

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0174.1
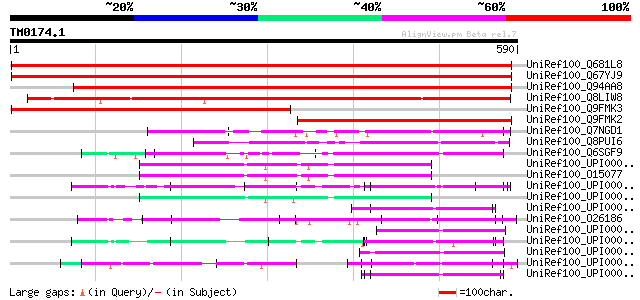
(590 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q681L8 Hypothetical protein At5g63200 [Arabidopsis tha... 777 0.0
UniRef100_Q67YJ9 Hypothetical protein At5g63200 [Arabidopsis tha... 774 0.0
UniRef100_Q94AA8 AT5g63200/MDC12_17 [Arabidopsis thaliana] 709 0.0
UniRef100_Q8LIW8 P0497A05.3 protein [Oryza sativa] 640 0.0
UniRef100_Q9FMK3 Arabidopsis thaliana genomic DNA, chromosome 5,... 416 e-114
UniRef100_Q9FMK2 Arabidopsis thaliana genomic DNA, chromosome 5,... 346 1e-93
UniRef100_Q7NGD1 Glr3240 protein [Gloeobacter violaceus] 94 1e-17
UniRef100_Q8PUI6 O-linked N-acetylglucosamine transferase [Metha... 87 2e-15
UniRef100_Q6SGF9 TPR domain/sulfotransferase domain protein [unc... 83 2e-14
UniRef100_UPI000036D04B UPI000036D04B UniRef100 entry 81 8e-14
UniRef100_O15077 KIAA0372 protein [Homo sapiens] 81 8e-14
UniRef100_UPI00002E2395 UPI00002E2395 UniRef100 entry 75 4e-12
UniRef100_UPI00001CECA3 UPI00001CECA3 UniRef100 entry 74 9e-12
UniRef100_UPI000026B4BA UPI000026B4BA UniRef100 entry 73 2e-11
UniRef100_O26186 O-linked GlcNAc transferase [Methanobacterium t... 72 5e-11
UniRef100_UPI00002E634F UPI00002E634F UniRef100 entry 71 1e-10
UniRef100_UPI00003165D7 UPI00003165D7 UniRef100 entry 70 1e-10
UniRef100_UPI0000328B13 UPI0000328B13 UniRef100 entry 70 2e-10
UniRef100_UPI000033283A UPI000033283A UniRef100 entry 69 4e-10
UniRef100_UPI000031E2B7 UPI000031E2B7 UniRef100 entry 69 4e-10
>UniRef100_Q681L8 Hypothetical protein At5g63200 [Arabidopsis thaliana]
Length = 649
Score = 777 bits (2006), Expect = 0.0
Identities = 389/583 (66%), Positives = 476/583 (80%), Gaps = 1/583 (0%)
Query: 3 RLTNDENSQDKSLLS-KDTDSTEGEGKKSHKLGKCRSRPSKTDSLDCGGDADVDQHVQGA 61
RL+N+E+ Q+ +L+ K+ + E E KK K+GKCRSR S DCG DAD D QG
Sbjct: 55 RLSNEESHQEGGILTCKEVEPGEVEAKKISKVGKCRSRSKIESSSDCGVDADGDLANQGV 114
Query: 62 PSSREEKVSSMKTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPE 121
P+SREEK+S++K GLIHVARKMPKNAHAHFILGLM QRL Q QKAI YEKAEEILL E
Sbjct: 115 PASREEKISNLKMGLIHVARKMPKNAHAHFILGLMFQRLGQSQKAIPEYEKAEEILLGCE 174
Query: 122 TEIERPDLLSLVQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVW 181
EI RP+LL LVQIHH QCL+L+ + S KELE EL+EILSKLK+S++ D+RQAAVW
Sbjct: 175 PEIARPELLLLVQIHHGQCLLLDGFGDTDSVKELEGEELEEILSKLKDSIKLDVRQAAVW 234
Query: 182 NTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILK 241
NTLG +LLK GR+ SAISVLSSLLA+ P+NYDCL NLG+AYLQ G++ELSAKCFQ+L+LK
Sbjct: 235 NTLGLMLLKAGRLMSAISVLSSLLALVPDNYDCLANLGVAYLQSGDMELSAKCFQDLVLK 294
Query: 242 DQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSA 301
D NHP AL+NYAA LLCK++S VAGAGA+ A DQ NVAKECLLAA+++D KSA
Sbjct: 295 DHNHPAALINYAAELLCKHSSTVAGAGANGGADASEDQKAPMNVAKECLLAALRSDPKSA 354
Query: 302 HIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSF 361
H W NLA ++ + GDHRSSSKCLEKAAKL+PNCM+TR+AVA R+K+AERSQD S+ LS+
Sbjct: 355 HAWVNLANSYYMMGDHRSSSKCLEKAAKLDPNCMATRFAVAVQRIKDAERSQDASDQLSW 414
Query: 362 GGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQ 421
GNEMAS+IR+G+S ++ P AWAGLAM HKAQHEI++A+ ++++ LTEMEERAV SLKQ
Sbjct: 415 AGNEMASVIREGESVPIDPPIAWAGLAMAHKAQHEIAAAFVADRNELTEMEERAVYSLKQ 474
Query: 422 AVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPS 481
AV EDP+D VRWHQLG+HSLC+QQ+K SQKYLKAAV R CSY WSNLG+SLQLS+E S
Sbjct: 475 AVTEDPEDAVRWHQLGLHSLCSQQYKLSQKYLKAAVGRSRECSYAWSNLGISLQLSDEHS 534
Query: 482 QAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLG 541
+AE+ YK+AL ++ + QAHAILSNLG YR +K+Y+ +KAMF+K+LEL+PGYAPA+NNLG
Sbjct: 535 EAEEVYKRALTVSKEDQAHAILSNLGNLYRQKKQYEVSKAMFSKALELKPGYAPAYNNLG 594
Query: 542 LVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKIC 584
LVFVAE EEAK CFEK+L++D LLDAA+SNL+K TMS++C
Sbjct: 595 LVFVAERRWEEAKSCFEKSLEADSLLDAAQSNLLKATTMSRLC 637
>UniRef100_Q67YJ9 Hypothetical protein At5g63200 [Arabidopsis thaliana]
Length = 649
Score = 774 bits (1998), Expect = 0.0
Identities = 388/583 (66%), Positives = 475/583 (80%), Gaps = 1/583 (0%)
Query: 3 RLTNDENSQDKSLLS-KDTDSTEGEGKKSHKLGKCRSRPSKTDSLDCGGDADVDQHVQGA 61
RL+N+E+ Q+ +L+ K+ + E E KK K+GKCRSR S DCG DAD D QG
Sbjct: 55 RLSNEESHQEGGILTCKEVEPGEVEAKKISKVGKCRSRSKIESSSDCGVDADGDLANQGV 114
Query: 62 PSSREEKVSSMKTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPE 121
P+SREEK+S++K GLIHVARKMPKNAHAHFILGLM QRL Q QKAI YEKAEEILL E
Sbjct: 115 PASREEKISNLKMGLIHVARKMPKNAHAHFILGLMFQRLGQSQKAIPEYEKAEEILLGCE 174
Query: 122 TEIERPDLLSLVQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVW 181
EI RP+LL LVQIHH QCL+L+ + S KELE EL+EILSKLK+S++ D+RQAAVW
Sbjct: 175 PEIARPELLLLVQIHHGQCLLLDGFGDTDSVKELEGEELEEILSKLKDSIKLDVRQAAVW 234
Query: 182 NTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILK 241
NTLG +LLK G + SAISVLSSLLA+ P+NYDCL NLG+AYLQ G++ELSAKCFQ+L+LK
Sbjct: 235 NTLGLMLLKAGCLMSAISVLSSLLALVPDNYDCLANLGVAYLQSGDMELSAKCFQDLVLK 294
Query: 242 DQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSA 301
D NHP AL+NYAA LLCK++S VAGAGA+ A DQ NVAKECLLAA+++D KSA
Sbjct: 295 DHNHPAALINYAAELLCKHSSTVAGAGANGGADASEDQKAPMNVAKECLLAALRSDPKSA 354
Query: 302 HIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSF 361
H W NLA ++ + GDHRSSSKCLEKAAKL+PNCM+TR+AVA R+K+AERSQD S+ LS+
Sbjct: 355 HAWVNLANSYYMMGDHRSSSKCLEKAAKLDPNCMATRFAVAVQRIKDAERSQDASDQLSW 414
Query: 362 GGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQ 421
GNEMAS+IR+G+S ++ P AWAGLAM HKAQHEI++A+ ++++ LTEMEERAV SLKQ
Sbjct: 415 AGNEMASVIREGESVPIDPPIAWAGLAMAHKAQHEIAAAFVADRNELTEMEERAVYSLKQ 474
Query: 422 AVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPS 481
AV EDP+D VRWHQLG+HSLC+QQ+K SQKYLKAAV R CSY WSNLG+SLQLS+E S
Sbjct: 475 AVTEDPEDAVRWHQLGLHSLCSQQYKLSQKYLKAAVGRSRECSYAWSNLGISLQLSDEHS 534
Query: 482 QAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLG 541
+AE+ YK+AL ++ + QAHAILSNLG YR +K+Y+ +KAMF+K+LEL+PGYAPA+NNLG
Sbjct: 535 EAEEVYKRALTVSKEDQAHAILSNLGNLYRQKKQYEVSKAMFSKALELKPGYAPAYNNLG 594
Query: 542 LVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSKIC 584
LVFVAE EEAK CFEK+L++D LLDAA+SNL+K TMS++C
Sbjct: 595 LVFVAERRWEEAKSCFEKSLEADSLLDAAQSNLLKATTMSRLC 637
>UniRef100_Q94AA8 AT5g63200/MDC12_17 [Arabidopsis thaliana]
Length = 523
Score = 709 bits (1831), Expect = 0.0
Identities = 352/510 (69%), Positives = 426/510 (83%)
Query: 75 GLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQ 134
GLIHVARKMPKNAHAHFILGLM QRL Q QKAI YEKAEEILL E EI RP+LL LVQ
Sbjct: 2 GLIHVARKMPKNAHAHFILGLMFQRLGQSQKAIPEYEKAEEILLGCEPEIARPELLLLVQ 61
Query: 135 IHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRV 194
IHH QCL+L+ + S KELE EL+EILSKLK+S++ D+RQAAVWNTLG +LLK G +
Sbjct: 62 IHHGQCLLLDGFGDTDSVKELEGEELEEILSKLKDSIKLDVRQAAVWNTLGLMLLKAGCL 121
Query: 195 QSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAA 254
SAISVLSSLLA+ P+NYDCL NLG+AYLQ G++ELSAKCFQ+L+LKD NHP AL+NYAA
Sbjct: 122 MSAISVLSSLLALVPDNYDCLANLGVAYLQSGDMELSAKCFQDLVLKDHNHPAALINYAA 181
Query: 255 LLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSIS 314
LLCK++S VAGAGA+ A DQ NVAKECLLAA+++D KSAH W NLA ++ +
Sbjct: 182 ELLCKHSSTVAGAGANGGADASEDQKAPMNVAKECLLAALRSDPKSAHAWVNLANSYYMM 241
Query: 315 GDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGD 374
GDHRSSSKCLEKAAKL+PNCM+TR+AVA R+K+AERSQD S+ LS+ GNEMAS+IR+G+
Sbjct: 242 GDHRSSSKCLEKAAKLDPNCMATRFAVAVQRIKDAERSQDASDQLSWAGNEMASVIREGE 301
Query: 375 SSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWH 434
S ++ P AWAGLAM HKAQHEI++A+ ++++ LTEMEERAV SLKQAV EDP+D VRWH
Sbjct: 302 SVPIDPPIAWAGLAMAHKAQHEIAAAFVADRNELTEMEERAVYSLKQAVTEDPEDAVRWH 361
Query: 435 QLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLA 494
QLG+HSLC+QQ+K SQKYLKAAV R CSY WSNLG+SLQLS+E S+AE+ YK+AL ++
Sbjct: 362 QLGLHSLCSQQYKLSQKYLKAAVGRSRECSYAWSNLGISLQLSDEHSEAEEVYKRALTVS 421
Query: 495 TKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAK 554
+ QAHAILSNLG YR +K+Y+ +KAMF+K+LEL+PGYAPA+NNLGLVFVAE EEAK
Sbjct: 422 KEDQAHAILSNLGNLYRQKKQYEVSKAMFSKALELKPGYAPAYNNLGLVFVAERRWEEAK 481
