61%), Gaps = 26/402 (6%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
M+L KS+ QT+V + + KH+ NLVFSP S++ +L ++A GS T +++ S
Sbjct: 1 MELGKSMENQTDVMVLLAKHVIPTVA-NGSNLVFSPMSINVLLCLIAAGSNCVTKEQILS 59
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYK 118
F+ S D+LN ++ +S L+D LS +W DKSL+ SFK L+ Y
Sbjct: 60 FIMLPSSDYLNAVLAKTVSVALNDGMERSDLHLSTAYGVWIDKSLSFKPSFKDLLENSYN 119
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTK--LIFANALYFKGVW 176
AT VDF TK +V EVN+W E TNGLI E+L +++ + + LI ANA+YFKG W
Sbjct: 120 ATCNQVDFATKPAEVINEVNAWAEVHTNGLIKEILSDDSIKTIRESMLILANAVYFKGAW 179
Query: 177 KHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFS 236
FD +T DFHLL+GT ++VPFM++ KK Q++ +DGF VLRL Y + + + F+
Sbjct: 180 SKKFDAKLTKSYDFHLLDGTMVKVPFMTNYKK-QYLEYYDGFKVLRLPYVEDQRQ---FA 235
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M I+LPN +DGLP+L+E+++S+ FL +PRQ++ F+IPKFK FE +AS+VLKE+
Sbjct: 236 MYIYLPNDRDGLPTLLEEISSKPRFLDNHIPRQRILTEAFKIPKFKFSFEFKASDVLKEM 295
Query: 297 GVVSPFSKSNANFTKMVVSP--------LDELCVESIHHKASIEVNEEGTEAGV--ACSM 346
G+ PF ++ + T+MV SP + L V ++ HKA IEV+EEGTEA SM
Sbjct: 296 GLTLPF--THGSLTEMVESPSIPENLCVAENLFVSNVFHKACIEVDEEGTEAAAVSVASM 353
Query: 347 CDVKLCVSARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
L + DFVADHPFLF +RE+ +G ILF+GQVL P
Sbjct: 354 TKDMLLMG-----DFVADHPFLFTVREEKSGVILFMGQVLDP 390
>UniRef100_Q41593 Serpin [Triticum aestivum]
Length = 398
Score = 303 bits (777), Expect = 4e-81
Identities = 171/390 (43%), Positives = 249/390 (63%), Gaps = 6/390 (1%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
D++ SI QT AL + + S + N FSP SL++ LS++A G+ +T D+L +
Sbjct: 8 DVRLSIAHQTRFALRLASTISSNPKSAASNAAFSPVSLYSALSLLAAGAGSATRDQLVAT 67
Query: 62 LRFDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYKA 119
L V+ L+ QV+ VL+D +T F N ++ D SL L SF+++ YKA
Sbjct: 68 LGTGKVEGLHALAEQVVQFVLADASSTGGSACRFANGVFVDASLLLKPSFQEIAVCKYKA 127
Query: 120 TLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWKHT 179
VDF+TK +V +VNSWVEK T+G I ++LP ++ TKL+ ANALYFKG W
Sbjct: 128 ETQSVDFQTKAAEVTTQVNSWVEKVTSGRIKDILPPGSIDNTTKLVLANALYFKGAWTEQ 187
Query: 180 FDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCI 239
FD T D F+LL+G+S++ PFMSS Q++ + DG VL+L YKQG D + +F M I
Sbjct: 188 FDSYGTKNDYFYLLDGSSVQTPFMSS-MDDQYLLSSDGLKVLKLPYKQGGDNR-QFFMYI 245
Query: 240 FLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVV 299
LP A GL SL EKL++E FL+ +PRQ+V +R+F++PKFKI F +EAS++LK LG+
Sbjct: 246 LLPEAPGGLSSLAEKLSAEPDFLERHIPRQRVALRQFKLPKFKISFGIEASDLLKCLGLQ 305
Query: 300 SPFSKSNANFTKMVVSPLDE-LCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTG 358
PF A+F++MV S + + L V S+ H+A +EVNE+GTEA + ++ V +
Sbjct: 306 LPFG-DEADFSEMVDSLMPQGLRVSSVFHQAFVEVNEQGTEAAASTAIKMVLQQARPPSV 364
Query: 359 IDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DF+ADHPFLFL+RED++G +LF+G V++P
