

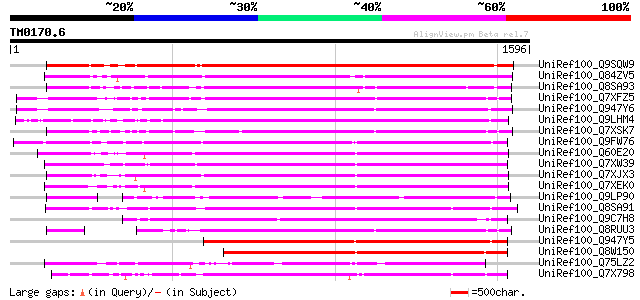
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0170.6
(1596 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SQW9 Putative retroelement pol polyprotein [Arabidop... 1177 0.0
UniRef100_Q84ZV5 Polyprotein [Glycine max] 1136 0.0
UniRef100_Q8SA93 Putative polyprotein [Zea mays] 1047 0.0
UniRef100_Q7XFZ5 Putative retroelement [Oryza sativa] 1045 0.0
UniRef100_Q947Y6 Putative retroelement [Oryza sativa] 1038 0.0
UniRef100_Q9LHM4 Retroelement pol polyprotein [Arabidopsis thali... 1036 0.0
UniRef100_Q7XSK7 OSJNBa0059D20.8 protein [Oryza sativa] 1028 0.0
UniRef100_Q9FW76 Putative gag-pol polyprotein [Oryza sativa] 983 0.0
UniRef100_Q60E20 Putative polyprotein [Oryza sativa] 971 0.0
UniRef100_Q7XW39 OSJNBb0062H02.17 protein [Oryza sativa] 961 0.0
UniRef100_Q7XJX3 OSJNBa0004L19.16 protein [Oryza sativa] 961 0.0
UniRef100_Q7XEK0 Hypothetical protein [Oryza sativa] 949 0.0
UniRef100_Q9LP90 T32E20.30 [Arabidopsis thaliana] 945 0.0
UniRef100_Q8SA91 Putative gag-pol polyprotein [Zea mays] 936 0.0
UniRef100_Q9C7H8 Gypsy/Ty3 element polyprotein, putative [Arabid... 916 0.0
UniRef100_Q8RUU3 Putative gag-pol polyprotein [Oryza sativa] 871 0.0
UniRef100_Q947Y5 Putative retroelement [Oryza sativa] 858 0.0
UniRef100_Q8W150 Polyprotein [Oryza sativa] 810 0.0
UniRef100_Q75LZ2 Putative gag-pol polyprotein [Oryza sativa] 764 0.0
UniRef100_Q7X798 OSJNBb0108J11.19 protein [Oryza sativa] 751 0.0
>UniRef100_Q9SQW9 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1661
Score = 1177 bits (3045), Expect = 0.0
Identities = 643/1453 (44%), Positives = 887/1453 (60%), Gaps = 72/1453 (4%)
Query: 113 KKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEEL 172
+ V+ P ++G + W+ R E F T E K+ A + G ++ ++ E+
Sbjct: 229 RTVDYPAYEGGNADDWLFRLEQCFLSNRTLEEEKLEKAVSCLTGASVTWWRCSKDREQIY 288
Query: 173 TWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYF 232
TW F++ + R+ + + L +RQ G VEEY FE LT +P + + F
Sbjct: 289 TWREFQEKFMLRFRPSRGSSAVDHLLNVRQTGTVEEYRERFEELTVDLPHVTSDILESAF 348
Query: 233 LHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGD---GVAGQARANRFGGNGSR 289
L+GL+ +R +V V VN A ++ + + +E + + V QAR N N
Sbjct: 349 LNGLRRSLRDQV---VRCRPVNLADIVEIAKLIESQERNAVSYQVRNQARTNTAPFNNQV 405
Query: 290 SGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSY 349
S G++ + R P D T+ +G
Sbjct: 406 S----------------TGSRVVDRAPTRQPFIPSRDTTRASGS---------------- 433
Query: 350 PQLMERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNEDEEETQ 409
E R C CG + + H C + L+ L + E E++E L E+NE E +
Sbjct: 434 ---GEARNSNPCRYCGDRWFQGHKCKPQKLKGLAITE---EVEEESPLIEELNEPLTEEE 487
Query: 410 GE------LSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRL 463
G+ +M+L L ++ ++MK+RG I VV+LVDSGAT NF+ LVR
Sbjct: 488 GDPEPAEGFKVMTLSSLNDESQ--EQSMKMRGYIGNTKVVLLVDSGATCNFISEALVREK 545
Query: 464 GWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLK 523
GW V T VK+G G ++ G+C + +E+ LFDLG D+VLG WL
Sbjct: 546 GWLVTQTRSFGVKVGGGRIIKSSGKCVDIPLEVQGIEFVQDYYLFDLGDLDLVLGFSWLA 605
Query: 524 TLGDTIVNWDTQLMSFWSDKKWITLQGMDT--RGE----HMEALQSITATGERKPGLLGT 577
LG+T NW +S+ + W++L G RG+ ME + T T LL
Sbjct: 606 GLGETRANWRDLRISWQIGRTWVSLYGDPDLCRGQISMRSMERVIKYTGTAY----LLEL 661
Query: 578 RNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNV 637
+ ++ KQ E ++ + +LL ++ VFQ Q LPP R REH IT+QEG PVN+
Sbjct: 662 ASLFESKKQ---EEQTALQPAIQRLLDQYQGVFQTPQLLPPVRNREHAITLQEGSSPVNI 718
Query: 638 RPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVT 697
RPYRY KNEIEK VREML+A IIR S S YSSPV+LVKKKD W CVDYRALN T
Sbjct: 719 RPYRYSFAQKNEIEKLVREMLNAQIIRPSVSPYSSPVLLVKKKDGGWRFCVDYRALNEAT 778
Query: 698 VPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMP 757
+PDK+PIPVIEELLDEL GA FSKLDLKSGY Q+R++ DV KTAF+THEGHYE++VMP
Sbjct: 779 IPDKYPIPVIEELLDELKGATVFSKLDLKSGYFQIRMKLSDVEKTAFKTHEGHYEFLVMP 838
Query: 758 FGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLT 817
FGL NAPSTFQS+MN++FRP LR+ VLVFFDDILVYS H+ HLE VL++L HQ
Sbjct: 839 FGLTNAPSTFQSVMNDLFRPYLRKFVLVFFDDILVYSPDMKTHLKHLETVLQLLHLHQFY 898
Query: 818 ANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKF 877
AN KKC F + + YLGH+ISEQGVA DP KV ++ WP PK+V +RGFLG TGYYR+F
Sbjct: 899 ANFKKCTFGSTRISYLGHIISEQGVATDPEKVEAMLQWPLPKSVTELRGFLGFTGYYRRF 958
Query: 878 IQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASG 937
++NYG+IARPL + KK++F+W + A AF+ LK ++ PVL LPDF QEF +E DASG
Sbjct: 959 VKNYGQIARPLRDQLKKNSFDWNEAATSAFQALKAAVSALPVLVLPDFQQEFTVETDASG 1018
Query: 938 IGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQ 997
+GIGA+L K IA+ S+A + +S YE+EL+A+ A+ W+ YL ++F++ TDQ
Sbjct: 1019 MGIGAVLSQNKRLIAFLSQAFSSQGRIRSVYERELLAIVKAVTKWKHYLSSKEFIIKTDQ 1078
Query: 998 RSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRPL-VSY 1056
RSL+ L QK V+ QQ WA+KL G + I YKPG N+ ADALSR L L ++
Sbjct: 1079 RSLRHLLEQKSVSTIQQRWASKLSGLKYRIEYKPGVDNKVADALSRRPPTEALSQLTITG 1138
Query: 1057 PLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPK 1116
P + + E Q+D +L QI+ A + V G + + +VI S IPK
Sbjct: 1139 PPTIDLTALKAEIQQDHELSQILKNWAQGDHHDSDFTVADGLIYRKGCLVIPVGSPFIPK 1198
Query: 1117 LLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLL 1176
+LE+FHT+P GGH G L+T++R+ + +YWRG+ K + ++ C +CQ +KY SPAGLL
Sbjct: 1199 MLEKFHTSPIGGHEGALKTFKRLTSEVYWRGLRKDVVNYIKGCQICQENKYSTLSPAGLL 1258
Query: 1177 QPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFA 1236
PLPIP+++W D+SL+F+ GLP S F I VVVDRL+KYSHFIPLKHP+TA++V E F
Sbjct: 1259 SPLPIPQQIWSDVSLDFVEGLPSSNRFNCILVVVDRLSKYSHFIPLKHPFTAKTVVEAFI 1318
Query: 1237 KEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLET 1296
+++++LHG P++++SDRD +F+S FW E+F+LQGT + S+AYHP++DGQTE+VNRCLE+
Sbjct: 1319 RDVVKLHGFPNTLVSDRDRIFLSGFWSELFKLQGTGLQKSTAYHPQTDGQTEVVNRCLES 1378
Query: 1297 YLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEA 1356
YLRCFA +P +W WL WAEYW+NTSYHSAT+ TPF+AVYGR+PPVL R+ T
Sbjct: 1379 YLRCFAGRRPTSWFQWLPWAEYWYNTSYHSATKTTPFQAVYGREPPVLLRYGDIPTNNAN 1438
Query: 1357 VEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANR 1416
VE+ L+DRD L +L+++L AQ +M+ A+ R+ FE+ EWV++K+R +RQ S+A+R
Sbjct: 1439 VEELLKDRDGMLVELRENLEIAQAQMKKAADKSRRDVAFEIDEWVYLKLRPYRQSSVAHR 1498
Query: 1417 VHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGLPEQL 1476
+ KL+ RYFGP+ ++ R+G VAY+L+LPE + IHPVFH+S LK+AV LP+ L
Sbjct: 1499 KNEKLSQRYFGPFKVLHRIGQVAYKLQLPEHSTIHPVFHVSQLKRAVPPSFTPQELPKIL 1558
Query: 1477 DGEELPSIQPALVLARRDILRQGESIS--QVLVQWQGKTPEEATWEDLATIRSEFPVFNL 1534
+ P +L +RQ + S +VLVQW G + E+TWE L T+ ++P F+L
Sbjct: 1559 SPTLEWNTGPEKLLD----IRQSNTNSGPEVLVQWSGLSTLESTWEPLLTLVQQYPDFDL 1614
Query: 1535 EDKVVSSEGSIVR 1547
EDKV GSI R
Sbjct: 1615 EDKVSLLRGSIDR 1627
>UniRef100_Q84ZV5 Polyprotein [Glycine max]
Length = 1552
Score = 1136 bits (2938), Expect = 0.0
Identities = 618/1462 (42%), Positives = 869/1462 (59%), Gaps = 71/1462 (4%)
Query: 106 NEFRQSVKKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSL 165
+ F+ K++ P FDGK++ WI +AE +F T ++ +A + ++ + +Y L
Sbjct: 91 SSFQVRSVKLDFPRFDGKNVMDWIFKAEQFFDYYATPDADRLIIASVHLDQDVVPWYQML 150
Query: 166 LATEEELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPE 225
TE +W+ F AL +G + L +L Q V EY F L ++ L
Sbjct: 151 QKTEPFSSWQAFTRALELDFGPSAYDCPRATLFKLNQSATVNEYYMQFTALVNRVDGLSA 210
Query: 226 KQYQGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFGG 285
+ F+ GL+EEI V+++ R L A+ K + ++
Sbjct: 211 EAILDCFVSGLQEEISRDVKAM-------EPRTLTKAVALAKLFE----------EKYTS 253
Query: 286 NGSRSGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKN-------TGDIRRAG 338
FS + T T P +N + D K T +
Sbjct: 254 PPKTKTFSNLARNF---------TSNTSATQKYPPTNQKNDNPKPNLPPLLPTPSTKPFN 304
Query: 339 PRDRGFNHLSYPQLMERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEE-DGEELDESKML 397
R++ +S ++ RR+K LC+ C + H CP++ + LL LEE D ++ DE M+
Sbjct: 305 LRNQNIKKISPAEIQLRREKNLCYFCDEKFSPAHKCPNRQVMLLQLEETDEDQTDEQVMV 364
Query: 398 AMEVNEDEEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDC 457
E N D++ L+ M + G+ T++ G + G+ V +LVD G++ NF+
Sbjct: 365 TEEANMDDDTHHLSLNAMR------GSNGVG-TIRFTGQVGGIAVKILVDGGSSDNFIQP 417
Query: 458 FLVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVL 517
+ + L V P + V +G+G A+G L + + ++ L + G D++L
Sbjct: 418 RVAQVLKLPVEPAPNLRVLVGNGQILSAEGIVQQLPLHIQGQEVKVPVYLLQISGADVIL 477
Query: 518 GIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQG------MDTRGEHMEALQSITATGERK 571
G WL TLG + ++ + F+ + K+ITLQG + H LQ+ + E
Sbjct: 478 GSTWLATLGPHVADYAALTLKFFQNDKFITLQGEGNSEATQAQLHHFRRLQNTKSIEECF 537
Query: 572 P-GLLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQE 630
L+ E K LP I+ EL LL+ + VF LPP R ++H I +++
Sbjct: 538 AIQLIQKEVPEDTLKDLPTNIDP----ELAILLHTYAQVFAVPASLPPQREQDHAIPLKQ 593
Query: 631 GKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDY 690
G GPV VRPYRYPH K++IEK ++EML GII+ S S +S P++LVKKKD +W C DY
Sbjct: 594 GSGPVKVRPYRYPHTQKDQIEKMIQEMLVQGIIQPSNSPFSLPILLVKKKDGSWRFCTDY 653
Query: 691 RALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGH 750
RALN++TV D FP+P ++ELLDELHGA +FSKLDL+SGYHQ+ V+ ED KTAFRTH GH
Sbjct: 654 RALNAITVKDSFPMPTVDELLDELHGAQYFSKLDLRSGYHQILVQPEDREKTAFRTHHGH 713
Query: 751 YEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEV 810
YE++VMPFGL NAP+TFQ LMN++F+ LR+ VLVFFDDIL+YS +W DH+ HLE VL+
Sbjct: 714 YEWLVMPFGLTNAPATFQCLMNKIFQFALRKFVLVFFDDILIYSASWKDHLKHLESVLQT 773
Query: 811 LQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGL 870
L+ HQL A KC F V+YLGH +S GV+++ KV +V WP P NVK +RGFLGL
Sbjct: 774 LKQHQLFARLSKCSFGDTEVDYLGHKVSGLGVSMENTKVQAVLDWPTPNNVKQLRGFLGL 833
Query: 871 TGYYRKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFR 930
TGYYR+FI++Y IA PLT+L +KD+F W +A+ AF LK+ +T APVL+LPDFSQ F
Sbjct: 834 TGYYRRFIKSYANIAGPLTDLLQKDSFLWNNEAEAAFVKLKKAMTEAPVLSLPDFSQPFI 893
Query: 931 LECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRK 990
LE DASGIG+GA+L HPIAYFSK L PR +SAY +EL+A+ A+ +R YLLG K
Sbjct: 894 LETDASGIGVGAVLGQNGHPIAYFSKKLAPRMQKQSAYTRELLAITEALSKFRHYLLGNK 953
Query: 991 FVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGEL 1050
F++ TDQRSLK + Q + T +QQ W K LGY+F+I YKPGK NQ ADALSR+
Sbjct: 954 FIIRTDQRSLKSLMDQSLQTPEQQAWLHKFLGYDFKIEYKPGKDNQAADALSRMFMLA-- 1011
Query: 1051 RPLVSYPLWEEQGVIADENQK-----DPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRV 1105
W E I E + DP L Q++ A Y V +G L ++DRV
Sbjct: 1012 --------WSEPHSIFLEELRARLISDPHLKQLMETYKQGADA-SHYTVREGLLYWKDRV 1062
Query: 1106 VISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRH 1165
VI + ++ K+L+E+H++P GGH+G RT R+ YW M + ++ ++ C +CQ+
Sbjct: 1063 VIPAEEEIVNKILQEYHSSPIGGHAGITRTLARLKAQFYWPKMQEDVKAYIQKCLICQQA 1122
Query: 1166 KYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHP 1225
K + PAGLLQPLPIP++VWED++++FI GLP S G I VV+DRLTKY+HFIPLK
Sbjct: 1123 KSNNTLPAGLLQPLPIPQQVWEDVAMDFITGLPNSFGLSVIMVVIDRLTKYAHFIPLKAD 1182
Query: 1226 YTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDG 1285
Y ++ VAE F I++LHG+P SI+SDRD +F S FW+ +F+LQGT MSSAYHP+SDG
Sbjct: 1183 YNSKVVAEAFMSHIVKLHGIPRSIVSDRDRVFTSTFWQHLFKLQGTTLAMSSAYHPQSDG 1242
Query: 1286 QTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLT 1345
Q+E++N+CLE YLRCF + PK W L WAE+W+NT+YH + TPF A+YGR+PP LT
Sbjct: 1243 QSEVLNKCLEMYLRCFTYEHPKGWVKALPWAEFWYNTAYHMSLGMTPFRALYGREPPTLT 1302
Query: 1346 RWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKI 1405
R V + L DRD L +LK +L AQ+ M+ QA+ KR F++G+ V VK+
Sbjct: 1303 RQACSIDDPAEVREQLTDRDALLAKLKINLTRAQQVMKRQADKKRLDVSFQIGDEVLVKL 1362
Query: 1406 RAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGN 1465
+ +RQ S R + KL+ RYFGP+ ++ ++G VAY+L+LP ARIHPVFH+S LK G
Sbjct: 1363 QPYRQHSAVLRKNQKLSMRYFGPFKVLAKIGDVAYKLELPSAARIHPVFHVSQLKPFNGT 1422
Query: 1466 YVEEPGLPEQLDGEEL-PSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLAT 1524
++P LP L E+ P +QP +LA R I+R I Q+LVQW+ +EATWED+
Sbjct: 1423 -AQDPYLPLPLTVTEMGPVMQPVKILASRIIIRGHNQIEQILVQWENGLQDEATWEDIED 1481
Query: 1525 IRSEFPVFNLEDKVV-SSEGSI 1545
I++ +P FNLEDKVV EG++
Sbjct: 1482 IKASYPTFNLEDKVVFKGEGNV 1503
>UniRef100_Q8SA93 Putative polyprotein [Zea mays]
Length = 2749
Score = 1047 bits (2708), Expect = 0.0
Identities = 601/1457 (41%), Positives = 823/1457 (56%), Gaps = 109/1457 (7%)
Query: 114 KVELPMFDGK-DLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEEL 172
K++ P +DG D W+++ E +F+ Q T + LA ++G +Y +L E
Sbjct: 472 KIDFPTYDGSVDPLNWLNQCEQFFRGQRTLVTDRTWLASYHLKGAAQTWYYALEQDEGMP 531
Query: 173 TWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYF 232
TW RF++ R+G G +L+ L V++Y F + KL +Q F
Sbjct: 532 TWGRFKEVCTLRFGPPVRGTRLSELARLPFTSTVQDYADRFNAMLGHTRKLDAQQKAELF 591
Query: 233 LHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFGGNGSRSGF 292
+ GL + IR V + + + RA E+ AR G SR G
Sbjct: 592 VGGLPDHIRADV---AIRDPQDLQSAMYLARAFEQRAAAQTTPPPAR----GFRQSRPGL 644
Query: 293 SGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQL 352