Query: 555 YCFEKALQSDPLLDAAKSNLVKVVTMSKIC 584
CFEK+L++D LLDAA+SNL+K TMS++C
Sbjct: 482 SCFEKSLEADSLLDAAQSNLLKATTMSRLC 511
>UniRef100_Q8LIW8 P0497A05.3 protein [Oryza sativa]
Length = 606
Score = 640 bits (1650), Expect = 0.0
Identities = 330/569 (57%), Positives = 427/569 (74%), Gaps = 12/569 (2%)
Query: 21 DSTEGEGKKSHKLGKCRSRPSKTDSLDCGGDADVDQHVQGAPSSREEKVSSMKTGLIHVA 80
DS+ L K + D+++C D DQH QGA +REEKVS++K L+HVA
Sbjct: 41 DSSTRSSNDEGSLAKSVITTKEPDTVEC---EDADQHCQGASVAREEKVSNLKAALVHVA 97
Query: 81 RKMPKNAHAHFILGLMHQRLNQPQ----KAILVYEKAEEILLRPETEIERPDLLSLVQIH 136
RKMPKNAHAHF+LGLM+QRL Q +AI YEK+ EILL+ E E+ RPDLLS V+IH
Sbjct: 98 RKMPKNAHAHFMLGLMYQRLAQLAAEIFQAIAAYEKSSEILLQDEEEVRRPDLLSSVRIH 157
Query: 137 HAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQS 196
HAQC IL++S ++ D+ELE EL EIL K+K SV+ D RQAAVWN LG +LL++G++QS
Sbjct: 158 HAQC-ILQTSMGDTFDEELESGELDEILVKMKSSVESDPRQAAVWNILGLVLLRSGQLQS 216
Query: 197 AISVLSSLLAIAPENYDCLGNLGIAYLQI---GNLELSAKCFQELILKDQNHPVALVNYA 253
AISVLSSL +AP+ D L NLG+AY+Q GNLEL+ KCFQEL++KDQNHP ALVNYA
Sbjct: 217 AISVLSSLTVVAPDYLDSLANLGVAYIQRFERGNLELATKCFQELVIKDQNHPAALVNYA 276
Query: 254 ALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSI 313
ALLLCKY S AG+G + S G+ Q VAKECLLAA+KAD K+A +W NLA A+ +
Sbjct: 277 ALLLCKYGSFAAGSGGNVSAGSCLHQKEGLAVAKECLLAAVKADPKAASVWVNLANAYYM 336
Query: 314 SGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDG 373
+G+HR+S +CLE+AAK EP+ M RYA+A HR+++A RSQ + L + NEMA+++++G
Sbjct: 337 AGEHRNSKRCLEQAAKHEPSHMPARYAIAVHRIRDAVRSQCSDDQLLWASNEMATVLKEG 396
Query: 374 DSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRW 433
D S V+ P AWAGLAM H+AQHEI++AY++EQ L+++EERA+ +LKQA+ EDPDD V+W
Sbjct: 397 DPSAVDAPIAWAGLAMAHRAQHEIAAAYDTEQINLSDVEERALYTLKQAIQEDPDDAVQW 456
Query: 434 HQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLL 493
HQLG++++CT QF S +LKAAVA CSY WSNLG++LQLS++ S E YK+AL+L
Sbjct: 457 HQLGLYNICTTQFSRSVNFLKAAVARSPDCSYVWSNLGIALQLSDD-SSCETVYKRALIL 515
Query: 494 ATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEA 553
++ QQ++AILSNLGI YR +Y+ A+ M +SLEL PG+APA NNLGLV +AEG EEA
Sbjct: 516 SSSQQSYAILSNLGILYRQHGRYELARRMLLRSLELCPGHAPANNNLGLVSIAEGRYEEA 575
Query: 554 KYCFEKALQSDPLLDAAKSNLVKVVTMSK 582
CFEK+LQSDPLLDAAKSNL KV+ +SK
Sbjct: 576 ISCFEKSLQSDPLLDAAKSNLAKVLALSK 604
>UniRef100_Q9FMK3 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MDC12
[Arabidopsis thaliana]
Length = 404
Score = 416 bits (1069), Expect = e-114
Identities = 213/325 (65%), Positives = 254/325 (77%), Gaps = 1/325 (0%)
Query: 3 RLTNDENSQDKSLLS-KDTDSTEGEGKKSHKLGKCRSRPSKTDSLDCGGDADVDQHVQGA 61
RL+N+E+ Q+ +L+ K+ + E E KK K+GKCRSR S DCG DAD D QG
Sbjct: 55 RLSNEESHQEGGILTCKEVEPGEVEAKKISKVGKCRSRSKIESSSDCGVDADGDLANQGV 114
Query: 62 PSSREEKVSSMKTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPE 121
P+SREEK+S++K GLIHVARKMPKNAHAHFILGLM QRL Q QKAI YEKAEEILL E
Sbjct: 115 PASREEKISNLKMGLIHVARKMPKNAHAHFILGLMFQRLGQSQKAIPEYEKAEEILLGCE 174
Query: 122 TEIERPDLLSLVQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVW 181
EI RP+LL LVQIHH QCL+L+ + S KELE EL+EILSKLK+S++ D+RQAAVW
Sbjct: 175 PEIARPELLLLVQIHHGQCLLLDGFGDTDSVKELEGEELEEILSKLKDSIKLDVRQAAVW 234
Query: 182 NTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILK 241
NTLG +LLK G + SAISVLSSLLA+ P+NYDCL NLG+AYLQ G++ELSAKCFQ+L+LK
Sbjct: 235 NTLGLMLLKAGCLMSAISVLSSLLALVPDNYDCLANLGVAYLQSGDMELSAKCFQDLVLK 294
Query: 242 DQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSA 301
D NHP AL+NYAA LLCK++S VAGAGA+ A DQ NVAKECLLAA+++D KSA
Sbjct: 295 DHNHPAALINYAAELLCKHSSTVAGAGANGGADASEDQKAPMNVAKECLLAALRSDPKSA 354
Query: 302 HIWGNLAYAFSISGDHRSSSKCLEK 326
H W NLA ++ + GDHRSSSKCLEK
Sbjct: 355 HAWVNLANSYYMMGDHRSSSKCLEK 379
>UniRef100_Q9FMK2 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MDC12
[Arabidopsis thaliana]
Length = 262
Score = 346 bits (888), Expect = 1e-93
Identities = 168/250 (67%), Positives = 213/250 (85%)
Query: 335 MSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQ 394
M+TR+AVA R+K+AERSQD S+ LS+ GNEMAS+IR+G+S ++ P AWAGLAM HKAQ
Sbjct: 1 MATRFAVAVQRIKDAERSQDASDQLSWAGNEMASVIREGESVPIDPPIAWAGLAMAHKAQ 60
Query: 395 HEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLK 454
HEI++A+ ++++ LTEMEERAV SLKQAV EDP+D VRWHQLG+HSLC+QQ+K SQKYLK
Sbjct: 61 HEIAAAFVADRNELTEMEERAVYSLKQAVTEDPEDAVRWHQLGLHSLCSQQYKLSQKYLK 120
Query: 455 AAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEK 514
AAV R CSY WSNLG+SLQLS+E S+AE+ YK+AL ++ + QAHAILSNLG YR +K
Sbjct: 121 AAVGRSRECSYAWSNLGISLQLSDEHSEAEEVYKRALTVSKEDQAHAILSNLGNLYRQKK 180
Query: 515 KYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
+Y+ +KAMF+K+LEL+PGYAPA+NNLGLVFVAE EEAK CFEK+L++D LLDAA+SNL
Sbjct: 181 QYEVSKAMFSKALELKPGYAPAYNNLGLVFVAERRWEEAKSCFEKSLEADSLLDAAQSNL 240
Query: 575 VKVVTMSKIC 584
+K TMS++C
Sbjct: 241 LKATTMSRLC 250
>UniRef100_Q7NGD1 Glr3240 protein [Gloeobacter violaceus]
Length = 433
Score = 93.6 bits (231), Expect = 1e-17
Identities = 100/426 (23%), Positives = 179/426 (41%), Gaps = 54/426 (12%)
Query: 161 KEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGI 220
K + +++ +++ R+ +WN LG L + GR AIS + + + D NL +
Sbjct: 58 KALEEQVRAALKAQPREPRLWNNLGVSLRRQGRNAEAISAYRQAVRLDSKYADAYSNLAV 117
Query: 221 AYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQV 280
A IG + + + + D+++PV +N A+V G G E A+ +
Sbjct: 118 ALTAIGRHDEALGAIRTALKLDRDNPVYRLN--------LATVYEGLGRD-GEAAIEYR- 167
Query: 281 MAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYA 340
A ++ +SA ++ LA G + + +KA LEP + R A
Sbjct: 168 -----------AYLQRAGESAAVYERLAKIQQKLGRNAQARTSRQKAVDLEPENGAYRLA 216
Query: 341 VATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSA 400
+ L+ G + A+I +L A AM ++ +++
Sbjct: 217 Y--------------GQALAAAGQDFAAI--------AQLEAAVRLKAMDARSWLLLATL 254
Query: 401 YESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACD 460
Y +Q + A +L++A A DP + LGV + + + L AV
Sbjct: 255 YNGQQQT-----DAAAAALREAAALDPGSARVQNDLGVTLSKLKLTDQAAEALNRAVTLA 309
Query: 461 RGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAK 520
+ WSNLGV L+ + ++AE+AY++A L + + I +NLG+ Y ++ Q A
Sbjct: 310 PSYAEAWSNLGVVLRGQGKMNEAEQAYQRA--LGIESRLPQIYNNLGVLYLESERVQEAV 367
Query: 521 AMFTKSLELQPGYAPAFNNLGLVFVAEG----LLEEAKYCFEKALQSDPLLDAAKSNLVK 576
F ++L L+PGY A NL + G ++ K E A L+ +++L K
Sbjct: 368 PQFQQALVLEPGYWEARRNLAMALGRLGKWGAAADQLKLALEAAQVPSALITQVQADLQK 427
Query: 577 VVTMSK 582
+ S+
Sbjct: 428 LQEASR 433
Score = 62.4 bits (150), Expect = 4e-08
Identities = 81/362 (22%), Positives = 139/362 (38%), Gaps = 48/362 (13%)
Query: 255 LLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSIS 314
L+LC A + A A A+ G V +E + AA+KA + +W NL +
Sbjct: 32 LVLCLAAGLFAAEIARAAPGPA---VPPPKALEEQVRAALKAQPREPRLWNNLGVSLRRQ 88
Query: 315 GDHRSSSKCLEKAAKLE-------PNCMSTRYAVATH-------RMKEAERSQDPSELLS 360
G + + +A +L+ N A+ H R +P L+
Sbjct: 89 GRNAEAISAYRQAVRLDSKYADAYSNLAVALTAIGRHDEALGAIRTALKLDRDNPVYRLN 148
Query: 361 FGGNEMASIIRDGDSSLV---------ELPTAWAGLAMVHKAQHEISSAYESEQHVLTEM 411
+ RDG++++ E + LA + + + A S Q +
Sbjct: 149 L-ATVYEGLGRDGEAAIEYRAYLQRAGESAAVYERLAKIQQKLGRNAQARTSRQKAVDLE 207
Query: 412 EER-------------------AVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKY 452
E A+ L+ AV D W L QQ +
Sbjct: 208 PENGAYRLAYGQALAAAGQDFAAIAQLEAAVRLKAMDARSWLLLATLYNGQQQTDAAAAA 267
Query: 453 LKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRH 512
L+ A A D G + ++LGV+L + QA +A +A+ LA SNLG+ R
Sbjct: 268 LREAAALDPGSARVQNDLGVTLSKLKLTDQAAEALNRAVTLAPSYA--EAWSNLGVVLRG 325
Query: 513 EKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKS 572
+ K A+ + ++L ++ +NNLG++++ ++EA F++AL +P A+
Sbjct: 326 QGKMNEAEQAYQRALGIESRLPQIYNNLGVLYLESERVQEAVPQFQQALVLEPGYWEARR 385
Query: 573 NL 574
NL
Sbjct: 386 NL 387
>UniRef100_Q8PUI6 O-linked N-acetylglucosamine transferase [Methanosarcina mazei]
Length = 412
Score = 86.7 bits (213), Expect = 2e-15
Identities = 86/369 (23%), Positives = 158/369 (42%), Gaps = 56/369 (15%)
