

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.5
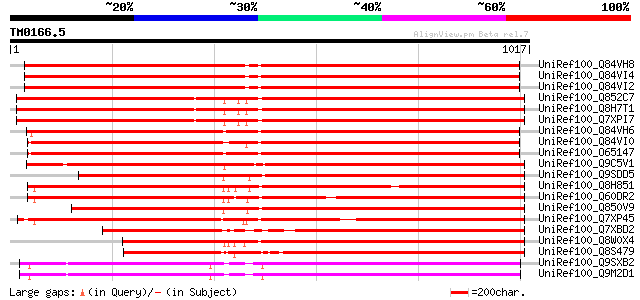
(1017 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84VH8 Gag-pol polyprotein [Glycine max] 915 0.0
UniRef100_Q84VI4 Gag-pol polyprotein [Glycine max] 914 0.0
UniRef100_Q84VI2 Gag-pol polyprotein [Glycine max] 909 0.0
UniRef100_Q852C7 Putative gag-pol polyprotein [Oryza sativa] 907 0.0
UniRef100_Q8H7T1 Putative Zea mays retrotransposon Opie-2 [Oryza... 906 0.0
UniRef100_Q7XPI7 OSJNBb0004A17.2 protein [Oryza sativa] 906 0.0
UniRef100_Q84VH6 Gag-pol polyprotein [Glycine max] 905 0.0
UniRef100_Q84VI0 Gag-pol polyprotein [Glycine max] 897 0.0
UniRef100_O65147 Gag-pol polyprotein [Glycine max] 893 0.0
UniRef100_Q9C5V1 Gag/pol polyprotein [Arabidopsis thaliana] 871 0.0
UniRef100_Q9SDD5 Similar to copia-type pol polyprotein [Oryza sa... 867 0.0
UniRef100_Q8H851 Putative Zea mays retrotransposon Opie-2 [Oryza... 855 0.0
UniRef100_Q60DR2 Putative polyprotein [Oryza sativa] 850 0.0
UniRef100_Q850V9 Putative polyprotein [Oryza sativa] 842 0.0
UniRef100_Q7XP45 OSJNBa0063G07.6 protein [Oryza sativa] 835 0.0
UniRef100_Q7XBD2 Retrotransposon Opie-2 [Zea mays] 785 0.0
UniRef100_Q8W0X4 Putative pol protein [Zea mays] 775 0.0
UniRef100_Q8S479 Putative pol protein [Zea mays] 726 0.0
UniRef100_Q9SXB2 T28P6.8 protein [Arabidopsis thaliana] 721 0.0
UniRef100_Q9M2D1 Copia-type polyprotein [Arabidopsis thaliana] 720 0.0
>UniRef100_Q84VH8 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 915 bits (2364), Expect = 0.0
Identities = 464/974 (47%), Positives = 648/974 (65%), Gaps = 14/974 (1%)
Query: 29 KSLEKGSIGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVL 88
K + G + +P +N VLLV+GL NL+SISQ+ ++G++V F ++ C V+ VL
Sbjct: 599 KIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVL 657
Query: 89 FSGKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLP 148
G R + + + S CL S DE IW +R GH LR + ++ +RG+P
Sbjct: 658 MKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIP 717
Query: 149 RLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVI 208
LK +C CQ GK K + +T+R LELLH+DL GP++ ES+GGK+Y V+
Sbjct: 718 NLKIEEGRICGECQIGKQVKMSHQKLRHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVV 777
Query: 209 VDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNS 268
VDD+SR+TWV F+R K ET +F ++Q+E + +RSDHG EFEN F E S
Sbjct: 778 VDDFSRFTWVNFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTS 837
Query: 269 QGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISI 328
+GI+H FS TPQQNG+VERKNRTLQE AR M+ + +LWAEA+NTA YIHN +++
Sbjct: 838 EGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTL 897
Query: 329 RPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGF 388
R T YE+ KGR+P + +FH FGS CY+L +EQ K D K+ FLGYS S+ +
Sbjct: 898 RRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAY 957
Query: 389 RIYNIIHQTVEESIQIRFDGKLGSEKSKLFE---RFADLSIDCSEANQPKNSSEDVAPEA 445
R++N +TV ESI + D + K + E D D +++ + +S+ E+
Sbjct: 958 RVFNSRTRTVMESINVVVDDLSPARKKDVEEDVRTLGDNVADAAKSGENAENSDSATDES 1017
Query: 446 EASEAAPTTSDQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIE 505
++ +S ++QK HP+ELIIG+ + V TRS E ++S VS IE
Sbjct: 1018 NINQPDKRSSTRIQKM------HPKELIIGDPNRGVTTRSR----EVEIVSNSCFVSKIE 1067
Query: 506 PKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTR 565
PK+V EAL D+ WI MQEEL+QF +N+VW L+ +P+G +VIGTKW+F+NK NE+G +TR
Sbjct: 1068 PKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITR 1127
Query: 566 NKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYIS 625
NKARLVAQGY+Q EG+D+ ETF+PVARLE+IRLL+ + L+QMDVKSAFLNGY++
Sbjct: 1128 NKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLN 1187
Query: 626 EEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLF 685
EEVYV+QP GF D +PDHVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF
Sbjct: 1188 EEVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLF 1247
Query: 686 CITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPG 745
+++I QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q
Sbjct: 1248 VKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMED 1307
Query: 746 VTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLT 805
++ Q +Y ++KKF M + + +TP L K+E + V Q LYR MIGSLLYLT
Sbjct: 1308 SIFLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLT 1367
Query: 806 ASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDAD 865
ASRPDI ++V +CAR+Q++P+ +HLT VKRILKY+ GT++ G+MY S L G+CDAD
Sbjct: 1368 ASRPDITYAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDAD 1427
Query: 866 FAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLE 925
+AG +RKSTSG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L+
Sbjct: 1428 WAGSADDRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLK 1487
Query: 926 DYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQ 985
+Y + + +YCDN +AI++SKNP+ HSR KHI++++HYIRD V ++L++VDT+ Q
Sbjct: 1488 EYNVEQDVMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQ 1547
Query: 986 WADIFTKPLAEDRF 999
ADIFTK L ++F
Sbjct: 1548 IADIFTKALDANQF 1561
>UniRef100_Q84VI4 Gag-pol polyprotein [Glycine max]
Length = 1574
Score = 914 bits (2361), Expect = 0.0
Identities = 464/974 (47%), Positives = 648/974 (65%), Gaps = 14/974 (1%)
Query: 29 KSLEKGSIGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVL 88
K + G + +P +N VLLV+GL NL+SISQ+ ++G++V F ++ C V+ VL
Sbjct: 597 KIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVL 655
Query: 89 FSGKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLP 148
G R + + + S CL S DE IW +R GH LR + ++ +RG+P
Sbjct: 656 MKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKIIDKGAVRGIP 715
Query: 149 RLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVI 208
LK +C CQ GK K + +T+R LELLH+DL GP++ ES+GGK+Y V+
Sbjct: 716 NLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVV 775
Query: 209 VDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNS 268
