

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
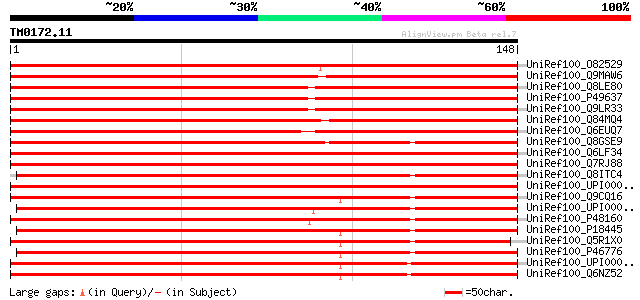
Query= TM0172.11
(148 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O82529 Ribosomal protein L27a [Petunia hybrida] 272 1e-72
UniRef100_Q9MAW6 60S ribosomal protein L27a [Panax ginseng] 265 2e-70
UniRef100_Q8LE80 60S ribosomal protein L27A [Arabidopsis thaliana] 261 3e-69
UniRef100_P49637 60S ribosomal protein L27a-3 [Arabidopsis thali... 260 4e-69
UniRef100_Q9LR33 60S ribosomal protein L27a-2 [Arabidopsis thali... 254 3e-67
UniRef100_Q84MQ4 Putative ribosomal protein L27a [Oryza sativa] 249 7e-66
UniRef100_Q6EUQ7 Putative 60S ribosomal protein L27a [Oryza sativa] 249 1e-65
UniRef100_Q8GSE9 Putative 60S ribosomal protein L27a [Oryza sativa] 241 2e-63
UniRef100_Q6LF34 Putative 60S ribosomal protein l27a [Plasmodium... 224 2e-58
UniRef100_Q7RJ88 Ribosomal protein L27a [Plasmodium yoelii yoelii] 223 5e-58
UniRef100_Q8ITC4 Ribosomal protein L22 [Aequipecten irradians] 222 2e-57
UniRef100_UPI000046BC1D UPI000046BC1D UniRef100 entry 221 2e-57
UniRef100_Q9CQ16 Mus musculus ES cells cDNA, RIKEN full-length e... 213 9e-55
UniRef100_UPI00003ADC0D UPI00003ADC0D UniRef100 entry 212 1e-54
UniRef100_P48160 60S ribosomal protein L27a [Dictyostelium disco... 211 2e-54
UniRef100_P18445 60S ribosomal protein L27a [Rattus norvegicus] 211 3e-54
UniRef100_Q5R1X0 Ribosomal protein L27a [Homo sapiens] 210 6e-54
UniRef100_P46776 60S ribosomal protein L27a [Homo sapiens] 210 6e-54
UniRef100_UPI000019475C UPI000019475C UniRef100 entry 209 8e-54
UniRef100_Q6NZ52 Hypothetical protein [Homo sapiens] 209 1e-53
>UniRef100_O82529 Ribosomal protein L27a [Petunia hybrida]
Length = 150
Score = 272 bits (696), Expect = 1e-72
Identities = 130/150 (86%), Positives = 138/150 (91%), Gaps = 2/150 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQ--AANDKSKAPVIDVTNYGYFKVLGKGVLPA 118
FHKLRN+F+CPTVNIDKLWSL+PQ+VKD+ A K AP+IDVT YGYFKVLGKGVLP
Sbjct: 61 FHKLRNKFYCPTVNIDKLWSLVPQDVKDKVAAGGAKGTAPLIDVTQYGYFKVLGKGVLPE 120
Query: 119 NQPVVVKAKLISKIAEKKIKENGGAVLLTA 148
NQ VVVKAKL+SK AEKKIKE GGAV+LTA
Sbjct: 121 NQAVVVKAKLVSKNAEKKIKEAGGAVVLTA 150
>UniRef100_Q9MAW6 60S ribosomal protein L27a [Panax ginseng]
Length = 146
Score = 265 bits (677), Expect = 2e-70
Identities = 124/148 (83%), Positives = 135/148 (90%), Gaps = 2/148 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKN KKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MTTRFKKNXKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FH+LRN+F+CP VNIDKLWSLLPQE K++A D APVIDVT +GYFKVLGKG LP++