Sbjct: 365 MDFIADHPFLFLVREDISGVVLFMGHVVNP 394
>UniRef100_O48706 Putative serpin [Arabidopsis thaliana]
Length = 389
Score = 302 bits (773), Expect = 1e-80
Identities = 179/397 (45%), Positives = 238/397 (59%), Gaps = 20/397 (5%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
M+L KSI Q V + K + + N+VFSP S++ +LS++A GS T +E+ S
Sbjct: 1 MELGKSIENQNNVVARLAKKVIETDVANGSNVVFSPMSINVLLSLIAAGSNPVTKEEILS 60
Query: 61 FLRFDSVDHLNTFFSQVLS--TVLSDTTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYK 118
FL S DHLN +++ T SD LS + +W DKS L SFK+L+ YK
Sbjct: 61 FLMSPSTDHLNAVLAKIADGGTERSD----LCLSTAHGVWIDKSSYLKPSFKELLENSYK 116
Query: 119 ATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTK-----LIFANALYFK 173
A+ + VDF TK +V EVN W + TNGLI ++L + + + LI ANA+YFK
Sbjct: 117 ASCSQVDFATKPVEVIDEVNIWADVHTNGLIKQILSRDCTDTIKEIRNSTLILANAVYFK 176
Query: 174 GVWKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKL 233
W FD +T +DFHLL+G +++VPFM S K Q++R +DGF VLRL Y + K
Sbjct: 177 AAWSRKFDAKLTKDNDFHLLDGNTVKVPFMMSYKD-QYLRGYDGFQVLRLPYVED---KR 232
Query: 234 RFSMCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVL 293
FSM I+LPN KDGL +L+EK+++E FL +P + V RIPK FE +AS VL
Sbjct: 233 HFSMYIYLPNDKDGLAALLEKISTEPGFLDSHIPLHRTPVDALRIPKLNFSFEFKASEVL 292
Query: 294 KELGVVSPFSKSNANFTKMVVSPL--DELCVESIHHKASIEVNEEGTEAGVACSMCDVKL 351
K++G+ SPF+ S N T+MV SP D+L V SI HKA IEV+EEGTEA +
Sbjct: 293 KDMGLTSPFT-SKGNLTEMVDSPSNGDKLHVSSIIHKACIEVDEEGTEAAAVSVAIMMPQ 351
Query: 352 CVSARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
C+ DFVADHPFLF +RED +G ILFIGQVL P
Sbjct: 352 CLMRNP--DFVADHPFLFTVREDNSGVILFIGQVLDP 386
>UniRef100_Q40076 Protein z-type serpin [Hordeum vulgare]
Length = 400
Score = 300 bits (768), Expect = 5e-80
Identities = 171/393 (43%), Positives = 246/393 (62%), Gaps = 10/393 (2%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
D++ SI QT AL + + S E N+ FSP SLH LS++ G+ G+T D+L +
Sbjct: 8 DVRLSIAHQTRFALRLASAISSNPERAAGNVAFSPLSLHVALSLITAGAGGATRDQLVAI 67
Query: 62 LR---FDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTH 116
L LN QV+ VL++ +T ++F N ++ D SL+L SF++L
Sbjct: 68 LGDGGAGDAKELNALAEQVVQFVLANESSTGGPRIAFANGIFVDASLSLKPSFEELAVCQ 127
Query: 117 YKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVW 176
YKA VDF+ K + +VNSWVE+ T GLI ++LP +V TKL+ NALYFKG W
Sbjct: 128 YKAKTQSVDFQHKTLEAVGQVNSWVEQVTTGLIKQILPPGSVDNTTKLVLGNALYFKGAW 187
Query: 177 KHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFS 236
FD S T D FHLL+G+SI+ FMSS KK Q+I + D VL+L Y +G DK+ +FS
Sbjct: 188 DQKFDESNTKCDSFHLLDGSSIQTQFMSSTKK-QYISSSDNLKVLKLPYAKGHDKR-QFS 245
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M I LP A+DGL SL ++L++E F++ +P+Q V V +F++PKFKI ++ EAS++L+ L
Sbjct: 246 MYILLPGAQDGLWSLAKRLSTEPEFIENHIPKQTVEVGRFQLPKFKISYQFEASSLLRAL 305
Query: 297 GVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSAR 356