RP + P + G A P F L+ +
Sbjct: 645 PAPP---------------------RPLTAPPTAAAQPAGTAAPARP----FRRLTPAEQ 679
Query: 353 MERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESK------MLAMEVNEDEE 406
ERR++GLCF C PY R HVCP RL LE D DE + +A+E E
Sbjct: 680 QERRRQGLCFNCDEPYVRGHVCP----RLFYLENDDYIDDEPQEEGADLQIALE-QEPPS 734
Query: 407 ETQGELSLMSLCEL-GMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGW 465
+ +SL L G++T P M L +ING +V LVDSG+T NF+ L+ RL
Sbjct: 735 RAAAIIPTVSLHALAGVRT---PNAMLLPVSINGHRLVALVDSGSTTNFMSVGLMSRL-- 789
Query: 466 EVVDTPRMTVKL----GDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEW 521
++ TP T+K+ GD Q R L++ ++ + C LG D++LG E+
Sbjct: 790 QLPSTPHPTIKVQVANGDNIPCQGMARSVDLRVGTEQFSIDCIG--LTLGTFDVILGFEF 847
Query: 522 LKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHMEALQSITATGERKPGLLGTRNEE 581
L+ LG + + D MSF + I G+ PG + +
Sbjct: 848 LRLLGPILWDCDRLSMSFTKGGRHIIWSGLGA------------------PGAVPPQPAA 889
Query: 582 KAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNVRPYR 641
P + LD LL +F++VF E QGLPP R +H I + G PV VRPYR
Sbjct: 890 CVVSSTPTQ------PLLDDLLRQFELVFAEPQGLPPARPYDHRIHLLPGAAPVAVRPYR 943
Query: 642 YPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDK 701
YP K+E+E+Q ML+ GIIR STS +S+PV+LV+K D +W C+DYRALN+ T DK
Sbjct: 944 YPQLQKDELERQCSAMLAQGIIRPSTSPFSAPVLLVRKPDNSWRFCIDYRALNAKTSKDK 1003
Query: 702 FPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLM 761
FPIPV++ELLDELHGA FF+KLDL+SGYHQVR+ DV KTAFRTHEGHYE++VMPFGL
Sbjct: 1004 FPIPVVDELLDELHGAHFFTKLDLRSGYHQVRMHPADVEKTAFRTHEGHYEFLVMPFGLS 1063
Query: 762 NAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKK 821
NAP+TFQ+LMN+V RP LR+ VLVFFDDIL+YSKTW++H+ H+ VL L+ HQL +
Sbjct: 1064 NAPATFQALMNDVLRPYLRKYVLVFFDDILIYSKTWAEHLQHISIVLHALRDHQLHLKRS 1123
Query: 822 KCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNY 881
KC F + V YLGH+IS GVA+D KV +V SWP P + +G+RGFLGL GYYRKFI+++
Sbjct: 1124 KCSFGARSVAYLGHVISAAGVAMDAAKVEAVSSWPAPHSARGLRGFLGLAGYYRKFIRDF 1183
Query: 882 GKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIG 941
G IA PLT L ++DAF W Q AF+ LK LTT PVL +P+F + F ++CDASG G G
Sbjct: 1184 GVIAAPLTRLLRRDAFTWDDDTQAAFQQLKTALTTGPVLQMPNFEKTFVVDCDASGTGFG 1243
Query: 942 AILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLK 1001
A+L G P+A+FS+ R+L +AYE+EL+ + A++HWRPYL GR F V TD SLK
Sbjct: 1244 AVLHQGAGPVAFFSRPFVTRHLKLAAYERELIGLVQAVRHWRPYLWGRHFAVRTDHYSLK 1303
Query: 1002 EFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRID------EAGELRPLVS 1055
L Q++ T Q W +KL G++FE+ Y+PG+ N ADALSR D AGEL +
Sbjct: 1304 YLLDQRLSTVPQHQWLSKLFGFDFEVEYRPGRLNVAADALSRRDAELLQPSAGELGAAAA 1363
Query: 1056 YPLWEEQGVIADENQK-------DPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVIS 1108
L D+ ++ +LCQ + + L +R+ G LL+ R+ +
Sbjct: 1364 LALSGPSFAFLDDIRRATATSPDSSRLCQQLQDGTLTAP----WRLEDGLLLHGSRIYVP 1419
Query: 1109 NKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYL 1168
N L + + H+ GH G +T R+ Y G + ++V C CQR+K
Sbjct: 1420 NHGDLRHQAILLAHS---AGHEGIQKTLHRLRAEFYVPGDRTLVADWVRTCTTCQRNKTE 1476
Query: 1169 ASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTA 1228
PAGLLQPL +P +VW DIS++FI GLP+ G I VVDR +KY+HFIPL HPYTA
Sbjct: 1477 TLQPAGLLQPLQVPSQVWADISMDFIEGLPKVGGKSVILTVVDRFSKYAHFIPLGHPYTA 1536
Query: 1229 RSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTE 1288
SVA F I+RLHG PSSI+SDRDP+F H W+++F+ G +MS+A+HP++DGQ+E
Sbjct: 1537 ASVARAFFDGIVRLHGFPSSIVSDRDPVFTGHVWRDLFKCAGVSLRMSTAFHPQTDGQSE 1596
Query: 1289 IVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWV 1348
+VN+ + YLRC D+P+ W WL WAEY +NTS+H+A + TPFE VYGR PP + +
Sbjct: 1597 VVNKVIAMYLRCVTGDRPRAWVDWLSWAEYCYNTSFHTALRATPFEVVYGRPPPPILPYQ 1656
Query: 1349 LGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAH 1408
G R A E+ L+DRD L +++Q LV AQ+ + ++ + E G+WV++++ H
Sbjct: 1657 AGSARTAAAEELLRDRDNILAEVRQRLVQAQQLSKRYYDAGHRDMELADGDWVWLRL-LH 1715
Query: 1409 RQV-SLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYV 1467
R V SL R KL RY GP+ ++ R+G VAYRL+LPEGAR+H VFH+ LLK+ G
Sbjct: 1716 RPVQSLEPRAKGKLGPRYAGPFRVLERIGKVAYRLELPEGARLHDVFHVGLLKRHKGEPP 1775
Query: 1468 EE-PGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIR 1526
E+ GLP +G LP+ P VL +Q +LVQWQG +PEEATWE L R
Sbjct: 1776 EQRAGLPPVQNGRLLPA--PLKVLR----AQQRRGTWHILVQWQGLSPEEATWEPLDDFR 1829
Query: 1527 SEFPVFNLEDKVVSSEG 1543
+P F LED++ + G
Sbjct: 1830 GLYPDFQLEDELFAHAG 1846
>UniRef100_Q7XFZ5 Putative retroelement [Oryza sativa]
Length = 1476
Score = 1045 bits (2703), Expect = 0.0
Identities = 608/1535 (39%), Positives = 839/1535 (54%), Gaps = 119/1535 (7%)
Query: 21 MEAKIDAMESELAAIRTALAAVTTAMKDLPTTLGTMVEKAVGKSVGIDVDSGDARPEREP 80
+++ ++ + LA+++++ A T A++D + + + + SG P+R P
Sbjct: 5 LKSTLETLAKTLASLQSSSEATTKAIEDNTQAIAALSVARTTEKASTE-SSGTPAPDRPP 63
Query: 81 REKTPESVGLRESGSDQPVLQGEALNEFRQSVKKVELPMFDGK-DLAGWISRAEIYFKVQ 139
+ PE P +DGK D +I+R E +F Q
Sbjct: 64 KHWRPE------------------------------FPKYDGKTDPLAFINRCESFFIQQ 93
Query: 140 ETSPEVKVSLAQLSMEGGTIHFYNSLLATEEELTWERFRDALLERYGGNGDGDVYEQLSE 199
P + +A +++ G +Y + E TWERF++ L RYG +LS
Sbjct: 94 HVIPAERTWMASYNLQDGAQLWYMHVQDNEGTPTWERFKELLNLRYGPPLRSVPLFELSA 153
Query: 200 LRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKVRSLVAMGGVNRARLL 259
R+ VE+Y F+ L + +L E Q F GL + +V+ L
Sbjct: 154 CRRTSTVEDYQDRFQALLPRAGRLEEAQQVQLFTGGLLPPLSLQVQQQKPASLEEAMSLA 213
Query: 260 VVTRAVEKEVKGDGVAGQARANRFGGNGSRSGFSGATKGNGTEWIWVKGNKETGQTVNRP 319
+E + F T G P
Sbjct: 214 RQFELMEPYL----------------------FPATTSARGV----------------LP 235
Query: 320 NSNPRGDQTKNTGDIRRAGP-----RDRGFNHLSYPQLMERRQKGLCFKCGGPYHRNHVC 374
PR +TG + + P R LS Q ERR+ GLC+ C Y R+H
Sbjct: 236 TPAPR----PSTGPVVKPAPATVTVEGRPVKRLSQAQQEERRRLGLCYNCDEKYSRSH-- 289
Query: 375 PDKHLRLLILEEDG--EELDESKMLAMEVNEDEEETQGELSLMSLCELGMKTGGIP--RT 430
+K + L E G EE D++ V +D EE E + SL + GIP +
Sbjct: 290 -NKVCKRLFFVEGGAIEEGDDT------VEDDTEEATVEAPVFSLHAVA----GIPLGKP 338
Query: 431 MKLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCA 490
+ L+ T+ +V LVD+G+THNF+ R G V PR+T + +G K G
Sbjct: 339 ILLQVTLGAASLVALVDTGSTHNFIGEDAALRTGLPVQPRPRLTATVANGEKVSCPGVLR 398
Query: 491 GLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWD--TQLMSFWSDKKWITL 548
I + + L G D+VLG +W+ LG TI WD T +SF + ++
Sbjct: 399 RAPITIQGMAFDVDLYVMPLAGYDMVLGTQWMAHLGTTIA-WDVTTGTVSFQHQGRTVSW 457
Query: 549 QGMDTRGEHMEALQSITATGERKPGLLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDV 608
Q + H A +TG G+ + A P LD LL FD
Sbjct: 458 QSLPP---HQRADVHAVSTGTSLVAATGSSSSTPAPTTEPAL--------LDGLLGSFDD 506
Query: 609 VFQEKQGLPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTS 668
VF E +GLPP RGR+H I + G PV VRPYRYP HK+E+E+Q M+ G+IR+STS
Sbjct: 507 VFAEPRGLPPPRGRDHAIHLLPGAPPVAVRPYRYPVAHKDELERQCAVMMEQGLIRRSTS 566
Query: 669 AYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSG 728
A+SSPV+LVKK D +W CVDYRALN++T+ D +PIPV++ELLDELHGA FF+KLDL+SG
Sbjct: 567 AFSSPVLLVKKADGSWRFCVDYRALNAITIKDAYPIPVVDELLDELHGAKFFTKLDLRSG 626
Query: 729 YHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFD 788
YHQVR+R EDV KTAFRTH+G YE++VMPFGL NAP+TFQ+LMN++ R LRR VLVFFD
Sbjct: 627 YHQVRMRAEDVAKTAFRTHDGLYEFLVMPFGLCNAPATFQALMNDILRIYLRRFVLVFFD 686
Query: 789 DILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNK 848
DIL+YS TW+DH+ H+ VL +L+ H+L + KC F + YLGH+I GV++DP K
Sbjct: 687 DILIYSNTWADHLRHIRAVLLLLRQHRLFVKRSKCAFGVSSISYLGHIIGATGVSMDPAK 746
Query: 849 VLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFE 908
V +V WP+P++ + VRGFLGL GYYRKF+ +YG IA PLT LTKK+ F W + AF
Sbjct: 747 VQAVVDWPQPRSARTVRGFLGLAGYYRKFVHDYGTIAAPLTALTKKEGFRWSDEVATAFH 806
Query: 909 DLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAY 968
LK +TTAPVLALPDF + F +ECDAS G GA+LL KHP+A+FS+ + PR+ A +AY
Sbjct: 807 ALKHAVTTAPVLALPDFVKPFVVECDASTHGFGAVLLQDKHPLAFFSRPVAPRHRALAAY 866
Query: 969 EKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEII 1028
E+EL+ + LAI+HWRPYL GR FVV TD SLK L Q++ T Q +W KLLG++F +
Sbjct: 867 ERELIGLVLAIRHWRPYLWGRAFVVRTDHYSLKYLLDQRLATIPQHHWVGKLLGFDFTVE 926
Query: 1029 YKPGKTNQGADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKLCQIVSEVALDPQA 1088
YK G +N ADALSR D +S P ++ + +P L I + ++
Sbjct: 927 YKSGASNVVADALSRRDTDEGAVLALSAPRFDYIERLRAAQTTEPALVAIRDAIQAGTRS 986
Query: 1089 HPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGM 1148
P + + G +++ R+ I S L+ ++L HT GH G RT R+ + + M
Sbjct: 987 AP-WALRDGMVMFDSRLYIPPSSPLLHEILAAIHT---DGHEGVQRTLHRLRRDFHSPAM 1042
Query: 1149 TKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFV 1208
+ +QEFV ACD CQR+K P GLL PLP+P VW DI L+F+ LPR G I
Sbjct: 1043 RRVVQEFVRACDTCQRNKSEHLHPGGLLLPLPVPTTVWADIGLDFVEALPRVGGKTVILT 1102
Query: 1209 VVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRL 1268
VVDR +KY HFIPL HPYTA SVA+ F +I+RLHG+P S++SDRDP+F S FW+E+ RL
Sbjct: 1103 VVDRFSKYCHFIPLAHPYTAESVAQAFYADIVRLHGIPQSMVSDRDPVFTSSFWRELMRL 1162
Query: 1269 QGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSAT 1328
GT+ M++A HP+SDGQTE N+ + YLRCF D+P+ W WL WAEY +NT+Y ++
Sbjct: 1163 TGTKMHMTTAIHPQSDGQTEAANKVIVMYLRCFTGDRPRQWVRWLPWAEYIYNTAYQTSL 1222
Query: 1329 QQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANS 1388
+ TPF VYGR PP++ + GETRV AV + + DRDE L ++ L AQ + +
Sbjct: 1223 RDTPFRVVYGRDPPIIRSYEPGETRVAAVARSMADRDEFLADVRYRLEQAQATHKKYYDK 1282
Query: 1389 KRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGA 1448
+ +EVG+ V +++R SL KL RYFGPY ++ + VA RL+LP A
Sbjct: 1283 GHRAVSYEVGDLVLLRLRHRAPASLPQVSKGKLKPRYFGPYRVVEVINPVAVRLELPPRA 1342
Query: 1449 RIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQ 1508
++H VFH+ LLKK VG P P L +I P R L +G + QVLV
Sbjct: 1343 KLHDVFHVGLLKKFVG---AAPPSPPALPAVHHGAIDPEPERVTRSRLARG--VRQVLVH 1397
Query: 1509 WQGKTPEEATWEDLATIRSEFPVFNLEDKVVSSEG 1543
W+G++ ATWEDL T + +P F LED++ EG
Sbjct: 1398 WKGESAASATWEDLDTFKERYPAFQLEDELALEEG 1432
>UniRef100_Q947Y6 Putative retroelement [Oryza sativa]
Length = 1461
Score = 1038 bits (2683), Expect = 0.0
Identities = 602/1545 (38%), Positives = 839/1545 (53%), Gaps = 147/1545 (9%)
Query: 21 MEAKIDAMESELAAIRTALAAVTTAMKDLPTTLGTMVEKAVGKSVGIDVDSGDARPEREP 80
++A I+A+ ++A + ++ A ++ + + + G GD P+R P
Sbjct: 5 LKASIEALSKDMALMHESIKANAASIAANAKAIAALATTTSSSTSGARPGFGDQPPDRPP 64
Query: 81 REKTPESVGLRESGSDQPVLQGEALNEFRQSVKKVELPMFDGK-DLAGWISRAEIYFKVQ 139
+ P+ P +DGK D +I+R E +F Q
Sbjct: 65 KHWRPD------------------------------FPHYDGKSDPLIFINRCESFFLQQ 94
Query: 140 ETSPEVKVSLAQLSMEGGTIHFYNSLLATEEEL-TWERFRDALLERYGGNGDGDVYEQLS 198
E KV +A ++ G +Y + E TW RF++ L RYG +LS
Sbjct: 95 RIMQEEKVWMASHNLLEGAQLWYMQVQEDERGTPTWTRFKELLNLRYGPPLRSAPLFELS 154
Query: 199 ELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKVRS------LVAMGG 252
R+ G VE+Y F+ L + +L E+Q F GL + +V+ AM
Sbjct: 155 SCRRTGTVEDYQDRFQALLPRAGRLDEEQRVQLFTGGLLPPLSLQVQMQNPQSLAAAMSL 214
Query: 253 VNRARLLVVTRAVEKEVKGDGVAGQARANRFGGNGSRSGFSGATKGNGTEWIWVKGNKET 312
+ L+ AV + G GV
Sbjct: 215 ARQFELIEQYTAVPAKAPGRGV-------------------------------------- 236
Query: 313 GQTVNRPNSNPRGDQTKNTGDIRRAGPR-----DRGFNHLSYPQLMERRQKGLCFKCGGP 367
+ P P+ G + A P +R L+ + ERR+ GLCF C
Sbjct: 237 ---LPAPAPRPQLALPAPAGAAKPAPPAATAADNRPVRRLNQAEQEERRRLGLCFNCDEK 293
Query: 368 YHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNEDEEETQGELSLMSLCELGMKTGGI 427
Y R H +K + L + E+ DE E+E E + SL + G
Sbjct: 294 YSRGH---NKVCKRLFFVDSVEDEDEEA--------PEDEVDAEAPVFSLHAVAGVAVGH 342
Query: 428 PRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPRMTVKLGDGYKSQAQG 487
P + LR + +V LVD+G+THNF+ R G V PRMT + +G K G
Sbjct: 343 P--ILLRVQLGATTLVALVDTGSTHNFIGESAAARTGLSVQPRPRMTATVANGEKVACPG 400
Query: 488 --RCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKW 545
R A + IE +H+ + L G DIVLG +W+ LG ++WD + D +
Sbjct: 401 VLRHAPITIEGMPFHVDL--YVMPLAGYDIVLGTQWMAKLGR--MSWDVTTRALTFDLEG 456
Query: 546 ITLQGMDTRGEHMEALQSITATGERKPGLLGTRNEEKAGKQLPGEINSSQLEELDKLLYR 605
T+ + A+++ +A GLL +
Sbjct: 457 RTICWQGAPNQDGPAVRAASADDSLLGGLLDS---------------------------- 488
Query: 606 FDVVFQEKQGLPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQ 665
F VF E GLPP RGR+H I +++G PV VRPYRYP HK+E+E+Q M+S GI+R+
Sbjct: 489 FADVFTEPTGLPPQRGRDHAIVLKQGTSPVAVRPYRYPAAHKDELERQCAAMISQGIVRR 548
Query: 666 STSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDL 725
S SA+SSPV+LVKK D +W CVDYRALN++TV D FPIPV++ELLDELHGA FFSKLDL
Sbjct: 549 SDSAFSSPVLLVKKADSSWRFCVDYRALNALTVKDAFPIPVVDELLDELHGARFFSKLDL 608
Query: 726 KSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLV 785
+SGYHQVR+R ED+HKTAFRTH+G YE++VMPFGL NAP+TFQ+LMN+V R LRR VLV
Sbjct: 609 RSGYHQVRMRPEDIHKTAFRTHDGLYEFLVMPFGLCNAPATFQALMNDVLRSFLRRFVLV 668
Query: 786 FFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVD 845
FFDDIL+YS TW+DH+ HL VL VL+ H+L + KC F V YLGH+IS GVA+D
Sbjct: 669 FFDDILIYSDTWADHLRHLRAVLTVLREHKLFIKRSKCAFGVDSVAYLGHVISAAGVAMD 728
Query: 846 PNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAFNWGKQAQQ 905