Sbjct: 61 FHRLRNKFYCPVVNIDKLWSLLPQEAKEKA--DAQNAPVIDVTQHGYFKVLGKGFLPSSH 118
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKAKL+SK AEKKIKE GGAV+LTA
Sbjct: 119 PIVVKAKLVSKTAEKKIKEAGGAVVLTA 146
>UniRef100_Q8LE80 60S ribosomal protein L27A [Arabidopsis thaliana]
Length = 146
Score = 261 bits (666), Expect = 3e-69
Identities = 123/148 (83%), Positives = 135/148 (91%), Gaps = 2/148 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKLRN+FFCP VN+DKLWSL+P++VK A + K+ P+IDVT +G+FKVLGKG LP N+
Sbjct: 61 FHKLRNKFFCPIVNLDKLWSLVPEDVK--AKSTKNNVPLIDVTQHGFFKVLGKGHLPENK 118
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P VVKAKLISK AEKKIKE GGAV+LTA
Sbjct: 119 PFVVKAKLISKTAEKKIKEAGGAVVLTA 146
>UniRef100_P49637 60S ribosomal protein L27a-3 [Arabidopsis thaliana]
Length = 146
Score = 260 bits (665), Expect = 4e-69
Identities = 123/148 (83%), Positives = 134/148 (90%), Gaps = 2/148 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKLRN+FFCP VN+DKLWSL+P++VK A + K P+IDVT +G+FKVLGKG LP N+
Sbjct: 61 FHKLRNKFFCPIVNLDKLWSLVPEDVK--AKSTKDNVPLIDVTQHGFFKVLGKGHLPENK 118
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P VVKAKLISK AEKKIKE GGAV+LTA
Sbjct: 119 PFVVKAKLISKTAEKKIKEAGGAVVLTA 146
>UniRef100_Q9LR33 60S ribosomal protein L27a-2 [Arabidopsis thaliana]
Length = 146
Score = 254 bits (649), Expect = 3e-67
Identities = 120/148 (81%), Positives = 131/148 (88%), Gaps = 2/148 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M T KKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MATALKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKLRN+FFCP VN+DKLWSL+P++VK A + K P+IDVT +G+FKVLGKG LP N+
Sbjct: 61 FHKLRNKFFCPIVNLDKLWSLVPEDVK--AKSSKDNVPLIDVTQHGFFKVLGKGHLPENK 118
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P VVKAKLISK AEKKIKE GGAV+LTA
Sbjct: 119 PFVVKAKLISKTAEKKIKEAGGAVVLTA 146
>UniRef100_Q84MQ4 Putative ribosomal protein L27a [Oryza sativa]
Length = 146
Score = 249 bits (637), Expect = 7e-66
Identities = 117/148 (79%), Positives = 130/148 (87%), Gaps = 2/148 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FHKL N+F CP VN+++LWS++P + +A KAPVIDVT +GY KVLGKG+LP +
Sbjct: 61 FHKLSNRFHCPAVNVERLWSMVPTDKAAEAG--AGKAPVIDVTQFGYTKVLGKGMLPPQR 118
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKAKLISK+AEKKIK GGAVLLTA
Sbjct: 119 PIVVKAKLISKVAEKKIKAAGGAVLLTA 146
>UniRef100_Q6EUQ7 Putative 60S ribosomal protein L27a [Oryza sativa]
Length = 144
Score = 249 bits (635), Expect = 1e-65
Identities = 117/148 (79%), Positives = 129/148 (87%), Gaps = 4/148 (2%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FH+L N+F CP VN+++LWS++P E A KAPVIDVT +GY KVLGKG+LP +