G+ PFS+ A+ ++MV S L + + HK+ +EVNEEGTEAG A V + + +
Sbjct: 306 GLQLPFSE-EADLSEMVDSS-QGLEISHVFHKSFVEVNEEGTEAGAATVAMGVAMSMPLK 363
Query: 357 TG-IDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DFVA+HPFLFLIRED+ G ++F+G V +P
Sbjct: 364 VDLVDFVANHPFLFLIREDIAGVVVFVGHVTNP 396
>UniRef100_Q40066 Protein zx [Hordeum vulgare]
Length = 398
Score = 299 bits (766), Expect = 8e-80
Identities = 178/394 (45%), Positives = 244/394 (61%), Gaps = 11/394 (2%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEK--NLVFSPFSLHAILSVMAVGSAGSTLDELF 59
D++ SI QT A+ + + S + N FSP SLH LS++A G+A +T D+L
Sbjct: 5 DIRLSIAHQTRFAVRLASAISSPSHAKGSSGNAAFSPLSLHVALSLVAAGAA-ATRDQLA 63
Query: 60 SFL---RFDSVDHLNTFFSQVLSTVLSDTTPSFLLSFVNEMWADKSLALSHSFKQLMTTH 116
+ L + L+ QV+ VL+D + + ++ D SL L SFK L+
Sbjct: 64 ATLGAAEKGDAEGLHALAEQVVQVVLADASGAGGPRSFANVFVDSSLKLKPSFKDLVVGK 123
Query: 117 YKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVW 176
YK VDF+TK +V +VNSWVEK T GLI E+LP +V T+L+ NALYFKG W
Sbjct: 124 YKGETQSVDFQTKAPEVAGQVNSWVEKITTGLIKEILPAGSVDSTTRLVLGNALYFKGSW 183
Query: 177 KHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFS 236
FD S T + FHLL+G+S++ PFMSS KK Q+I ++D VL+L Y+QG DK+ +FS
Sbjct: 184 TEKFDASKTKDEKFHLLDGSSVQTPFMSSTKK-QYISSYDSLKVLKLPYQQGGDKR-QFS 241
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M I LP A+DGL +L KL++E F++ +P QKV V +F++PKFKI F EAS++LK L
Sbjct: 242 MYILLPEAQDGLWNLANKLSTEPEFMEKHMPMQKVPVGQFKLPKFKISFGFEASDMLKGL 301
Query: 297 GVVSPFSKSNANFTKMVVSP-LDELCVESIHHKASIEVNEEGTEAGV-ACSMCDVKLCVS 354
G+ PFS S A+ ++MV SP L V S+ HK+ +EVNEEGTEA + ++
Sbjct: 302 GLQLPFS-SEADLSEMVDSPAARSLYVSSVFHKSFVEVNEEGTEAAARTARVVTLRSLPV 360
Query: 355 ARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DFVADHPFLFLIREDLTG +LF+G V +P
Sbjct: 361 EPVKVDFVADHPFLFLIREDLTGVVLFVGHVFNP 394
>UniRef100_P06293 Protein Z [Hordeum vulgare]
Length = 399
Score = 295 bits (754), Expect = 2e-78
Identities = 172/393 (43%), Positives = 246/393 (61%), Gaps = 11/393 (2%)
Query: 2 DLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFSF 61
D++ SI QT AL + + S E N+ FSP SLH LS++ G+A +T D+L +
Sbjct: 8 DVRLSIAHQTRFALRLRSAISSNPERAAGNVAFSPLSLHVALSLITAGAA-ATRDQLVAI 66
Query: 62 LR---FDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTH 116
L LN QV+ VL++ +T ++F N ++ D SL+L SF++L
Sbjct: 67 LGDGGAGDAKELNALAEQVVQFVLANESSTGGPRIAFANGIFVDASLSLKPSFEELAVCQ 126
Query: 117 YKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVW 176
YKA VDF+ K + +VNSWVE+ T GLI ++LP +V TKLI NALYFKG W
Sbjct: 127 YKAKTQSVDFQHKTLEAVGQVNSWVEQVTTGLIKQILPPGSVDNTTKLILGNALYFKGAW 186
Query: 177 KHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFS 236
FD S T D FHLL+G+SI+ FMSS KK Q+I + D VL+L Y +G DK+ +FS
Sbjct: 187 DQKFDESNTKCDSFHLLDGSSIQTQFMSSTKK-QYISSSDNLKVLKLPYAKGHDKR-QFS 244
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M I LP A+DGL SL ++L++E F++ +P+Q V V +F++PKFKI ++ EAS++L+ L