P KV +++ WP+P++ + VRGFLGL GYYRKF+ NYG IA PLT L KK+ F W + A
Sbjct: 729 PAKVQAIREWPQPRSARAVRGFLGLAGYYRKFVHNYGTIAAPLTALLKKEGFAWTEAATA 788
Query: 906 AFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAK 965
AF+ LK +++AP+LA+PDF++ F +ECDAS G GA+L+ HP+A+FS+ + PR+ A
Sbjct: 789 AFDALKAAVSSAPILAMPDFTKAFTVECDASSHGFGAVLIQDGHPLAFFSRPVAPRHRAL 848
Query: 966 SAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNF 1025
+AYE+EL+ + LA++HWRPYL GR F V TD SLK L Q++ T Q +W KLLG++F
Sbjct: 849 AAYERELIGLVLAVRHWRPYLWGRHFTVKTDHYSLKYLLDQRLSTIPQHHWVGKLLGFDF 908
Query: 1026 EIIYKPGKTNQGADALSRIDEAGELRPLV-SYPLWEEQGVIADENQKDPKLCQIVSEVAL 1084
+ YKPG N ADALSR D + LV S P ++ + DP L + +E+
Sbjct: 909 TVEYKPGAANTVADALSRRDTTEDASVLVLSAPRFDFIERLRQAQDVDPALVALQAEIRS 968
Query: 1085 DPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLY 1144
+A P + + G +L+ R+ + S L+ ++L H GH G RT R+ + +
Sbjct: 969 GTRAGP-WSMADGMVLFAGRLYLPPASPLLQEVLRAVH---EEGHEGVQRTLHRLRRDFH 1024
Query: 1145 WRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFE 1204
+ M +Q+FV C+VCQR+K PAGLL PLP+P+ VW D++L+F+ LPR RG
Sbjct: 1025 FPNMKSVVQDFVRTCEVCQRYKAEHLQPAGLLLPLPVPQGVWTDVALDFVEALPRVRGKS 1084
Query: 1205 AIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKE 1264
I VVDR +KY HFIPL HPY+A SVA++F EI+RLHGVP S++SDRDP+F S FW E
Sbjct: 1085 VILTVVDRFSKYCHFIPLAHPYSAESVAQVFFAEIVRLHGVPQSMVSDRDPVFTSAFWSE 1144
Query: 1265 MFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSY 1324
+ RL GT+ M++A+HP+SDGQ+E NR + YLRC D+P+ W WL WAE+ FNT+Y
Sbjct: 1145 LMRLVGTKLHMTTAFHPQSDGQSEAANRVIIMYLRCLTGDRPRQWLRWLPWAEFVFNTAY 1204
Query: 1325 HSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRA 1384
++ + TPF VYGR PP + + G+TRV AV K +++R E L ++ L AQ +
Sbjct: 1205 QTSLRDTPFRVVYGRDPPSIRSYEPGDTRVAAVAKSMEERSEFLEDIRYRLEQAQAIQKK 1264
Query: 1385 QANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKL 1444
+ + F+VG+WV +++R SL+ V KL RYFGPY I + VA RL L
Sbjct: 1265 YYDKSHRAVSFQVGDWVLLRLRQRAPASLSLAVSGKLKPRYFGPYRIAEMINEVAARLAL 1324
Query: 1445 PEGARIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRDIL---RQGES 1501
P GAR+H VFHI LLKK G + P LP++ V + + R
Sbjct: 1325 PAGARLHDVFHIGLLKKWHGAPPDAP--------PPLPNVHHGAVACEPERVIKARLARG 1376
Query: 1502 ISQVLVQWQGKTPEEATWEDLATIRSEFPVFNLEDKVVSSEGSIV 1546
+ QVLVQW+G + ATWED + +P LED++ G V
Sbjct: 1377 VRQVLVQWKGTSAASATWEDREPFFARYPALQLEDELPLDGGGDV 1421
>UniRef100_Q9LHM4 Retroelement pol polyprotein [Arabidopsis thaliana]
Length = 1499
Score = 1036 bits (2680), Expect = 0.0
Identities = 597/1530 (39%), Positives = 887/1530 (57%), Gaps = 73/1530 (4%)
Query: 18 VTQMEAKIDAMESELAAIRTALAAVTTAMKDLPTTLGTMVEKAVGKSVGIDVDSGDARPE 77
VT++E + E+ L AV + +T G MV++ +D + +
Sbjct: 26 VTRLETTVAEQHKEMMKQFADLYAVLSR-----STAGKMVDE----QSTLDRSAPRSSQS 76
Query: 78 REPREKTPESVGLRESGSDQPVLQGEALNEFRQSVK--KVELPMFDGKDLAGWISRAEIY 135
E R P+ R++ Q ++ + N + + K++ P FDG L W+ + E +
Sbjct: 77 MENRSGYPDPY--RDARHQQ--VRSDHFNAYNNLTRLGKIDFPRFDGTRLKEWLFKVEEF 132
Query: 136 FKVQETSPEVKVSLAQLSMEG--GTIH--FYNSLLATEEELTWERFRDALLERYGGNGDG 191
F V T ++KV +A + + T H F S + E W+ + L ER+ + D
Sbjct: 133 FGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLEVLYDWKGYVKLLKERFEDDCD- 191
Query: 192 DVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKVRSLVAMG 251
D +L L++ + +Y FE + ++ L E+ +L GL+ + + VR
Sbjct: 192 DPMAELKHLQETDGIIDYHQKFELIKTRV-NLSEEYLVSVYLAGLRTDTQMHVRMFQPQ- 249
Query: 252 GVNRARLLVVTRAVEKEVKGDGVAGQARANRFGGNGSRSGFSGATKGNGTEWIWVKGNKE 311
L + + EK NR G G++ K +G +
Sbjct: 250 --TVRHCLFLGKTYEKAHPKKPANTTWSTNRSAPTG---GYNKYQK---------EGESK 295
Query: 312 TGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQLMERRQKGLCFKCGGPYHRN 371
T N+ N P Q K +S ++ +RR KGLC+ C Y
Sbjct: 296 TDHYGNKGNFKPVSQQPKK----------------MSQQEMSDRRSKGLCYFCDEKYTPE 339
Query: 372 HVCPDKHLRLLILEEDGEELDESKMLAMEVNEDEEETQGELSLMSLCELGMKTGGIPRTM 431
H K +L ++ D EE ++++ E+ D++E ++S+ ++ + +TM
Sbjct: 340 HYLVHKKTQLFRMDVD-EEFEDARE---ELVNDDDEHMPQISVNAVSGIAGY-----KTM 390
Query: 432 KLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPRMT-VKLGDGYKSQAQGRCA 490
+++GT + + +L+DSG+THNF+D +LG +V DT +T V + DG K + +G+
Sbjct: 391 RVKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKV-DTAGLTRVSVADGRKLRVEGKVT 449
Query: 491 GLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQG 550
++ + L L G D+VLG++WL+TLG + M F + + + L G
Sbjct: 450 DFSWKLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHG 509
Query: 551 MDTRGEHMEALQSITATGERKPGL----LGTRNEEKAGKQLPGEINSSQLEE---LDKLL 603
+ + Q + E + L + +E G+ +S+L E ++++L
Sbjct: 510 LTSGSVREVKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVL 569
Query: 604 YRFDVVFQEKQGLPPGRGRE-HCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGI 662
+ +F E LPP R + H I + EG PVN RPYRY H KNEI+K V ++L+ G
Sbjct: 570 NEYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGT 629
Query: 663 IRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSK 722
++ S+S Y+SPV+LVKKKD TW +CVDYR LN +TV D FPIP+IE+L+DEL GAV FSK
Sbjct: 630 VQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSK 689
Query: 723 LDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRG 782
+DL++GYHQVR+ +D+ KTAF+TH GH+EY+VMPFGL NAP+TFQ LMN +F+P LR+
Sbjct: 690 IDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKF 749
Query: 783 VLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGV 842
VLVFFDDILVYS + +H HL++V EV++ ++L A KC FA VEYLGH IS QG+
Sbjct: 750 VLVFFDDILVYSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGI 809
Query: 843 AVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAFNWGKQ 902
DP K+ +VK WP+P +K +RGFLGL GYYR+F++++G IA PL LTK DAF W
Sbjct: 810 ETDPAKIKAVKEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALTKTDAFEWTAV 869
Query: 903 AQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRN 962
AQQAFEDLK L APVL+LP F ++F +E DA G GIGA+L+ HP+AY S+ L +
Sbjct: 870 AQQAFEDLKAALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQ 929
Query: 963 LAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLG 1022
L S YEKEL+AV A++ WR YLL F++ TDQRSLK L Q++ T QQ W KLL
Sbjct: 930 LHLSIYEKELLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLE 989
Query: 1023 YNFEIIYKPGKTNQGADALSRIDEAGELRPLVSYPLWEEQGVIADENQKDPKLCQIVSEV 1082
+++EI Y+ GK N ADALSR++ + L ++ + I D +L I++ +
Sbjct: 990 FDYEIQYRQGKENVVADALSRVEGSEVLHMAMTVVECDLLKDIQAGYANDSQLQDIITAL 1049
Query: 1083 ALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATN 1142
DP + + Q L + ++V+ ++ +L H + GGHSG T++R+
Sbjct: 1050 QRDPDSKKYFSWSQNILRRKSKIVVPANDNIKNTILLWLHGSGVGGHSGRDVTHQRVKGL 1109
Query: 1143 LYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRG 1202
YW+GM K IQ ++ +C CQ+ K ++ GLLQPLPIP+ +W ++S++FI GLP S G
Sbjct: 1110 FYWKGMIKDIQAYIRSCGTCQQCKSDPAASPGLLQPLPIPDTIWSEVSMDFIEGLPVSGG 1169
Query: 1203 FEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFW 1262
I VVVDRL+K +HFI L HPY+A +VA + + +LHG P+SI+SDRD +F S FW
Sbjct: 1170 KTVIMVVVDRLSKAAHFIALSHPYSALTVAHAYLDNVFKLHGCPTSIVSDRDVVFTSEFW 1229
Query: 1263 KEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNT 1322
+E F LQG K++SAYHP+SDGQTE+VNRCLETYLRC D+P+ W+ WL AEYW+NT
Sbjct: 1230 REFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRCMCHDRPQLWSKWLALAEYWYNT 1289
Query: 1323 SYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERM 1382
+YHS+++ TPFE VYG+ PPV ++ GE++V V + LQ+R++ L LK HL+ AQ RM
Sbjct: 1290 NYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARSLQEREDMLLFLKFHLMRAQHRM 1349
Query: 1383 RAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRL 1442
+ A+ R +EFE+G++V+VK++ +RQ S+ R + KL+ +YFGPY II R G VAY+L
Sbjct: 1350 KQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKL 1409
Query: 1443 KLPEGARIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLARRDILRQGESI 1502
LP +++HPVFH+S LK VGN LP + +++ P V+ R+ + RQG+++
Sbjct: 1410 ALPSYSQVHPVFHVSQLKVLVGNVSTTVHLPSVM--QDVFEKVPEKVVERKMVNRQGKAV 1467
Query: 1503 SQVLVQWQGKTPEEATWEDLATIRSEFPVF 1532
++VLV+W + EEATWE L ++ FP F
Sbjct: 1468 TKVLVKWSNEPLEEATWEFLFDLQKTFPEF 1497
>UniRef100_Q7XSK7 OSJNBa0059D20.8 protein [Oryza sativa]
Length = 1463
Score = 1028 bits (2659), Expect = 0.0
Identities = 588/1441 (40%), Positives = 825/1441 (56%), Gaps = 91/1441 (6%)
Query: 113 KKVELPMFDGK-DLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEE 171
KK + P +DG D ++++ E YF+ T E +V +A ++ T +Y LL E
Sbjct: 58 KKWDFPRYDGTTDPLLFLNKFEAYFRHHRTMAEERVGMASYHLDDVTQTWYTQLLEDEGT 117
Query: 172 LTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGY 231
TW RF++ + R+G +LSE R+ G VEEY F+ L + +L E Q
Sbjct: 118 PTWGRFKELVNLRFGPPLRSAPLFELSECRRTGTVEEYSNRFQALLPRAGRLDESQRVQL 177
Query: 232 FLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFGGNGSRSG 291
F GL + VR ++ L ++ ++V+ +R R+
Sbjct: 178 FTGGLLPPLSHAVR-------IHHPETLAAAMSLARQVE------LMERDRPAPPPLRAP 224
Query: 292 FSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQ 351
G Q + P Q ++ + RR P +
Sbjct: 225 PRGLLPAPAPRLAL----PAPAQQLALPAPPAAAPQGRDAANPRRLTPEE---------- 270
Query: 352 LMERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNEDEEETQGE 411
+ ER ++GLCF C + R H ++ R L DG E+D+ +A+E + E
Sbjct: 271 MAERCRQGLCFNCNEKFTRGH---NRFCRRLFFV-DGVEIDD---VAIEGDAAAAAGDTE 323
Query: 412 LSLMSLCELGMKTGGIP--RTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVD 469
+ SL + G+P T++L+ T+ ++ L+D G+TH+F+ RR G +
Sbjct: 324 APVFSLHAVA----GVPIADTIQLQVTVGDASLLALLDGGSTHSFIGEEAARRAGLPIQS 379
Query: 470 TPRMTVKLGDGYKSQAQG--RCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGD 527
+PRMT + +G + G R A I +H + L G D+VLG WL TLG
Sbjct: 380 SPRMTAIVANGERVACPGVIRDAAFTINGSTFHTDLF--VMPLAGFDVVLGTRWLGTLGP 437
Query: 528 TIVNWDTQLMSFWSDKKWITLQGM-DTRGEHMEALQSITATGERKPGLLGTRNEEKAGKQ 586
+ ++ ++ M+F D + +G+ T H+ L + + T
Sbjct: 438 IVWDFTSRSMAFQRDGQRFAWKGVASTSTTHLRTLAAASGT------------------- 478
Query: 587 LPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNVRPYRYPHHH 646
LD+LL ++ VF E GLPP RGR+H I ++ PV VRPYRYP H
Sbjct: 479 -----------LLDELLVAYEDVFGEPTGLPPPRGRDHAIVLKPSSAPVAVRPYRYPAAH 527
Query: 647 KNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPV 706
K+E+E+Q M+ G++R+S S +SSPV+LVKK D +W CVDYRALN++TV D FPIPV
Sbjct: 528 KDELERQCAAMIEQGVVRRSDSPFSSPVLLVKKADGSWRFCVDYRALNALTVKDAFPIPV 587
Query: 707 IEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPST 766
+ DELHGA FF+KLDL+SGYHQVR+R EDVHKTAFRTH+G YE++VMPFGL NAP+T
Sbjct: 588 V----DELHGARFFTKLDLRSGYHQVRMRPEDVHKTAFRTHDGLYEFLVMPFGLCNAPAT 643
Query: 767 FQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFA 826
FQ+LMN+V RP LRR VLVFFDDIL+YS+TW+DH+ HL VL VL+ H+L + KC F
Sbjct: 644 FQALMNDVLRPFLRRFVLVFFDDILIYSETWTDHLRHLRTVLSVLRQHRLFVKRSKCTFG 703
Query: 827 TKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIAR 886
+ V YLGH+ISE GVA+DP KV ++ W P++ + VR FLGL GYYRKF+ NYG IA
Sbjct: 704 SPSVSYLGHVISEAGVAMDPAKVQAIHEWLVPRSARAVRSFLGLAGYYRKFVHNYGTIAA 763
Query: 887 PLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLY 946
PLT LTKKD F+W + AF+ LK +T+APVLA+PDF++ F +E DAS G GA+L+
Sbjct: 764 PLTALTKKDGFSWTEDTAAAFDALKAAVTSAPVLAMPDFAKPFTVEGDASTHGFGAVLVQ 823
Query: 947 GKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQ 1006
HP+A+FS+ + R+ A +AYE+EL+ + A++HWRPYL GR+FVV TD SLK L Q
Sbjct: 824 DGHPVAFFSRPVVLRHRALAAYERELIGLVHAVRHWRPYLWGRRFVVKTDHYSLKYLLDQ 883
Query: 1007 KIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRID-EAGELRPLVSYPLWEEQGVI 1065
++ T Q +W KLLG++F + YKPG N ADALSR D E G + L S P ++ +
Sbjct: 884 RLATIPQHHWVGKLLGFDFAVEYKPGAANTVADALSRRDTEEGAILAL-SAPRFDFISKL 942
Query: 1066 ADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTP 1125
D ++DP L + EV+ + P + +V L Y + I S L +++E H
Sbjct: 943 HDAQRQDPALTALRDEVSAGTRTGP-WALVDDLLQYNSWLYIPPASPLAREIIEATH--- 998
Query: 1126 HGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERV 1185
GH G RT R+ + M + +Q++V +C VCQR+K SPAGLL PLP+P+ V
Sbjct: 999 EDGHEGVKRTMHRLRREFHIPNMKQLVQDWVRSCAVCQRYKSEHLSPAGLLLPLPVPQGV 1058
Query: 1186 WEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGV 1245
W DI+L+FI LPR RG I VVDR +KY HFIPL HPY+A SVA+ F EI+ LHGV
Sbjct: 1059 WTDIALDFIEALPRVRGKSVILTVVDRFSKYCHFIPLAHPYSAESVAQAFFAEIVHLHGV 1118
Query: 1246 PSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQ 1305
P S++SDRDP+F S FW+E+ RL GT+ M++A+HP+SDGQ+E NR + YLRC D+
Sbjct: 1119 PQSMVSDRDPIFTSTFWRELMRLMGTKLHMTTAFHPQSDGQSEAANRVIIMYLRCLTGDR 1178
Query: 1306 PKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRD 1365
P+ W WL WAE+ FNT+Y S+ + TPF VYGR PP + + G+TRV AV K +++R
Sbjct: 1179 PRQWLRWLPWAEFIFNTAYQSSLRDTPFRVVYGRDPPSIRSYEAGDTRVAAVAKSMEERA 1238
Query: 1366 EALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARY 1425
E L ++ L AQ + + +H ++VG+W +++R SL KL R+
Sbjct: 1239 EFLFDIRYRLEQAQAVQKLHYDKHHRHVAYQVGDWALLRLRQRPTTSLPQSGTGKLKPRF 1298
Query: 1426 FGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPSIQ 1485
+GPY I + VA RL+LP GAR+H VFHI LLKK G PG P L +I