Query: 215 LGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEG 274
L G+ L++G + F++ I KD + L N AA L
Sbjct: 68 LNECGLDLLRLGKYNEAIIAFEKAIDKDPGNIYLLNNKAAAL------------------ 109
Query: 275 ALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNC 334
+ + A + A+K + + A +W N+A+++S G++ + + KA L+P+
Sbjct: 110 ---ESLGRFEEALKLYQEAVKINSEDADLWNNMAFSYSQIGEYEKAVEAYGKALDLKPDY 166
Query: 335 MSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQ 394
+ Y A + + +A R ++ + I+ +S+ E AWAG +
Sbjct: 167 PNAWYGKALN-LSQAGRYEEAVDAYD--------IVLKENSNYKE---AWAGKGIA---- 210
Query: 395 HEISSAYESEQHVLTEME--ERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKY 452
L +M + A+ + +A+ DP+ W+ GV FK + K
Sbjct: 211 -------------LGQMGNYDEAIIAYDKALEIDPEFLEAWYYKGVDLDSLGSFKQALKA 257
Query: 453 LKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRH 512
+ AV D W+N+G+ L+ E +A A+++A+ + ++ + N G
Sbjct: 258 YEKAVEIDPENDDAWNNMGIDLENLERYDEAINAFEKAIEINSENSD--VWYNKGFTLSQ 315
Query: 513 EKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKS 572
+++ A + K+++L P Y A+++LG V EEA +EKAL+ DP +AA S
Sbjct: 316 VQRFDEAVEAYRKAVQLDPEYLEAYSSLGFVLAQLKRFEEALDIYEKALKLDP--EAADS 373
Query: 573 NLVKVVTMS 581
K V +S
Sbjct: 374 WFGKAVCLS 382
Score = 38.9 bits (89), Expect = 0.43
Identities = 39/175 (22%), Positives = 77/175 (43%), Gaps = 15/175 (8%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETE-------IERPDLLSLVQIH 136
P+ A + G+ L ++A+ YEKA EI PE + I+ +L +
Sbjct: 232 PEFLEAWYYKGVDLDSLGSFKQALKAYEKAVEI--DPENDDAWNNMGIDLENLERYDEAI 289
Query: 137 HAQCLILESSSENSS---DKELEPHELK---EILSKLKESVQFDIRQAAVWNTLGFILLK 190
+A +E +SENS +K +++ E + +++VQ D +++LGF+L +
Sbjct: 290 NAFEKAIEINSENSDVWYNKGFTLSQVQRFDEAVEAYRKAVQLDPEYLEAYSSLGFVLAQ 349
Query: 191 TGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNH 245
R + A+ + L + PE D + +G E + +++ + D +
Sbjct: 350 LKRFEEALDIYEKALKLDPEAADSWFGKAVCLSYLGREEEAEDAYRKAVEIDPRY 404
Score = 35.8 bits (81), Expect = 3.6
Identities = 34/164 (20%), Positives = 67/164 (40%), Gaps = 19/164 (11%)
Query: 94 GLMHQRLNQPQKAILVYEKAEEILLRPE-------TEIERPDLLSLVQIHHAQCLILESS 146
G+ ++ +AI+ Y+KA EI PE ++ L S Q A +E
Sbjct: 208 GIALGQMGNYDEAIIAYDKALEI--DPEFLEAWYYKGVDLDSLGSFKQALKAYEKAVEID 265
Query: 147 SENSSD--------KELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAI 198
EN + LE ++ E ++ +++++ + + VW GF L + R A+
Sbjct: 266 PENDDAWNNMGIDLENLERYD--EAINAFEKAIEINSENSDVWYNKGFTLSQVQRFDEAV 323
Query: 199 SVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKD 242
+ + PE + +LG Q+ E + +++ + D
Sbjct: 324 EAYRKAVQLDPEYLEAYSSLGFVLAQLKRFEEALDIYEKALKLD 367
>UniRef100_Q6SGF9 TPR domain/sulfotransferase domain protein [uncultured bacterium
560]
Length = 723
Score = 83.2 bits (204), Expect = 2e-14
Identities = 98/419 (23%), Positives = 173/419 (40%), Gaps = 56/419 (13%)
Query: 159 ELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNL 218
++ E + +K + + ++N G G++ A+ LAI P+ + NL
Sbjct: 26 KINEAIDAIKVLTKDFPNEPLLYNISGVCYKTIGQLDVAVKSFEKALAIKPDYTEVNYNL 85
Query: 219 GIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALAD 278
G+ + ++G L+ + KC+++++ + H A N G + E +
Sbjct: 86 GLTFQELGQLDAAVKCYEDVLAVNPEHAEAHNNL---------------GVTLKE---LE 127
Query: 279 QVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTR 338
Q+ AA + E +A IK D AH NL A G + K EKA ++P+ T
Sbjct: 128 QLDAAVKSYEKAIA-IKPDYAEAH--NNLGNALKELGQLDVAVKSYEKAIAIKPDFAETH 184
Query: 339 YAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEIS 398
N + + ++ L +L A Q + S
Sbjct: 185 -------------------------NNLGNALQ----GLGQLDEAVKSYEQAIAIQSDFS 215
Query: 399 SAYESEQHVLTEMEER--AVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAA 456
+AY + +VL E+ + AV S ++A+A PD + LG Q + K + A
Sbjct: 216 NAYYNLGNVLRELGQLDDAVKSYEKAIAIKPDYDEAHNNLGNALQGLGQLDEAVKSYEQA 275
Query: 457 VACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQ-QAHAILSNLGIFYRHEKK 515
+A S + NLG L+ + A ++YK+A+++ +AH +NLGI + +
Sbjct: 276 IAIQSDFSNAYYNLGNVLRELGQVDTAVRSYKKAIVIKPDYAKAH---NNLGIALQDLGQ 332
Query: 516 YQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
A K++ + P +A A +NLG+ G ++ A +EKA+ P A NL
Sbjct: 333 MDTAVKNLEKAIAITPDFAEAHHNLGIALQDLGQIDAAVKGYEKAIAIKPDYAEAYHNL 391
Score = 63.5 bits (153), Expect = 2e-08
Identities = 90/407 (22%), Positives = 158/407 (38%), Gaps = 54/407 (13%)
Query: 169 ESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNL-GIAYLQIGN 227
E++Q + + V + + L +G++ AI + L P N L N+ G+ Y IG
Sbjct: 4 ENIQLALSREQVDSVI--TLYSSGKINEAIDAIKVLTKDFP-NEPLLYNISGVCYKTIGQ 60
Query: 228 LELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAK 287
L+++ K F++ + ++ VNY + G ++ + A
Sbjct: 61 LDVAVKSFEKALAIKPDYTE--VNY-------------------NLGLTFQELGQLDAAV 99
Query: 288 ECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMK 347
+C + + + A NL ++ K EKA ++P+ YA A + +
Sbjct: 100 KCYEDVLAVNPEHAEAHNNLGVTLKELEQLDAAVKSYEKAIAIKPD-----YAEAHNNLG 154
Query: 348 EAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHV 407
A + ++ + +I D + L A GL +
Sbjct: 155 NALKELGQLDVAVKSYEKAIAIKPDFAETHNNLGNALQGLGQL----------------- 197
Query: 408 LTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTW 467
+ AV S +QA+A D ++ LG Q + K + A+A
Sbjct: 198 -----DEAVKSYEQAIAIQSDFSNAYYNLGNVLRELGQLDDAVKSYEKAIAIKPDYDEAH 252
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSL 527
+NLG +LQ + +A K+Y+QA+ A + NLG R + A + K++
Sbjct: 253 NNLGNALQGLGQLDEAVKSYEQAI--AIQSDFSNAYYNLGNVLRELGQVDTAVRSYKKAI 310
Query: 528 ELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
++P YA A NNLG+ G ++ A EKA+ P A NL
Sbjct: 311 VIKPDYAKAHNNLGIALQDLGQMDTAVKNLEKAIAITPDFAEAHHNL 357
Score = 49.3 bits (116), Expect = 3e-04
Identities = 69/302 (22%), Positives = 123/302 (39%), Gaps = 39/302 (12%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPE---------TEIERPDLLSLVQ 134
P++A AH LG+ + L Q A+ YEKA I ++P+ ++ L +
Sbjct: 110 PEHAEAHNNLGVTLKELEQLDAAVKSYEKA--IAIKPDYAEAHNNLGNALKELGQLDVAV 167
Query: 135 IHHAQCLILE---SSSENSSDKELEP-HELKEILSKLKESVQFDIRQAAVWNTLGFILLK 190
+ + + ++ + + N+ L+ +L E + ++++ + + LG +L +
Sbjct: 168 KSYEKAIAIKPDFAETHNNLGNALQGLGQLDEAVKSYEQAIAIQSDFSNAYYNLGNVLRE 227
Query: 191 TGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALV 250
G++ A+ +AI P+ + NLG A +G L+ + K +++ I + A
Sbjct: 228 LGQLDDAVKSYEKAIAIKPDYDEAHNNLGNALQGLGQLDEAVKSYEQAIAIQSDFSNAYY 287
Query: 251 NY-----------AALLLCKYASVVAGAGASASEG---ALADQVMAANVAKECLLA-AIK 295
N A+ K A V+ A A AL D K A AI
Sbjct: 288 NLGNVLRELGQVDTAVRSYKKAIVIKPDYAKAHNNLGIALQDLGQMDTAVKNLEKAIAIT 347
Query: 296 ADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAER--SQ 353
D AH NL A G ++ K EKA ++P+ YA A H + ++ +
Sbjct: 348 PDFAEAH--HNLGIALQDLGQIDAAVKGYEKAIAIKPD-----YAEAYHNLSYLKKYTAN 400
Query: 354 DP 355
DP
Sbjct: 401 DP 402
Score = 47.0 bits (110), Expect = 0.002
Identities = 85/387 (21%), Positives = 146/387 (36%), Gaps = 71/387 (18%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHAQCLIL 143
P ++ LGL Q L Q A+ YE + + + PE HA+
Sbjct: 76 PDYTEVNYNLGLTFQELGQLDAAVKCYE--DVLAVNPE---------------HAEA--- 115
Query: 144 ESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSS 203
++ + KELE +L + ++++ A N LG L + G++ A+
Sbjct: 116 -HNNLGVTLKELE--QLDAAVKSYEKAIAIKPDYAEAHNNLGNALKELGQLDVAVKSYEK 172
Query: 204 LLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASV 263
+AI P+ + NLG A +G L+ + K +++ I + A N +L
Sbjct: 173 AIAIKPDFAETHNNLGNALQGLGQLDEAVKSYEQAIAIQSDFSNAYYNLGNVL------- 225
Query: 264 VAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKC 323
G L D V + A AIK D AH NL A G + K
Sbjct: 226 -------RELGQLDDAVKSYEKA-----IAIKPDYDEAH--NNLGNALQGLGQLDEAVKS 271
Query: 324 LEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTA 383
E+A ++ + + Y + + L G ++ + +R ++V P
Sbjct: 272 YEQAIAIQSDFSNAYYNLG-------------NVLRELG--QVDTAVRSYKKAIVIKPD- 315
Query: 384 WAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCT 443
KA + + A + L +M + AV +L++A+A PD H LG+
Sbjct: 316 ------YAKAHNNLGIALQD----LGQM-DTAVKNLEKAIAITPDFAEAHHNLGIALQDL 364
Query: 444 QQFKTSQKYLKAAVACDRGCSYTWSNL 470