Sbjct: 245 MYILLPGAQDGLWSLAKRLSTEPEFIENHIPKQTVEVGRFQLPKFKISYQFEASSLLRAL 304
Query: 297 GVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSAR 356
G+ PFS+ A+ ++MV S L + + HK+ +EVNEEGTEAG A V + + +
Sbjct: 305 GLQLPFSE-EADLSEMVDSS-QGLEISHVFHKSFVEVNEEGTEAGAATVAMGVAMSMPLK 362
Query: 357 TG-IDFVADHPFLFLIREDLTGTILFIGQVLHP 388
+DFVA+HPFLFLIRED+ G ++F+G V +P
Sbjct: 363 VDLVDFVANHPFLFLIREDIAGVVVFVGHVTNP 395
>UniRef100_Q9SH52 F22C12.22 [Arabidopsis thaliana]
Length = 651
Score = 288 bits (736), Expect = 2e-76
Identities = 169/398 (42%), Positives = 239/398 (59%), Gaps = 33/398 (8%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLD-ELF 59
MD++++++ QT VA+ ++ H+ S ++ N++FSP S+++ +++ A G G + ++
Sbjct: 1 MDVREAMKNQTHVAMILSGHVLSSAP-KDSNVIFSPASINSAITMHAAGPGGDLVSGQIL 59
Query: 60 SFLRFDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTHY 117
SFLR S+D L T F ++ S V +D T ++ N +W DKSL FK L +
Sbjct: 60 SFLRSSSIDELKTVFRELASVVYADRSATGGPKITAANGLWIDKSLPTDPKFKDLFENFF 119
Query: 118 KATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWK 177
KA VDFR++ ++V +EVNSWVE TN LI +LLP +V LT I+ANAL FKG WK
Sbjct: 120 KAVYVPVDFRSEAEEVRKEVNSWVEHHTNNLIKDLLPDGSVTSLTNKIYANALSFKGAWK 179
Query: 178 HTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLR-FS 236
F+ T +DF+L+NGTS+ VPFMSS + Q++R +DGF VLRL Y++G D R FS
Sbjct: 180 RPFEKYYTRDNDFYLVNGTSVSVPFMSS-YENQYVRAYDGFKVLRLPYQRGSDDTNRKFS 238
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M +LP+ KDGL L+EK+AS FL +P + + KFRIPKFKI F ++VL L
Sbjct: 239 MYFYLPDKKDGLDDLLEKMASTPGFLDSHIPTYRDELEKFRIPKFKIEFGFSVTSVLDRL 298
Query: 297 GVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACS--MCDVKL-CV 353
G+ S S++HKA +E++EEG EA A + C L V
Sbjct: 299 GLRS----------------------MSMYHKACVEIDEEGAEAAAATADGDCGCSLDFV 336
Query: 354 SARTGIDFVADHPFLFLIREDLTGTILFIGQVLHPEGA 391
IDFVADHPFLFLIRE+ TGT+LF+ + EGA
Sbjct: 337 EPPKKIDFVADHPFLFLIREEKTGTVLFV--EMEDEGA 372
Score = 156 bits (395), Expect = 9e-37
Identities = 124/392 (31%), Positives = 177/392 (44%), Gaps = 115/392 (29%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDELFS 60
M+ + +++ Q EVA+ ++ HLFS N VFSP S+ A+ ++MA G
Sbjct: 367 MEDEGAMKKQNEVAMILSWHLFSTVAKHSNN-VFSPASITAVFTMMASGPG--------- 416
Query: 61 FLRFDSVDHLNTFFSQVLSTVLSDTTPSFLLSFVNEMWADKSLALSHSFKQLMTTHYKAT 120
S+++SD SFL S S+ +S +++TT +
Sbjct: 417 ------------------SSLISDQILSFLGS--------SSIDELNSVFRVITTVFADA 450
Query: 121 LALVD----FRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVW 176
D F K ++V VNSW +
Sbjct: 451 FGFGDECFYFIFKAEEVPINVNSWASRH-------------------------------- 478
Query: 177 KHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFS 236
T++ V MSS K Q+I +DGF VL+L ++QG D FS
Sbjct: 479 -------------------TNVSVSLMSSYKD-QYIEAYDGFKVLKLPFRQGNDTSRNFS 518