Sbjct: 1299 YGPYRITELINDVAVRLELPAGARLHDVFHIGLLKKFHG---PPPGAPPALPPLHHGAIA 1355
Query: 1486 PALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFPVFNLEDKVVSSEGSI 1545
P A R L +G + Q LVQW+G++P ATWED+ +R+++P LED++ EG
Sbjct: 1356 PEPERAVRFRLARG--VRQALVQWKGESPASATWEDIEVLRAKYPALQLEDELSLEEGGD 1413
Query: 1546 V 1546
V
Sbjct: 1414 V 1414
>UniRef100_Q9FW76 Putative gag-pol polyprotein [Oryza sativa]
Length = 1608
Score = 983 bits (2540), Expect = 0.0
Identities = 576/1536 (37%), Positives = 856/1536 (55%), Gaps = 62/1536 (4%)
Query: 11 KKTTRVAVTQMEAKIDAMESELAAIRTALAAVTTAMKDLPT--TLGTMVEKAVGKSVGID 68
KK+ V + +E K++ + S+LA + + + ++++ P+ V AV ++
Sbjct: 43 KKSLEVRLPAVEKKVEILSSDLATLNHKVQQLESSIQRQPSGDEFAGKVTTAVLPKEELN 102
Query: 69 VDSGD-ARPEREPREKTPESV---GLRESGSDQPVLQGEALNEFRQSVKKVELPMFDGKD 124
S + +R+P +P+ GL + + G A S+ + P F+G +
Sbjct: 103 YHSTPFIKGQRDPEFASPDQQNQDGLTFQSGNSGLGGGIA-----GSIPPMSCPQFNGDN 157
Query: 125 LAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEELTWERFRDALLER 184
W S E+YF + P+ V +A L+ G + S+ + TW D + R
Sbjct: 158 PQLWKSNCEVYFDIYGIHPQNWVKIATLNFCGNAAFWLQSVRSQLAGATWFELCDRVCGR 217
Query: 185 YGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGYFLHGLKEEIRGKV 244
+ + + Q + Q V +Y+ F+ + Q+ YF+ + ++ +
Sbjct: 218 FARDRKQALIRQWIHITQTSSVADYVDRFDSIMHQLMAYGGSNDPAYFVTKFVDGLKDHI 277
Query: 245 RSLVAMGGVNRARLL--VVTRAVEKEVKGDGVAGQARANRFGGNGSRSGFSGATKGNGTE 302
R +V V R + L T A+ +E +GV Q+ +N+ K T
Sbjct: 278 RVVVM---VQRPQDLDSACTVALLQEEALEGV--QSVSNK--------------KNETTT 318
Query: 303 WIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQLMERRQKGLCF 362
++ K + + S P + G G D+ SY RR KGLCF
Sbjct: 319 FLKTKPSHNLTSPTFQARSIPFTNIEDKRGVEFSKGRDDKVSALRSY-----RRSKGLCF 373
Query: 363 KCGGPYHRNHVCPDKHLRLLILEE--DGEELDESKMLAMEVNEDEEETQGELSLMSLCEL 420
CG + R+H C ++L ++EE + + D + E + EE SLM++
Sbjct: 374 VCGEKWGRDHKCATT-VQLHVVEELINALKTDPEENCNSEGAPESEED----SLMAISFQ 428
Query: 421 GMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPRMTVKLGDG 480
+ ++++LRG + +++LVDSG+TH+F+D L +L + V++ DG
Sbjct: 429 ALNGTDSSKSIRLRGWVQNTELLMLVDSGSTHSFIDAKLGAQLCGLQKLNQAIKVQVADG 488
Query: 481 YKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFW 540
+ + +L LG D +LG++WL+ V+W + ++F
Sbjct: 489 SQLFCDSFLPNCSWWSQGHSFTSDFRLLPLGSYDAILGMDWLEQFSPMQVDWVHKWIAFQ 548
Query: 541 SDKKWITLQGMDTRGEHMEALQSITATGERKPGLLGTRNEEKAGKQLPGEINSSQLEELD 600
+ + LQG+ + + + G K G + + L ++ E +
Sbjct: 549 HHGQAVQLQGIHPQLSTCFPISNDQLQGMSKKGAVMCLVHLNVAETLTA---TTVPEIVQ 605
Query: 601 KLLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSA 660
+L F +F E LPP R +H I + EG PVN+RPYRY K+EIE+QV EML +
Sbjct: 606 PILNEFQEIFSEPTELPPKRNCDHHIPLVEGAKPVNLRPYRYKPALKDEIERQVAEMLRS 665
Query: 661 GIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFF 720
G+I+ S+S +SSP +LVKKKD TW +C+DYR LN VTV K+P+PVI+ELLDEL G+ +F
Sbjct: 666 GVIQPSSSPFSSPALLVKKKDGTWRLCIDYRQLNDVTVKSKYPVPVIDELLDELAGSKWF 725
Query: 721 SKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLR 780
SKLDL++GYHQ+R+ E D +KTAF+TH GHYEY VM FGL AP+TF S MNE P+LR
Sbjct: 726 SKLDLRAGYHQIRMAEGDEYKTAFQTHSGHYEYKVMSFGLTGAPATFLSAMNETLSPVLR 785
Query: 781 RGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQ 840
+ LVFFDDIL+YS T H+ H+ VL++L HQ KC FA + + YLGH+I
Sbjct: 786 KFALVFFDDILIYSPTLELHLQHVRTVLQLLSAHQWKVKLSKCSFAQQEISYLGHVIGAA 845
Query: 841 GVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKD-AFNW 899
GVA DP K+ V SWP+P +K +RGFLGL GYYRKF++++G I++PLT+L KK F W
Sbjct: 846 GVATDPAKIQDVVSWPQPTTIKKLRGFLGLAGYYRKFVRHFGLISKPLTQLLKKGIPFKW 905
Query: 900 GKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALG 959
+ + AF+ LK+ L APVLALPDFS+ F +E DAS +GIGA+L KHPIAY S+ALG
Sbjct: 906 TPEIESAFQQLKQALVAAPVLALPDFSKHFTIETDASDVGIGAVLSQEKHPIAYLSRALG 965
Query: 960 PRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAK 1019
P+ S YEKE MA+ LA++HWRPYL +F++ TD SL Q++ T QQ K
Sbjct: 966 PKTRGLSTYEKEYMAIILAVEHWRPYLQQGEFIILTDHHSLMHLTEQRLHTPWQQKAFTK 1025
Query: 1020 LLGYNFEIIYKPGKTNQGADALSR----IDEAGELRPLVSYPLWEEQGVIADENQKDPKL 1075
LLG ++I Y+ G +N ADALSR I E + + P W ++ + Q D +
Sbjct: 1026 LLGLQYKICYRKGVSNAAADALSRRESPISEVAAISECI--PSWMQE--LMQGYQLDGQS 1081
Query: 1076 CQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRT 1135
Q+++E+A+ P + Y++ QG L Y+ ++ + N ++L KL+ E H TP GGHSGF T
Sbjct: 1082 KQLLAELAISPNSRKDYQLCQGILKYKGKIWVGNNTALQHKLVNELHATPLGGHSGFPVT 1141
Query: 1136 YRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFII 1195
YR++ + W GM K I+E + +C VC + K + GLLQPLP+P W+ ISL+FI
Sbjct: 1142 YRKVKSLFAWPGMKKLIKEQLQSCQVCLQAKPDRARYPGLLQPLPVPAGAWQTISLDFIE 1201
Query: 1196 GLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDP 1255
GLPRS + I VVVD+ +KYSHFIPL HP+ A VA+ F K I +LHG+P +IISDRD
Sbjct: 1202 GLPRSSHYNCILVVVDKFSKYSHFIPLSHPFNAGGVAQEFMKNIYKLHGLPRAIISDRDK 1261
Query: 1256 LFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHW 1315
+F S FW ++F GT MSSAYHP+SDGQTE VN+CLE YLRCF P W+SWL+
Sbjct: 1262 IFTSQFWDQLFSKFGTDLHMSSAYHPQSDGQTERVNQCLEIYLRCFVHAAPHKWSSWLYL 1321
Query: 1316 AEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHL 1375
AE+W+NTS+HS +TPFE +YG P L + ++ + + +R + L+QHL
Sbjct: 1322 AEFWYNTSFHSTLNKTPFEVLYGYTPSHFGIG-LDDCQIADLHEWHTERKFMQQLLQQHL 1380
Query: 1376 VAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRV 1435
AQ++M+ QA+ KR ++F VG+WV++K++ + Q +A R KLA RY+GP+ ++ RV
Sbjct: 1381 NRAQQQMKHQADKKRSFRQFAVGDWVYLKLQPYVQTFVAQRACHKLAFRYYGPFQVMSRV 1440
Query: 1436 GAVAYRLKLPEGARIHPVFHISLLKKAVG-NYVEEPGLPEQLDGEELPSIQPALVLARRD 1494
G VAY ++LP + IHPVFH+S LK AVG + + LP L ++ P L +R
Sbjct: 1441 GTVAYHIQLPATSSIHPVFHVSQLKAAVGFSKKVQDELPTSLGALQV----PFQFLDKRL 1496
Query: 1495 ILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFP 1530
+ + S+ Q+L W +P E+TWED+ + + FP
Sbjct: 1497 VKKGNRSVLQLLTHWYHSSPSESTWEDMEDLFARFP 1532
>UniRef100_Q60E20 Putative polyprotein [Oryza sativa]
Length = 1475
Score = 971 bits (2509), Expect = 0.0
Identities = 553/1478 (37%), Positives = 821/1478 (55%), Gaps = 97/1478 (6%)
Query: 86 ESVGLRESGSDQPVLQGEAL--NEFRQSVKK---VELPMFDGKDLAGWISRAEIYFKVQE 140
++ G+R + + G+A N++ ++V K +E+P+F G+D W+ + E +F++
Sbjct: 64 QAQGVRMLDQQRNIYLGQAGEGNQYAEAVLKGPRLEIPLFSGEDPIDWLKQCEKFFEITG 123
Query: 141 TSPEVKVSLAQLSMEGGTIHFYNSLLATEEELTWERFRDALLERYGGNGDGDVYEQLSEL 200
T + V+LA + G + ++ + + + W ++ + R+ + + E +
Sbjct: 124 TPLDQWVNLAVAHLNGRALKWFGGIGLPWQVIAWPQWCSMVCTRFSAASEHEAIELFQNV 183
Query: 201 RQQGI-VEEYITDFEYLTAQI----PKLPEKQYQGYFLHGLKEEIRGKVRSLVAMGGVNR 255
+Q G VE+YI FE + P L E+ F+ GL+ +I+ V G
Sbjct: 184 KQFGTTVEQYIDKFEDYVDLVKRDHPYLQEQYLTSCFIGGLRADIKYDVCGQKPQG---- 239
Query: 256 ARLLVVTRAVEKEVKGDGVAGQARANRFGGNGSRSGFSGATKGNGTEWIWVKGNKETGQT 315
LL A A A + GN +R+ GN
Sbjct: 240 --LLETYWA-------------ANARKMVGNFNRNRNQNPLGGNQGR------------- 271
Query: 316 VNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQLMERRQKGLCFKCGGPYHRNHVCP 375
N+N RG Q + GD RR++ C+ C P+ H C
Sbjct: 272 ----NANHRG-QNRGEGD---------------------RREEKKCWFCKEPWFPRHQCK 305
Query: 376 DKH-LRLLILEEDGEELDESKMLAMEVNEDEEETQGELS--------LMSLCELGMKTGG 426
K + L+ E+DG+E E+ + E +E + S LM + + ++
Sbjct: 306 IKQAIHALLEEDDGQEDKETSNTGGDEEEKKETEESATSENESPTEELMYISQTAVQGTS 365
Query: 427 IPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGWEVVDTPRMTVKLGDGYKSQAQ 486
P T + ING V LVDSG+T F+D R + + +T V + G + +
Sbjct: 366 RPDTFSVLIKINGRTAVGLVDSGSTTTFMDQDYALRNYYPLKNTDTKKVVVAGGGELKTD 425
Query: 487 GRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWI 546
+ E+ +L L G DI+LG +W+ ++ +++ K I
Sbjct: 426 VMVPDISYEIQGECFTNQFKLLPLKGYDIILGADWIYNYSPISLDLKQRILGITKGNKVI 485
Query: 547 TLQGMDTRGEHMEALQSITATGER-----KPGLLGTRNEEKAGKQLPGEINSSQLEELDK 601
LQ +H + +G+R K G LG + + E E++
Sbjct: 486 LLQDFTKPNKHFQI------SGKRLEKMLKKGALGMVIQVNVMSETVEEEGHVIPEDISD 539
Query: 602 LLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAG 661
++ +F V +E +GLPP R +H I +Q G P N+RPYR PH+ K +E + E++ +
Sbjct: 540 IIQQFPAVLKEPKGLPPKRECDHVINLQSGAVPPNIRPYRVPHYQKEAMENIINELIESK 599
Query: 662 IIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFS 721
I+ S S YSSP ++V+KKD +W MCVDYR LN+ TV +KFP+P+IE+LLDEL+GA FS
Sbjct: 600 EIQTSDSPYSSPAVMVRKKDGSWRMCVDYRQLNAQTVKNKFPMPIIEDLLDELNGARIFS 659
Query: 722 KLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRR 781
KLDL+SGYHQ+R+ E+DVHKTAFRTH GHYEY VMPFGL N P+TFQSLMN V P LRR
Sbjct: 660 KLDLRSGYHQIRMAEKDVHKTAFRTHLGHYEYQVMPFGLTNDPATFQSLMNHVLAPFLRR 719
Query: 782 GVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQG 841
VLVFFDDIL+YSKT ++H+ H++ V++ LQ + L KKC F V YLGH+IS+ G
Sbjct: 720 FVLVFFDDILIYSKTRAEHLEHVKLVMQALQDNHLVIKLKKCAFGLASVSYLGHVISQDG 779
Query: 842 VAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAFNWGK 901
VA DP KV +K+WP PK+V VR FLG+TGYYR+FIQ YG I RP+ ++ KK+ F WG
Sbjct: 780 VATDPKKVGKIKNWPTPKDVTDVRKFLGMTGYYRRFIQGYGTICRPIHDMLKKNGFQWGA 839
Query: 902 QAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPR 961
AFE LK KL T+PVLALPDF Q F +E DA G+GIGA+L+ G PIA+FSKALGP+
Sbjct: 840 DQTTAFETLKHKLRTSPVLALPDFDQAFTIEADACGVGIGAVLMQGGRPIAFFSKALGPK 899
Query: 962 NLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLL 1021
+S YEKE MA+ A++ WR Y+LG K ++ TDQ+SLK + Q++V G Q KL+
Sbjct: 900 AAGQSIYEKEAMAILEALKKWRHYVLGSKLIIKTDQQSLKFMMGQRLVEGIQHKLLLKLM 959
Query: 1022 GYNFEIIYKPGKTNQGADALSRIDE----AGELRPL-VSYPLWEEQGVIADENQKDPKLC 1076
Y++ I YK GK N ADALSR+ + A P+ V P W I + D +
Sbjct: 960 EYDYTIEYKSGKENLVADALSRLPQKEAVADRCHPMTVVIPEWIVD--IQRSYENDVQAH 1017
Query: 1077 QIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTY 1136
+I+S + Y++ G L Y+ R+ + + + +L+ +H++ GGHSG T+
Sbjct: 1018 KILSLIGTAADPDREYKLEAGLLKYKGRIYVGEATDIRRQLITTYHSSSFGGHSGMRATH 1077
Query: 1137 RRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIG 1196
RI YW GM ++ F+ C CQ K GLL PL IP+ W I+++FI G
Sbjct: 1078 HRIKMLFYWHGMRGEVERFIRECPTCQITKSEHVHIPGLLNPLEIPDMAWTHITMDFIEG 1137
Query: 1197 LPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPL 1256
LP+S+G + I VVVDRLTKY+HFI L HPY V E F I +LHG+P II+DRD +
Sbjct: 1138 LPKSQGKDVILVVVDRLTKYAHFIALAHPYDVEQVVEAFMNNIHKLHGMPMVIITDRDRI 1197
Query: 1257 FVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWA 1316
F S ++E+F+ + + S+AYHP+ DGQTE VN+CLE+YLR +P W SWL A
Sbjct: 1198 FTSSLFQEIFKAMKVKLRFSTAYHPQMDGQTERVNQCLESYLRNMTFQEPHKWYSWLALA 1257
Query: 1317 EYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLV 1376
E+W+NT++H+A Q TPF+A+YG PP + + + E +++++ L +LK L
Sbjct: 1258 EWWYNTTFHTAIQMTPFKAMYGYSPPQINEFSVPCNISEEARVTIEEKEAILNKLKNSLA 1317
Query: 1377 AAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVG 1436
AQ RM+ A+ R + EVG+ V++K++ +RQ + R KL ++++GP+ ++ ++G
Sbjct: 1318 DAQHRMKYFADKNRTERNLEVGDMVYLKLKPYRQSAFGIRGSLKLRSKFYGPFKVLQKIG 1377
Query: 1437 AVAYRLKLPEGARIHPVFHISLLKKAVGNY-VEEPGLPEQLDGEELPSIQPALVLARRDI 1495
+AY+L+LP+ A+IHPVFH+S LKK +G + + LP ++ + +P VL RR +
Sbjct: 1378 QLAYKLQLPDDAQIHPVFHVSQLKKHLGKHAIPMSNLPSVGPDGQIKT-EPLAVLQRRMV 1436
Query: 1496 LRQGESISQVLVQWQGKTPEEATWEDLATIRSEFPVFN 1533
R+G +++Q L+ WQ +P EATWED + I++ FP FN
Sbjct: 1437 PRKGVAVTQWLILWQNLSPAEATWEDASVIQAMFPSFN 1474
>UniRef100_Q7XW39 OSJNBb0062H02.17 protein [Oryza sativa]
Length = 1629
Score = 961 bits (2484), Expect = 0.0
Identities = 544/1438 (37%), Positives = 796/1438 (54%), Gaps = 47/1438 (3%)
Query: 108 FRQSVKKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLA 167
+ V K++ P FDG D W + E YF V T P + V +A + G + S A
Sbjct: 172 YSHRVPKLDFPKFDGTDPQDWRMKCEHYFDVNNTYPGLWVRVAIIYFSGRAASWLRSTKA 231
Query: 168 TEEELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQ 227
WE F AL +++ + + Q+ +RQ G V EY F+ L ++
Sbjct: 232 HVRFPNWEDFCAALSDKFDRDQHELLIRQMDGIRQSGTVWEYYEQFDELMNKLLVYDPVV 291
Query: 228 YQGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEV--KGDGVAGQARANRFGG 285
Y H E + K+R++V + + +++EV D V G+ G
Sbjct: 292 NMHYLTHRFTEGLYRKIRNVVLLQRPRDLESALAVALLQEEVLETADEVTGKEVKKSEGN 351
Query: 286 NGSRSGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRG------DQTKNTGDIRRAGP 339
+ RS N + P G + K ++RR+
Sbjct: 352 SLGRS----------------VANLRGAYPLPTPPMRSGGINMGIKSEEKKESEVRRSSG 395
Query: 340 RDRGFNHLSYPQLMERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAM 399
+ + L +RR +GLC+ C + H C + ++L ++E L ES +
Sbjct: 396 TNERLSSLK----AQRRAQGLCYICAEKWSPTHKCSNT-VQLHAVQELFTVLHESVEDGL 450
Query: 400 EVNED-EEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCF 458
+ E+T +SL ++ G +TGG M++ G I G +++LVDSG++ +F+
Sbjct: 451 STTDHVVEQTLMAVSLQAV--QGTETGG---CMRMLGQIQGKEILILVDSGSSASFISKR 505
Query: 459 LVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLG 518
+ L + + V + G K + + + + ++ +L D++LG
Sbjct: 506 VASSLMGVLEQPVHVQVMVAGGAKLHCCSEILNCEWTIQGHVFFTNLKVLELNNYDMILG 565
Query: 519 IEWLKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHMEALQS--ITATGERKPGLLG 576
++WL V+W T+ + I L G+ + E + S + +R
Sbjct: 566 MDWLMQHSPMTVDWTTKSLIIAYAGTQIQLYGVRSDTEQCAHISSKQLRELNDR----TA 621