Q + K + A+A + + NL
Sbjct: 365 GQIDAAVKGYEKAIAIKPDYAEAYHNL 391
>UniRef100_UPI000036D04B UPI000036D04B UniRef100 entry
Length = 1322
Score = 81.3 bits (199), Expect = 8e-14
Identities = 89/362 (24%), Positives = 152/362 (41%), Gaps = 33/362 (9%)
Query: 152 DKELEPHELKEILSKLKESVQFDIRQAAVWNTL--GFILLKTGRVQSAISVLSSLLAIAP 209
D +E +++ L+ L Q A W L G LK G+ A++ L + L P
Sbjct: 286 DLSVELEDMEMALAILTTVTQKASAGTAKWAWLRRGLYYLKAGQHSQAVADLQAALRADP 345
Query: 210 ENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAAL--LLCKYASVVAGA 267
++++C +LG AYL G + K F + + ++ AA+ +L KY VA
Sbjct: 346 KDFNCWESLGEAYLSRGGYTTALKSFTKASELNPESTYSVFKVAAIQQILGKYKEAVAQY 405
Query: 268 GASASEGALADQVMAANVAKECLLAAIKA------DVKSAHIWGNLAYAFSISGDHRSSS 321
+ D V A EC L KA D K+ F+ + HR+
Sbjct: 406 QMIIKK--TEDYVPALKGLGECHLMMAKAALVDYLDGKAVDYIEKALEYFTCALQHRADV 463
Query: 322 KCLEKAAKLEPNCMSTRYAVATHRM-----------KEAERSQDPSELLSFGGNEMASII 370
CL K A C+ YAV+ ++ KE ++ +ELL GG +
Sbjct: 464 SCLWKLAGDACTCL---YAVSPSKVNVHVLGVLLGQKEGKQVLKKNELLHLGGRCYGRAL 520
Query: 371 RDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDP 430
+ L+ W L + + Q + + S + L E+ E+++ LK+AV D ++
Sbjct: 521 K-----LMSTSNTWCDLGINYYRQAQHLAETGSNMNDLKELLEKSLHCLKKAVRLDSNNH 575
Query: 431 VRWHQLGVHSLCT--QQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYK 488
+ W+ LGV + + + +Q ++ ++ + W+NLGV +E QA +A+K
Sbjct: 576 LYWNALGVVACYSGIGNYALAQHCFIKSIQSEQINAVAWTNLGVLYLTNENIEQAHEAFK 635
Query: 489 QA 490
A
Sbjct: 636 MA 637
Score = 39.3 bits (90), Expect = 0.33
Identities = 27/104 (25%), Positives = 51/104 (48%), Gaps = 6/104 (5%)
Query: 158 HELKEILSK----LKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYD 213
++LKE+L K LK++V+ D WN LG + +G A++ + +I E +
Sbjct: 551 NDLKELLEKSLHCLKKAVRLDSNNHLYWNALGVVACYSGIGNYALAQHCFIKSIQSEQIN 610
Query: 214 CLG--NLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAAL 255
+ NLG+ YL N+E + + F+ D ++ + + A +
Sbjct: 611 AVAWTNLGVLYLTNENIEQAHEAFKMAQSLDPSYLMCWIGQALI 654
>UniRef100_O15077 KIAA0372 protein [Homo sapiens]
Length = 1568
Score = 81.3 bits (199), Expect = 8e-14
Identities = 90/362 (24%), Positives = 152/362 (41%), Gaps = 33/362 (9%)
Query: 152 DKELEPHELKEILSKLKESVQFDIRQAAVWNTL--GFILLKTGRVQSAISVLSSLLAIAP 209
D +E +++ L+ L Q A W L G LK G+ A++ L + L P
Sbjct: 540 DLSVELEDMEMALAILTTVTQKASAGTAKWAWLRRGLYYLKAGQHSQAVADLQAALRADP 599
Query: 210 ENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAAL--LLCKYASVVAGA 267
++++C +LG AYL G + K F + + ++ AA+ +L KY VA
Sbjct: 600 KDFNCWESLGEAYLSRGGYTTALKSFTKASELNPESIYSVFKVAAIQQILGKYKEAVAQY 659
Query: 268 GASASEGALADQVMAANVAKECLLAAIKA------DVKSAHIWGNLAYAFSISGDHRSSS 321
+ D V A EC L KA D K+ F+ + HR+
Sbjct: 660 QMIIKK--KEDYVPALKGLGECHLMMAKAALVDYLDGKAVDYIEKALEYFTCALQHRADV 717
Query: 322 KCLEKAAKLEPNCMSTRYAVATHRM-----------KEAERSQDPSELLSFGGNEMASII 370
CL K A C+ YAVA ++ KE ++ +ELL GG +
Sbjct: 718 SCLWKLAGDACTCL---YAVAPSKVNVHVLGVLLGQKEGKQVLKKNELLHLGGRCYGRAL 774
Query: 371 RDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDP 430
+ L+ W L + + Q + + S + L E+ E+++ LK+AV D ++
Sbjct: 775 K-----LMSTSNTWCDLGINYYRQAQHLAETGSNMNDLKELLEKSLHCLKKAVRLDSNNH 829
Query: 431 VRWHQLGVHSLCT--QQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYK 488
+ W+ LGV + + + +Q ++ ++ + W+NLGV +E QA +A+K
Sbjct: 830 LYWNALGVVACYSGIGNYALAQHCFIKSIQSEQINAVAWTNLGVLYLTNENIEQAHEAFK 889
Query: 489 QA 490
A
Sbjct: 890 MA 891
Score = 39.3 bits (90), Expect = 0.33
Identities = 27/104 (25%), Positives = 51/104 (48%), Gaps = 6/104 (5%)
Query: 158 HELKEILSK----LKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYD 213
++LKE+L K LK++V+ D WN LG + +G A++ + +I E +
Sbjct: 805 NDLKELLEKSLHCLKKAVRLDSNNHLYWNALGVVACYSGIGNYALAQHCFIKSIQSEQIN 864
Query: 214 CLG--NLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAAL 255
+ NLG+ YL N+E + + F+ D ++ + + A +
Sbjct: 865 AVAWTNLGVLYLTNENIEQAHEAFKMAQSLDPSYLMCWIGQALI 908
Score = 37.0 bits (84), Expect = 1.6
Identities = 25/87 (28%), Positives = 42/87 (47%), Gaps = 1/87 (1%)
Query: 444 QQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQ-QAHAI 502
+++K + K+ K + ++ W +GV+ E+P QA+ AYK+A L Q A
Sbjct: 24 KEYKEALKHCKTVLKQEKNNYNAWVFIGVAAAELEQPDQAQSAYKKAAELEPDQLLAWQG 83
Query: 503 LSNLGIFYRHEKKYQRAKAMFTKSLEL 529
L+NL Y H ++ K L+L
Sbjct: 84 LANLYEKYNHINAKDDLPGVYQKLLDL 110
>UniRef100_UPI00002E2395 UPI00002E2395 UniRef100 entry
Length = 326
Score = 75.5 bits (184), Expect = 4e-12
Identities = 71/301 (23%), Positives = 128/301 (41%), Gaps = 29/301 (9%)
Query: 274 GALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN 333
GA++ + VA + I+ H + NL A G++ + L+KA +L+P+
Sbjct: 52 GAISFEKGQKEVAIKHFRKVIELRPHHPHAYNNLGAALIDIGEYEEAKSNLKKAIELQPD 111
Query: 334 CMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKA 393
YA A + + + + N+ SI E A+ L +V
Sbjct: 112 -----YAEAYNNLGNVYKEMEEY-------NQAISIYEKAIELNPEYYEAYNNLGVVLGK 159
Query: 394 QHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYL 453
+ YE + VL KQ + PD + LG C ++++ S+ L
Sbjct: 160 NEQ----YEEAEKVL-----------KQVIKSQPDFVEALNNLGSILTCMEKYEESKAIL 204
Query: 454 KAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHE 513
++ + SNLG + E QA YK+A+ + K + L+N+G+
Sbjct: 205 SRSIEIYPSNAEALSNLGDVYRKLHEYEQAVDYYKRAIQI--KPNYYEALNNMGLALTEL 262
Query: 514 KKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSN 573
KKY AK + +++++ P +A A+NNLG+ + E+A +++A+Q P A +N
Sbjct: 263 KKYSEAKEILKQAIKINPKFAEAYNNLGIYYEKLNEYEQAVDHYKRAIQIKPNYYEALNN 322
Query: 574 L 574
+
Sbjct: 323 M 323
Score = 65.5 bits (158), Expect = 4e-09
Identities = 49/172 (28%), Positives = 87/172 (50%), Gaps = 6/172 (3%)
Query: 413 ERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGV 472
E A +LK+A+ PD ++ LG +++ + + A+ + ++NLGV
Sbjct: 96 EEAKSNLKKAIELQPDYAEAYNNLGNVYKEMEEYNQAISIYEKAIELNPEYYEAYNNLGV 155
Query: 473 SLQLSEEPSQAEKAYKQALLLATKQQAHAI--LSNLGIFYRHEKKYQRAKAMFTKSLELQ 530
L +E+ +AEK KQ + K Q + L+NLG +KY+ +KA+ ++S+E+
Sbjct: 156 VLGKNEQYEEAEKVLKQVI----KSQPDFVEALNNLGSILTCMEKYEESKAILSRSIEIY 211
Query: 531 PGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSK 582
P A A +NLG V+ E+A +++A+Q P A +N+ +T K
Sbjct: 212 PSNAEALSNLGDVYRKLHEYEQAVDYYKRAIQIKPNYYEALNNMGLALTELK 263
Score = 65.1 bits (157), Expect = 6e-09
Identities = 78/358 (21%), Positives = 143/358 (39%), Gaps = 56/358 (15%)
Query: 188 LLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPV 247
L G+ +S S L+ P + +G + G E++ K F+++I +HP
Sbjct: 21 LYHQGKFDDVLSRSSQLIKEYPHTFVLHNIIGAISFEKGQKEVAIKHFRKVIELRPHHPH 80
Query: 248 ALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNL 307
A N A L+ + AK L AI+ A + NL
Sbjct: 81 AYNNLGAALI---------------------DIGEYEEAKSNLKKAIELQPDYAEAYNNL 119
Query: 308 AYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMA 367
+ ++ + EKA +L P Y A + + + G NE
Sbjct: 120 GNVYKEMEEYNQAISIYEKAIELNPE-----YYEAYNNLG-----------VVLGKNE-- 161
Query: 368 SIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEME--ERAVCSLKQAVAE 425
+ A L V K+Q + A + +LT ME E + L +++
Sbjct: 162 -----------QYEEAEKVLKQVIKSQPDFVEALNNLGSILTCMEKYEESKAILSRSIEI 210
Query: 426 DPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEK 485
P + LG +++ + Y K A+ +N+G++L ++ S+A++
Sbjct: 211 YPSNAEALSNLGDVYRKLHEYEQAVDYYKRAIQIKPNYYEALNNMGLALTELKKYSEAKE 270
Query: 486 AYKQALLLATK-QQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGL 542
KQA+ + K +A+ +NLGI+Y +Y++A + ++++++P Y A NN+GL
Sbjct: 271 ILKQAIKINPKFAEAY---NNLGIYYEKLNEYEQAVDHYKRAIQIKPNYYEALNNMGL 325
Score = 61.6 bits (148), Expect = 6e-08
Identities = 45/159 (28%), Positives = 81/159 (50%), Gaps = 2/159 (1%)
Query: 421 QAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEP 480
Q + E P V + +G S Q + + K+ + + + ++NLG +L E
Sbjct: 36 QLIKEYPHTFVLHNIIGAISFEKGQKEVAIKHFRKVIELRPHHPHAYNNLGAALIDIGEY 95
Query: 481 SQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNL 540
+A+ K+A+ L A A +NLG Y+ ++Y +A +++ K++EL P Y A+NNL
Sbjct: 96 EEAKSNLKKAIELQP-DYAEAY-NNLGNVYKEMEEYNQAISIYEKAIELNPEYYEAYNNL 153