Sbjct: 61 FHRLSNRFHCPAVNVERLWSMVPAE----AGAGAGKAPVIDVTQFGYTKVLGKGMLPPER 116
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKAKLISK+AEKKIK GGAVLLTA
Sbjct: 117 PIVVKAKLISKVAEKKIKAAGGAVLLTA 144
>UniRef100_Q8GSE9 Putative 60S ribosomal protein L27a [Oryza sativa]
Length = 146
Score = 241 bits (616), Expect = 2e-63
Identities = 113/148 (76%), Positives = 131/148 (88%), Gaps = 2/148 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
MTT +KNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY
Sbjct: 1 MTTSLRKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
FH+LRN+F+ P VN+++LWS++P E +AA KAP++DVT +GYFKVLGKG+LP +
Sbjct: 61 FHRLRNKFYSPAVNVERLWSMVPAEQAAEAAG-AGKAPLLDVTQFGYFKVLGKGLLP-EK 118
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKAKLISK+AEKKIK GGAV+LTA
Sbjct: 119 PIVVKAKLISKVAEKKIKAAGGAVVLTA 146
>UniRef100_Q6LF34 Putative 60S ribosomal protein l27a [Plasmodium falciparum]
Length = 148
Score = 224 bits (572), Expect = 2e-58
Identities = 103/148 (69%), Positives = 120/148 (80%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M TRFKKNRKKRGHVSAGHGR+GKHRKHPGGRG AGG+HH RI FDKYHPGYFGKVGMR+
Sbjct: 1 MATRFKKNRKKRGHVSAGHGRVGKHRKHPGGRGKAGGLHHMRINFDKYHPGYFGKVGMRH 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
+ L+N+ +CPT+N+DKLW LLP+E K + + +K APVIDVT GYFKVLG G L NQ
Sbjct: 61 LNLLKNRTYCPTINVDKLWGLLPEEKKKEFSENKDIAPVIDVTRKGYFKVLGNGKLKHNQ 120
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKA+ S +AEKKIK GG +L A
Sbjct: 121 PIVVKARYFSSVAEKKIKAVGGQCILVA 148
>UniRef100_Q7RJ88 Ribosomal protein L27a [Plasmodium yoelii yoelii]
Length = 148
Score = 223 bits (569), Expect = 5e-58
Identities = 106/148 (71%), Positives = 120/148 (80%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M TRFKKNRKKRGHVSAGHGRIGKHRKHPGGRG AGGMHH RI FDKYHPGYFGKVGMR+
Sbjct: 1 MATRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGKAGGMHHMRINFDKYHPGYFGKVGMRH 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
+ L+N+ +C T+NIDKLW LLP+E K + A +++ APVIDVT GYFKVLG G L NQ
Sbjct: 61 LNLLKNRKYCRTINIDKLWGLLPEEKKQEFAKNENIAPVIDVTRVGYFKVLGNGNLEHNQ 120
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKA+ S IAEKKIK GG +L A
Sbjct: 121 PIVVKARYFSSIAEKKIKAVGGQCILVA 148
>UniRef100_Q8ITC4 Ribosomal protein L22 [Aequipecten irradians]
Length = 152
Score = 222 bits (565), Expect = 2e-57
Identities = 103/146 (70%), Positives = 119/146 (80%), Gaps = 1/146 (0%)
Query: 3 TRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRYFH 62
T+ KK RK RGHVS GHGRIGKHRKHPGGRGNAGG HHHRI FDKYHPGYFGKVGMRY+H