Query: 577 TRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVN 636
N + E E + +L F VF E +GLPP R +H I + G GPVN
Sbjct: 622 VSNLVQFCSVFALEYQEQIPEVVQTVLTEFSSVFDEPKGLPPIRQFDHTIPLLPGAGPVN 681
Query: 637 VRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSV 696
VRPYRY KNEIE QV+EMLS GII+ S+S +SSPV+LVKKKD +W CVDYR LN++
Sbjct: 682 VRPYRYTPIQKNEIESQVQEMLSKGIIQPSSSPFSSPVLLVKKKDGSWRFCVDYRHLNAI 741
Query: 697 TVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVM 756
TV +K+P+PVI+ELLDEL GA +FSKLDL+SGYHQ+R+ +D HKTAF+TH GH+E+ V+
Sbjct: 742 TVKNKYPLPVIDELLDELAGAQWFSKLDLRSGYHQIRMHPDDEHKTAFQTHHGHFEFRVL 801
Query: 757 PFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQL 816
PFGL +AP+TFQ +MN V +LRR VLVF DDIL+YSK+ +H+ HL+ V ++L HQL
Sbjct: 802 PFGLTSAPATFQGVMNSVLATLLRRCVLVFVDDILIYSKSLEEHVQHLKTVFQILLKHQL 861
Query: 817 TANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRK 876
+ KC FA + + YLGH+I GV+ DP K+ ++ WP P +VK +R FLGL+GYYRK
Sbjct: 862 KVKRTKCSFAQQELAYLGHIIQPNGVSTDPEKIQVIQHWPAPTSVKELRSFLGLSGYYRK 921
Query: 877 FIQNYGKIARPLTELTKK-DAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDA 935
F++NYG +++PLT L +K + W + + AF+ LK+ L TA VLA+PDF F +E DA
Sbjct: 922 FVRNYGILSKPLTNLLRKGQLYIWTAETEDAFQALKQALITALVLAMPDFQTPFVVETDA 981
Query: 936 SGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVST 995
S GIGA+L+ HP+A+ S+ALG R+ S YEKE +A+ LA+ HWRPYL +F + T
Sbjct: 982 SDKGIGAVLMQNNHPLAFLSRALGLRHPGLSTYEKESLAIMLAVDHWRPYLQHDEFFIRT 1041
Query: 996 DQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRI--DEAGELRPL 1053
D RSL Q++ T Q KLLG ++II+K G N ADALSR + EL L
Sbjct: 1042 DHRSLAFLTEQRLTTPWQHKALTKLLGLRYKIIFKKGIDNSAADALSRYPGSDRVELSAL 1101
Query: 1054 -VSYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSS 1112
V+ P W I DP C V + ++ A P + + G L +Q+R+ + +
Sbjct: 1102 SVAVPEWIND--IVAGYSSDPDACSKVQTLCINSGAVPNFSLRNGVLYFQNRLWVGHNVD 1159
Query: 1113 LIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSP 1172
+ ++L HT GGHSG TY+R+ W + T+ ++V AC VCQ+ K
Sbjct: 1160 VQQRILANLHTAAVGGHSGIQVTYQRVKQLFAWPRLRATVVQYVQACSVCQQAKSEHVKY 1219
Query: 1173 AGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVA 1232
G+LQPLP+P+ W+ +SL+F+ GLP+S F I VVVD+ +KYSHF+PL HP++A VA
Sbjct: 1220 PGMLQPLPVPDHAWQIVSLDFVEGLPKSASFNCILVVVDKFSKYSHFVPLTHPFSALDVA 1279
Query: 1233 EIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNR 1292
E + + I RLHG+P S+ISDRD +F S W +FRL GTQ +MSS+YHP++DGQTE VN+
Sbjct: 1280 EAYMQHIHRLHGLPQSLISDRDRIFTSTLWTTLFRLAGTQLRMSSSYHPQTDGQTERVNQ 1339
Query: 1293 CLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGET 1352
CLET+LRCF P W+ WL AEYW+NTS+HSA TPFE +YG KP
Sbjct: 1340 CLETFLRCFVHACPSQWSRWLALAEYWYNTSFHSALGTTPFEVLYGHKPRYFGLSASAAC 1399
Query: 1353 RVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVS 1412
R + + + L +R++ ++ HL+ AQ RM+ QA+ R + F VG+WV++K++ Q S
Sbjct: 1400 RSDDLVEWLHEREKMQALIRDHLLRAQTRMKQQADQHRSERSFAVGDWVYLKLQPFVQQS 1459
Query: 1413 LANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGL 1472
+ R + KL+ R++GP+ ++ +VG VAYRL LP + IHPV H+S LKKA+ +
Sbjct: 1460 VVTRANRKLSFRFYGPFQVLDKVGTVAYRLDLPSSSLIHPVVHVSQLKKALAPTEQVHSP 1519
Query: 1473 PEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFP 1530
LD + PA +L RR I + + + Q+ V+W G P TWE+ +R FP
Sbjct: 1520 LPVLDPTNATHVCPAQILDRRFIRKGSKLVEQIQVRWTGDAPAATTWENPQELRRRFP 1577
>UniRef100_Q7XJX3 OSJNBa0004L19.16 protein [Oryza sativa]
Length = 1586
Score = 961 bits (2484), Expect = 0.0
Identities = 530/1442 (36%), Positives = 813/1442 (55%), Gaps = 85/1442 (5%)
Query: 114 KVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEELT 173
++E+P+F G+D W+ + E ++++ T E V+LA ++G + +Y + + +T
Sbjct: 207 RLEIPLFSGEDPIDWLKQCEKFYEISGTPAEQWVNLAIAHLQGKAMKWYRGIGIPWQLIT 266
Query: 174 WERFRDALLERYGGNGDGDVYEQLSELRQQG-IVEEYITDFE----YLTAQIPKLPEKQY 228
W ++ + R+ + E ++Q VE+YI FE + + P L E+
Sbjct: 267 WPQWCAMVSTRFSAADTHEAVELFQNVKQYNQTVEQYIDKFEEYVDLVRREHPYLQEQYL 326
Query: 229 QGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFGGNGS 288
F+ GL+ +I+ V G + T+ E+ A AR N N +
Sbjct: 327 NSCFIGGLRGDIKHDVCGHKPQGLLES---YWYTKNYER-------AANARKNLLNFNRN 376
Query: 289 R-SGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHL 347
R +G +G + R PR++
Sbjct: 377 RFQNQAGPIQGRNV--------------------------------VNRGQPREQ----- 399
Query: 348 SYPQLMERRQKGLCFKCGGPYHRNHVCP-DKHLRLLILE--------EDGEELDESKMLA 398
+E++++ C+ C P+ H C K L L++E E+GE +
Sbjct: 400 -----VEKKEERKCWFCKEPWFPKHQCKVKKALNALLMEGEEGKDEGEEGELTGNQEDCK 454
Query: 399 MEVNEDEEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCF 458
+E E + + + LM + + P T + ING V LVDSG+T F+D
Sbjct: 455 LEKEEAPPDDENQEELMFVSHNAVYGTTRPDTFSVIIQINGRRAVGLVDSGSTSTFMDQD 514
Query: 459 LVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLG 518
R +V T V + G + +++ + L ++ + L G D++LG
Sbjct: 515 YAVRNHCPLVSTDAKKVVVAGGGELKSEVQVPELVYQIQGETFSNKFNIIPLKGYDVILG 574
Query: 519 IEWLKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHMEALQSITATGERKPGL---- 574
+W+ ++ + + +K + +Q G+H+ RK GL
Sbjct: 575 ADWIYKYSPITLDLKKRELGITKGEKTVVIQDFTRPGKHLWVDSKKVDQILRKGGLGCLF 634
Query: 575 -LGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKG 633
+ EE+ ++P E++ ++L F V ++ +GLPP R +H IT++ G
Sbjct: 635 QITRVKEEETSHEIP--------EDIKEILQEFPAVLKDPKGLPPRRNCDHVITLKSGAE 686
Query: 634 PVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRAL 693
P N+RPYR PH+ K +EK + E++ + I+ S YSSP ++V+KKD +W +CVDYR L
Sbjct: 687 PPNLRPYRVPHYQKEAMEKIIAELIESKEIQVSDIPYSSPAVMVRKKDGSWRLCVDYRQL 746
Query: 694 NSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEY 753
N+ TV +KFP+P+IE+LLDEL+GA FSKLDL+SGYHQ+R+ +D+ KTAFRTH GHYEY
Sbjct: 747 NAQTVKNKFPMPIIEDLLDELNGAKVFSKLDLRSGYHQIRMATQDIPKTAFRTHLGHYEY 806
Query: 754 MVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQH 813
VMPFGL NAP+TFQSLMN+V P LR+ VLVFFDDIL+YSK W++H H+ +V++VL+
Sbjct: 807 QVMPFGLTNAPTTFQSLMNQVLAPFLRKYVLVFFDDILIYSKDWAEHKEHIRQVMKVLEE 866
Query: 814 HQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGY 873
++L KKC F V YLGH+IS+ GVA DP KV + ++P PK+V +R FLG+TGY
Sbjct: 867 NKLVVKLKKCAFGLPSVTYLGHIISQDGVATDPKKVEKIATYPTPKSVTDLRKFLGMTGY 926
Query: 874 YRKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLEC 933
YR+FI+NYG + RPL ++ KK+ F W ++ +AFE LK + T+PVL+LPDF++EF +E
Sbjct: 927 YRRFIKNYGIVCRPLHDMLKKEGFQWEREQTEAFETLKTHMCTSPVLSLPDFTKEFVIEA 986
Query: 934 DASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVV 993
DA G GIGA+L+ P+AYFSK LGP+ A+S YEKE MA+ A++ WR Y+LG + ++
Sbjct: 987 DACGNGIGAVLMQSGRPLAYFSKTLGPKAAAQSIYEKEAMAILEALKKWRHYVLGSRLII 1046
Query: 994 STDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRPL 1053
TDQ+SLK ++Q++V G Q KL+ Y++ I YK GK N ADALSRI A + + +
Sbjct: 1047 KTDQQSLKFMMNQRLVEGIQHKLLLKLMEYDYSIEYKAGKENLVADALSRIPPAEQCQAI 1106
Query: 1054 VS-YPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSS 1112
+ P W I + D + +I+S + + Y G L Y+ R+ + +
Sbjct: 1107 TTVIPEWVRD--IQRSYEGDVQAHKILSLIGTEGDTDGSYSQEAGLLRYKGRIYVGENTE 1164
Query: 1113 LIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSP 1172
+ +L+ +H++ GGHSG TY RI + YW G+ K ++ F+ C +CQ K
Sbjct: 1165 IREELIRSYHSSAFGGHSGMRATYHRIKSLFYWPGLKKAVEGFIRECPICQVTKAEHIHI 1224
Query: 1173 AGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVA 1232
GLL PL +P+ W I+++F+ GLP+S G + I VVVDRLTKY+HFI + HPYT V
Sbjct: 1225 PGLLDPLEVPDMAWAHITMDFVEGLPKSNGKDVILVVVDRLTKYAHFIAMAHPYTVEQVV 1284
Query: 1233 EIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNR 1292
E+F I RLHG+P +II+DRD +F S ++E+F+ + K S++YHP++DGQTE VN+
Sbjct: 1285 ELFMNNIHRLHGMPMAIITDRDRIFTSQLFQEIFKSMKVRLKFSTSYHPQTDGQTERVNQ 1344
Query: 1293 CLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGET 1352
CLE+YLR +P W SWL AE+W+NT+YH++ Q TPF+A+YG PP + + +
Sbjct: 1345 CLESYLRSMTFQEPTRWHSWLALAEWWYNTTYHTSIQMTPFQALYGYPPPQINEFSVPCN 1404
Query: 1353 RVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVS 1412
E ++ +D +++LK L AQ R++ A+ R + VG+ V++K++ +RQ +
Sbjct: 1405 VSEEARVTIEQKDAIIQKLKYSLTEAQRRIKHYADRNRSERTLAVGDMVYLKLQPYRQTA 1464
Query: 1413 LANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGN-YVEEPG 1471
R KL ++++GP+ I+ +VG VAY+L+LPEG+ IHPVFH+S LKK +G+ V
Sbjct: 1465 FGIRGSLKLRSKFYGPFKIMEKVGRVAYKLQLPEGSNIHPVFHVSQLKKHIGSRAVPMAN 1524
Query: 1472 LPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFPV 1531
LP ++ + +P VL RR I R G +++Q LV W +P EATWED + I+S FP
Sbjct: 1525 LPSVGPDGQIKT-EPVAVLKRRMIPRGGVAVTQWLVLWHNLSPSEATWEDASMIQSMFPS 1583
Query: 1532 FN 1533
FN
Sbjct: 1584 FN 1585
>UniRef100_Q7XEK0 Hypothetical protein [Oryza sativa]
Length = 1611
Score = 949 bits (2453), Expect = 0.0
Identities = 542/1456 (37%), Positives = 812/1456 (55%), Gaps = 95/1456 (6%)
Query: 106 NEFRQSVKK---VELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFY 162
N++ ++V K +E+ +F G+D W+ + E +F++ T + V+LA + G ++
Sbjct: 220 NQYAEAVIKGPRLEISLFTGEDPVDWLKQCEKFFEITGTPVDQWVNLAVAHLYGRAAKWF 279
Query: 163 NSLLATEEELTWERFRDALLERYGGNGDGDVYEQLSELRQQGI-VEEYITDFEYLTAQI- 220
+ + +TW ++ + R+ + E ++Q G+ VE+YI FE +
Sbjct: 280 RGVGLPWQVITWPQWCAMVCTRFSTANTHEAVELFQNVKQYGMTVEQYIDKFEEYMDLVR 339
Query: 221 ---PKLPEKQYQGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQ 277
P L E + F+ GL+ +I+ +V G G
Sbjct: 340 RDHPYLQEPYFTSCFISGLRGDIK-------------------------HDVCGQKPQGL 374
Query: 278 ARANRFGGNGSRSGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRA 337
+ + N ++ S N NR G+ KN +
Sbjct: 375 LESYWYAKNYEKAANSRKAAAN----------------FNRNRLQTGGNTGKNV--YNKG 416
Query: 338 GPRDRGFNHLSYPQLMERRQKGLCFKCGGPYHRNHVCPDKHL--RLLILEEDGEELDESK 395
PR G +++++ C+ C P+ H C K LL+ E+ E++E
Sbjct: 417 QPRQEG----------DKKEEKKCWFCKEPWFPRHQCKVKQAIHALLVENEESVEVEED- 465
Query: 396 MLAMEVNEDEEETQGEL----------SLMSLCELGMKTGGIPRTMKLRGTINGVPVVVL 445
++E E + E QGE LMS+ + + P T + +NG V L
Sbjct: 466 --SVEEEEIKGEKQGEKLPEQTENVQEELMSISQSAVYGLTRPDTFSVMIKVNGKKAVGL 523
Query: 446 VDSGATHNFVDCFLVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSP 505
VDSG+T F+D + + +T V + G + +++ G++ E+ S
Sbjct: 524 VDSGSTTTFMDSKFAIKSQCTLENTKMRKVIVAGGGELKSELIVPGMEYEIQGESFTNSF 583
Query: 506 QLFDLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHMEALQSIT 565
L L DI+LG +W+ ++ + M + + +Q G++ +
Sbjct: 584 NLLSLERYDIILGADWIFKYSPITLDLRKREMKITKGGRELEIQDFTKPGKYFQV----- 638
Query: 566 ATGERKPGLLGTRNEEKAGKQLPGEINSSQLE-----ELDKLLYRFDVVFQEKQGLPPGR 620
+K G + + Q+ + S +E ++ +L F V +E +GLPP R
Sbjct: 639 --SNKKMGKMIKKGALGCVIQINAITDQSNVEVGIPKDIQIVLQSFPKVLKEPKGLPPRR 696
Query: 621 GREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKK 680
+H I ++ G P N+RPYR PH K +E + E+ IR S S Y SP ++V+KK
Sbjct: 697 SCDHVINLKVGSEPPNLRPYRVPHFQKGAMEDIITELFRTQEIRISDSPYVSPAVMVRKK 756
Query: 681 DQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVH 740
D +W +CVDYR LN+ T+ +KFP+P+IE+LLDELHGA FSKLDL+SGYHQ+R+ E D+
Sbjct: 757 DGSWRLCVDYRQLNAQTIKNKFPMPIIEDLLDELHGAKVFSKLDLRSGYHQIRMAEGDIP 816
Query: 741 KTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDH 800
KTAFRTH GHYEY VMPFGL NAP+TFQ+LMN+V P LR+ VLVFF DIL+YSKT S+H
Sbjct: 817 KTAFRTHLGHYEYNVMPFGLTNAPATFQALMNQVLAPFLRKFVLVFFADILIYSKTQSEH 876
Query: 801 MLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKN 860
+ H++ V++ L +QL KKC+F V YLGH+IS +GV+ DP K+ +K+ PKN
Sbjct: 877 LEHIKLVMQALSANQLVVRLKKCEFGLDRVSYLGHIISSEGVSTDPKKISDIKNRKPPKN 936
Query: 861 VKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVL 920
V VR FLG+ GYYR+FI+ YG I RPL +L KKD F WG Q+AFE LKEK+ +PVL
Sbjct: 937 VTEVREFLGMAGYYRRFIKGYGVICRPLHDLLKKDGFKWGDTQQEAFELLKEKMCNSPVL 996
Query: 921 ALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQ 980
ALPDFSQ F +E DA GIGIGA+L+ P+AYFSKALGP+ A+S YEKE +A+ A++
Sbjct: 997 ALPDFSQPFVIETDACGIGIGAVLMQKGRPLAYFSKALGPKAAAQSVYEKEAIAILEALK 1056
Query: 981 HWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADA 1040
WR Y+LG ++ TDQ+SLK + Q++V G Q KL+ +++ I YK GK N ADA
Sbjct: 1057 KWRHYILGGSLIIKTDQQSLKFMMSQRLVEGIQHKLLLKLMEFDYVIEYKSGKENLVADA 1116
Query: 1041 LSRIDEAGELRPL---VSYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQG 1097
LSR E + L V P W + I ++D +I+S + D Y++ G
Sbjct: 1117 LSRSPNLKEEQCLPITVVVPEWVQD--IKRSYEEDIFAHKILSLIETDGDPERHYKLESG 1174
Query: 1098 TLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV* 1157
L Y+ R+ + + + LLE +H + GGHSG TY RI YW G+ K ++ ++
Sbjct: 1175 LLKYKGRIYVGETTEIRMLLLEAYHASYFGGHSGIRATYHRIKQLFYWPGLKKQVEHYIR 1234
Query: 1158 ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYS 1217
C CQ K GLL PL +P+ W I+++FI GLP+S+G + I VVVDRLTKY+
Sbjct: 1235 ECPTCQITKAEHIHIPGLLNPLEVPDMAWTHITMDFIEGLPKSQGKDVILVVVDRLTKYA 1294
Query: 1218 HFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSS 1277
HF+ L HPYT V +IF I +LHG+P I++DRD +F S+F++E+F+ Q + + S+
Sbjct: 1295 HFLALSHPYTVEQVVQIFMDNIHKLHGMPMVIVTDRDRVFTSNFFQEIFKTQKVKLRFST 1354
Query: 1278 AYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVY 1337
A+HP++DGQTE VN+CLE+YLR +P+ W SWL AE+W+NT+YH++ Q TPF+A+Y