Query: 541 GLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVT 579
G+V EEA+ ++ ++S P A +NL ++T
Sbjct: 154 GVVLGKNEQYEEAEKVLKQVIKSQPDFVEALNNLGSILT 192
Score = 54.7 bits (130), Expect = 8e-06
Identities = 65/263 (24%), Positives = 108/263 (40%), Gaps = 48/263 (18%)
Query: 73 KTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSL 132
K+ L P A A+ LG +++ + + +AI +YEKA E L PE E + L +
Sbjct: 99 KSNLKKAIELQPDYAEAYNNLGNVYKEMEEYNQAISIYEKAIE--LNPEY-YEAYNNLGV 155
Query: 133 VQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTG 192
V +N +E E LK+++ + V+ N LG IL
Sbjct: 156 VL------------GKNEQYEEAEK-VLKQVIKSQPDFVE-------ALNNLGSILTCME 195
Query: 193 RVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNY 252
+ + + ++LS + I P N + L NLG Y ++ E + ++ I N+ AL N
Sbjct: 196 KYEESKAILSRSIEIYPSNAEALSNLGDVYRKLHEYEQAVDYYKRAIQIKPNYYEALNNM 255
Query: 253 AALL--LCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYA 310
L L KY+ AKE L AIK + K A + NL
Sbjct: 256 GLALTELKKYSE-----------------------AKEILKQAIKINPKFAEAYNNLGIY 292
Query: 311 FSISGDHRSSSKCLEKAAKLEPN 333
+ ++ + ++A +++PN
Sbjct: 293 YEKLNEYEQAVDHYKRAIQIKPN 315
>UniRef100_UPI00001CECA3 UPI00001CECA3 UniRef100 entry
Length = 1526
Score = 74.3 bits (181), Expect = 9e-12
Identities = 86/359 (23%), Positives = 146/359 (39%), Gaps = 27/359 (7%)
Query: 152 DKELEPHELKEILSKLKESVQFDIRQAAVWNTL--GFILLKTGRVQSAISVLSSLLAIAP 209
D +E + + L+ L Q AA W L G LK G A++ L + L P
Sbjct: 623 DLSVELEDTETALAILTAVTQKASSGAAKWAWLRRGLYYLKAGHHSQAVADLQAALRADP 682
Query: 210 ENYDCLGNLGIAYLQIGNLELSAKCFQEL--ILKDQNHPVALVNYAALLLCKYASVVAGA 267
++ +C +LG AYL G+ + K F + D + V V +L +Y+ +A
Sbjct: 683 KDCNCWESLGEAYLSRGSYTTALKSFTRASELSPDSTYSVFKVAAIQQILGRYSEAIAQY 742
Query: 268 GASASEGALADQVMAANVAKECLLAAIKA------DVKSAHIWGNLAYAFSISGDHRSSS 321
+ D V A EC L KA D K+ F+ + HR+
Sbjct: 743 QLIIKKE--EDYVPALKGLGECHLMLGKAALVDFLDGKAVDYVEQALGYFTRALQHRADV 800
Query: 322 KCLEKAA--------KLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRDG 373
CL K + P ++ R + A +E + ELL GG ++
Sbjct: 801 SCLWKLMGDACTCLHPVSPAKVNVRVSGALLGQQEGQEVLRKDELLHLGGRCYGRALK-- 858
Query: 374 DSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERAVCSLKQAVAEDPDDPVRW 433
L+ W L + + Q + + S + LTE+ E+++ LK+AV D ++ + W
Sbjct: 859 ---LMSTSNTWCDLGINYYRQAQHLAETGSSMNDLTELLEKSLHCLKKAVRLDSNNHLYW 915
Query: 434 HQLGVHSLC--TQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQA 490
+ LGV + + +Q ++ ++ + W+NLGV +E QA +A++ A
Sbjct: 916 NALGVVTCYHGVGNYALAQHCFIKSIQAEQINAAAWTNLGVLYLATENIEQAHEAFQMA 974
Score = 37.4 bits (85), Expect = 1.2
Identities = 27/104 (25%), Positives = 48/104 (45%), Gaps = 6/104 (5%)
Query: 158 HELKEILSK----LKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYD 213
++L E+L K LK++V+ D WN LG + G A++ + +I E +
Sbjct: 888 NDLTELLEKSLHCLKKAVRLDSNNHLYWNALGVVTCYHGVGNYALAQHCFIKSIQAEQIN 947
Query: 214 CLG--NLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAAL 255
NLG+ YL N+E + + FQ D ++ + + A +
Sbjct: 948 AAAWTNLGVLYLATENIEQAHEAFQMAQSLDPSYLLCWIGQALI 991
>UniRef100_UPI000026B4BA UPI000026B4BA UniRef100 entry
Length = 321
Score = 73.2 bits (178), Expect = 2e-11
Identities = 54/166 (32%), Positives = 84/166 (50%), Gaps = 4/166 (2%)
Query: 398 SSAYESEQHVLTEME--ERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKA 455
S AY + + L E+ + A S KQA+A PD + LG + +Q
Sbjct: 28 SEAYNNLGNNLQELGRLDEAETSYKQAIALKPDSAEAHNNLGNALRKRGRLDEAQTSYTH 87
Query: 456 AVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKK 515
A+A S NLGV+LQ + AE +Y QA++L K + +NLG + +
Sbjct: 88 AIALTSDLSEIHYNLGVTLQDLGKLEDAEASYTQAIVL--KSKFTKAYNNLGNTLKELGR 145
Query: 516 YQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKAL 561
A+A +T+++ L P YA +NNLG+V G LEEA+ C+++A+
Sbjct: 146 IAEAEARYTQAIALNPDYAEIYNNLGVVLQDIGKLEEAESCYKQAI 191
Score = 49.7 bits (117), Expect = 2e-04
Identities = 36/145 (24%), Positives = 71/145 (48%), Gaps = 2/145 (1%)
Query: 421 QAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEP 480
QA+A +P ++ LG + + ++ K A+A + +NLG +L+
Sbjct: 19 QAIALNPHYSEAYNNLGNNLQELGRLDEAETSYKQAIALKPDSAEAHNNLGNALRKRGRL 78
Query: 481 SQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNL 540
+A+ +Y A+ L + I NLG+ + K + A+A +T+++ L+ + A+NNL
Sbjct: 79 DEAQTSYTHAIALTS--DLSEIHYNLGVTLQDLGKLEDAEASYTQAIVLKSKFTKAYNNL 136
Query: 541 GLVFVAEGLLEEAKYCFEKALQSDP 565
G G + EA+ + +A+ +P
Sbjct: 137 GNTLKELGRIAEAEARYTQAIALNP 161
Score = 45.8 bits (107), Expect = 0.003
Identities = 35/120 (29%), Positives = 58/120 (48%), Gaps = 4/120 (3%)
Query: 456 AVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQ-QAHAILSNLGIFYRHEK 514
A+A + S ++NLG +LQ +AE +YKQA+ L +AH +NLG R
Sbjct: 20 AIALNPHYSEAYNNLGNNLQELGRLDEAETSYKQAIALKPDSAEAH---NNLGNALRKRG 76
Query: 515 KYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
+ A+ +T ++ L + NLG+ G LE+A+ + +A+ A +NL
Sbjct: 77 RLDEAQTSYTHAIALTSDLSEIHYNLGVTLQDLGKLEDAEASYTQAIVLKSKFTKAYNNL 136
Score = 39.3 bits (90), Expect = 0.33
Identities = 19/81 (23%), Positives = 41/81 (50%)
Query: 159 ELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNL 218
+L++ + +++ + +N LG L + GR+ A + + +A+ P+ + NL
Sbjct: 111 KLEDAEASYTQAIVLKSKFTKAYNNLGNTLKELGRIAEAEARYTQAIALNPDYAEIYNNL 170
Query: 219 GIAYLQIGNLELSAKCFQELI 239
G+ IG LE + C+++ I
Sbjct: 171 GVVLQDIGKLEEAESCYKQAI 191
Score = 36.2 bits (82), Expect = 2.8
Identities = 46/204 (22%), Positives = 76/204 (36%), Gaps = 29/204 (14%)
Query: 133 VQIHHAQCLILE---SSSENSSDKEL-EPHELKEILSKLKESVQFDIRQAAVWNTLGFIL 188
++ H Q + L S + N+ L E L E + K+++ A N LG L
Sbjct: 13 IEASHTQAIALNPHYSEAYNNLGNNLQELGRLDEAETSYKQAIALKPDSAEAHNNLGNAL 72
Query: 189 LKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLE-LSAKCFQELILKDQNHPV 247
K GR+ A + + +A+ + + NLG+ +G LE A Q ++LK
Sbjct: 73 RKRGRLDEAQTSYTHAIALTSDLSEIHYNLGVTLQDLGKLEDAEASYTQAIVLKS----- 127
Query: 248 ALVNYAALLLCKYASVVAGAGASASE-GALADQVMAANVAKECLLAAIKADVKSAHIWGN 306
K+ G + E G +A+ A+ AI + A I+ N
Sbjct: 128 -----------KFTKAYNNLGNTLKELGRIAE-------AEARYTQAIALNPDYAEIYNN 169
Query: 307 LAYAFSISGDHRSSSKCLEKAAKL 330
L G + C ++A L
Sbjct: 170 LGVVLQDIGKLEEAESCYKQAITL 193
Score = 35.8 bits (81), Expect = 3.6
Identities = 19/60 (31%), Positives = 34/60 (56%)
Query: 515 KYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
++ +A T+++ L P Y+ A+NNLG G L+EA+ +++A+ P A +NL
Sbjct: 9 RWGEIEASHTQAIALNPHYSEAYNNLGNNLQELGRLDEAETSYKQAIALKPDSAEAHNNL 68
>UniRef100_O26186 O-linked GlcNAc transferase [Methanobacterium thermoautotrophicum]
Length = 379
Score = 72.0 bits (175), Expect = 5e-11
Identities = 64/266 (24%), Positives = 118/266 (44%), Gaps = 9/266 (3%)
Query: 315 GDHRSSSKCLEKAAKLEPNCMSTRY--AVATHRMKEAERSQDPSELLSFGGNEMASIIRD 372
G ++ + K KA K PN + A+ ++K E++ + E + ++A +
Sbjct: 30 GKYKEALKEFRKALKARPNNPEILHYNAITLLKLKRPEKALECYEKILKNNPKLAEAWNN 89
Query: 373 GDSSLVELPTAWAGLAMVHKA---QHEISSAYESEQHVLTEME--ERAVCSLKQAVAEDP 427
L EL L KA + + A+ ++ VL E+ + A+ ++A+ +P
Sbjct: 90 KGLVLKELGRYDEALECYEKALKINPKYAGAWNNKALVLKELGRYDEALECYEKALQINP 149
Query: 428 DDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAY 487
W+ G + +++K + K + A+ + W G++L + +A K Y
Sbjct: 150 KLADAWYNKGSVLIYLKKYKKALKCFEKAIELNPKNYRAWGTKGITLHNLKIYEEALKCY 209
Query: 488 KQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAE 547
+ L L Q +N G+ + +Y + + K+L++ P A A+NN G+V
Sbjct: 210 DKVLQL--NPQDDKAWNNKGLVFNELGRYDESLECYEKALQINPKLAEAWNNKGVVLSEL 267
Query: 548 GLLEEAKYCFEKALQSDPLLDAAKSN 573
G EEA C+EKAL+ DP D +N
Sbjct: 268 GRYEEALECYEKALEIDPEDDKTWNN 293
Score = 68.2 bits (165), Expect = 7e-10
Identities = 82/383 (21%), Positives = 145/383 (37%), Gaps = 62/383 (16%)
Query: 189 LKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVA 248
LK G+ + A+ L P N + L I L++ E + +C+++++ +N+P
Sbjct: 27 LKQGKYKEALKEFRKALKARPNNPEILHYNAITLLKLKRPEKALECYEKIL---KNNPKL 83
Query: 249 LVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLA 308
A A ++G + ++ + A EC A+K + K A W N A
Sbjct: 84 ------------------AEAWNNKGLVLKELGRYDEALECYEKALKINPKYAGAWNNKA 125
Query: 309 YAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMAS 368