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M +LP+ KDGL +L+EK+AS FL +P QKV+V +F IPKFKI F AS L
Sbjct: 519 MHFYLPDEKDGLDNLVEKMASSVGFLDSHIPSQKVKVGEFGIPKFKIEFGFSASRAFNRL 578
Query: 297 GVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSAR 356
G LDE+ +++ KA +E++EEG EA A ++ C +
Sbjct: 579 G-------------------LDEM---ALYQKACVEIDEEGAEAIAATAVVGGFGCAFVK 616
Query: 357 TGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
IDFVADHPFLF+IRED TGT+LF+GQ+ P
Sbjct: 617 R-IDFVADHPFLFMIREDKTGTVLFVGQIFDP 647
>UniRef100_Q9ZQR6 Putative serpin protein [Arabidopsis thaliana]
Length = 407
Score = 283 bits (724), Expect = 6e-75
Identities = 171/401 (42%), Positives = 237/401 (58%), Gaps = 31/401 (7%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGSTLDE-LF 59
+D+Q++++ Q EV+L + + S + N VFSP S++A+L+V A + TL +
Sbjct: 29 IDMQEAMKNQNEVSLLLVGKVISAVA-KNSNCVFSPASINAVLTVTAANTDNKTLRSFIL 87
Query: 60 SFLRFDSVDHLNTFFSQVLSTVLSD--TTPSFLLSFVNEMWADKSLALSHSFKQLMTTHY 117
SFL+ S + N F ++ S V D T ++ VN +W ++SL+ + ++ L +
Sbjct: 88 SFLKSSSTEETNAIFHELASVVFKDGSETGGPKIAAVNGVWMEQSLSCNPDWEDLFLNFF 147
Query: 118 KATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKLTKLIFANALYFKGVWK 177
KA+ A VDFR K ++V +VN+W + TN LI E+LP +V LT I+ NALYFKG W+
Sbjct: 148 KASFAKVDFRHKAEEVRLDVNTWASRHTNDLIKEILPRGSVTSLTNWIYGNALYFKGAWE 207
Query: 178 HTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLR-FS 236
FD SMT FHLLNG S+ VPFM S +K QFI +DGF VLRL Y+QGRD R FS
Sbjct: 208 KAFDKSMTRDKPFHLLNGKSVSVPFMRSYEK-QFIEAYDGFKVLRLPYRQGRDDTNREFS 266
Query: 237 MCIFLPNAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKEL 296
M ++LP+ K L +L+E++ S FL +P +V V FRIPKFKI F EAS+V +
Sbjct: 267 MYLYLPDKKGELDNLLERITSNPGFLDSHIPEYRVDVGDFRIPKFKIEFGFEASSVFNDF 326
Query: 297 GVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVK-LCV-S 354
EL V S+H KA IE++EEGTEA A ++ V C+
Sbjct: 327 ----------------------ELNV-SLHQKALIEIDEEGTEAAAATTVVVVTGSCLWE 363
Query: 355 ARTGIDFVADHPFLFLIREDLTGTILFIGQVLHPEGAAMSL 395
+ IDFVADHPFLFLIRED TGT+LF GQ+ P + +L
Sbjct: 364 PKKKIDFVADHPFLFLIREDKTGTLLFAGQIFDPSELSSAL 404
>UniRef100_O82283 Putative serpin [Arabidopsis thaliana]
Length = 374
Score = 280 bits (715), Expect = 7e-74
Identities = 164/395 (41%), Positives = 230/395 (57%), Gaps = 45/395 (11%)
Query: 1 MDLQKSIRCQTEVALSITKHLFSKEEYQEKNLVFSPFSLHAILSVMAVGSAGST--LDEL 58
MDL++S+ Q ++ L +T L ++ ILS++A S G T D++
Sbjct: 15 MDLKESVGNQNDIVLRLTAPL-----------------INVILSIIAASSPGDTDTADKI 57
Query: 59 FSFLRFDSVDHLNTFFSQVLSTVLSDTTPSF--LLSFVNEMWADKSLALSHSFKQLMTTH 116
S L+ S D L+ S++++TVL+D+T S +S N +W +K+L + SFK L+
Sbjct: 58 VSLLQASSTDKLHAVSSEIVTTVLADSTASGGPTISAANGLWIEKTLNVEPSFKDLLLNS 117
Query: 117 YKATLALVDFRTKGDQVCREVNSWVEKETNGLITELLPTNAVH-KLTKLIFANALYFKGV 175
YKA VDFRTK D+V REVNSWVEK+TNGLIT LLP+N LT IFANAL+F G
Sbjct: 118 YKAAFNRVDFRTKADEVNREVNSWVEKQTNGLITNLLPSNPKSAPLTDHIFANALFFNGR 177