Sbjct: 1355 AHHPQTDGQTERVNQCLESYLRSMTFQEPQKWFSWLALAEWWYNTTYHTSIQMTPFQALY 1414
Query: 1338 GRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEV 1397
G PP +T + + E L+D+ L++LK + AQ R++ A+ R + E+
Sbjct: 1415 GYPPPQITEFAIPCNMSEEARVTLEDKALILQKLKSSIGEAQRRIKFYADKGRSERTLEL 1474
Query: 1398 GEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHIS 1457
G+ V++K++ +RQV++ R KL ++Y+GP+ +I ++GAVAY+L+LP+GA IHPVFH+S
Sbjct: 1475 GDMVYLKLQPYRQVAMGIRGSLKLRSKYYGPFKVIEKMGAVAYKLQLPDGAGIHPVFHVS 1534
Query: 1458 LLKKAVG-NYVEEPGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEE 1516
LKK +G + P LP + + +PA VL RR I R E ++Q L+ W+ TP E
Sbjct: 1535 QLKKHLGARAIPMPNLP-AIGPDGQIKTEPAAVLQRRMIPRHNEPVTQWLILWENLTPAE 1593
Query: 1517 ATWEDLATIRSEFPVF 1532
ATWED + I++ FP F
Sbjct: 1594 ATWEDASYIQAAFPNF 1609
>UniRef100_Q9LP90 T32E20.30 [Arabidopsis thaliana]
Length = 1397
Score = 945 bits (2443), Expect = 0.0
Identities = 520/1205 (43%), Positives = 722/1205 (59%), Gaps = 124/1205 (10%)
Query: 347 LSYPQLMERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNEDEE 406
L +L E+R+KG+CF+C GP+ + H CP+K LR+L + ++ +M +E N EE
Sbjct: 290 LQLVKLDEKRRKGICFRCDGPWSKEHKCPNKELRVLTV------INGFEMEVLESNSVEE 343
Query: 407 ETQGELSLMSLCELGMKTG-GIPR--TMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRL 463
E S+ EL + G+P T+K++G+I G + F
Sbjct: 344 EFHD--SVAQFAELSFSSYMGLPSYTTIKMKGSIC---------KGEWCHTQFYF----- 387
Query: 464 GWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMG-----EYHLRCSPQLFDLGGPDIVLG 518
P ++LG G Q G C + + + E L DLG D++LG
Sbjct: 388 -------PNFHIRLGTGITVQGLGLCDKVTMTLPVGCGQELELTTHFITLDLGPVDVILG 440
Query: 519 IEWLKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHMEALQSITATGERKPGLLGTR 578
I WL+TLGD VNW+ +SF + +TL+G +L+S + +
Sbjct: 441 IAWLRTLGDCKVNWERHELSFLYHGRTVTLRGDPELDTFKMSLKSFSTKFRLQ------- 493
Query: 579 NEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNVR 638
K+L +NS Q +GLPP +G EH I++ G ++VR
Sbjct: 494 -----NKELEVSLNSHQ----------------NLKGLPPIKGNEHAISLLPGTRAISVR 532
Query: 639 PYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTV 698
PYRYPH HK +E V EML GIIR S S +SSPV+LVKKKDQ+W CVDYRALN T+
Sbjct: 533 PYRYPHAHKEAMEGLVSEMLDNGIIRASKSPFSSPVLLVKKKDQSWRFCVDYRALNRATI 592
Query: 699 PDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPF 758
P+KFPIP+I++LLDELHGA+ FSKLDL++GYHQ+R++ ED+ KT FRTH+GH+E++VMPF
Sbjct: 593 PNKFPIPMIDQLLDELHGAIIFSKLDLRAGYHQIRMKVEDIEKTTFRTHDGHFEFLVMPF 652
Query: 759 GLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTA 818
GL NAP+TFQS MN++ RP LR+ VLVFFDDIL+YS+ +H HL VL+VL+ HQ A
Sbjct: 653 GLSNAPATFQSSMNDMLRPFLRKFVLVFFDDILIYSRNEQEHEEHLAMVLKVLEEHQFYA 712
Query: 819 NKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFI 878
N+KK T QGV+ DP K +++ W P++VK +RGFLGLTGYYR+F+
Sbjct: 713 NRKKPYHIT------------QGVSTDPTKTVAMTKWVTPQSVKELRGFLGLTGYYRRFL 760
Query: 879 QNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGI 938
+ YG +ARPLTEL KKD+F W + AQ+AF+ LK ++TAPVLALPDF +
Sbjct: 761 KGYGTLARPLTELLKKDSFVWSESAQEAFDALKRAMSTAPVLALPDFGK----------- 809
Query: 939 GIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQR 998
++G L + K YE+ELMA+ L+IQ W+ YL+GR+FV+ TDQ+
Sbjct: 810 ------VHG----------LTSKEQLKPVYERELMAIVLSIQKWKHYLMGRRFVLHTDQK 853
Query: 999 SLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSR-IDEAGELRPLV--- 1054
SLK Q+ V+ D Q W KLL Y F+I+YK G N+ AD LSR + G ++
Sbjct: 854 SLKFLQEQREVSMDYQKWLTKLLHYEFDILYKLGVDNKAADGLSRMVQPTGSFSSMLLMA 913
Query: 1055 -SYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSL 1113
+ P + + +E + L +V E Q Y V +G L + R++I S
Sbjct: 914 FTVPTVLQLHDLYEEIDSNAHLQHLVKECLSAKQGTSAYTVKEGRLWKKQRLIIPKDSKF 973
Query: 1114 IPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPA 1173
+P +L E+H+ GGHSG L+T +RI + +W GM K IQ+FV C++CQR KY SPA
Sbjct: 974 LPLILAEYHSGLLGGHSGVLKTMKRIQQSFHWEGMMKDIQKFVAKCEMCQRQKYSTLSPA 1033
Query: 1174 GLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAE 1233
GLLQPLPIP +VWEDISL+F+ GLP DRL+KY HFI LKHP+ A VA
Sbjct: 1034 GLLQPLPIPTQVWEDISLDFVEGLP------------DRLSKYGHFIGLKHPFNAVDVAR 1081
Query: 1234 IFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRC 1293
IF E+++LHG P+SI+SDRD F+S FWK+ F+L GT+ K S+A+HP++DGQTE++NR
Sbjct: 1082 IFIHEVVKLHGFPASIVSDRDNTFLSSFWKDCFKLSGTKLKYSTAFHPQTDGQTEVLNRT 1141
Query: 1294 LETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETR 1353
LETYLRCFA+ PKTW +L AE W+N+S+H+ + TPF+ +YGR PP + R+ T+
Sbjct: 1142 LETYLRCFASAHPKTWFQYLPRAELWYNSSFHTTIKTTPFKVLYGRDPPPIMRFEANSTK 1201
Query: 1354 VEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSL 1413
+E L+ RD L +K+HLV AQ+ M+ + R+ EF+ VF+K+R +RQ S+
Sbjct: 1202 NCELEGLLKQRDLMLADIKEHLVNAQQLMKNNDDKHRREVEFDTRNRVFLKLRPYRQNSV 1261
Query: 1414 ANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGLP 1473
RV KLAA+YFGP+ I+ R+G VAYRLKLPEG++IH VFH+S LK+ +G++ + LP
Sbjct: 1262 TKRVCQKLAAKYFGPFEIMERIGKVAYRLKLPEGSKIHLVFHVSQLKQVLGDHHQVIPLP 1321
Query: 1474 EQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFPVFN 1533
E L + + P VL R + + + LV WQG E TWE ++ +FP
Sbjct: 1322 EVLTADNEFVVVPEAVLETR---YNEDGLLEALVHWQGLPVHEDTWEIAKDLKKQFPGLA 1378
Query: 1534 LEDKV 1538
L+DK+
Sbjct: 1379 LQDKL 1383
Score = 85.5 bits (210), Expect = 1e-14
Identities = 50/165 (30%), Positives = 94/165 (56%), Gaps = 6/165 (3%)
Query: 112 VKKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEE 171
+K VE+PMFDG + GWI+R E +F+ + +++L +S+ G + +YN ++ +
Sbjct: 127 LKNVEMPMFDGSGIYGWIARVERFFRSGGYNEAEQLALVSVSVSGEALSWYNWAISRGDF 186
Query: 172 LTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQGY 231
++W + + L+ R+G + L ++Q G V EY+ FE L++Q+ L +++ +G
Sbjct: 187 VSWLKLKSGLMLRFGNLKLRGPSQSLFCIKQTGSVAEYVQRFEDLSSQVGGLDDQKLEGI 246
Query: 232 FLHGLKEEIR-----GKVRSLVAMGGVNRA-RLLVVTRAVEKEVK 270
FL+GL E++ K ++L M V R+ V+ R V+KE++
Sbjct: 247 FLNGLTGEMQELVHMHKPQNLPEMVAVARSMETSVMRRVVQKELQ 291
>UniRef100_Q8SA91 Putative gag-pol polyprotein [Zea mays]
Length = 2396
Score = 936 bits (2419), Expect = 0.0
Identities = 540/1471 (36%), Positives = 822/1471 (55%), Gaps = 96/1471 (6%)
Query: 111 SVKKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEE 170
++ +V P FDG + W + E YF + + P + + +A + G + S+
Sbjct: 171 NLPRVNFPQFDGDNPQLWKTLCENYFDMYDVEPYMWIRVATMHFIGRAASWLQSVGRRVC 230
Query: 171 ELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDF----EYLTAQIPKLPEK 226
L+W F L +R+G + QL + Q G V EY+ F ++L+A
Sbjct: 231 MLSWSEFCRQLQDRFGREQHESLIRQLFHIHQSGTVAEYVEQFSILVDHLSAYEANADPL 290
Query: 227 QYQGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFGGN 286
Y F+ GL+++I+ + V R L A + + R R +
Sbjct: 291 YYTMRFIDGLRDDIKAVIM-------VQRPSNL--DTACSLALVQEEATTARRWRRSEPS 341
Query: 287 GSRSG-FSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFN 345
S +G +G T+W NK++ Q+ + D+ ++ RRA
Sbjct: 342 SSHAGPKTGVQLSASTKWT---SNKDSTQSASH------SDKLESLRRFRRA-------- 384
Query: 346 HLSYPQLMERRQKGLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNEDE 405
KGLC KC ++ H C +L +EE L + ++ +++ E
Sbjct: 385 ------------KGLCDKCAEKWNPGHKCAAT-AQLHAMEEVWSLLVDEEVPESDLSPPE 431
Query: 406 EETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRLGW 465
+ +S TG +T+KL G+I P+++L+DSG++H F++ L L
Sbjct: 432 PAPEQLFVTISKSAWTGSTGR--QTLKLNGSIQNHPLLILIDSGSSHTFLNDQLRPHLQG 489
Query: 466 EVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLKTL 525
+ V++ +G + + ++ L D+V+G++WL++
Sbjct: 490 VTSMASTLQVQVANGAMVTCHYKLLQAQWQIQNCSFTSDVSFLPLPYYDMVVGMDWLESF 549
Query: 526 GDTIVNWDTQLMSFWSDKKWITLQG------MDTRGE--HMEALQSITATGERKPGLLGT 577
V+W + + + LQG DT E ME+ S++++ + P +
Sbjct: 550 SPMRVDWAQKWLIIPYQGSSVLLQGNTAGVPADTVIELLFMESASSVSSSPDSHPAI--- 606
Query: 578 RNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNV 637
LL +F VF E QGLPP R +H I + EG PV+V
Sbjct: 607 ----------------------QALLQQFSSVFAEPQGLPPSRDCDHAIPLVEGAQPVSV 644
Query: 638 RPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVT 697
RPYRYP K++IEKQV+EML G+I++S S+++SPV+LVKKKD TW CVDYR LN++T
Sbjct: 645 RPYRYPPALKDKIEKQVQEMLHQGVIQKSNSSFASPVLLVKKKDMTWRFCVDYRYLNALT 704
Query: 698 VPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMP 757
+ K+P+PV ++L+DEL + +FSKLDL++GYHQ+ ++ + +KTAF+TH GHYE+ VM
Sbjct: 705 LKSKYPVPVFDQLIDELAHSKWFSKLDLRAGYHQILLKPGEEYKTAFQTHVGHYEFRVMA 764
Query: 758 FGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLT 817
FGL AP+TF S MNE +P+LR+ LVFFDDIL+YSK++ +H+LHL++VL++L
Sbjct: 765 FGLTGAPNTFLSAMNETLKPVLRKCALVFFDDILIYSKSFEEHLLHLQKVLQLLLSDNWK 824
Query: 818 ANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKF 877
KC+FA YLGH+ISEQGV+ P+K+ ++ SW P + K +R FLGL G+YRKF
Sbjct: 825 VKLSKCEFAKTNTAYLGHIISEQGVSTYPSKIQAISSWAVPTSAKELRCFLGLAGFYRKF 884
Query: 878 IQNYGKIARPLTELTKKDA-FNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDAS 936
++++G I+RPL +L KK F W +AFE LK+ L TAPVLALPDFSQ F + DAS
Sbjct: 885 VKHFGIISRPLFDLLKKHTLFVWTVDHSKAFEVLKQALVTAPVLALPDFSQPFCIHTDAS 944
Query: 937 GIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTD 996
G+GA+L+ HP+A+ SKALGP+N S YEKE MA+ LAI WR YL +F++ TD
Sbjct: 945 YYGVGAVLMQSGHPLAFLSKALGPKNQGLSTYEKEYMAIILAIAQWRSYLQLAEFIIYTD 1004
Query: 997 QRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSR-IDEAGELRPLV- 1054
RSL + Q++ T QQ KL G ++I+Y+ G N ADALSR + E +
Sbjct: 1005 HRSLAQLNEQRLHTIWQQKMYTKLAGLQYKIVYRKGVDNGAADALSRKVQEDSHCCAISH 1064
Query: 1055 SYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLI 1114
S P W ++ + + KDP Q+++++ L+ + + QG + +++R+ + L
Sbjct: 1065 SVPTWLQE--VVEGYDKDPTSKQLLAQLILNSADKAPFSLHQGIIRHKNRIWLGGNLQLQ 1122
Query: 1115 PKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAG 1174
K+L+ H T GGHSG TY ++ YW GM + ++V +C VCQ+ K + G
Sbjct: 1123 QKVLQAMHDTAVGGHSGAPATYHKVKQMFYWPGMRADVLQYVQSCTVCQQSKPDRAKYPG 1182
Query: 1175 LLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEI 1234
LLQPL +P + W ISL+FI GLPRS + I VVVD+ +KY HF+PL HP+TA VA +
Sbjct: 1183 LLQPLEVPPQAWHTISLDFIEGLPRSAHYNCILVVVDKFSKYGHFLPLLHPFTAAKVARV 1242
Query: 1235 FAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCL 1294
F + +LHG+P +IISDRD +F S FW+++F++ GT MSS+YHP+SDGQTE +N+CL
Sbjct: 1243 FLDNVYKLHGLPVNIISDRDRIFTSSFWQQLFQITGTNLSMSSSYHPQSDGQTERLNQCL 1302
Query: 1295 ETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKP---PVLTRWVLGE 1351
ET+LRC+ P W++WL AEYW+NT+ HS +TPFE +YG P +L V+ +
Sbjct: 1303 ETFLRCYVHTCPSRWSAWLSVAEYWYNTTVHSTLGRTPFEVLYGHTPRHFGILVDTVVPQ 1362
Query: 1352 TRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQV 1411
+E L++R+ + +K HL AQ+RM+ QA+ +R + F VG+WV++K++ + Q
Sbjct: 1363 PELETW---LKERELMTKVIKLHLHRAQDRMKRQADKQRSERVFSVGDWVYLKLQPYIQS 1419
Query: 1412 SLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVG-NYVEEP 1470
S+A R + KL+ ++FGP+ I R+G+VAYRL LP + IHP+FH+S LK+ +G + P
Sbjct: 1420 SVATRSNHKLSFKFFGPFQITDRLGSVAYRLALPASSSIHPIFHVSQLKRVIGRDQRASP 1479
Query: 1471 GLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFP 1530
LP+ + P P +L RR I R GE I+QV V W G T + ATWED+ +R+ FP
Sbjct: 1480 QLPQDVG----PIQVPTRILQRRFIDRGGELIAQVKVVWSGMTEDLATWEDVEALRARFP 1535
Query: 1531 VFNLEDKV-VSSEGSIVRKDNNHNGLLAQDD 1560
+ D+ +G++ + + LA +
Sbjct: 1536 KALIWDQAGARGQGNVSKASSEDKKKLAASE 1566
>UniRef100_Q9C7H8 Gypsy/Ty3 element polyprotein, putative [Arabidopsis thaliana]
Length = 1447
Score = 916 bits (2367), Expect = 0.0
Identities = 509/1195 (42%), Positives = 718/1195 (59%), Gaps = 54/1195 (4%)
Query: 348 SYPQLMERRQK----GLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNE 403
SY + E QK GLC+ C + H K +L ++ D E D A+EV
Sbjct: 289 SYRPVKEVEQKSDHLGLCYFCDEKFTPEHYLVHKKTQLFRMDVDEEFED-----AVEVLS 343
Query: 404 DEEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRRL 463
D++ Q + +S+ + +G +TM ++GT++ + +L+DSG+THNF+D + +L
Sbjct: 344 DDDHEQKPMPQISVNAVSGISGY--KTMGVKGTVDKRDLFILIDSGSTHNFIDSTVAAKL 401
Query: 464 GWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEWLK 523
G V V + DG K G+ G ++ + L L G D+VLG++WL+
Sbjct: 402 GCHVESAGLTKVAVADGRKLNVDGQIKGFTWKLQSTTFQSDILLIPLQGVDMVLGVQWLE 461
Query: 524 TLGDTIVNWDTQLMSFWSDKKWITLQGMDT---RGEHMEALQSITATGERKPGLLGTRNE 580
TLG + M F+ + + L G+ T R LQ T + + ++ R
Sbjct: 462 TLGRISWEFKKLEMQFFYKNQRVWLHGIITGSVRDIKAHKLQK-TQADQIQLAMVCVREV 520
Query: 581 EKAGKQLPGEIN---SSQLEE--LDKLLYRFDVVFQEKQGLPPGRGR-EHCITIQEGKGP 634
+Q G I+ S +EE + ++ F VF E LPP R + +H I + EG P
Sbjct: 521 VSDEEQEIGSISALTSDVVEESVVQNIVEEFPDVFAEPTDLPPFREKHDHKIKLLEGANP 580
Query: 635 VNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALN 694
VN RPYRY H K+EI+K V++M+ +G I+ S+S ++SPV+LVKKKD TW +CVDY LN
Sbjct: 581 VNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSPFASPVVLVKKKDGTWRLCVDYTELN 640
Query: 695 SVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYM 754
+TV D+F IP+IE+L+DEL G+V FSK+DL++GYHQVR+ +D+ KTAF+TH GH+EY+
Sbjct: 641 GMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYL 700
Query: 755 VMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHH 814