VDD+SR+TWVKF+R K ET +F ++Q+E + +RSDHG EFEN E S
Sbjct: 776 VDDFSRFTWVKFIREKSETFEVFKELSLRLQREKDCVIKRIRSDHGREFENSRLTEFCTS 835
Query: 269 QGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISI 328
+GI+H FS TPQQNG+VERKNRTLQE AR M+ + +LWAEA+NTA YIHN +++
Sbjct: 836 EGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTL 895
Query: 329 RPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGF 388
R T YE+ KGR+P + +FH FGS CY+L +EQ K D K+ FLGYS S+ +
Sbjct: 896 RRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAY 955
Query: 389 RIYNIIHQTVEESIQIRFDGKLGSEKSKLFERF---ADLSIDCSEANQPKNSSEDVAPEA 445
R++N +TV ESI + D + K + E D D +++ + +S+ E+
Sbjct: 956 RVFNSRTRTVMESINVVVDDLSPARKKDVEEDVRTSGDNVADAAKSGENAENSDSATDES 1015
Query: 446 EASEAAPTTSDQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIE 505
++ +S ++QK HP+ELIIG+ + V TRS E ++S VS IE
Sbjct: 1016 NINQPDKRSSTRIQKM------HPKELIIGDPNRGVTTRSR----EVEIVSNSCFVSKIE 1065
Query: 506 PKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTR 565
PK+V EAL D+ WI MQEEL+QF +N+VW L+ +P+G +VIGTKW+F+NK NE+G +TR
Sbjct: 1066 PKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITR 1125
Query: 566 NKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYIS 625
NKARLVAQGY+Q EG+D+ ETF+PVARLE+IRLL+ + L+QMDVKSAFLNGY++
Sbjct: 1126 NKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLN 1185
Query: 626 EEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLF 685
EEVYV+QP GF D +PDHVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF
Sbjct: 1186 EEVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLF 1245
Query: 686 CITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPG 745
+++I QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q
Sbjct: 1246 VKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMED 1305
Query: 746 VTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLT 805
++ Q +Y ++KKF M + + +TP L K+E + V Q LYR MIGSLLYLT
Sbjct: 1306 SIFLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLT 1365
Query: 806 ASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDAD 865
ASRPDI ++V +CAR+Q++P+ +HLT VKRILKY+ GT++ G+MY S L G+CDAD
Sbjct: 1366 ASRPDITYAVGVCARYQANPKISHLTQVKRILKYVNGTSDYGIMYCHCSNPMLVGYCDAD 1425
Query: 866 FAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLE 925
+AG +RKSTSG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L+
Sbjct: 1426 WAGSADDRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLK 1485
Query: 926 DYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQ 985
+Y + + +YCDN +AI++SKNP+ HSR KHI++++HYIRD V ++L++VDT+ Q
Sbjct: 1486 EYNVEQDVMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQ 1545
Query: 986 WADIFTKPLAEDRF 999
ADIFTK L ++F
Sbjct: 1546 IADIFTKALDANQF 1559
>UniRef100_Q84VI2 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 909 bits (2350), Expect = 0.0
Identities = 462/974 (47%), Positives = 647/974 (65%), Gaps = 14/974 (1%)
Query: 29 KSLEKGSIGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVL 88
K + G + +P +N VLLV+GL NL+SISQ+ ++G++V F ++ C V+ VL
Sbjct: 599 KIIGMGKLVHDGLPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVL 657
Query: 89 FSGKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLP 148
G R + + + S CL S DE IW +R GH LR + ++ + +RG+P
Sbjct: 658 MKGSRSKDNCYLWTPQETSYSSTCLSSKEDEVRIWHQRFGHLHLRGMKKILDKSAVRGIP 717
Query: 149 RLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVI 208
LK +C CQ GK K + +T+R LELLH+DL GP++ ES+GGK+Y V+
Sbjct: 718 NLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAYVV 777
Query: 209 VDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNS 268
VDD+SR+TWV F+R K T +F ++Q+E + +RSDHG EFEN F E S
Sbjct: 778 VDDFSRFTWVNFIREKSGTFEVFKKLSLRLQREKDCVIKRIRSDHGREFENSRFTEFCTS 837
Query: 269 QGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISI 328
+GI+H FS TPQQNG+VERKNRTLQE AR M+ + +LWAEA+NTA YIHN +++
Sbjct: 838 EGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTL 897
Query: 329 RPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGF 388
R T YE+ KGR+P + +FH FGS CY+L +EQ K D K+ FLGYS S+ +
Sbjct: 898 RRGTPTTLYEIWKGRKPSVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAY 957
Query: 389 RIYNIIHQTVEESIQIRFDGKLGSEKSKLFERF---ADLSIDCSEANQPKNSSEDVAPEA 445
R++N +TV ESI + D + K + E D D +++ + +S+ E+
Sbjct: 958 RVFNSRTRTVMESINVVVDDLSPARKKDVEEDVRTSGDNVADAAKSGENAENSDSATDES 1017
Query: 446 EASEAAPTTSDQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIE 505
++ +S ++QK HP+ELIIG+ + V TRS E ++S VS IE
Sbjct: 1018 NINQPDKRSSTRIQKM------HPKELIIGDPNRGVTTRSR----EVEIVSNSCFVSKIE 1067
Query: 506 PKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTR 565
PK+V EAL D+ WI MQEEL+QF +N+VW L+ +P+G +VIGTKW+F+NK NE+G +TR
Sbjct: 1068 PKNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITR 1127
Query: 566 NKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYIS 625
NKARLVAQGY+Q EG+D+ ETF+PVARLE+IRLL+ + L+QMDVKSAFLNGY++
Sbjct: 1128 NKARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLN 1187
Query: 626 EEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLF 685
EEVYV+QP GF D +PDHVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF
Sbjct: 1188 EEVYVEQPKGFADPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLF 1247
Query: 686 CITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPG 745
+++I QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q
Sbjct: 1248 VKQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMED 1307
Query: 746 VTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLT 805
++ Q +Y ++KKF M + + +TP L K+E + V QK YR MIGSLLYLT
Sbjct: 1308 SIFLSQSRYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQKPYRSMIGSLLYLT 1367
Query: 806 ASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDAD 865
ASRPDI ++V +CAR+Q++P+ +HL VKRILKY+ GT++ G+MY S L G+CDAD
Sbjct: 1368 ASRPDITYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSSSMLVGYCDAD 1427