Sbjct: 8 TKLKKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGQHHHRINFDKYHPGYFGKVGMRYYH 67
Query: 63 KLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQPV 122
K +N+FFCPTVN+DKLWSL+ + +++ A+ K KAPVIDV GY+KVLGKG LP QPV
Sbjct: 68 KTQNKFFCPTVNLDKLWSLVSDQTREKYASSKDKAPVIDVVRAGYYKVLGKGHLP-KQPV 126
Query: 123 VVKAKLISKIAEKKIKENGGAVLLTA 148
+VKA+ S+ AE KIK GGA +L A
Sbjct: 127 IVKARFFSRSAELKIKAAGGACVLVA 152
>UniRef100_UPI000046BC1D UPI000046BC1D UniRef100 entry
Length = 148
Score = 221 bits (564), Expect = 2e-57
Identities = 104/148 (70%), Positives = 120/148 (80%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M TRFKKNRKKRGHVSAGHGRIGKHRKHPGGRG AGGMHH RI FDKYHPGYFGKVGMR+
Sbjct: 1 MATRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGKAGGMHHMRINFDKYHPGYFGKVGMRH 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQ 120
+ L+N+ +C T+N+DKLW LLP+E K + A +++ APVIDVT GYFKVLG G L NQ
Sbjct: 61 LNLLKNRKYCRTINVDKLWGLLPEEKKKEFAKNENIAPVIDVTRVGYFKVLGNGHLEHNQ 120
Query: 121 PVVVKAKLISKIAEKKIKENGGAVLLTA 148
P+VVKA+ S +AEKKIK GG +L A
Sbjct: 121 PIVVKARYFSSMAEKKIKAVGGQCILVA 148
>UniRef100_Q9CQ16 Mus musculus ES cells cDNA, RIKEN full-length enriched library,
clone:2400004P05 product:ribosomal protein L27a, full
insert sequence [Mus musculus]
Length = 148
Score = 213 bits (541), Expect = 9e-55
Identities = 102/149 (68%), Positives = 120/149 (80%), Gaps = 2/149 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M +R +K RK RGHVS GHGRIGKHRKHPGGRGNAGGMHHHRI FDKYHPGYFGKVGMR+
Sbjct: 1 MPSRLRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGMHHHRINFDKYHPGYFGKVGMRH 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKS-KAPVIDVTNYGYFKVLGKGVLPAN 119
+H RNQ FCPTVN+DKLW+L+ ++ + AA +K+ AP+IDV GY+KVLGKG LP
Sbjct: 61 YHLKRNQSFCPTVNLDKLWTLVSEQTRVNAAKNKTGVAPIIDVVRSGYYKVLGKGKLP-K 119
Query: 120 QPVVVKAKLISKIAEKKIKENGGAVLLTA 148
QPV+VKAK S+ AE+KIK GGA +L A
Sbjct: 120 QPVIVKAKFFSRRAEEKIKGVGGACVLVA 148
>UniRef100_UPI00003ADC0D UPI00003ADC0D UniRef100 entry
Length = 148
Score = 212 bits (540), Expect = 1e-54
Identities = 102/147 (69%), Positives = 118/147 (79%), Gaps = 2/147 (1%)
Query: 3 TRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRYFH 62
+R +K RK RGHVS GHGRIGKHRKHPGGRGNAGGMHHHRI FDKYHPGYFGKVGMR++H
Sbjct: 3 SRLRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGMHHHRINFDKYHPGYFGKVGMRHYH 62
Query: 63 KLRNQFFCPTVNIDKLWSLLPQEVK-DQAANDKSKAPVIDVTNYGYFKVLGKGVLPANQP 121
RNQ FCPTVN+DKLW+L+ ++ + + A N APVIDV GY+KVLGKG LP QP
Sbjct: 63 LKRNQKFCPTVNLDKLWTLVTEQTRLNYAKNQAGLAPVIDVVRSGYYKVLGKGKLP-KQP 121
Query: 122 VVVKAKLISKIAEKKIKENGGAVLLTA 148