VM FGL NAP+TFQSLMN VFR LR+ VLVFFDDIL+YS + +H HL V EV++ H
Sbjct: 701 VMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMRLH 760
Query: 815 QLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYY 874
+L A K E+LGH IS + + DP K+ +VK WP P VK VRGFLG GYY
Sbjct: 761 KLFAKGSK--------EHLGHFISAREIETDPAKIQAVKEWPTPTTVKQVRGFLGFAGYY 812
Query: 875 RKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECD 934
R+F++N+G IA PL LTK D F W +AQ AF+ LK L APVLALP F ++F +E D
Sbjct: 813 RRFVRNFGVIAGPLHALTKTDGFCWSLEAQSAFDTLKAVLCNAPVLALPVFDKQFMVETD 872
Query: 935 ASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVS 994
A G GI A+L+ HP+AY S+ L + L S YEKEL+A A++ WR YLL F++
Sbjct: 873 ACGQGIRAVLMQKGHPLAYISRQLKGKQLHLSIYEKELLAFIFAVRKWRHYLLPSHFIIK 932
Query: 995 TDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRPLV 1054
TDQRSLK L Q++ T QQ W KLL +++EI Y+ GK N ADALSR++ + L +
Sbjct: 933 TDQRSLKYLLEQRLNTPVQQQWLPKLLEFDYEIQYRQGKENLVADALSRVEGSEVLHMAL 992
Query: 1055 SYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLI 1114
S + I + D L I+S + P A Y Q L + ++V+ N +
Sbjct: 993 SIVECDFLKEIQVAYESDGVLKDIISALQQHPDAKKHYSWSQDILRRKSKIVVPNDVEIT 1052
Query: 1115 PKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAG 1174
KLL+ H + GG SG +++R+ + YW+GM K IQ F+ +C CQ+ K ++ G
Sbjct: 1053 NKLLQWLHCSGMGGRSGRDASHQRVKSLFYWKGMVKDIQAFIRSCGTCQQCKSDNAAYPG 1112
Query: 1175 LLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEI 1234
LLQPLPIP+++W D+S++FI GLP S G I VVVDRL+K +HF+ L HPY+A +VA+
Sbjct: 1113 LLQPLPIPDKIWCDVSMDFIEGLPNSGGKSVIMVVVDRLSKAAHFVALAHPYSALTVAQA 1172
Query: 1235 FAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCL 1294
F + + HG P+SI+SDRD LF S FWKE F+LQG + +MSSAYHP+SDGQTE+VNRCL
Sbjct: 1173 FLDNVYKHHGCPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCL 1232
Query: 1295 ETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRV 1354
E YLRC +P W WL AEYW+NT+YHS++Q TPFE VYG+ PP+ ++ G+++V
Sbjct: 1233 ENYLRCMCHARPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKV 1292
Query: 1355 EAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLA 1414
V + LQ+R+ L LK HL+ AQ RM+ A+ R + F++G++V+VK++ +RQ S+
Sbjct: 1293 AVVARSLQERENMLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVV 1352
Query: 1415 NRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGLPE 1474
RV+ KL+ +YFGPY II + G V VGN LP
Sbjct: 1353 LRVNQKLSPKYFGPYKIIEKCGEV-----------------------MVGNVTTSTQLPS 1389
Query: 1475 QLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEF 1529
L ++ P +L R+ + RQG + + VLV+W G+ EEATW+ L + +F
Sbjct: 1390 VL--PDIFEKAPEYILERKLVKRQGRAATMVLVKWIGEPVEEATWKFLFDRQQKF 1442
Score = 49.3 bits (116), Expect = 0.001
Identities = 35/136 (25%), Positives = 63/136 (45%), Gaps = 6/136 (4%)
Query: 114 KVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSME--GGTIH--FYNSLLATE 169
K++ P FDG + W+ + E +F V T E+KV + + + T H F S + +
Sbjct: 116 KIDFPRFDGSRINEWLFKVEEFFGVDFTPEEMKVKMVAIHFDSHAATWHHSFIQSGIGLD 175
Query: 170 EELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQ 229
W + L +R+ D D +L +L++ + EY FE + ++ L E+
Sbjct: 176 VFFNWPEYVKLLKDRFEDACD-DPMAELKKLQETDGIVEYHQQFELIKVRL-NLSEEYLV 233
Query: 230 GYFLHGLKEEIRGKVR 245
+L GL+ + + VR
Sbjct: 234 SVYLAGLRTDTQMHVR 249
>UniRef100_Q8RUU3 Putative gag-pol polyprotein [Oryza sativa]
Length = 1338
Score = 871 bits (2251), Expect = 0.0
Identities = 479/1155 (41%), Positives = 658/1155 (56%), Gaps = 69/1155 (5%)
Query: 391 LDESKMLAMEVNEDEEET-QGELSLMSLCELGMKTG-GIPRTMKLRGTINGVPVVVLVDS 448
L E + L + +++ T Q + + L TG M + + + L+DS
Sbjct: 220 LSEDQQLGEDDTDNDSATDQADAAATLRISLHAATGVRASDAMHITAHLGDTDLYTLIDS 279
Query: 449 GATHNFVDCFLVRRLGWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLF 508
G H F+ R+G PRM GL + + P +F
Sbjct: 280 GLMHTFLSQDTAARVGR--APQPRM-----------------GLNVTVANGDKVACPGVF 320
Query: 509 DLGGPDIVLGIEWLKTLGDTIVNWDTQLMSFWSDKKWITLQGMDTRGEHMEALQSITATG 568
PD+ L I + D + ++ MSFW +TL G+
Sbjct: 321 ----PDMPLQIAGEEFATD-VYDFTVLTMSFWHR---VTLHGLP---------------- 356
Query: 569 ERKPGLLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITI 628
G Q P + + LD LL F VF E GLPP R R H I +
Sbjct: 357 ---------------GHQRPRALACAPAALLDSLLEEFADVFTEPTGLPPARDRSHRIQL 401
Query: 629 QEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCV 688
G PV VRPYRYP HK+E+E+Q R M G+I +STSA+SSPV+LVKK D +W CV
Sbjct: 402 LPGTAPVAVRPYRYPVRHKDELERQCRVMEENGLIHRSTSAFSSPVLLVKKADGSWRFCV 461
Query: 689 DYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHE 748
+YRALN TV DK+PIPV++ELLDELHGA FSKLDL+SGYHQVR+ +D+ KTAFRTH+
Sbjct: 462 NYRALNERTVKDKYPIPVVDELLDELHGAAIFSKLDLRSGYHQVRMHPDDIDKTAFRTHD 521
Query: 749 GHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVL 808
G YE++V+PFGL NA +TFQSLMN+V RP LRR VLVFFDDILVYS TW+ H+ HL V
Sbjct: 522 GLYEFLVIPFGLTNALATFQSLMNDVLRPFLRRFVLVFFDDILVYSPTWTSHLQHLRTVF 581
Query: 809 EVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFL 868
L QL KC F V YLGH+IS+ G A+D K+ +V WP P++ K +RGFL
Sbjct: 582 TALWAAQLFVKHTKCSFGDPSVAYLGHIISQHGFAMDAAKIQAVAEWPRPRSPKELRGFL 641
Query: 869 GLTGYYRKFIQNYGKIARPLTELTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQE 928
GL YYRKFIQ++G +A PLT+L +KD+F W AF+ LK LTT PVL+LPDF++
Sbjct: 642 GLASYYRKFIQDFGSVAAPLTQLLRKDSFAWAPATDDAFQRLKLALTTTPVLSLPDFNRP 701
Query: 929 FRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLG 988
F +ECDASG G GA+L G+ PIAYFS+ + R+ A +AYE+EL+++ A++HWRPYL G
Sbjct: 702 FVVECDASGTGFGAVLHQGEDPIAYFSRPIATRHHALAAYERELISLVQAVRHWRPYLWG 761
Query: 989 RKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAG 1048
R+F+V TD SLK L Q++ T Q +W +KLLG++F + YKPGK N ADALS
Sbjct: 762 RQFIVKTDHYSLKFLLDQRLSTIPQHHWVSKLLGFDFVVEYKPGKQNAAADALSCRAAPD 821
Query: 1049 ELRPLVSYPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVIS 1108
++S P ++ I DP L + E+ + P + V+ G + Y+ R+ I
Sbjct: 822 SQAFVLSTPTFDLLKDIRTAGDTDPALQALRDEINSGTRTTP-WAVIDGLVTYKRRIYIP 880
Query: 1109 NKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYL 1168
S + ++ H GH G +T R+ + + + + + + C CQR+K
Sbjct: 881 PGSPWVSVVVAAAH---DDGHEGIQKTLHRLRRDFHTPDDRRVVHDHIQGCLTCQRNKTD 937
Query: 1169 ASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTA 1228
PAGLL PLP+P +W D++++F+ GLPR G I VVDR +KY+H I L H YTA
Sbjct: 938 HLHPAGLLLPLPVPSAIWSDVAMDFVEGLPRVGGKSVILTVVDRFSKYAHLIALAHSYTA 997
Query: 1229 RSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTE 1288
+VA F +I+RLHGVP SI+SDRDP+F S FW +F T+ S+A+HP+SDGQ++
Sbjct: 998 ETVARAFFVDIVRLHGVPESIVSDRDPVFTSAFWTALFTATCTKLHRSTAFHPQSDGQSK 1057
Query: 1289 IVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWV 1348
VN+ + LRC D+ + W WL WAEY +NTS+H+A + TPF+ VYGR PP + +
Sbjct: 1058 AVNKAIAMCLRCMTGDRSRQWLRWLPWAEYIYNTSFHAALRDTPFKLVYGRDPPSIRAYD 1117
Query: 1349 LGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAH 1408
E RV AV + +++RD L ++ L AQ+ + + K + FEVG WV++++R
Sbjct: 1118 ASELRVAAVAQSIEERDAFLADVRLRLEQAQQYAKRYYDQKHREVSFEVGAWVWLRVRHR 1177
Query: 1409 RQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVE 1468
SL V KL R++GPY ++ + VAYRL LP G R+H VFH+ LLK VG
Sbjct: 1178 VPASLPEAVKGKLRPRFYGPYRVVAVINEVAYRLALPPGTRLHDVFHVGLLKPFVG---V 1234
Query: 1469 EPGLPEQLDGEELPSIQPALVLARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSE 1528
P P L + + QP R L +G + Q+LVQW+G +WEDL R+
Sbjct: 1235 SPSAPLALPPIQHGAAQPVPRQVLRARLARG--VRQLLVQWEGLPASATSWEDLDDFRNR 1292
Query: 1529 FPVFNLEDKVVSSEG 1543
+P F L D+++ G
Sbjct: 1293 YPSFQLADELLIEGG 1307
Score = 52.8 bits (125), Expect = 9e-05
Identities = 36/118 (30%), Positives = 55/118 (46%), Gaps = 1/118 (0%)
Query: 114 KVELPMFDGK-DLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATEEEL 172
K++ P FDG+ D +++R E +F+ Q T + KV LA + G +Y L E
Sbjct: 110 KLDFPKFDGRGDPLPFLNRCEQFFRGQRTPEDNKVWLASYHLLDGAQQWYTRLERDHEPP 169
Query: 173 TWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQYQG 230
+W RF + L RYG +L+ R+ V++Y F L + L E Q G
Sbjct: 170 SWHRFSELLNMRYGPPLHSTPLGELAACRRTTTVDDYAERFLDLLTRTGYLSEDQQLG 227
>UniRef100_Q947Y5 Putative retroelement [Oryza sativa]
Length = 1923
Score = 858 bits (2216), Expect = 0.0
Identities = 437/941 (46%), Positives = 609/941 (64%), Gaps = 14/941 (1%)
Query: 597 EELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKGPVNVRPYRYPHHHKNEIEKQVRE 656
+ + +L RF VFQE +PP R +H I + EG PVN+RPYR+ K+EIE+QV E
Sbjct: 38 DSIKHVLDRFQEVFQEPTEMPPVRNCDHKIPLMEGASPVNLRPYRHTPALKDEIERQVTE 97
Query: 657 MLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHG 716
ML +G+I+ S SA+SSP +LVKKKD TW +C+DY+ LN++T+ K+P+PVI+ELLDEL G
Sbjct: 98 MLQSGVIQNSNSAFSSPALLVKKKDGTWRLCIDYKHLNAITIKGKYPLPVIDELLDELSG 157
Query: 717 AVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFR 776
A +FSKLDL++GYHQ+R++ + HKTAF+TH GHYEY VM FGL AP+TFQ MN+
Sbjct: 158 AKYFSKLDLRAGYHQIRLQPGEEHKTAFQTHSGHYEYRVMSFGLTGAPATFQKAMNDTLA 217
Query: 777 PMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHL 836
+LR+ LVFFDDIL+YS H+ HLE+VL++LQ Q KC FA + + YLGH+
Sbjct: 218 TVLRKFTLVFFDDILIYSPDLPSHIQHLEQVLQLLQAQQWKVKLSKCSFAQQQLAYLGHI 277
Query: 837 ISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDA 896
I + GV DP+K+ V W P++VK +RGFLGL GYYRKF++N+G I +PLT+L KK
Sbjct: 278 IGKDGVTTDPSKIADVLHWKIPQSVKQLRGFLGLAGYYRKFVRNFGTINKPLTQLLKKGV 337
Query: 897 -FNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFS 955
F W Q +AF LK+ L +APVLALPDFS+ F +E DA +GIGA+L +HPIA+ S
Sbjct: 338 PFKWTAQMDEAFNALKQALVSAPVLALPDFSKTFTVETDACDMGIGAVLSQDRHPIAFVS 397
Query: 956 KALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQN 1015
KALGP+ S YEKE +A+ LA+ WR YL +FV+ TD +L Q++ T Q
Sbjct: 398 KALGPKTRGLSTYEKEYLAILLAVDQWRSYLQHDEFVILTDHHNLMHITDQRLHTPLQHK 457
Query: 1016 WAAKLLGYNFEIIYKPGKTNQGADALSRID-EAGELRPLVS--YPLWEEQGVIADENQKD 1072
KL+G +++ Y+ G +N ADALSR D E + VS PLW + + D
Sbjct: 458 AFTKLMGLQYKVCYRRGTSNAAADALSRRDEETNDQLWAVSECQPLW--LTAVVKGYETD 515
Query: 1073 PKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGF 1132
+ Q+++E+AL P A + +VQG L Y+ ++ I + SL +L+ H +P GGHSGF
Sbjct: 516 EQAQQLLTELALHPTAREHFHLVQGVLRYKGKIWIGHNLSLQQQLVTALHASPIGGHSGF 575
Query: 1133 LRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLN 1192
TY+R+ W M K IQ++V C +CQ+ K + GLLQPLPIPE W+ +SL+
Sbjct: 576 PVTYQRVKALFAWPQMKKMIQQWVKNCTICQQAKPDLAKYPGLLQPLPIPEGAWQVVSLD 635
Query: 1193 FIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISD 1252
FI GLP+S + I VVVD+ ++Y+HF+PL HP++A VA + K I +LHG+P +ISD
Sbjct: 636 FIEGLPKSERYNCILVVVDKFSRYAHFVPLSHPFSALDVAVSYMKNIYKLHGMPKVLISD 695
Query: 1253 RDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASW 1312
RD +F S W+ +F G +SSAYHP+SDGQT+ VN+CLE +LRCF P W +W
Sbjct: 696 RDKIFTSKLWEFLFLKSGIALHLSSAYHPQSDGQTKRVNQCLEMFLRCFTNAAPFKWVTW 755
Query: 1313 LHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGET-RVEAVEKDLQDRDEALRQL 1371
LH A YW+NT +HSA +TPFE +YG P L V ET + + L +R L
Sbjct: 756 LHLAGYWYNTCFHSALNKTPFEVLYGHNP--LQLGVTMETCAIPDLAVWLHERKLMAELL 813
Query: 1372 KQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPI 1431
+QHL Q++M+ A+ R +EF VG+WVF+K++ + Q S+A+R KLA +++GP+ I
Sbjct: 814 QQHLHRVQQKMKFHADKNRSFREFAVGDWVFLKLQPYVQKSVASRACHKLAFKFYGPFQI 873
Query: 1432 IGRVGAVAYRLKLPEGARIHPVFHISLLKKAVG-NYVEEPGLPEQLDGEELPSIQPALVL 1490
+ RVG VAY+L+LP+ + IHPVFH+S LK A G + + LP+ L ++ P +L
Sbjct: 874 LARVGTVAYKLQLPDDSTIHPVFHVSQLKVAHGFKHQVQSRLPKFLKS----TVYPLQIL 929
Query: 1491 ARRDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFPV 1531
+R I + ++SQVLV W E+ATWED ++ FP+
Sbjct: 930 DQRLIRKGNRTVSQVLVYWSDSVAEDATWEDREDLQQRFPM 970
>UniRef100_Q8W150 Polyprotein [Oryza sativa]
Length = 933
Score = 810 bits (2091), Expect = 0.0
Identities = 404/878 (46%), Positives = 574/878 (65%), Gaps = 10/878 (1%)
Query: 657 MLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRALNSVTVPDKFPIPVIEELLDELHG 716
ML GII+ S+S +SSP +LVKKKD +W +C+DYR LN++T +P+P+I+ELLDEL G
Sbjct: 1 MLQNGIIQHSSSPFSSPALLVKKKDGSWRVCIDYRQLNAITKKGTYPMPIIDELLDELAG 60
Query: 717 AVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEYMVMPFGLMNAPSTFQSLMNEVFR 776
A FSKLDL++GYHQ+R+ E + KTAF+TH GHYEY VM FGL AP+TFQ MN+ R
Sbjct: 61 AKIFSKLDLRAGYHQIRMAEGEEFKTAFQTHSGHYEYKVMSFGLTGAPATFQGAMNDTLR 120
Query: 777 PMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQHHQLTANKKKCQFATKFVEYLGHL 836
P+LR+ LVFFDDIL+YS + H+ HL++VL++L HQ KC FA + YLGH+
Sbjct: 121 PLLRKCALVFFDDILIYSPDMNSHLDHLKQVLQLLDTHQWKVKLSKCDFAQTQISYLGHI 180
Query: 837 ISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGYYRKFIQNYGKIARPLTELTKKDA 896
IS QGV+ DP+K+ S+ W P +K +RGFLGL GYYRKF++++G +++PLT+L KKDA
Sbjct: 181 ISGQGVSTDPSKIQSIVDWAVPTTLKKLRGFLGLAGYYRKFVKDFGTLSKPLTQLLKKDA 240
Query: 897 -FNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLECDASGIGIGAILLYGKHPIAYFS 955
F W + QAF+ LK LT+ PVLALP+F Q F +E DAS IGIGA+L +HP+A+ S
Sbjct: 241 PFVWSAEVNQAFQALKHALTSTPVLALPNFQQGFTIETDASDIGIGAVLSQNQHPVAFVS 300
Query: 956 KALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFVVSTDQRSLKEFLHQKIVTGDQQN 1015