Query: 866 FAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLE 925
+AG +RKSTSG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L+
Sbjct: 1428 WAGSADDRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLK 1487
Query: 926 DYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQ 985
+Y + + +YCDN +AI++SKNP+ HSR KHI++++HYIRD V ++L++VDT+ Q
Sbjct: 1488 EYNVEQDVMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLKHVDTEEQ 1547
Query: 986 WADIFTKPLAEDRF 999
ADIFTK L ++F
Sbjct: 1548 IADIFTKALDANQF 1561
>UniRef100_Q852C7 Putative gag-pol polyprotein [Oryza sativa]
Length = 1969
Score = 907 bits (2343), Expect = 0.0
Identities = 478/1022 (46%), Positives = 657/1022 (63%), Gaps = 35/1022 (3%)
Query: 13 RLSQEETLALEETRREKSLEKGSIGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIF 72
R S +ET+ + + + KG+I ++ DV LV L +NLLS+SQ+ ++ +V F
Sbjct: 776 RASGDETITFGDASSGRVMAKGTIKVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRF 835
Query: 73 NQTGCKAVSQTNGSVLFSGKRKNYIYKIKSSELLSQKVKCLM-SVNDEQWIWLRRLGHAS 131
+ + + + V F R ++ +CL+ S N + + W RRLGH
Sbjct: 836 KKDRSRVLDASESPV-FDISRVGRVFFANFDSSAPGPSRCLIASENRDLFFWHRRLGHIG 894
Query: 132 LRKISQLSKLNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLF 191
+S++S ++LIRGLP+LK + +C C+ GK T K +V T P +LLH+D
Sbjct: 895 FDHLSRISGMDLIRGLPKLKVQKDLVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMDTV 954
Query: 192 GPVKTESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRS 251
GP + +S+GGK Y LV+VDD+SR++WV FL K+ET F + + EF ++ +RS
Sbjct: 955 GPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETFGFFQSLARSLALEFPGALRAIRS 1014
Query: 252 DHGGEFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHL 311
D+G EF+N AFE +S G+ H FS P PQQNGVVERKNRTL EMARTM+ E + +
Sbjct: 1015 DNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKF 1074
Query: 312 WAEAINTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDS 371
W EAI+ A +I N + +R IL KTPYEL GR+P +S+ FG C++L + L KF+S
Sbjct: 1075 WTEAISAACFISNRVFLRTILHKTPYELRFGRRPKVSHLRVFGCKCFVLKS-GNLDKFES 1133
Query: 372 KALKCYFLGYSERSKGFRIYNIIHQTVEESIQIRFDGKLGSEKSKLF-----ERFADLSI 426
++L FLGY+ S+ +R+Y + + E+ ++ FD + ++ F D
Sbjct: 1134 RSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDEASPGARPEISGVPDESIFVDEDS 1193
Query: 427 DCSEANQPKNSSEDVAPEAEA--------SEAAPTTSDQLQKK-----------KRIAVS 467
D + + + P E S APTTS ++ + I
Sbjct: 1194 DDDDDDSIPPPLDSTPPVQETGSPSTTSPSGDAPTTSSSAAEEIDGGTSGPTAPRHIQNR 1253
Query: 468 HPEELIIGNKDAPV-RTRSMLKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEEL 526
HP + +IG V R RS L V+ EPK+V AL D+ W+ M EEL
Sbjct: 1254 HPPDSMIGGLGERVTRNRSY-------ELVNSAFVASFEPKNVCHALSDENWVNAMHEEL 1306
Query: 527 DQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTET 586
+ F +N VW+L+ P GF+VIGTKWVF+NKL E G + RNKARLVAQG++Q EG+D+ ET
Sbjct: 1307 ENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEET 1366
Query: 587 FSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVY 646
F+PVARLEAIR+L++F+ + L QMDVKSAFLNG I EEVYVKQPPGFE+ K+P+HV+
Sbjct: 1367 FAPVARLEAIRILLAFAASKGFKLFQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVF 1426
Query: 647 KLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFG 706
KL+K+LYGLKQAPRAWYERL +FLLQN F G D TLF + D L+VQIYVDDIIFG
Sbjct: 1427 KLEKALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFG 1486
Query: 707 SANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSD 766
++ +L +FS +M EFEMSMMGEL +FLG+QI Q ++HQ KY+ ELLKKF+M+D
Sbjct: 1487 GSSHALVAQFSDVMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMAD 1546
Query: 767 CNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPR 826
C TPM T L +E +V Q+ YR MIGSLLYLTASRPDI FSV LCARFQ+ PR
Sbjct: 1547 CKPIATPMATTSSLGPDEDGEEVDQREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPR 1606
Query: 827 ETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSN 886
+H AVKRI +Y+K T G+ Y +S ++ F DADFAG +++RKSTSG+CHFLG++
Sbjct: 1607 TSHRQAVKRIFRYIKSTLEYGIWYSCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTS 1666
Query: 887 LVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISL 946
LV+WSS++Q+++A STAEAEYV+AA+ C+Q +WM + L+DYGLS VP+ CDNT+AI++
Sbjct: 1667 LVSWSSRKQSSVAQSTAEAEYVAAASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINI 1726
Query: 947 SKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILENL 1006
+KNP+ HSR KHIE++YH++RD+V+KGT+ LE+V+++ Q ADIFTKPL RF F+ L
Sbjct: 1727 AKNPVQHSRTKHIEIRYHFLRDNVEKGTIVLEFVESEKQLADIFTKPLDRSRFEFLRSEL 1786
Query: 1007 NM 1008
+
Sbjct: 1787 GV 1788
>UniRef100_Q8H7T1 Putative Zea mays retrotransposon Opie-2 [Oryza sativa]
Length = 2145
Score = 906 bits (2341), Expect = 0.0
Identities = 477/1022 (46%), Positives = 657/1022 (63%), Gaps = 35/1022 (3%)
Query: 13 RLSQEETLALEETRREKSLEKGSIGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIF 72
R S +ET+ + + + KG+I ++ DV LV L +NLLS+SQ+ ++ +V F
Sbjct: 776 RASSDETITFGDASSGRVMAKGTIKVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRF 835
Query: 73 NQTGCKAVSQTNGSVLFSGKRKNYIYKIKSSELLSQKVKCLM-SVNDEQWIWLRRLGHAS 131
+ + + + V F R ++ +CL+ S N + + W RRLGH
Sbjct: 836 KKDRSRVLDASESPV-FDISRVGRVFFANFDSSAPGPSRCLIASENRDLFFWHRRLGHIG 894
Query: 132 LRKISQLSKLNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLF 191
+S++S ++LIRGLP+LK + +C C+ GK T K +V T P +LLH+D
Sbjct: 895 FDHLSRISGMDLIRGLPKLKVPKDLVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMDTV 954
Query: 192 GPVKTESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRS 251
GP + +S+GGK Y LV+VDD+SR++WV FL K+ET F + + EF ++ +RS
Sbjct: 955 GPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETFGFFQSLARSLALEFPGALRAIRS 1014
Query: 252 DHGGEFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHL 311
D+G EF+N AFE +S G+ H FS P PQQNGVVERKNRTL EMARTM+ E + +
Sbjct: 1015 DNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKF 1074
Query: 312 WAEAINTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDS 371