G + + +C EKA ++ P Y
Sbjct: 126 LVLKELGRYDEALECYEKALQINPKLADAWY----------------------------- 156
Query: 369 IIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYES------EQHVLTEMEERAVCSLKQA 422
+ S L+ L L KA Y + H L EE C K
Sbjct: 157 ---NKGSVLIYLKKYKKALKCFEKAIELNPKNYRAWGTKGITLHNLKIYEEALKCYDK-V 212
Query: 423 VAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQ 482
+ +P D W+ G+ ++ S + + A+ + + W+N GV L +
Sbjct: 213 LQLNPQDDKAWNNKGLVFNELGRYDESLECYEKALQINPKLAEAWNNKGVVLSELGRYEE 272
Query: 483 AEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGL 542
A + Y++AL + + +N G+ KY+ A F K+LE+ P +A A+ G+
Sbjct: 273 ALECYEKALEIDPEDDK--TWNNKGLVLEELGKYKDALECFQKALEINPEFADAWKWKGI 330
Query: 543 VFVAEGLLEEAKYCFEKALQSDP 565
+ EE+ C++KAL+ +P
Sbjct: 331 ILEDLKKPEESLKCYKKALKLNP 353
Score = 60.5 bits (145), Expect = 1e-07
Identities = 71/355 (20%), Positives = 150/355 (42%), Gaps = 36/355 (10%)
Query: 155 LEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDC 214
L+ + KE L + +++++ + + LLK R + A+ +L P+ +
Sbjct: 27 LKQGKYKEALKEFRKALKARPNNPEILHYNAITLLKLKRPEKALECYEKILKNNPKLAEA 86
Query: 215 LGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEG 274
N G+ ++G + + +C+++ + + + AGA ++
Sbjct: 87 WNNKGLVLKELGRYDEALECYEKALKINPKY---------------------AGAWNNKA 125
Query: 275 ALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEP-- 332
+ ++ + A EC A++ + K A W N ++ + KC EKA +L P
Sbjct: 126 LVLKELGRYDEALECYEKALQINPKLADAWYNKGSVLIYLKKYKKALKCFEKAIELNPKN 185
Query: 333 -NCMSTRYAVATHRMK---EAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLA 388
T+ + H +K EA + D ++L + + G EL L
Sbjct: 186 YRAWGTK-GITLHNLKIYEEALKCYD--KVLQLNPQDDKAWNNKG-LVFNELGRYDESLE 241
Query: 389 MVHKA---QHEISSAYESEQHVLTEM--EERAVCSLKQAVAEDPDDPVRWHQLGVHSLCT 443
KA +++ A+ ++ VL+E+ E A+ ++A+ DP+D W+ G+
Sbjct: 242 CYEKALQINPKLAEAWNNKGVVLSELGRYEEALECYEKALEIDPEDDKTWNNKGLVLEEL 301
Query: 444 QQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQ 498
++K + + + A+ + + W G+ L+ ++P ++ K YK+AL L +Q
Sbjct: 302 GKYKDALECFQKALEINPEFADAWKWKGIILEDLKKPEESLKCYKKALKLNPPKQ 356
Score = 58.2 bits (139), Expect = 7e-07
Identities = 45/158 (28%), Positives = 72/158 (45%), Gaps = 3/158 (1%)
Query: 433 WHQLGVHSLCTQ-QFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQAL 491
WH G S Q ++K + K + A+ ++L + P +A + Y++
Sbjct: 18 WHLAGGRSSLKQGKYKEALKEFRKALKARPNNPEILHYNAITLLKLKRPEKALECYEK-- 75
Query: 492 LLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLE 551
+L + +N G+ + +Y A + K+L++ P YA A+NN LV G +
Sbjct: 76 ILKNNPKLAEAWNNKGLVLKELGRYDEALECYEKALKINPKYAGAWNNKALVLKELGRYD 135
Query: 552 EAKYCFEKALQSDPLLDAAKSNLVKVVTMSKICKGLLK 589
EA C+EKALQ +P L A N V+ K K LK
Sbjct: 136 EALECYEKALQINPKLADAWYNKGSVLIYLKKYKKALK 173
Score = 52.8 bits (125), Expect = 3e-05
Identities = 52/254 (20%), Positives = 104/254 (40%), Gaps = 44/254 (17%)
Query: 79 VARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHA 138
+ + PK A A GL+ + L + +A+ YEKA + + P+ +
Sbjct: 76 ILKNNPKLAEAWNNKGLVLKELGRYDEALECYEKA--LKINPK----------YAGAWNN 123
Query: 139 QCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAI 198
+ L+L KEL ++ E L ++++Q + + A W G +L+ + + A+
Sbjct: 124 KALVL---------KELGRYD--EALECYEKALQINPKLADAWYNKGSVLIYLKKYKKAL 172
Query: 199 SVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLC 258
+ + P+NY G GI + E + KC+ +++ + A N
Sbjct: 173 KCFEKAIELNPKNYRAWGTKGITLHNLKIYEEALKCYDKVLQLNPQDDKAWNN------- 225
Query: 259 KYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHR 318
+G + +++ + + EC A++ + K A W N S G +
Sbjct: 226 --------------KGLVFNELGRYDESLECYEKALQINPKLAEAWNNKGVVLSELGRYE 271
Query: 319 SSSKCLEKAAKLEP 332
+ +C EKA +++P
Sbjct: 272 EALECYEKALEIDP 285
Score = 45.4 bits (106), Expect = 0.005
Identities = 56/282 (19%), Positives = 106/282 (36%), Gaps = 34/282 (12%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLR-PETEIERPDLLSLVQIHHAQCLI 142
PK A A L+ + L + +A+ YEKA +I + + + +L ++ +
Sbjct: 115 PKYAGAWNNKALVLKELGRYDEALECYEKALQINPKLADAWYNKGSVLIYLKKYKKALKC 174
Query: 143 LESSSENS-------SDKELEPHELK---EILSKLKESVQFDIRQAAVWNTLGFILLKTG 192
E + E + K + H LK E L + +Q + + WN G + + G
Sbjct: 175 FEKAIELNPKNYRAWGTKGITLHNLKIYEEALKCYDKVLQLNPQDDKAWNNKGLVFNELG 234
Query: 193 RVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNY 252
R ++ L I P+ + N G+ ++G E + +C+++ + D N
Sbjct: 235 RYDESLECYEKALQINPKLAEAWNNKGVVLSELGRYEEALECYEKALEIDPEDDKTWNN- 293
Query: 253 AALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFS 312
+G + +++ A EC A++ + + A W
Sbjct: 294 --------------------KGLVLEELGKYKDALECFQKALEINPEFADAWKWKGIILE 333
Query: 313 ISGDHRSSSKCLEKAAKLEPNCMSTRYAVA--THRMKEAERS 352
S KC +KA KL P +T T + ++ +RS
Sbjct: 334 DLKKPEESLKCYKKALKLNPPKQNTMVHARKNTTKTRQTQRS 375
>UniRef100_UPI00002E634F UPI00002E634F UniRef100 entry
Length = 284
Score = 70.9 bits (172), Expect = 1e-10
Identities = 43/152 (28%), Positives = 78/152 (51%), Gaps = 4/152 (2%)
Query: 427 PDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKA 486
P +P ++ LG L ++K +QK LK A+ + + ++NLG ++ QA K
Sbjct: 3 PKNPQAYNNLGTTLLDLNEYKKAQKALKKAIKLEPAYAEAYNNLGNIYNKIKKYDQAIKE 62
Query: 487 YKQALLLATKQ-QAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFV 545
YK+A++L + +AH +NLGI + +K++ A+ K++ + P + AFN +
Sbjct: 63 YKKAIILNPEYYEAH---NNLGIVLGNNEKFEEAEKSLAKAISINPHFPEAFNTQATFLI 119
Query: 546 AEGLLEEAKYCFEKALQSDPLLDAAKSNLVKV 577
G E+A ++A++ P AA NL +
Sbjct: 120 KSGNFEKALVKIKRAIELKPNFSAAYINLALI 151
Score = 47.8 bits (112), Expect = 0.001
Identities = 46/182 (25%), Positives = 74/182 (40%), Gaps = 21/182 (11%)
Query: 155 LEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDC 214
L+ +E K+ LK++++ + A +N LG I K + AI + + PE Y+
Sbjct: 17 LDLNEYKKAQKALKKAIKLEPAYAEAYNNLGNIYNKIKKYDQAIKEYKKAIILNPEYYEA 76
Query: 215 LGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEG 274
NLGI E + K + I + + P A A L+ G
Sbjct: 77 HNNLGIVLGNNEKFEEAEKSLAKAISINPHFPEAFNTQATFLI--------------KSG 122
Query: 275 ALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNC 334
++ A E +K + +A+I NLA + G+H SS+ LE A L+P
Sbjct: 123 NFEKALVKIKRAIE-----LKPNFSAAYI--NLALIYDEFGNHNLSSEYLEIAKDLDPYL 175
Query: 335 MS 336
S
Sbjct: 176 FS 177
Score = 42.4 bits (98), Expect = 0.039
Identities = 38/170 (22%), Positives = 77/170 (44%), Gaps = 4/170 (2%)
Query: 400 AYESEQHVLTEMEE--RAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAV 457
AY + L ++ E +A +LK+A+ +P ++ LG +++ + K K A+
Sbjct: 8 AYNNLGTTLLDLNEYKKAQKALKKAIKLEPAYAEAYNNLGNIYNKIKKYDQAIKEYKKAI 67
Query: 458 ACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQ 517
+ +NLG+ L +E+ +AEK+ +A ++ + F ++
Sbjct: 68 ILNPEYYEAHNNLGIVLGNNEKFEEAEKSLAKA--ISINPHFPEAFNTQATFLIKSGNFE 125
Query: 518 RAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLL 567
+A +++EL+P ++ A+ NL L++ G + E A DP L
Sbjct: 126 KALVKIKRAIELKPNFSAAYINLALIYDEFGNHNLSSEYLEIAKDLDPYL 175
Score = 37.0 bits (84), Expect = 1.6
Identities = 35/159 (22%), Positives = 69/159 (43%), Gaps = 25/159 (15%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSLVQIHHAQCLIL 143
P A A+ LG ++ ++ + +AI Y+KA I+L PE + H+ ++L
Sbjct: 37 PAYAEAYNNLGNIYNKIKKYDQAIKEYKKA--IILNPE----------YYEAHNNLGIVL 84
Query: 144 ESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSS 203
++ + +E L +++ + +NT L+K+G + A+ +
Sbjct: 85 GNNEK-----------FEEAEKSLAKAISINPHFPEAFNTQATFLIKSGNFEKALVKIKR 133
Query: 204 LLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKD 242
+ + P NL + Y + GN LS++ + I KD
Sbjct: 134 AIELKPNFSAAYINLALIYDEFGNHNLSSEYLE--IAKD 170
>UniRef100_UPI00003165D7 UPI00003165D7 UniRef100 entry
Length = 344
Score = 70.5 bits (171), Expect = 1e-10
Identities = 42/163 (25%), Positives = 84/163 (50%), Gaps = 2/163 (1%)
Query: 412 EERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLG 471
+E A+ ++ + P P ++ LG + +++ ++ LK A+ + ++NLG
Sbjct: 10 KEVAIKHFRKVIELRPHHPHAYNNLGAALIDVGEYEEAKSNLKKAIELQPDYAEAYNNLG 69
Query: 472 VSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQP 531