Query: 176 WKHTFDPSMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRF 235
W FDPS+T DFHLL+GT + VPFM+ ++ ++GF V+ L Y++GR+ F
Sbjct: 178 WDSQFDPSLTKDSDFHLLDGTKVRVPFMTG-ASCRYTHVYEGFKVINLQYRRGREDSRSF 236
Query: 236 SMCIFLPNAKDGLPSLIEKLASESCFLKGK--LPRQKVRVRKFRIPKFKICFELEASNVL 293
SM I+LP+ KDGLPS++E+LAS FLK LP +++ +IP+FK F EAS L
Sbjct: 237 SMQIYLPDEKDGLPSMLERLASTRGFLKDNEVLPSHSAVIKELKIPRFKFDFAFEASEAL 296
Query: 294 KELGVVSPFSKSNANFTKMVVSPLDELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCV 353
K G+V P S I HK+ IEV+E G++A A + +
Sbjct: 297 KGFGLVVPLS--------------------MIMHKSCIEVDEVGSKAAAAAAFRGIGCRR 336
Query: 354 SARTGIDFVADHPFLFLIREDLTGTILFIGQVLHP 388
DFVADHPFLF+++E +G +LF+GQV+ P
Sbjct: 337 PPPEKHDFVADHPFLFIVKEYRSGLVLFLGQVMDP 371
>UniRef100_Q5PP54 At2g25240 [Arabidopsis thaliana]
Length = 324
Score = 277 bits (709), Expect = 3e-73
Identities = 159/328 (48%), Positives = 210/328 (63%), Gaps = 15/328 (4%)
Query: 66 SVDHLNTFFSQVLSTVLSDTTPSFL-LSFVNEMWADKSLALSHSFKQLMTTHYKATLALV 124
S DHLN +Q++ T S L LS N +W DK +L SFK L+ YKAT + V
Sbjct: 4 STDHLNLVLAQIID---GGTEKSDLRLSIANGVWIDKFFSLKLSFKDLLENSYKATCSQV 60
Query: 125 DFRTKGDQVCREVNSWVEKETNGLITELLPTNAVHKL--TKLIFANALYFKGVWKHTFDP 182
DF +K +V EVN+W E TNGLI ++L +++ + + L+ ANA+YFKG W FD
Sbjct: 61 DFASKPSEVIDEVNTWAEVHTNGLIKQILSRDSIDTIRSSTLVLANAVYFKGAWSSKFDA 120
Query: 183 SMTFVDDFHLLNGTSIEVPFMSSEKKTQFIRTFDGFNVLRLSYKQGRDKKLRFSMCIFLP 242
+MT +DFHLL+GTS++VPFM++ + Q++R++DGF VLRL Y + + + FSM I+LP
Sbjct: 121 NMTKKNDFHLLDGTSVKVPFMTNYED-QYLRSYDGFKVLRLPYIEDQRQ---FSMYIYLP 176
Query: 243 NAKDGLPSLIEKLASESCFLKGKLPRQKVRVRKFRIPKFKICFELEASNVLKELGVVSPF 302
N K+GL L+EK+ SE F +P + V FRIPKFK FE AS VLK++G+ SPF
Sbjct: 177 NDKEGLAPLLEKIGSEPSFFDNHIPLHCISVGAFRIPKFKFSFEFNASEVLKDMGLTSPF 236
Query: 303 SKSNANFTKMVVSPL--DELCVESIHHKASIEVNEEGTEAGVACSMCDVKLCVSARTGID 360
+ + T+MV SP D+L V SI HKA IEV+EEGTEA A V C S R D
Sbjct: 237 N-NGGGLTEMVDSPSNGDDLYVSSILHKACIEVDEEGTEA--AAVSVGVVSCTSFRRNPD 293
Query: 361 FVADHPFLFLIREDLTGTILFIGQVLHP 388
FVAD PFLF +RED +G ILF+GQVL P
Sbjct: 294 FVADRPFLFTVREDKSGVILFMGQVLDP 321
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.136 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 634,066,588
Number of Sequences: 2790947
Number of extensions: 25614314
Number of successful extensions: 69599
Number of sequences better than 10.0: 1006
Number of HSP's better than 10.0 without gapping: 786
Number of HSP's successfully gapped in prelim test: 220
Number of HSP's that attempted gapping in prelim test: 65612
Number of HSP's gapped (non-prelim): 1128
length of query: 402
length of database: 848,049,833
effective HSP length: 130
effective length of query: 272
effective length of database: 485,226,723
effective search space: 131981668656
effective search space used: 131981668656
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0175.7