W EAI+ A +I N + +R IL KTPYEL GR+P +S+ FG C++L + L KF+S
Sbjct: 1075 WTEAISAACFISNRVFLRTILHKTPYELRFGRRPKVSHLRVFGCKCFVLKS-GNLDKFES 1133
Query: 372 KALKCYFLGYSERSKGFRIYNIIHQTVEESIQIRFDGKLGSEKSKLF-----ERFADLSI 426
++L FLGY+ S+ +R+Y + + E+ ++ FD + ++ F D
Sbjct: 1134 RSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDEASPGARPEISGVPDESIFVDEDS 1193
Query: 427 DCSEANQPKNSSEDVAPEAEA--------SEAAPTTSDQLQKK-----------KRIAVS 467
D + + + P E S APTTS ++ + I
Sbjct: 1194 DDDDDDSIPPPLDSTPPVQETGSPSTTSPSGDAPTTSSSAAEEIDGGTSGPTAPRHIQNR 1253
Query: 468 HPEELIIGNKDAPV-RTRSMLKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEEL 526
HP + +IG V R RS L V+ EPK+V AL D+ W+ M EEL
Sbjct: 1254 HPPDSMIGGLGERVTRNRSY-------ELVNSAFVASFEPKNVCHALSDENWVNAMHEEL 1306
Query: 527 DQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTET 586
+ F +N VW+L+ P GF+VIGTKWVF+NKL E G + RNKARLVAQG++Q EG+D+ ET
Sbjct: 1307 ENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEET 1366
Query: 587 FSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVY 646
F+PVARLEAIR+L++F+ + L QMDVKSAFLNG I EEVYVKQPPGFE+ K+P+HV+
Sbjct: 1367 FAPVARLEAIRILLAFAASKGFKLFQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVF 1426
Query: 647 KLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFG 706
KL+K+LYGLKQAPRAWYERL +FLLQN F G D TLF + D L+VQIYVDDIIFG
Sbjct: 1427 KLEKALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFG 1486
Query: 707 SANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSD 766
++ +L +FS +M EFEMSMMGEL +FLG+QI Q ++HQ KY+ ELLKKF+M+D
Sbjct: 1487 GSSHALVAQFSDVMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMAD 1546
Query: 767 CNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPR 826
C TPM T L +E +V Q+ YR MIGSLLYLTASRPDI FSV LCARFQ+ PR
Sbjct: 1547 CKPIATPMATTSSLGPDEDGEEVDQREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPR 1606
Query: 827 ETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSN 886
+H AVKR+ +Y+K T G+ Y +S ++ F DADFAG +++RKSTSG+CHFLG++
Sbjct: 1607 TSHRQAVKRMFRYIKSTLEYGIWYSCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTS 1666
Query: 887 LVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISL 946
LV+WSS++Q+++A STAEAEYV+AA+ C+Q +WM + L+DYGLS VP+ CDNT+AI++
Sbjct: 1667 LVSWSSRKQSSVAQSTAEAEYVAAASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINI 1726
Query: 947 SKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILENL 1006
+KNP+ HSR KHIE++YH++RD+V+KGT+ LE+V+++ Q ADIFTKPL RF F+ L
Sbjct: 1727 AKNPVQHSRTKHIEIRYHFLRDNVEKGTIVLEFVESEKQLADIFTKPLDRSRFEFLRSEL 1786
Query: 1007 NM 1008
+
Sbjct: 1787 GV 1788
>UniRef100_Q7XPI7 OSJNBb0004A17.2 protein [Oryza sativa]
Length = 1877
Score = 906 bits (2341), Expect = 0.0
Identities = 477/1022 (46%), Positives = 657/1022 (63%), Gaps = 35/1022 (3%)
Query: 13 RLSQEETLALEETRREKSLEKGSIGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIF 72
R S +ET+ + + + KG+I ++ DV LV L +NLLS+SQ+ ++ +V F
Sbjct: 858 RASSDETITFGDASSGRVMAKGTIKVNDKFMLKDVALVSKLKYNLLSVSQLCDENLEVRF 917
Query: 73 NQTGCKAVSQTNGSVLFSGKRKNYIYKIKSSELLSQKVKCLM-SVNDEQWIWLRRLGHAS 131
+ + + + V F R ++ +CL+ S N + + W RRLGH
Sbjct: 918 KKDRSRVLDASESPV-FDISRVGRVFFANFDSSAPGPSRCLIASENRDLFFWHRRLGHIG 976
Query: 132 LRKISQLSKLNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLF 191
+S++S ++LIRGLP+LK + +C C+ GK T K +V T P +LLH+D
Sbjct: 977 FDHLSRISGMDLIRGLPKLKVPKDLVCAPCRHGKMTSSSHKPVTMVMTDGPGQLLHMDTV 1036
Query: 192 GPVKTESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRS 251
GP + +S+GGK Y LV+VDD+SR++WV FL K+ET F + + EF ++ +RS
Sbjct: 1037 GPARVQSVGGKWYVLVVVDDFSRYSWVYFLESKEETFGFFQSLARSLALEFPGALRAIRS 1096
Query: 252 DHGGEFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHL 311
D+G EF+N AFE +S G+ H FS P PQQNGVVERKNRTL EMARTM+ E + +
Sbjct: 1097 DNGSEFKNSAFESFCDSSGVEHQFSSPYVPQQNGVVERKNRTLVEMARTMLDEFTTPRKF 1156
Query: 312 WAEAINTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDS 371
W EAI+ A +I N + +R IL KTPYEL GR+P +S+ FG C++L + L KF+S
Sbjct: 1157 WTEAISAACFISNRVFLRTILHKTPYELRFGRRPKVSHLRVFGCKCFVLKS-GNLDKFES 1215
Query: 372 KALKCYFLGYSERSKGFRIYNIIHQTVEESIQIRFDGKLGSEKSKLF-----ERFADLSI 426
++L FLGY+ S+ +R+Y + + E+ ++ FD + ++ F D
Sbjct: 1216 RSLDGIFLGYATHSRAYRVYVLSTNKIVETCEVTFDEASPGARPEISGVPDESIFVDEDS 1275
Query: 427 DCSEANQPKNSSEDVAPEAEA--------SEAAPTTSDQLQKK-----------KRIAVS 467
D + + + P E S APTTS ++ + I
Sbjct: 1276 DDDDDDSIPPPLDSTPPVQETGSPSTTSPSGDAPTTSSSAAEEIDGGTSGPTAPRHIQNR 1335
Query: 468 HPEELIIGNKDAPV-RTRSMLKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEEL 526
HP + +IG V R RS L V+ EPK+V AL D+ W+ M EEL
Sbjct: 1336 HPPDSMIGGLGERVTRNRSY-------ELVNSAFVASFEPKNVCHALSDENWVNAMHEEL 1388
Query: 527 DQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTET 586
+ F +N VW+L+ P GF+VIGTKWVF+NKL E G + RNKARLVAQG++Q EG+D+ ET
Sbjct: 1389 ENFERNKVWSLVEPPLGFNVIGTKWVFKNKLGEDGSIVRNKARLVAQGFTQVEGLDFEET 1448
Query: 587 FSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVY 646
F+PVARLEAIR+L++F+ + L QMDVKSAFLNG I EEVYVKQPPGFE+ K+P+HV+
Sbjct: 1449 FAPVARLEAIRILLAFAASKGFKLFQMDVKSAFLNGVIEEEVYVKQPPGFENPKFPNHVF 1508
Query: 647 KLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFG 706
KL+K+LYGLKQAPRAWYERL +FLLQN F G D TLF + D L+VQIYVDDIIFG
Sbjct: 1509 KLEKALYGLKQAPRAWYERLKTFLLQNGFEMGAVDKTLFTLHSGIDFLLVQIYVDDIIFG 1568
Query: 707 SANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSD 766
++ +L +FS +M EFEMSMMGEL +FLG+QI Q ++HQ KY+ ELLKKF+M+D
Sbjct: 1569 GSSHALVAQFSDVMSREFEMSMMGELTFFLGLQIKQTKEGIFVHQTKYSKELLKKFDMAD 1628
Query: 767 CNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPR 826
C TPM T L +E +V Q+ YR MIGSLLYLTASRPDI FSV LCARFQ+ PR
Sbjct: 1629 CKPIATPMATTSSLGPDEDGEEVDQREYRSMIGSLLYLTASRPDIHFSVCLCARFQASPR 1688
Query: 827 ETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSN 886
+H AVKR+ +Y+K T G+ Y +S ++ F DADFAG +++RKSTSG+CHFLG++
Sbjct: 1689 TSHRQAVKRMFRYIKSTLEYGIWYSCSSALSVRAFSDADFAGCKIDRKSTSGTCHFLGTS 1748