V+VKAK S+ AE+KIKE GGA +L A
Sbjct: 122 VIVKAKFFSRRAEEKIKEVGGACVLVA 148
>UniRef100_P48160 60S ribosomal protein L27a [Dictyostelium discoideum]
Length = 148
Score = 211 bits (538), Expect = 2e-54
Identities = 98/149 (65%), Positives = 121/149 (80%), Gaps = 2/149 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M TRF K+RK RG V AG+GR+GKHRKHPGGRGNAGG+ HHRI FDKYHPGYFGK+GMR+
Sbjct: 1 MPTRFSKHRKSRGDVCAGYGRVGKHRKHPGGRGNAGGLTHHRINFDKYHPGYFGKLGMRH 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEV-KDQAANDKSKAPVIDVTNYGYFKVLGKGVLPAN 119
FH LRNQ+ CPTV+++K+W+L+P+ V K AA + APV+DVT G+FKVLG G+LP
Sbjct: 61 FHLLRNQYHCPTVSLEKIWTLVPESVRKSLAAKNDGTAPVVDVTQKGFFKVLGHGILP-T 119
Query: 120 QPVVVKAKLISKIAEKKIKENGGAVLLTA 148
QP++VKA+ SK+AEKKIK GGA +L A
Sbjct: 120 QPIIVKARYFSKVAEKKIKAVGGACILVA 148
>UniRef100_P18445 60S ribosomal protein L27a [Rattus norvegicus]
Length = 147
Score = 211 bits (537), Expect = 3e-54
Identities = 101/147 (68%), Positives = 119/147 (80%), Gaps = 2/147 (1%)
Query: 3 TRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRYFH 62
+R +K RK RGHVS GHGRIGKHRKHPGGRGNAGGMHHHRI FDKYHPGYFGKVGMR++H
Sbjct: 2 SRLRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGMHHHRINFDKYHPGYFGKVGMRHYH 61
Query: 63 KLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKS-KAPVIDVTNYGYFKVLGKGVLPANQP 121
RNQ FCPTVN+DKLW+L+ ++ + AA +K+ AP+IDV GY+KVLGKG LP QP
Sbjct: 62 LKRNQSFCPTVNLDKLWTLVSEQTRVNAAKNKNGVAPIIDVVRSGYYKVLGKGKLP-KQP 120
Query: 122 VVVKAKLISKIAEKKIKENGGAVLLTA 148
V+VKAK S+ AE+KIK GGA +L A
Sbjct: 121 VIVKAKFFSRRAEEKIKGVGGACVLVA 147
>UniRef100_Q5R1X0 Ribosomal protein L27a [Homo sapiens]
Length = 148
Score = 210 bits (534), Expect = 6e-54
Identities = 99/147 (67%), Positives = 119/147 (80%), Gaps = 2/147 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M +R +K RK RGHVS GHGRIGKHRKHPGGRGNAGG+HHHRI FDKYHPGYFGKVGM++
Sbjct: 1 MPSRLRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGLHHHRINFDKYHPGYFGKVGMKH 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKS-KAPVIDVTNYGYFKVLGKGVLPAN 119
+H RNQ FCPTVN+DKLW+L+ ++ + AA +K+ AP+IDV GY+KVLGKG LP
Sbjct: 61 YHLKRNQSFCPTVNLDKLWTLVSEQTRVNAAKNKTGAAPIIDVVRSGYYKVLGKGKLP-K 119
Query: 120 QPVVVKAKLISKIAEKKIKENGGAVLL 146
QPV+VKAK S+ AE+KIK GGA +L
Sbjct: 120 QPVIVKAKFFSRRAEEKIKSVGGACVL 146
>UniRef100_P46776 60S ribosomal protein L27a [Homo sapiens]
Length = 147
Score = 210 bits (534), Expect = 6e-54
Identities = 99/147 (67%), Positives = 119/147 (80%), Gaps = 2/147 (1%)
Query: 3 TRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRYFH 62
+R +K RK RGHVS GHGRIGKHRKHPGGRGNAGG+HHHRI FDKYHPGYFGKVGM+++H