+ EE +QA Y++A+ L + + +NLG+ + ++Y+ A + K++ ++P
Sbjct: 70 NVYKEMEEYNQAISVYEKAIEL--NPEYYEAYNNLGVVLGNNEQYKEAIEAYNKAISIKP 127
Query: 532 GYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
YA AF N+G+ + LEEA ++KA+ P D A N+
Sbjct: 128 DYAEAFYNMGVTLQGQDKLEEAIASYKKAVSIKPDYDEAYFNI 170
Score = 61.2 bits (147), Expect = 8e-08
Identities = 59/281 (20%), Positives = 113/281 (39%), Gaps = 33/281 (11%)
Query: 302 HIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSF 361
H + NL A G++ + L+KA +L+P+ YA A + + + +
Sbjct: 29 HAYNNLGAALIDVGEYEEAKSNLKKAIELQPD-----YAEAYNNLGNVYKEMEEY----- 78
Query: 362 GGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLT------------ 409
N+ S+ E A+ L +V + A E+ ++
Sbjct: 79 --NQAISVYEKAIELNPEYYEAYNNLGVVLGNNEQYKEAIEAYNKAISIKPDYAEAFYNM 136
Query: 410 -------EMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRG 462
+ E A+ S K+AV+ PD + +GV + S + + + +
Sbjct: 137 GVTLQGQDKLEEAIASYKKAVSIKPDYDEAYFNIGVGLQDKGNLEESIQAYNSTIEINPN 196
Query: 463 CSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAM 522
+ + N G +L+ +A KAY AL + + +GI + K + +
Sbjct: 197 NADAFYNKGNALKQLGNLEEAMKAYNNALFINPNDAE--VYCTMGIVLNQQGKVEESIDA 254
Query: 523 FTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQS 563
+ K+L L+P YA A+N++ + +G LEEA + KA+++
Sbjct: 255 YNKALSLKPDYAKAYNHMAIALHHQGKLEEAIEGYNKAIEA 295
Score = 57.8 bits (138), Expect = 9e-07
Identities = 37/153 (24%), Positives = 77/153 (50%), Gaps = 2/153 (1%)
Query: 413 ERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGV 472
E A +LK+A+ PD ++ LG +++ + + A+ + ++NLGV
Sbjct: 45 EEAKSNLKKAIELQPDYAEAYNNLGNVYKEMEEYNQAISVYEKAIELNPEYYEAYNNLGV 104
Query: 473 SLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPG 532
L +E+ +A +AY +A+ + K N+G+ + + K + A A + K++ ++P
Sbjct: 105 VLGNNEQYKEAIEAYNKAISI--KPDYAEAFYNMGVTLQGQDKLEEAIASYKKAVSIKPD 162
Query: 533 YAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDP 565
Y A+ N+G+ +G LEE+ + ++ +P
Sbjct: 163 YDEAYFNIGVGLQDKGNLEESIQAYNSTIEINP 195
Score = 50.8 bits (120), Expect = 1e-04
Identities = 74/386 (19%), Positives = 143/386 (36%), Gaps = 59/386 (15%)
Query: 188 LLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPV 247
L G+ + AI ++ + P + NLG A + +G E + ++ I ++
Sbjct: 4 LFSKGQKEVAIKHFRKVIELRPHHPHAYNNLGAALIDVGEYEEAKSNLKKAIELQPDYAE 63
Query: 248 ALVNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNL 307
A N G + ++ N A AI+ + + + NL
Sbjct: 64 AYNNL---------------------GNVYKEMEEYNQAISVYEKAIELNPEYYEAYNNL 102
Query: 308 AYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMA 367
+ ++ + + KA ++P+ YA A + M + QD E +
Sbjct: 103 GVVLGNNEQYKEAIEAYNKAISIKPD-----YAEAFYNMGVTLQGQDKLEEAIASYKKAV 157
Query: 368 SIIRDGDSSLVELPTAWAGLAMVHKAQHEIS-SAYESEQHVLTEMEERAVCSLKQAVAED 426
SI D D + + G+ + K E S AY S + +
Sbjct: 158 SIKPDYDEAYFNI-----GVGLQDKGNLEESIQAYNSTIEI------------------N 194
Query: 427 PDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKA 486
P++ ++ G + + K A+ + + + +G+ L + ++ A
Sbjct: 195 PNNADAFYNKGNALKQLGNLEEAMKAYNNALFINPNDAEVYCTMGIVLNQQGKVEESIDA 254
Query: 487 YKQALLLATKQQAHAILSNLGIFYRHEKK-------YQRAKAMFTKSLELQPGYAPAFNN 539
Y +AL L K +++ I H+ K Y +A F+K++ QP YA A+ +
Sbjct: 255 YNKALSL--KPDYAKAYNHMAIALHHQGKLEEAIEGYNKAIEAFSKNISNQPDYAEAYKH 312
Query: 540 LGLVFVAEGLLEEAKYCFEKALQSDP 565
LG+ +G L++A F +A+ P
Sbjct: 313 LGIGSKDQGQLKKAIEAFTQAISHQP 338
Score = 48.1 bits (113), Expect = 7e-04
Identities = 72/343 (20%), Positives = 126/343 (35%), Gaps = 49/343 (14%)
Query: 73 KTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDLLSL 132
K+ L P A A+ LG +++ + + +AI VYEKA E L PE E + L +
Sbjct: 48 KSNLKKAIELQPDYAEAYNNLGNVYKEMEEYNQAISVYEKAIE--LNPEY-YEAYNNLGV 104
Query: 133 VQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLKTG 192
V ++ Q KE + +++ A + +G L
Sbjct: 105 VLGNNEQ--------------------YKEAIEAYNKAISIKPDYAEAFYNMGVTLQGQD 144
Query: 193 RVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALVNY 252
+++ AI+ ++I P+ + N+G+ GNLE S + + I + N+ A N
Sbjct: 145 KLEEAIASYKKAVSIKPDYDEAYFNIGVGLQDKGNLEESIQAYNSTIEINPNNADAFYN- 203
Query: 253 AALLLCKYASVVAGAGASASEGALADQVMAANVAKECLLAAIKADVKSAHIWGNLAYAFS 312
A G L + + A N A+ + A ++ + +
Sbjct: 204 -------------KGNALKQLGNLEEAMKAYN-------NALFINPNDAEVYCTMGIVLN 243
Query: 313 ISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHRMKEAERSQDPSELLSFGGNEMASIIRD 372
G S KA L+P+ YA A + M A Q E G N+
Sbjct: 244 QQGKVEESIDAYNKALSLKPD-----YAKAYNHMAIALHHQGKLEEAIEGYNKAIEAFSK 298
Query: 373 GDSSLVELPTAWAGLAMVHKAQHEISSAYESEQHVLTEMEERA 415
S+ + A+ L + K Q ++ A E+ ++ + A
Sbjct: 299 NISNQPDYAEAYKHLGIGSKDQGQLKKAIEAFTQAISHQPDYA 341
Score = 40.0 bits (92), Expect = 0.19
Identities = 22/55 (40%), Positives = 31/55 (56%)
Query: 523 FTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKV 577
F K +EL+P + A+NNLG + G EEAK +KA++ P A +NL V
Sbjct: 17 FRKVIELRPHHPHAYNNLGAALIDVGEYEEAKSNLKKAIELQPDYAEAYNNLGNV 71
Score = 39.7 bits (91), Expect = 0.25
Identities = 37/175 (21%), Positives = 64/175 (36%), Gaps = 21/175 (12%)
Query: 159 ELKEILSKLKESVQFDIRQAAVWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNL 218
E +E S LK++++ A +N LG + + AISV + + PE Y+ NL
Sbjct: 43 EYEEAKSNLKKAIELQPDYAEAYNNLGNVYKEMEEYNQAISVYEKAIELNPEYYEAYNNL 102
Query: 219 GIAYLQIGNLELSAKCFQELILKDQNHPVALVNYAALLLCKYASVVAGAGASASEGALAD 278
G+ + + + + + I ++ A N L G E A+A
Sbjct: 103 GVVLGNNEQYKEAIEAYNKAISIKPDYAEAFYNMGVTL----------QGQDKLEEAIAS 152
Query: 279 QVMAANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN 333
A +IK D A+ N+ G+ S + ++ PN
Sbjct: 153 YKKA---------VSIKPDYDEAYF--NIGVGLQDKGNLEESIQAYNSTIEINPN 196
>UniRef100_UPI0000328B13 UPI0000328B13 UniRef100 entry
Length = 275
Score = 70.1 bits (170), Expect = 2e-10
Identities = 55/170 (32%), Positives = 85/170 (49%), Gaps = 8/170 (4%)
Query: 408 LTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTW 467
L + E AV K E P++ W LG T +F S K + AVA + +
Sbjct: 14 LVDAENLAVSLTK----EFPNNNFSWKILGAIFKATGRFAESIKANQTAVALNPKDAGAH 69
Query: 468 SNLGVSLQLSEEPSQAEKAYKQALLLATK-QQAHAILSNLGIFYRHEKKYQRAKAMFTKS 526
SNLGV LQ + +AE++ KQA++L +AH NLGI K + A+ +TK+
Sbjct: 70 SNLGVVLQEQGKLDEAEESCKQAIILKPDLAEAHY---NLGITLIELGKIKEAEESYTKA 126
Query: 527 LELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVK 576
L+P +A A NL F+ G L++A+ + KA+ P + A + L++
Sbjct: 127 TTLKPNFAEAHYNLANTFIKLGRLDQAELSYRKAIALKPNFEKAHNKLLR 176
Score = 37.0 bits (84), Expect = 1.6
Identities = 21/62 (33%), Positives = 30/62 (47%), Gaps = 1/62 (1%)
Query: 181 WNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNL-ELSAKCFQELI 239
W LG I TGR +I + +A+ P++ NLG+ + G L E C Q +I
Sbjct: 35 WKILGAIFKATGRFAESIKANQTAVALNPKDAGAHSNLGVVLQEQGKLDEAEESCKQAII 94
Query: 240 LK 241
LK
Sbjct: 95 LK 96
>UniRef100_UPI000033283A UPI000033283A UniRef100 entry
Length = 257
Score = 68.9 bits (167), Expect = 4e-10
Identities = 50/183 (27%), Positives = 91/183 (49%), Gaps = 4/183 (2%)
Query: 394 QHEISSAYESEQHVLTEMEE--RAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQK 451
Q + + AY + +V EMEE +A+ ++A+ +P+ ++ LG +Q++ ++K
Sbjct: 5 QPDYAEAYNNLGNVYKEMEEYNQAISVYEKAIELNPEYYEAYNNLGAALGKNEQYEEAEK 64
Query: 452 YLKAAVACDRGCSYTWSNLGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYR 511
LK + +NLG S+Q S E + K + A + LSNLG +R
Sbjct: 65 VLKQVIKSQPKFVEALNNLG-SIQTSMEKYEEAKVILNRSIEIHSSNAES-LSNLGNVFR 122
Query: 512 HEKKYQRAKAMFTKSLELQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAK 571
+Y+RA + ++++++P Y A NN+GL EAK ++A++ +P A
Sbjct: 123 KLHEYERAVDHYKRAIQIKPNYYEALNNMGLALTELKKYNEAKNILKQAIKVNPKFAEAY 182
Query: 572 SNL 574
+NL
Sbjct: 183 NNL 185
Score = 62.4 bits (150), Expect = 4e-08
Identities = 41/152 (26%), Positives = 74/152 (47%), Gaps = 2/152 (1%)
Query: 410 EMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSN 469
E E A LKQ + P + LG ++++ ++ L ++ + + SN
Sbjct: 57 EQYEEAEKVLKQVIKSQPKFVEALNNLGSIQTSMEKYEEAKVILNRSIEIHSSNAESLSN 116
Query: 470 LGVSLQLSEEPSQAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLEL 529
LG + E +A YK+A+ + K + L+N+G+ KKY AK + +++++