Query: 887 LVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISL 946
LV+WSS++Q+++A STAEAEYV+AA+ C+Q +WM + L+DYGLS VP+ CDNT+AI++
Sbjct: 1749 LVSWSSRKQSSVAQSTAEAEYVAAASACSQVLWMISTLKDYGLSFSGVPLLCDNTSAINI 1808
Query: 947 SKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILENL 1006
+KNP+ HSR KHIE++YH++RD+V+KGT+ LE+V+++ Q ADIFTKPL RF F+ L
Sbjct: 1809 AKNPVQHSRTKHIEIRYHFLRDNVEKGTIVLEFVESEKQLADIFTKPLDRSRFEFLRSEL 1868
Query: 1007 NM 1008
+
Sbjct: 1869 GV 1870
>UniRef100_Q84VH6 Gag-pol polyprotein [Glycine max]
Length = 1577
Score = 905 bits (2338), Expect = 0.0
Identities = 465/973 (47%), Positives = 640/973 (64%), Gaps = 14/973 (1%)
Query: 33 KGSI-GDGKI-----PVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGS 86
KG I G GK+ P +N VLLV+GL NL+SISQ+ ++G++V F ++ C V+
Sbjct: 598 KGKITGMGKLVHEGLPSLNKVLLVKGLTVNLISISQLCDEGFNVNFTKSEC-LVTNEKSE 656
Query: 87 VLFSGKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRG 146
VL G R + + + S CL S DE IW +R GH LR + ++ +RG
Sbjct: 657 VLMKGSRSKDNCYLWTPQESSHSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRG 716
Query: 147 LPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGL 206
+P LK +C CQ GK K + +T+R LELLH+DL GP++ ES+GGK+Y
Sbjct: 717 IPNLKIEEGRICGECQIGKQVKMSHQKLQHQTTSRVLELLHMDLMGPMQVESLGGKRYAY 776
Query: 207 VIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELF 266
V+VDD+SR+TWV F+R K +T +F ++Q+E + +RSDHG EFEN F E
Sbjct: 777 VVVDDFSRFTWVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFC 836
Query: 267 NSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNII 326
S+GI+H FS TPQQNG+VERKNRTLQE AR M+ + +LWAEA+NTA YIHN +
Sbjct: 837 TSEGITHEFSAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRV 896
Query: 327 SIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSK 386
++R T YE+ KGR+P + +FH FGS CY+L +EQ K D K+ FLGYS S+
Sbjct: 897 TLRRGTPTTLYEIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSR 956
Query: 387 GFRIYNIIHQTVEESIQIRFDGKLGSEKSKLFERFADLSIDCSEANQPKNSSEDVAPEAE 446
+R++N +TV ESI + D + K + E D+ S+E+
Sbjct: 957 AYRVFNSRTRTVMESINVVVDDLTPARKKDVEE---DVRTSGDNVADTAKSAENAENSDS 1013
Query: 447 ASEAAPTTSDQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIEP 506
A++ + RI HP+ELIIG+ + V TRS E ++S VS IEP
Sbjct: 1014 ATDEPNINQPDKRPSIRIQKMHPKELIIGDPNRGVTTRSR----EIEIVSNSCFVSKIEP 1069
Query: 507 KSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRN 566
K+V EAL D+ WI MQEEL+QF +N+VW L+ +P+G +VIGTKW+F+NK NE+G +TRN
Sbjct: 1070 KNVKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRN 1129
Query: 567 KARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISE 626
KARLVAQGY+Q EG+D+ ETF+PVARLE+IRLL+ + L+QMDVKSAFLNGY++E
Sbjct: 1130 KARLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNE 1189
Query: 627 EVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFC 686
E YV+QP GF D +PDHVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF
Sbjct: 1190 EAYVEQPKGFVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFV 1249
Query: 687 ITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGV 746
+++I QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q
Sbjct: 1250 KQDAENLMIAQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDS 1309
Query: 747 TYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTA 806
++ Q KY ++KKF M + + +TP L K+E + V Q LYR MIGSLLYLTA
Sbjct: 1310 IFLSQSKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTA 1369
Query: 807 SRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADF 866
SRPDI ++V +CAR+Q++P+ +HL VKRILKY+ GT++ G+MY S L G+CDAD+
Sbjct: 1370 SRPDITYAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSGSMLVGYCDADW 1429
Query: 867 AGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLED 926
AG +RKSTSG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L++
Sbjct: 1430 AGSADDRKSTSGGCFYLGNNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKE 1489
Query: 927 YGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQW 986
Y + + +YCDN +AI++SKNP+ HSR KHI++++HYIR+ V ++LE+VDT+ Q
Sbjct: 1490 YNVEQDVMTLYCDNMSAINISKNPVQHSRTKHIDIRHHYIRELVDDKVITLEHVDTEEQI 1549
Query: 987 ADIFTKPLAEDRF 999
ADIFTK L +F
Sbjct: 1550 ADIFTKALDAKQF 1562
>UniRef100_Q84VI0 Gag-pol polyprotein [Glycine max]
Length = 1576
Score = 897 bits (2318), Expect = 0.0
Identities = 460/971 (47%), Positives = 640/971 (65%), Gaps = 23/971 (2%)
Query: 36 IGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVLFSGKRKN 95
+ DG +P +N VLLV+GL NL+SISQ+ ++G++V F ++ C V+ VL G R
Sbjct: 607 VHDG-LPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSK 664
Query: 96 YIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLPRLKYSSE 155
+ + + S CL S DE IW +R GH LR + ++ +RG+P LK
Sbjct: 665 DNCYLWTPQETSYSSTCLSSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEG 724
Query: 156 ALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVIVDDYSRW 215
+C CQ GK K + +T+ LELLH+DL GP++ ES+GGK+Y V+VDD+SR+
Sbjct: 725 RICGECQIGKQVKMSHQKLQHQTTSMVLELLHMDLMGPMQVESLGGKRYAYVVVDDFSRF 784
Query: 216 TWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNSQGISHNF 275
TWV F+R K +T +F ++Q+E + +RSDHG EFEN F E S+GI+H F
Sbjct: 785 TWVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEF 844
Query: 276 SCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISIRPILEKT 335
S TPQQNG+VERKNRTLQE R M+ + +LWAEA+NTA YIHN +++R T
Sbjct: 845 SAAITPQQNGIVERKNRTLQEATRVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTT 904
Query: 336 PYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIH 395
YE+ KGR+P + +FH FGS CY+L +EQ K D K+ FLGYS S+ +R++N
Sbjct: 905 LYEIWKGRKPTVKHFHIFGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRT 964
Query: 396 QTVEESIQIRFDGKLGSEKSKLFERFADLSIDCSEANQPKNSSEDVAPEAEASEAAPTTS 455
+TV ESI + D + K + E + +++ D A AE +E + +T+
Sbjct: 965 RTVMESINVVVDDLTPARKKDVEE----------DVRTSEDNVADTAKSAENAEKSDSTT 1014
Query: 456 DQLQKKK-------RIAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIEPKS 508