KALGPR S YEKE +A+ +A+ HWRPYL ++F++ TD SL Q++ T QQ
Sbjct: 301 KALGPRTQGLSTYEKECLAIMMAVDHWRPYLQFQEFLIITDHHSLMHLTEQRLHTPWQQK 360
Query: 1016 WAAKLLGYNFEIIYKPGKTNQGADALSR--IDEAGELRPL-VSYPLWEEQGVIADENQKD 1072
KL G F+I+Y+ GK N ADALSR +E E + P+W + I +D
Sbjct: 361 AFTKLSGLQFQIVYRKGKHNAAADALSRHVPEETSEFLGISTCSPVWLQD--ILHGYDQD 418
Query: 1073 PKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGF 1132
P +++ +A++P ++P Y + +G + ++ +V + N S++ +++ H +P GGHSGF
Sbjct: 419 PLALSLLTGLAVNPSSYPHYSLSKGLIKHKGKVWVGNNSNIQQQIISALHDSPLGGHSGF 478
Query: 1133 LRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLN 1192
TY+RI + W M T+Q+ + +C VC + K S GLLQPLP+P+ W+ IS++
Sbjct: 479 PVTYKRIKSLFSWPHMKLTVQKQLASCAVCLQAKPDRSKYPGLLQPLPVPDGAWQIISMD 538
Query: 1193 FIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISD 1252
FI GLP+S + I VVVD+ +KY+HF+PL HP++A VA++F + +LHG+P IISD
Sbjct: 539 FIEGLPKSYHQDCILVVVDKFSKYAHFMPLSHPFSALDVAKVFMLNVYKLHGLPQIIISD 598
Query: 1253 RDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASW 1312
RD +F S W+++F GT+ +SSAYHP+SDGQTE VN+CLE +LRCF P W+ W
Sbjct: 599 RDKIFTSALWEQLFLRSGTKLHLSSAYHPQSDGQTERVNQCLEIFLRCFVHATPAKWSLW 658
Query: 1313 LHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRWVLGETRVEAVEKDLQDRDEALRQLK 1372
LH AE+W+N+SYHSA +TPFE +YG PP + ++ L +R + L+
Sbjct: 659 LHLAEFWYNSSYHSALNKTPFEVLYG-YPPSHFGIRADACVISDLDSWLSERHLMTQLLR 717
Query: 1373 QHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPII 1432
QHL AQ+ M+ QA+ KR + F+VG+WVF+K++ + Q S+A R + KL+ RYFGPY I+
Sbjct: 718 QHLNRAQQVMKTQADKKRSFRSFQVGDWVFLKLQPYVQSSVAKRANHKLSFRYFGPYQIL 777
Query: 1433 GRVGAVAYRLKLPEGARIHPVFHISLLKKAVGNYVEEPGLPEQLDGEELPSIQPALVLAR 1492
+VG+VAY+L+LP + +HPVFH+S LK G + + QL P +L
Sbjct: 778 SKVGSVAYKLQLPADSMVHPVFHVSQLK---GTQNFKHSIQSQLPNITDHIQYPVQILDT 834
Query: 1493 RDILRQGESISQVLVQWQGKTPEEATWEDLATIRSEFP 1530
R + + + Q+LV W EATWED ++ FP
Sbjct: 835 RIQKKGNKVVRQILVCWSNLPAVEATWEDEEELKQRFP 872
>UniRef100_Q75LZ2 Putative gag-pol polyprotein [Oryza sativa]
Length = 1377
Score = 764 bits (1973), Expect = 0.0
Identities = 477/1376 (34%), Positives = 712/1376 (51%), Gaps = 170/1376 (12%)
Query: 108 FRQSVKKVELPMFDGKDLAGWISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLA 167
F + K++ P FDG D W R E YF V T P + V +A + G + S A
Sbjct: 146 FNHRLPKLDFPQFDGSDPQSWRMRCEHYFGVYGTHPNLWVRIATIYFMGRAASWLRSSRA 205
Query: 168 TEEELTWERFRDALLERYGGNGDGDVYEQLSELRQQGIVEEYITDFEYLTAQIPKLPEKQ 227
E + W RF +A+ ++ N + QL +++Q G V E+ F+ L Q+
Sbjct: 206 HEVFVDWGRFYEAMNRKFDRNQHRQLIRQLDQIKQTGSVSEFYERFDDLMNQLLTYDPMY 265
Query: 228 YQGYFLHGLKEEIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFGGNG 287
+H E +R +R V + +V +++E+
Sbjct: 266 NTLNLVHRFIEGLRFDIREAVLLHCPEDLESALVLALLQEEL------------------ 307
Query: 288 SRSGFSGATKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHL 347
++ N + +V G+++ P + +T+ ++ G +RG N
Sbjct: 308 --------SERNQEQRKFVDGSQKFKNAYPLPLPPTKVLKTEE----KKGGELNRGTNLS 355
Query: 348 SYPQLME--RRQKGLCFKCGGPYHRNHVCPDKHLRLLILEEDGEELDESKMLAMEVNEDE 405
+ RR +GLC+ C + H C ++L ++E + NED+
Sbjct: 356 DKVSALRSFRRAQGLCYLCAEKWSPTHKCSGT-VQLHAVQE------LFALFPDNANEDQ 408
Query: 406 EETQGE---LSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNFVDCFLVRR 462
E SL+++ ++ T++L+G I G+ +++LVDSG+ +F+ L R
Sbjct: 409 ASISSEDSDSSLLAMSVHAVQGTDSQSTIRLQGKIQGLNILMLVDSGSLASFISDQLADR 468
Query: 463 L-GWEVVDTPRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGGPDIVLGIEW 521
L G + + P ++VK+ +G Q + + + ++ LGG D +LG++W
Sbjct: 469 LTGVQFLAHP-LSVKVANGQVLHCQKEMPSAEWFVSGQKFLTTFKVVALGGYDAILGMDW 527
Query: 522 LKTLGDTIVNWDTQLMSFWSDKKWITLQGMDT--------RGEHMEALQSITATGERKPG 573
L ++W + + + L+G+ + +H++ L SI
Sbjct: 528 LTKYSPMQIDWQLKSIHLTVMGHEVLLRGVQSDTSDCALISSQHLQHLHSI--------- 578
Query: 574 LLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCITIQEGKG 633
+ A P +Q +E++K Q K L G IQ
Sbjct: 579 ------DSVAVNVRPYRYTPAQKDEIEK---------QVKDMLQKG-------IIQPSAS 616
Query: 634 PVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MCVDYRAL 693
P + S +++ KK D TW CVDYR L
Sbjct: 617 PFS----------------------------------SPVLLVKKK-DGTWRFCVDYRHL 641
Query: 694 NSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTHEGHYEY 753
N++TV +K+P+P+I+EL+DEL GA +FSKLDL+SGYHQ+R+ + KTAF+TH GH+E+
Sbjct: 642 NAITVKNKYPLPIIDELMDELAGACWFSKLDLRSGYHQIRMAVGEEAKTAFKTHNGHFEF 701
Query: 754 MVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERVLEVLQH 813
V+PFGL +AP+TFQ +MN V LR+ VLVF DDILVYS+T +H HL +V E L+
Sbjct: 702 KVLPFGLTSAPATFQGVMNTVLADQLRQNVLVFVDDILVYSRTLEEHKNHLRQVFETLR- 760
Query: 814 HQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGFLGLTGY 873
H+IS GV+ DP K+ V+ WP P +VK VR FLGL GY
Sbjct: 761 ---------------------HIISADGVSTDPEKIQVVRQWPVPVSVKDVRSFLGLAGY 799
Query: 874 YRKFIQNYGKIARPLTE-LTKKDAFNWGKQAQQAFEDLKEKLTTAPVLALPDFSQEFRLE 932
YRKF++++G I++PLT+ L K F W + Q+AF+ LK+ L +APVL +PDF + F +E
Sbjct: 800 YRKFVRHFGIISKPLTKLLCKGQPFIWTQHHQEAFDTLKQSLISAPVLVMPDFQKMFVVE 859
Query: 933 CDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYLLGRKFV 992
DAS GIGA+L+ +H +A+ SKALG R S YEKE +A+ LA+ HWRPYL F+
Sbjct: 860 TDASDRGIGAVLMQDQHSVAFLSKALGHRTQVLSTYEKESLAIILAVDHWRPYLQHDDFL 919
Query: 993 VSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSRIDEAGELRP 1052
+ TD RSL +Q + T Q KLLG + I+YK G N ADALS G P
Sbjct: 920 IRTDHRSLAFLDNQLLTTSWQYKAMTKLLGLRYRIVYKKGLENGAADALSHRSSDG--LP 977
Query: 1053 LVS-----YPLWEEQGVIADENQKDPKLCQIVSEVALDPQAHPGYRVVQGTLLYQDRVVI 1107
++S P W + + + DP+ Q++ + + G L ++ RV +
Sbjct: 978 ILSALSVGLPDWLQD--VVSGYKSDPEALQLLHSFKEGTCHSAHFELQNGILYFKKRVWV 1035
Query: 1108 SNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIATNLYWRGMTKTIQEFV*ACDVCQRHKY 1167
N L ++L HT GGHSG IQ+FV +C VCQ+ K
Sbjct: 1036 GNNRQLQQQVLANLHTAALGGHSG--------------------IQDFVQSCAVCQQAKP 1075
Query: 1168 LASSPAGLLQPLPIPERVWEDISLNFIIGLPRSRGFEAIFVVVDRLTKYSHFIPLKHPYT 1227
GLLQP+ IPE W+ IS++FI GLPRS + +I V+VD+ TK++HF+PL HP+T
Sbjct: 1076 EHVPYPGLLQPVQIPEHAWQVISMDFIEGLPRSASYNSILVIVDKFTKFAHFLPLSHPFT 1135
Query: 1228 ARSVAEIFAKEIIRLHGVPSSIISDRDPLFVSHFWKEMFRLQGTQFKMSSAYHPESDGQT 1287
A SVA++F I +HG+P +IISDRD +F S FWKE+FR GTQ +MSS+YHP++DGQT
Sbjct: 1136 AVSVAQLFMDRIHSIHGLPQAIISDRDRVFTSIFWKELFRRSGTQLQMSSSYHPQTDGQT 1195
Query: 1288 EIVNRCLETYLRCFAADQPKTWASWLHWAEYWFNTSYHSATQQTPFEAVYGRKPPVLTRW 1347
E VN+CLE +LRCF P W+ WL A++W+NTS+HS TPFEA++G KP
Sbjct: 1196 ERVNQCLEMFLRCFVHACPSRWSKWLSLAQFWYNTSFHSTLMLTPFEAMFGHKPRHFGLS 1255
Query: 1348 VLGETRVEAVEKDLQDRDEALRQLKQHLVAAQERMRAQANSKRKHQEFEVGEWVFVKIRA 1407
V A++ +QDR + HLV AQ+RM++QA+SKR + F VG+WVF+K++
Sbjct: 1256 VDSVGVPSALDSWMQDRHNMQSLIYHHLVRAQQRMKSQADSKRTDRVFAVGDWVFMKLQP 1315
Query: 1408 HRQVSLANRVHAKLAARYFGPYPIIGRVGAVAYRLKLPEGARIHPVFHISLLKKAV 1463
+ Q S+ R + KLA +++GP+ I+ RVG+VAY+L+LP + IHPV H+S LKKAV
Sbjct: 1316 YVQQSVMTRANQKLAFKFYGPFQILQRVGSVAYKLQLPATSLIHPVVHVSQLKKAV 1371
>UniRef100_Q7X798 OSJNBb0108J11.19 protein [Oryza sativa]
Length = 1516
Score = 751 bits (1939), Expect = 0.0
Identities = 513/1475 (34%), Positives = 763/1475 (50%), Gaps = 149/1475 (10%)
Query: 128 WISRAEIYFKVQETSPEVKVSLAQLSMEGGTIHFYNSLLATE---EELTWERFRDALLER 184
W+ E + + + + KV+ A ++G ++++ +AT E+TW F + R
Sbjct: 104 WLRAIEKKLNLLQCNDQEKVAFATHQLQGPASAWWDNYVATRLDATEVTWAEFCQSF--R 161
Query: 185 YGGNGDGDVYEQLSELR--QQGI--VEEYITDFEYLTAQIPK--LPEKQYQGYFLHGLKE 238
+G + +Q E R QQG V EY+ +F L P+ + + Q FL GL +
Sbjct: 162 KAQIPEGIMAQQKREFRALQQGTMSVTEYLHEFNRLARYAPEDVRTDAEKQEKFLFGLDD 221
Query: 239 EIRGKVRSLVAMGGVNRARLLVVTRAVEKEVKGDGVAGQARANRFG---GNGSRSGFSGA 295
E+ ++ S GV +V +A+ ++ + + + + +A + GN + F
Sbjct: 222 ELTNQLIS-----GVYEDFEKLVDKAIRQDEQRNKMDRKRKAAQISFPQGNNQKPRFMTG 276
Query: 296 TKGNGTEWIWVKGNKETGQTVNRPNSNPRGDQTKNTGDIRRAGPRDRGFNHLSYPQLM-- 353
G + I + +RP D N G + + NH S P +
Sbjct: 277 QLGGPSTLIVCQ---------HRPYHPSNFDNDYNGGSHNNS----KQHNHNSTPPPLMA 323
Query: 354 -------------ERRQKGL------CFKCGGPYHRNHVCPDKHLRLLILEEDGEELDES 394
E+ +KG CF CG H CP L G ++
Sbjct: 324 PDQSDPPAVSVQSEQPKKGAVGKPGPCFNCGKHGHFAGKCPK-------LNRTGPRFIQA 376
Query: 395 KMLAMEVNEDEEETQGELSLMSLCELGMKTGGIPRTMKLRGTINGVPVVVLVDSGATHNF 454
+ A V+ +E + E+ L G P +N P VL DSGA+H+F
Sbjct: 377 R--ANHVSAEEAQAAPEVVL----------GTFP--------VNSYPATVLFDSGASHSF 416
Query: 455 VDCFLVRRLGWEVVDT--PRMTVKLGDGYKSQAQGRCAGLKIEMGEYHLRCSPQLFDLGG 512
+ G VV+ P LG+ ++ C + IE+ + L +
Sbjct: 417 ISKRFAGTHGLSVVELKIPMQVHTLGNDMRTAHY--CPSVTIEIKRSPFLSNLILLESKD 474
Query: 513 PDIVLGIEWLKTLGDTIVNWDTQLMSFWSDK-KWITLQGMDTRGE----HMEALQSITAT 567
D++LG++WL T +++ ++ ++ +DK + IT + ++ + A++ T T
Sbjct: 475 LDVILGMDWL-TRNKGVIDCASRTITLTNDKGEKITFRSPASQKSVASLNQAAIEGQTET 533
Query: 568 GERKPGLLGTRNEEKAGKQLPGEINSSQLEELDKLLYRFDVVFQEKQGLPPGRGREHCIT 627
E+ P +LE++ + +V ++ +PP R E I
Sbjct: 534 VEKSP---------------------RKLEDIPIVQEYPEVFPEDLTTMPPKREIEFRID 572
Query: 628 IQEGKGPVNVRPYRYPHHHKNEIEKQVREMLSAGIIRQSTSAYSSPVILVKKKDQTW*MC 687
+ G P+ RPYR + E++KQV E L G IR STS + +PVI V+KKD+T MC
Sbjct: 573 LAPGTAPIYKRPYRMAANELAEVKKQVDEQLQKGYIRPSTSPWGAPVIFVEKKDKTKRMC 632
Query: 688 VDYRALNSVTVPDKFPIPVIEELLDELHGAVFFSKLDLKSGYHQVRVREEDVHKTAFRTH 747
VDYRALN VT+ +K+P+P I++L D+L GA FSK+DL+SGYHQ+R+REED+ KTAF T
Sbjct: 633 VDYRALNEVTIKNKYPLPRIDDLFDQLKGAKVFSKIDLRSGYHQLRIREEDIPKTAFTTR 692
Query: 748 EGHYEYMVMPFGLMNAPSTFQSLMNEVFRPMLRRGVLVFFDDILVYSKTWSDHMLHLERV 807
G +E VM FGL+NAP+ F +LMN+VF L + V+VF DDILVYSK+ +H HL V
Sbjct: 693 YGLFECTVMSFGLINAPAFFMNLMNKVFMEFLDKFVVVFIDDILVYSKSEEEHEQHLRLV 752
Query: 808 LEVLQHHQLTANKKKCQFATKFVEYLGHLISEQGVAVDPNKVLSVKSWPEPKNVKGVRGF 867
LE L+ HQL A KC F V++LGH+IS QGVAVDP+ V SV W PK V +R F
Sbjct: 753 LEKLKEHQLYAKFSKCDFWLSEVKFLGHVISAQGVAVDPSNVESVTKWTPPKTVSQIRSF 812
Query: 868 LGLTGYYRKFIQNYGKIARPLTELTKKD-AFNWGKQAQQAFEDLKEKLTTAPVLALPDFS 926
LGL GYYR+FI+N+ KIARP+T+L KKD F W + Q+FE+LK+KL +APVL LPD +
Sbjct: 813 LGLAGYYRRFIENFSKIARPMTQLLKKDEKFKWSAECNQSFEELKKKLVSAPVLILPDQT 872
Query: 927 QEFRLECDASGIGIGAILLYGKHPIAYFSKALGPRNLAKSAYEKELMAVALAIQHWRPYL 986
++F++ CDAS G+G +L+ +AY S+ L P ++ EL AV A++ WR YL
Sbjct: 873 KDFQVYCDASRQGLGCVLMQEGRVVAYASRQLRPHETNYPTHDLELAAVVHALKIWRHYL 932
Query: 987 LGRKFVVSTDQRSLKEFLHQKIVTGDQQNWAAKLLGYNFEIIYKPGKTNQGADALSR--- 1043
+G + V TD +SLK Q + Q+ W + Y+ I Y PGK N ADALSR
Sbjct: 933 IGNRCEVYTDHKSLKYIFTQPDLNLRQRRWLELIKDYDMGIHYHPGKANVVADALSRKSY 992
Query: 1044 -----------------------IDEAGELRPLVSYPLWEEQGVIADENQKDPKLCQIVS 1080
I E G + L + P +Q A N DP++ ++
Sbjct: 993 CNALCTEDMCGKLQQDLEHLNLGIVEHGYVAALEARPTLVDQVRAAQVN--DPEIAELKK 1050
Query: 1081 EVALDPQAHPGYRVVQGTLLYQDRVVISNKSSLIPKLLEEFHTTPHGGHSGFLRTYRRIA 1140
+ + +A + GT+ +R+ + + L +L E H T + H G + Y+ +
Sbjct: 1051 NMRVG-KARDFHEDENGTIWLGERLCVPDDKELKDLILTEAHQTQYSIHPGSTKMYQDLK 1109
Query: 1141 TNLYWRGMTKTIQEFV*ACDVCQRHKYLASSPAGLLQPLPIPERVWEDISLNFIIGLPR- 1199
+W M + I EFV CDVCQR K PAGLLQPL IPE WE+I ++FI GLPR
Sbjct: 1110 EKFWWVSMRREIAEFVALCDVCQRVKAEHQRPAGLLQPLQIPEWKWEEIGMDFITGLPRT 1169
Query: 1200 SRGFEAIFVVVDRLTKYSHFIPLKHPYTARSVAEIFAKEIIRLHGVPSSIISDRDPLFVS 1259
S G ++I+VVVDRLTK +HFIP+ YT + +AE++ I+ LHGVP I+SDR F S
Sbjct: 1170 SSGHDSIWVVVDRLTKVAHFIPVHTTYTGKRLAELYLSRIMCLHGVPKKIVSDRGSQFTS 1229
Query: 1260 HFWKEMFRLQGTQFKMSSAYHPESDGQTEIVNRCLETYLRCFAADQPKTWASWLHWAEYW 1319
FW+++ GT+ S+AYHP++DGQTE VN+ LE LR A D W L +AE+
Sbjct: 1230 KFWQKLQEELGTRLNFSTAYHPQTDGQTERVNQILEDMLRACALDFGGAWDKSLPYAEFS 1289
Query: 1320 FNTSYHSATQQTPFEAVYGRKPPVLTRW-VLGETRVEAVEKDLQDRDEALRQLKQHLVAA 1378
+N SY S+ Q PFEA+YGRK W GE ++ E L + +E +R +++ L A
Sbjct: 1290 YNNSYQSSLQMAPFEALYGRKCRTPLFWDQTGERQLFGTEV-LTEAEEKVRTVRERLRIA 1348
Query: 1379 QERMRAQANSKRKHQEFEVGEWVFVKIRAHRQVSLANRVHAKLAARYFGPYPIIGRVGAV 1438
Q R ++ A+++R+ FE G++V++++ R V + KLA R+ GPY I+ R G V
Sbjct: 1349 QSRQKSYADNRRRELTFEAGDYVYLRVTPLRGVH-RFQTKGKLAPRFVGPYRILERRGEV 1407
Query: 1439 AYRLKLPEG-ARIHPVFHISLLKKAVGNYVEEPGLPEQLD-GEELPSIQ-PALVLARRDI 1495
AY+L+LP IH VFH+S LKK + EE E +D E+L ++ P +L +
Sbjct: 1408 AYQLELPSNMVGIHDVFHVSQLKKCL-RVPEEQASSEHIDIQEDLTYVEKPICILETSER 1466
Query: 1496 LRQGESISQVLVQWQGKTPEEATWEDLATIRSEFP 1530
+ + I VQW + EEATWE +++ P
Sbjct: 1467 RTRNKVIKFCKVQWSHHSEEEATWEREDELKAAHP 1501
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,747,972,915
Number of Sequences: 2790947
Number of extensions: 122694509
Number of successful extensions: 396749
Number of sequences better than 10.0: 29511
Number of HSP's better than 10.0 without gapping: 2845
Number of HSP's successfully gapped in prelim test: 26666
Number of HSP's that attempted gapping in prelim test: 344371
Number of HSP's gapped (non-prelim): 46231
length of query: 1596
length of database: 848,049,833
effective HSP length: 141
effective length of query: 1455
effective length of database: 454,526,306
effective search space: 661335775230
effective search space used: 661335775230
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 82 (36.2 bits)
Lotus: description of TM0170.6