Sbjct: 117 LGNVFRKLHEYERAVDHYKRAIQI--KPNYYEALNNMGLALTELKKYNEAKNILKQAIKV 174
Query: 530 QPGYAPAFNNLGLVFVAEGLLEEAKYCFEKAL 561
P +A A+NNLG V+ +A CF+KA+
Sbjct: 175 NPKFAEAYNNLGYVYNKIQNFHKAIKCFKKAI 206
Score = 55.8 bits (133), Expect = 3e-06
Identities = 44/172 (25%), Positives = 83/172 (47%), Gaps = 5/172 (2%)
Query: 422 AVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEEPS 481
A+ PD ++ LG +++ + + A+ + ++NLG +L +E+
Sbjct: 1 AIELQPDYAEAYNNLGNVYKEMEEYNQAISVYEKAIELNPEYYEAYNNLGAALGKNEQYE 60
Query: 482 QAEKAYKQALLLATKQQAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFNNLG 541
+AEK KQ ++ ++ + L+NLG +KY+ AK + +S+E+ A + +NLG
Sbjct: 61 EAEKVLKQ--VIKSQPKFVEALNNLGSIQTSMEKYEEAKVILNRSIEIHSSNAESLSNLG 118
Query: 542 LVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNLVKVVTMSK---ICKGLLKE 590
VF E A +++A+Q P A +N+ +T K K +LK+
Sbjct: 119 NVFRKLHEYERAVDHYKRAIQIKPNYYEALNNMGLALTELKKYNEAKNILKQ 170
Score = 52.0 bits (123), Expect = 5e-05
Identities = 63/267 (23%), Positives = 110/267 (40%), Gaps = 37/267 (13%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEI-------------LLRPETEIERPDLL 130
P A A+ LG +++ + + +AI VYEKA E+ L + E + +
Sbjct: 6 PDYAEAYNNLGNVYKEMEEYNQAISVYEKAIELNPEYYEAYNNLGAALGKNEQYEEAEKV 65
Query: 131 SLVQIHHAQCLILESSSE-NSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILL 189
L Q+ +Q +E+ + S +E +E +++ L S++ A + LG +
Sbjct: 66 -LKQVIKSQPKFVEALNNLGSIQTSMEKYEEAKVI--LNRSIEIHSSNAESLSNLGNVFR 122
Query: 190 KTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVAL 249
K + A+ + I P Y+ L N+G+A + LK N +
Sbjct: 123 KLHEYERAVDHYKRAIQIKPNYYEALNNMGLALTE---------------LKKYNEAKNI 167
Query: 250 VNYAALLLCKYASVVAGAGASASEGALADQVMAANVAKECLL---AAIKADVKSAHIWGN 306
+ A + K+A G ++ + + A K+ +L AIK D A I N
Sbjct: 168 LKQAIKVNPKFAEAYNNLGYVYNK--IQNFHKAIKCFKKAILYYKKAIKFDKNLAGIHNN 225
Query: 307 LAYAFSISGDHRSSSKCLEKAAKLEPN 333
L+YA S+ GD+ +S + A L N
Sbjct: 226 LSYALSLIGDNSNSIHHQKIAVSLNTN 252
Score = 48.1 bits (113), Expect = 7e-04
Identities = 41/181 (22%), Positives = 72/181 (39%), Gaps = 23/181 (12%)
Query: 60 GAPSSREEKVSSMKTGLIHVARKMPKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLR 119
GA + E+ + L V + PK A LG + + + YE+A+ IL R
Sbjct: 50 GAALGKNEQYEEAEKVLKQVIKSQPKFVEALNNLGSIQTSMEK-------YEEAKVILNR 102
Query: 120 PETEIERPDLLSLVQIHHAQCLILESSSENSSDKELEPHELKEILSKLKESVQFDIRQAA 179
++IH + S N + + HE + + K ++Q
Sbjct: 103 S------------IEIHSSNA----ESLSNLGNVFRKLHEYERAVDHYKRAIQIKPNYYE 146
Query: 180 VWNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELI 239
N +G L + + A ++L + + P+ + NLG Y +I N + KCF++ I
Sbjct: 147 ALNNMGLALTELKKYNEAKNILKQAIKVNPKFAEAYNNLGYVYNKIQNFHKAIKCFKKAI 206
Query: 240 L 240
L
Sbjct: 207 L 207
>UniRef100_UPI000031E2B7 UPI000031E2B7 UniRef100 entry
Length = 320
Score = 68.9 bits (167), Expect = 4e-10
Identities = 49/156 (31%), Positives = 80/156 (50%), Gaps = 4/156 (2%)
Query: 420 KQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGVSLQLSEE 479
++AV P D LG+ + ++ K A+A + SNLG++L+
Sbjct: 66 QKAVKLSPQDAEAHSNLGITLKELGRLDEAETSYKQAIALKPDYAEAHSNLGITLKELGR 125
Query: 480 PSQAEKAYKQALLLATKQ-QAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQPGYAPAFN 538
+AE +YKQA+ L +AH SNLGI + + A+ + +++ L+P YA A +
Sbjct: 126 LDEAETSYKQAIALKPDYAEAH---SNLGITLKELGRLDEAETSYKQAIALKPDYAEAHS 182
Query: 539 NLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
NLG +F A G L+EA+ +++A+ P A SNL
Sbjct: 183 NLGNMFKALGRLDEAETSYKQAIALKPDYAEAHSNL 218
Score = 65.5 bits (158), Expect = 4e-09
Identities = 48/160 (30%), Positives = 79/160 (49%), Gaps = 4/160 (2%)
Query: 413 ERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSNLGV 472
+ A S KQA+A PD LG+ + ++ K A+A + SNLG+
Sbjct: 93 DEAETSYKQAIALKPDYAEAHSNLGITLKELGRLDEAETSYKQAIALKPDYAEAHSNLGI 152
Query: 473 SLQLSEEPSQAEKAYKQALLLATKQ-QAHAILSNLGIFYRHEKKYQRAKAMFTKSLELQP 531
+L+ +AE +YKQA+ L +AH SNLG ++ + A+ + +++ L+P
Sbjct: 153 TLKELGRLDEAETSYKQAIALKPDYAEAH---SNLGNMFKALGRLDEAETSYKQAIALKP 209
Query: 532 GYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAK 571
YA A +NLG G L+EA+ +++A+ P AK
Sbjct: 210 DYAEAHSNLGNTLQELGRLDEAETSYKQAIALKPDYAEAK 249
Score = 63.9 bits (154), Expect = 1e-08
Identities = 52/166 (31%), Positives = 81/166 (48%), Gaps = 8/166 (4%)
Query: 410 EMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSYTWSN 469
E E+ AV K E P W LG L + + + K + AV + SN
Sbjct: 26 EAEKLAVSITK----EFPKHQFGWKALGALFLQSGRNFEAVKVNQKAVKLSPQDAEAHSN 81
Query: 470 LGVSLQLSEEPSQAEKAYKQALLLATK-QQAHAILSNLGIFYRHEKKYQRAKAMFTKSLE 528
LG++L+ +AE +YKQA+ L +AH SNLGI + + A+ + +++
Sbjct: 82 LGITLKELGRLDEAETSYKQAIALKPDYAEAH---SNLGITLKELGRLDEAETSYKQAIA 138
Query: 529 LQPGYAPAFNNLGLVFVAEGLLEEAKYCFEKALQSDPLLDAAKSNL 574
L+P YA A +NLG+ G L+EA+ +++A+ P A SNL
Sbjct: 139 LKPDYAEAHSNLGITLKELGRLDEAETSYKQAIALKPDYAEAHSNL 184
Score = 42.0 bits (97), Expect = 0.051
Identities = 61/252 (24%), Positives = 96/252 (37%), Gaps = 31/252 (12%)
Query: 286 AKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPNCMSTRYAVATHR 345
A++ ++ K K W L F SG + + K +KA KL P
Sbjct: 27 AEKLAVSITKEFPKHQFGWKALGALFLQSGRNFEAVKVNQKAVKLSP------------- 73
Query: 346 MKEAERSQDPSELLSFGGNEMASIIRDGDSSLVELPTAWAGLAMVHKAQHEISSAYESEQ 405
++AE + L G L E T++ + E S
Sbjct: 74 -QDAEAHSNLGITLKELGR------------LDEAETSYKQAIALKPDYAEAHSNLGITL 120
Query: 406 HVLTEMEERAVCSLKQAVAEDPDDPVRWHQLGVHSLCTQQFKTSQKYLKAAVACDRGCSY 465
L ++E A S KQA+A PD LG+ + ++ K A+A +
Sbjct: 121 KELGRLDE-AETSYKQAIALKPDYAEAHSNLGITLKELGRLDEAETSYKQAIALKPDYAE 179
Query: 466 TWSNLGVSLQLSEEPSQAEKAYKQALLLATK-QQAHAILSNLGIFYRHEKKYQRAKAMFT 524
SNLG + +AE +YKQA+ L +AH SNLG + + A+ +
Sbjct: 180 AHSNLGNMFKALGRLDEAETSYKQAIALKPDYAEAH---SNLGNTLQELGRLDEAETSYK 236
Query: 525 KSLELQPGYAPA 536
+++ L+P YA A
Sbjct: 237 QAIALKPDYAEA 248
Score = 40.4 bits (93), Expect = 0.15
Identities = 39/172 (22%), Positives = 69/172 (39%), Gaps = 25/172 (14%)
Query: 181 WNTLGFILLKTGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELIL 240
W LG + L++GR A+ V + ++P++ + NLGI ++G L+ + +++ I
Sbjct: 45 WKALGALFLQSGRNFEAVKVNQKAVKLSPQDAEAHSNLGITLKELGRLDEAETSYKQAIA 104
Query: 241 KDQNHPVALVN-------------------YAALLLCKYASVVAGAGASASEGALADQVM 281
++ A N A L YA + G + E D+
Sbjct: 105 LKPDYAEAHSNLGITLKELGRLDEAETSYKQAIALKPDYAEAHSNLGITLKELGRLDE-- 162
Query: 282 AANVAKECLLAAIKADVKSAHIWGNLAYAFSISGDHRSSSKCLEKAAKLEPN 333
A K+ + A+K D AH NL F G + ++A L+P+
Sbjct: 163 AETSYKQAI--ALKPDYAEAH--SNLGNMFKALGRLDEAETSYKQAIALKPD 210
Score = 37.0 bits (84), Expect = 1.6
Identities = 40/185 (21%), Positives = 77/185 (41%), Gaps = 15/185 (8%)
Query: 84 PKNAHAHFILGLMHQRLNQPQKAILVYEKAEEILLRPETEIERPDL---------LSLVQ 134
P++A AH LG+ + L + +A Y++A I L+P+ +L L +
Sbjct: 73 PQDAEAHSNLGITLKELGRLDEAETSYKQA--IALKPDYAEAHSNLGITLKELGRLDEAE 130
Query: 135 IHHAQCLILE----SSSENSSDKELEPHELKEILSKLKESVQFDIRQAAVWNTLGFILLK 190
+ Q + L+ + N E L E + K+++ A + LG +
Sbjct: 131 TSYKQAIALKPDYAEAHSNLGITLKELGRLDEAETSYKQAIALKPDYAEAHSNLGNMFKA 190
Query: 191 TGRVQSAISVLSSLLAIAPENYDCLGNLGIAYLQIGNLELSAKCFQELILKDQNHPVALV 250
GR+ A + +A+ P+ + NLG ++G L+ + +++ I ++ A
Sbjct: 191 LGRLDEAETSYKQAIALKPDYAEAHSNLGNTLQELGRLDEAETSYKQAIALKPDYAEAKH 250
Query: 251 NYAAL 255
AAL
Sbjct: 251 LLAAL 255
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.129 0.364
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 899,132,336
Number of Sequences: 2790947
Number of extensions: 34922676
Number of successful extensions: 120819
Number of sequences better than 10.0: 2091
Number of HSP's better than 10.0 without gapping: 1282
Number of HSP's successfully gapped in prelim test: 823
Number of HSP's that attempted gapping in prelim test: 107719
Number of HSP's gapped (non-prelim): 10723
length of query: 590
length of database: 848,049,833
effective HSP length: 133
effective length of query: 457
effective length of database: 476,853,882
effective search space: 217922224074
effective search space used: 217922224074
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0174.1