D+ + RI P+ELIIG+ + V TRS E ++S VS IEPK+
Sbjct: 1015 DEPNINQPDKSPFIRIQKMQPKELIIGDPNRGVTTRSR----EIEIVSNSCFVSKIEPKN 1070
Query: 509 VDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKA 568
V EAL D+ WI MQEEL+QF +N+VW L+ +P+G +VIGTKW+F+NK NE+G +TRNKA
Sbjct: 1071 VKEALTDEFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKA 1130
Query: 569 RLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEV 628
RLVAQGY+Q EG+D+ ETF+PVARLE+IRLL+ + L+QMDVKSAFLNGY++EE
Sbjct: 1131 RLVAQGYTQIEGVDFDETFAPVARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEA 1190
Query: 629 YVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCIT 688
YV+QP GF D + DHVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF
Sbjct: 1191 YVEQPKGFVDPTHLDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQ 1250
Query: 689 YKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTY 748
+++I QIYVDDI+FG + + + F MQ+EFEMS++GEL YFLG+Q+ Q +
Sbjct: 1251 DAENLMIAQIYVDDIVFGGMSNEMLRHFVPQMQSEFEMSLVGELHYFLGLQVKQMEDSIF 1310
Query: 749 IHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASR 808
+ Q KY ++KKF M + + +TP L K+E + V Q LYR MIGSLLYLTASR
Sbjct: 1311 LSQSKYAKNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQNLYRSMIGSLLYLTASR 1370
Query: 809 PDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAG 868
PDI F+V +CAR+Q++P+ +HL VKRILKY+ GT++ G+MY S+ L G+CDAD+AG
Sbjct: 1371 PDITFAVGVCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAG 1430
Query: 869 DRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYG 928
+RK TSG C +LG+NL++W SK+QN ++LSTAEAEY++A + C+Q +WMK L++Y
Sbjct: 1431 SADDRKCTSGGCFYLGTNLISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYN 1490
Query: 929 LSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWAD 988
+ + +YCDN +AI++SKNP+ H+R KHI++++HYIRD V ++LE+VDT+ Q AD
Sbjct: 1491 VEQDVMTLYCDNMSAINISKNPVQHNRTKHIDIRHHYIRDLVDDKIITLEHVDTEEQVAD 1550
Query: 989 IFTKPLAEDRF 999
IFTK L ++F
Sbjct: 1551 IFTKALDANQF 1561
>UniRef100_O65147 Gag-pol polyprotein [Glycine max]
Length = 1550
Score = 893 bits (2308), Expect = 0.0
Identities = 457/964 (47%), Positives = 633/964 (65%), Gaps = 9/964 (0%)
Query: 36 IGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVLFSGKRKN 95
+ DG +P +N VLLV+GL NL+SISQ+ ++G++V F ++ C V+ VL G R
Sbjct: 581 VHDG-LPSLNKVLLVKGLTANLISISQLCDEGFNVNFTKSEC-LVTNEKSEVLMKGSRSK 638
Query: 96 YIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLPRLKYSSE 155
+ + + S CL S DE IW +R GH LR + ++ +RG+P LK
Sbjct: 639 DNCYLWTPQETSYSSTCLFSKEDEVKIWHQRFGHLHLRGMKKIIDKGAVRGIPNLKIEEG 698
Query: 156 ALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVIVDDYSRW 215
+C CQ GK K + +T+R LELLH+DL GP++ ES+G K+Y V+VDD+SR+
Sbjct: 699 RICGECQIGKQVKMSNQKLQHQTTSRVLELLHMDLMGPMQVESLGRKRYAYVVVDDFSRF 758
Query: 216 TWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNSQGISHNF 275
TWV F+R K +T +F ++Q+E + +RSDHG EFEN F E S+GI+H F
Sbjct: 759 TWVNFIREKSDTFEVFKELSLRLQREKDCVIKRIRSDHGREFENSKFTEFCTSEGITHEF 818
Query: 276 SCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISIRPILEKT 335
S TPQQNG+VERKNRTLQE AR M+ + +LWAEA+NTA YIHN +++R T
Sbjct: 819 SAAITPQQNGIVERKNRTLQEAARVMLHAKELPYNLWAEAMNTACYIHNRVTLRRGTPTT 878
Query: 336 PYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIH 395
YE+ KGR+P + +FH GS CY+L +EQ K D K+ FLGYS S+ +R++N
Sbjct: 879 LYEIWKGRKPTVKHFHICGSPCYILADREQRRKMDPKSDAGIFLGYSTNSRAYRVFNSRT 938
Query: 396 QTVEESIQIRFDGKLGSEKSKLFERFADLSIDCSEANQPKNSSEDVAPEAEASEAAPTTS 455
+TV ESI + D + K + E D+ S+E+ A++
Sbjct: 939 RTVMESINVVVDDLTPARKKDVEE---DVRTSGDNVADTAKSAENAENSDSATDEPNINQ 995
Query: 456 DQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIEPKSVDEALED 515
+ RI HP+ELIIG+ + V TRS E ++S VS IEPK+V EAL D
Sbjct: 996 PDKRPSIRIQKMHPKELIIGDPNRGVTTRSR----EIEIISNSCFVSKIEPKNVKEALTD 1051
Query: 516 KGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGY 575
+ WI MQEEL+QF +N+VW L+ +P+G +VIGTKW+F+NK NE+G +TRNKARLVAQGY
Sbjct: 1052 EFWINAMQEELEQFKRNEVWELVPRPEGTNVIGTKWIFKNKTNEEGVITRNKARLVAQGY 1111
Query: 576 SQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPG 635
+Q EG+D+ ETF+P ARLE+IRLL+ + L+QMDVKSAFLNGY++EE YV+QP G
Sbjct: 1112 TQIEGVDFDETFAPGARLESIRLLLGVACILKFKLYQMDVKSAFLNGYLNEEAYVEQPKG 1171
Query: 636 FEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILI 695
F D +PDHVY+LKK+LYGLKQAPRAWYERL+ FL Q + +G D TLF +++I
Sbjct: 1172 FVDPTHPDHVYRLKKALYGLKQAPRAWYERLTEFLTQQGYRKGGIDKTLFVKQDAENLMI 1231
Query: 696 VQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYT 755
QIYVDDI+FG + + + F + MQ+EFEMS++GEL YFLG+Q+ Q ++ Q KY
Sbjct: 1232 AQIYVDDIVFGGMSNEMLRHFVQQMQSEFEMSLVGELTYFLGLQVKQMEDSIFLSQSKYA 1291
Query: 756 LELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSV 815
++KKF M + + +TP L K+E + V Q LYR MIGSLLYLTASRPDI ++V
Sbjct: 1292 KNIVKKFGMENASHKRTPAPTHLKLSKDEAGTSVDQSLYRSMIGSLLYLTASRPDITYAV 1351
Query: 816 HLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKS 875
CAR+Q++P+ +HL VKRILKY+ GT++ G+MY S+ L G+CDAD+AG +RKS
Sbjct: 1352 GGCARYQANPKISHLNQVKRILKYVNGTSDYGIMYCHCSDSMLVGYCDADWAGSVDDRKS 1411
Query: 876 TSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVP 935
T G C +LG+N ++W SK+QN ++LSTAEAEY++A + C+Q +WMK L++Y + +
Sbjct: 1412 TFGGCFYLGTNFISWFSKKQNCVSLSTAEAEYIAAGSSCSQLVWMKQMLKEYNVEQDVMT 1471
Query: 936 IYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLA 995
+YCDN +AI++SKNP+ HSR KHI++++HYIRD V ++LE+VDT+ Q ADIFTK L
Sbjct: 1472 LYCDNLSAINISKNPVQHSRTKHIDIRHHYIRDLVDDKVITLEHVDTEEQIADIFTKALD 1531
Query: 996 EDRF 999
++F
Sbjct: 1532 ANQF 1535
>UniRef100_Q9C5V1 Gag/pol polyprotein [Arabidopsis thaliana]
Length = 1643
Score = 871 bits (2250), Expect = 0.0
Identities = 446/995 (44%), Positives = 642/995 (63%), Gaps = 28/995 (2%)
Query: 33 KGSIGDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVLFSGK 92
KG + + + P + +V VEGL NL+S+SQ+ ++G V FN+ C A ++ N + L +
Sbjct: 655 KGDLTETEKPHLTNVYFVEGLTANLISVSQLCDEGLTVSFNKVKCWATNERNQNTLTGVR 714