Sbjct: 2 SRLRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGLHHHRINFDKYHPGYFGKVGMKHYH 61
Query: 63 KLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKS-KAPVIDVTNYGYFKVLGKGVLPANQP 121
RNQ FCPTVN+DKLW+L+ ++ + AA +K+ AP+IDV GY+KVLGKG LP QP
Sbjct: 62 LKRNQSFCPTVNLDKLWTLVSEQTRVNAAKNKTGAAPIIDVVRSGYYKVLGKGKLP-KQP 120
Query: 122 VVVKAKLISKIAEKKIKENGGAVLLTA 148
V+VKAK S+ AE+KIK GGA +L A
Sbjct: 121 VIVKAKFFSRRAEEKIKSVGGACVLVA 147
>UniRef100_UPI000019475C UPI000019475C UniRef100 entry
Length = 148
Score = 209 bits (533), Expect = 8e-54
Identities = 101/149 (67%), Positives = 119/149 (79%), Gaps = 2/149 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M +R +K RK RGHVS GHGRIGKHRKHPGGRGNAGGMHHHRI FDKYHPGYFGKVGMR+
Sbjct: 1 MPSRLRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGMHHHRINFDKYHPGYFGKVGMRH 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKS-KAPVIDVTNYGYFKVLGKGVLPAN 119
+H RNQ FCPTVN+DKLW+L+ ++ + AA +K+ AP+IDV GY+KVLGKG L
Sbjct: 61 YHLKRNQSFCPTVNLDKLWTLVSEQTRVNAAKNKTGVAPIIDVVRSGYYKVLGKGKL-LK 119
Query: 120 QPVVVKAKLISKIAEKKIKENGGAVLLTA 148
QPV+VKAK S+ AE+KIK GGA +L A
Sbjct: 120 QPVIVKAKFFSRRAEEKIKGVGGACVLVA 148
>UniRef100_Q6NZ52 Hypothetical protein [Homo sapiens]
Length = 148
Score = 209 bits (532), Expect = 1e-53
Identities = 100/149 (67%), Positives = 121/149 (81%), Gaps = 2/149 (1%)
Query: 1 MTTRFKKNRKKRGHVSAGHGRIGKHRKHPGGRGNAGGMHHHRILFDKYHPGYFGKVGMRY 60
M +R +K RK RGHVS GHGRIGKHRKHPGGRGNAGG+HHHRI FDKYHPGYFGKVGM++
Sbjct: 1 MPSRLRKTRKLRGHVSHGHGRIGKHRKHPGGRGNAGGLHHHRINFDKYHPGYFGKVGMKH 60
Query: 61 FHKLRNQFFCPTVNIDKLWSLLPQEVKDQAANDKS-KAPVIDVTNYGYFKVLGKGVLPAN 119
++ RNQ FCPTVN+DKLW+L+ ++ + AA +K+ AP+IDV GY+KVLGKG L A
Sbjct: 61 YNLKRNQSFCPTVNLDKLWTLVSEQTRVNAAKNKTGAAPIIDVVRSGYYKVLGKGKL-AK 119
Query: 120 QPVVVKAKLISKIAEKKIKENGGAVLLTA 148
QPV+VKAKL S+ AE+KIK GGA +L A
Sbjct: 120 QPVIVKAKLFSRRAEEKIKSVGGACVLVA 148
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 292,094,649
Number of Sequences: 2790947
Number of extensions: 13097366
Number of successful extensions: 30479
Number of sequences better than 10.0: 244
Number of HSP's better than 10.0 without gapping: 145
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 29870
Number of HSP's gapped (non-prelim): 321
length of query: 148
length of database: 848,049,833
effective HSP length: 124
effective length of query: 24
effective length of database: 501,972,405
effective search space: 12047337720
effective search space used: 12047337720
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Lotus: description of TM0172.11