Query: 93 RKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLPRLKY 152
N Y + ++ CL + ++ +W +RLGH + R +S+L ++RG+P LK+
Sbjct: 715 TGNNCYMWEEPKI------CLRAEKEDPVLWHQRLGHMNARSMSKLVNKEMVRGVPELKH 768
Query: 153 SSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVIVDDY 212
+ +C AC +GK + K + TT+ L+L+H+DL GP++TESI GK+Y V+VDD+
Sbjct: 769 IEKIVCGACNQGKQIRVQHKRVEGIQTTQVLDLIHMDLMGPMQTESIAGKRYVFVLVDDF 828
Query: 213 SRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNSQGIS 272
SR+ WV+F+R K ET F Q++ E + + +RSD GGEF N+AF SQGI
Sbjct: 829 SRYAWVRFIREKSETANSFKILALQLKNEKKMGIKQIRSDRGGEFMNEAFNSFCESQGIF 888
Query: 273 HNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISIRPIL 332
H +S PRTPQ NGVVERKNRTLQEMAR M+ + + + WAEAI+TA Y+ N + +R
Sbjct: 889 HQYSAPRTPQSNGVVERKNRTLQEMARAMIHGNGVPEKFWAEAISTACYVINRVYVRLGS 948
Query: 333 EKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGFRIYN 392
+KTPYE+ KG++P++SYF FG CY++N K+QLGKFDS++ + +FLGY+ S +R++N
Sbjct: 949 DKTPYEIWKGKKPNLSYFRVFGCVCYIMNDKDQLGKFDSRSEEGFFLGYATNSLAYRVWN 1008
Query: 393 IIHQTVEESIQIRFDGKLGSEKSKLF---------------ERFADLSIDCSEANQPKNS 437
+EES+ + FD E + E D D + N+
Sbjct: 1009 KQRGKIEESMNVVFDDGSMPELQIIVRNRNEPQTSISNNHGEERNDNQFDNGDINKSGEE 1068
Query: 438 SEDVAPEAEASE---AAPTTSDQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKPSEETL 494
S++ P A+ + D ++ V + G K R + EE +
Sbjct: 1069 SDEEVPPAQVHRDHASKDIIGDPSGERVTRGVKQDYRQLAGIKQKH-RVMASFACFEEIM 1127
Query: 495 LSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFR 554
S VS++EPK+V EALED WIL M+EEL++F+++ VW L+ +P +VIGTKW+F+
Sbjct: 1128 FSC--FVSIVEPKNVKEALEDHFWILAMEEELEEFSRHQVWDLVPRPPQVNVIGTKWIFK 1185
Query: 555 NKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMD 614
NK +E G +TRNKARLVAQGY+Q EG+D+ ETF+PVARLE IR L+ + LHQMD
Sbjct: 1186 NKFDEVGNITRNKARLVAQGYTQVEGLDFDETFAPVARLECIRFLLGTACGMGFKLHQMD 1245
Query: 615 VKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNE 674
VK AFLNG I EEVYV+QP GFE+ ++P++VYKLKK+LYGLKQAPRAWYERL++FL+
Sbjct: 1246 VKCAFLNGIIEEEVYVEQPKGFENLEFPEYVYKLKKALYGLKQAPRAWYERLTTFLIVQG 1305
Query: 675 FVRGKGDNTLFCITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKY 734
+ RG D TLF + I+I+QIYVDDI+FG + L K F K M EF MSM+GELKY
Sbjct: 1306 YTRGSVDKTLFVKNDVHGIIIIQIYVDDIVFGGTSDKLVKTFVKTMTTEFRMSMVGELKY 1365
Query: 735 FLGIQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLY 794
FLG+QI+Q I Q Y L+K+F M + TPM T L K+E KV +KLY
Sbjct: 1366 FLGLQINQTDEGITISQSTYAQNLVKRFGMCSSKPAPTPMSTTTKLFKDEKGVKVDEKLY 1425
Query: 795 RGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTS 854
RGMIGSLLYLTA+RPD+ SV LCAR+QS+P+ +HL AVKRI+KY+ GT N GL Y + +
Sbjct: 1426 RGMIGSLLYLTATRPDLCLSVGLCARYQSNPKASHLLAVKRIIKYVSGTINYGLNYTRDT 1485
Query: 855 EYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCC 914
L G+CDAD+ G+ +R+ST+G FLGSNL++W SK+QN ++LS+ ++EY++ +CC
Sbjct: 1486 SLVLVGYCDADWGGNLDDRRSTTGGVFFLGSNLISWHSKKQNCVSLSSTQSEYIALGSCC 1545
Query: 915 TQTIWMKNHLEDYGLSL-KKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKG 973
TQ +WM+ DYG++ + + CDN +AI++SKNP+ HS KHI +++H++R+ V++
Sbjct: 1546 TQLLWMRQMGLDYGMTFPDPLLVKCDNESAIAISKNPVQHSVTKHIAIRHHFVRELVEEK 1605
Query: 974 TLSLEYVDTDHQWADIFTKPLAEDRFLFILENLNM 1008
+++E+V T+ Q DIFTKPL + F+ + ++L +
Sbjct: 1606 QITVEHVPTEIQLVDIFTKPLDLNTFVNLQKSLGI 1640
>UniRef100_Q9SDD5 Similar to copia-type pol polyprotein [Oryza sativa]
Length = 940
Score = 867 bits (2241), Expect = 0.0
Identities = 452/905 (49%), Positives = 595/905 (64%), Gaps = 37/905 (4%)
Query: 136 SQLSKLNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVK 195
SQ SK I GL +++ + +C ACQ GK KNV++TTRPLELLH+DLFGP+
Sbjct: 34 SQTSKARHILGLTNIQFEKDRVCSACQAGKQIGAHHPVKNVMTTTRPLELLHMDLFGPIA 93
Query: 196 TESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGG 255
SIGG KYGLVIVDD+S +TWV FL K ET IF F + Q EF + +RSD+
Sbjct: 94 YLSIGGNKYGLVIVDDFSCFTWVFFLHDKSETQAIFKKFARRAQNEFDLKIKNIRSDNVK 153
Query: 256 EFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEA 315
EF+N E + +GI H FS P +PQQNGV ERKNRTL E+ARTM+ E + WAEA
Sbjct: 154 EFKNTCIESFLDEEGIKHEFSAPYSPQQNGVAERKNRTLIEIARTMLDEYKTSDRFWAEA 213
Query: 316 INTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALK 375
+NT + N + + +L+KTPYEL G +P++SYF FGS CY+LN K + KF K
Sbjct: 214 VNTVCHDINRLYLHRLLKKTPYELLTGNKPNVSYFRVFGSKCYILNKKARSSKFAPKVDG 273
Query: 376 CYFLGYSERSKGFRIYNIIHQTVEESIQIRFDGKLGSEKSKL--------------FERF 421
+ LGY +R++N VE + + FD GS+ ++ +R
Sbjct: 274 GFLLGYGSNECAYRVFNKTSGIVEIAPDVTFDETNGSQVEQVDSHVLGEEEDPREAIKRL 333
Query: 422 ADLSIDCSEANQPKNSSEDVAPEAEASEAAPTTS--DQLQKKKRIAVS------------ 467
A + E Q +SS V P P+TS DQ ++ +++ S
Sbjct: 334 ALGDVRPREPQQGASSSTQVEPPTSTQANDPSTSSLDQGEEGEQVPPSSINLAHPRIHQS 393
Query: 468 ----HPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQ 523
HP + I+G+ + V TRS + E VS +EP V+EAL D W++ MQ
Sbjct: 394 IQRDHPTDNILGDINKGVSTRSHIANFCEHY----SFVSSLEPLRVEEALNDPDWVMAMQ 449
Query: 524 EELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDY 583
EEL+ FT+N+VWTL+ + + +VIGTKW+FRNK +E G V RNKARLVAQG++Q EG+D+
Sbjct: 450 EELNNFTRNEVWTLVERSRQ-NVIGTKWIFRNKQDEAGVVIRNKARLVAQGFTQIEGLDF 508
Query: 584 TETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPD 643
ETF+PVARLE+IR+L++F+ N N L+QMDVKSAFLNG I+E VYV+QPPGF+D KYP+
Sbjct: 509 GETFAPVARLESIRILLTFATNLNFKLYQMDVKSAFLNGLINELVYVEQPPGFKDPKYPN 568
Query: 644 HVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDI 703
HVYKL K+LY LKQAPRAWYE L +FL++N F GK D+TLF + NDI + QIYVDDI
Sbjct: 569 HVYKLHKALYELKQAPRAWYECLRNFLVKNGFEIGKADSTLFTKRHDNDIFVCQIYVDDI 628
Query: 704 IFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFN 763
IFGS N S +EFS++M FEMSMMGELK+FLG+QI Q T+I Q KY ++LKKF
Sbjct: 629 IFGSTNKSFSEEFSRMMTKRFEMSMMGELKFFLGLQIKQLKEGTFICQTKYLKDMLKKFG 688
Query: 764 MSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQS 823
M + TPM L+ E V QK+YR +IGSLLYL ASRPDI+ SV +CARFQ+
Sbjct: 689 MENAKPIHTPMPSNGHLDLNEQGKDVDQKVYRSIIGSLLYLCASRPDIMLSVCMCARFQA 748
Query: 824 DPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFL 883
P+E HL AVKRIL+YL T NLGL Y K + + L G+ DAD+AG +V+RKSTSG+C FL
Sbjct: 749 APKECHLVAVKRILRYLVHTPNLGLWYPKGARFDLIGYADADYAGCKVDRKSTSGTCQFL 808
Query: 884 GSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAA 943
G +LV+WSSK+QN++ALSTAEAEY+S +CC Q IWMK L DYGL++ K+P+ CDN +A
Sbjct: 809 GRSLVSWSSKKQNSVALSTAEAEYISTGSCCAQLIWMKQTLRDYGLNVSKIPLLCDNESA 868
Query: 944 ISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFIL 1003
I ++ NP+ HSR KHI++++H++RDH +G + +++V D Q ADIFTKPL E RF +
Sbjct: 869 IKIANNPVQHSRTKHIDIRHHFLRDHSTRGDIDIQHVRIDKQLADIFTKPLDEARFCELR 928
Query: 1004 ENLNM 1008
LN+
Sbjct: 929 SELNI 933
>UniRef100_Q8H851 Putative Zea mays retrotransposon Opie-2 [Oryza sativa]
Length = 2011
Score = 855 bits (2210), Expect = 0.0
Identities = 461/1001 (46%), Positives = 622/1001 (62%), Gaps = 46/1001 (4%)
Query: 36 IGDGKIPVIND-----VLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVLFS 90
IG GKI + ND V LV+ L NLLS++QI + F S + S +F
Sbjct: 599 IGLGKIAISNDLSIDNVSLVKSLNFNLLSVAQICDLVLSCAFFPQEVIVSSLLDKSCVFK 658
Query: 91 GKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLPRL 150
G R +Y + + CL++ W+W RRL H + ++S+LSK +L+ GL +
Sbjct: 659 GFRYGNLYLGDFNSSEANLKTCLVAKTSLGWLWHRRLAHVGMNQLSKLSKRDLVVGLKDV 718
Query: 151 KYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVIVD 210
K+ + LC ACQ GK K+++ST+RPLELLH++ FGP +SIGG + LVIVD
Sbjct: 719 KFEKDKLCSACQAGKQVACSHPTKSIMSTSRPLELLHMEFFGPTTYKSIGGNSHCLVIVD 778
Query: 211 DYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNSQG 270
DYSR+TW+ FL K +F F + Q EF +++ +RSD+G +F+N E+ + G
Sbjct: 779 DYSRYTWMFFLHDKSIVAELFKKFAKRGQNEFNCTLVKIRSDNGSKFKNTNIEDYCDDLG 838
Query: 271 ISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISIRP 330
I H S +PQQNGVVE KNRTL EMARTM+ E ++ WAEAINTA + N + +
Sbjct: 839 IKHELSATYSPQQNGVVEMKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHR 898
Query: 331 ILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGFRI 390
+L+KT YEL GR+P+++YF FG CY+ +L KF+S+ + + LGY+ SK +R+
Sbjct: 899 LLKKTSYELIVGRKPNVAYFRVFGCKCYIYRKGVRLTKFESRCDEGFLLGYASNSKAYRV 958
Query: 391 YNIIHQTVEESIQIRFDGKLGSEKSK----------LFERFADLSID----CSEANQPKN 436
YN VEE+ ++FD GS++ L ++SI ++P
Sbjct: 959 YNKNKGIVEETADVQFDETNGSQEGHENLDDVGDEGLMRAMKNMSIGDVKPIEVEDKPST 1018
Query: 437 SSED-----VAPEAEASEAAPTTSDQLQKKKRIAVS----HPEELIIGNKDAPVRTRSML 487
S++D P E + L RI + HP + ++G+ V+TRS +
Sbjct: 1019 STQDEPSTSATPSQAQVEVEEEKAQDLPMPPRIHTALSKDHPIDQVLGDISKGVQTRSRV 1078
Query: 488 KPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHVI 547
++ VS +EPK VDEAL D WI M +EL+ F +N VWTL+ + + +VI
Sbjct: 1079 A----SICEHYSFVSCLEPKHVDEALCDPDWINAMHKELNNFARNKVWTLVERLRDHNVI 1134
Query: 548 GTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNHN 607
GTKWVFRNK +E G V RNKAR VAQG++Q EG+D+ ETF+PV RLEAI +L++F+ N
Sbjct: 1135 GTKWVFRNKQDENGLVVRNKARFVAQGFTQVEGLDFGETFAPVTRLEAICILLAFASCFN 1194
Query: 608 ITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERLS 667
I L QMDVKSAFLNG I+E V+V+QPPGFED KYP+HVYKL K+LYGLKQAPRAWYERL
Sbjct: 1195 IKLFQMDVKSAFLNGEIAELVFVEQPPGFEDPKYPNHVYKLSKALYGLKQAPRAWYERLR 1254
Query: 668 SFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEMS 727
FLL +F GK D TLF +D + QIYVDDIIFG N CKEF +M EFEMS
Sbjct: 1255 DFLLSKDFKIGKVDTTLFTKIIGDDFFVCQIYVDDIIFGCTNEVFCKEFGDMMSREFEMS 1314
Query: 728 MMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVSS 787
M+GEL +F G+QI Q T F + D KTPM L+ +E
Sbjct: 1315 MIGELSFFHGLQIKQLKDGT--------------FGLEDAKPIKTPMATNGHLDLDEGGK 1360
Query: 788 KVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNLG 847
V KLYR MIGSLLYLTASRPDI+FSV +CARFQ+ P+E HL AVKRIL+YLK ++ +G
Sbjct: 1361 PVDLKLYRSMIGSLLYLTASRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTIG 1420
Query: 848 LMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAEY 907
L Y K +++ L G+ D+D+AG +V+RKSTSGSC LG +LV+WSSK+QN +AL AEAEY
Sbjct: 1421 LWYPKGAKFKLVGYSDSDYAGCKVDRKSTSGSCQMLGRSLVSWSSKKQNFVALFIAEAEY 1480
Query: 908 VSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYIR 967
VSA +CC Q +WMK L DYG+S K P+ C+N +AI ++ NP+ HSR KHI++++H++R
Sbjct: 1481 VSAGSCCAQLLWMKQILLDYGISFTKTPLLCENDSAIKIANNPVQHSRTKHIDIRHHFLR 1540
Query: 968 DHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILENLNM 1008
DHV K + + ++ T+ Q ADIFTKPL E RF + LN+
Sbjct: 1541 DHVAKCDIVISHIRTEDQLADIFTKPLDETRFCKLRNELNL 1581
>UniRef100_Q60DR2 Putative polyprotein [Oryza sativa]
Length = 1577
Score = 850 bits (2196), Expect = 0.0
Identities = 456/1002 (45%), Positives = 621/1002 (61%), Gaps = 50/1002 (4%)
Query: 36 IGDGKIPVIND-----VLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVLFS 90
IG GKI + ND V LV+ L NLLS++QI + F S + S +F
Sbjct: 590 IGLGKIAISNDLSIDNVSLVKSLNFNLLSVAQICDLSLSCAFFPQEVIVSSLLDKSCVFK 649
Query: 91 GKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKISQLSKLNLIRGLPRL 150
G R +Y + + + CL++ W+W RRL H + ++S+ SK +L+ GL +
Sbjct: 650 GFRYGNLYLVDFNSSEANLKTCLVAKTSLGWLWHRRLAHVGMNQLSKFSKRDLVMGLKDV 709
Query: 151 KYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVKTESIGGKKYGLVIVD 210
K+ + LC ACQ GK K+++ST++PLELLH+DLF P +SIGG + LVIVD
Sbjct: 710 KFEKDKLCSACQAGKQVACSHPTKSIMSTSKPLELLHMDLFDPTTYKSIGGNSHCLVIVD 769
Query: 211 DYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNSQG 270
DYSR+TWV FL K +F F + Q EF +++ +RS+ G EF+N E+ + G
Sbjct: 770 DYSRYTWVFFLHDKSIVADLFKKFAKRAQNEFSCTLVKIRSNIGSEFKNTNIEDYCDDLG 829
Query: 271 ISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISIRP 330
I H +PQQNGVVERKNRTL EMARTM+ E ++ WAEAINTA + N + +
Sbjct: 830 IKHELFATYSPQQNGVVERKNRTLIEMARTMLDEYGVSDSFWAEAINTACHATNRLYLHR 889
Query: 331 ILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGFRI 390
+L+KT YE+ GR+P+I+YF FG CY+ +L KF+S+ + + LGY+ +SK +R+
Sbjct: 890 VLKKTSYEVIVGRKPNIAYFRVFGCKCYIHRKGVRLTKFESRCDEGFLLGYASKSKAYRV 949
Query: 391 YNIIHQTVEESIQIRFDGKLGSEKSK----------LFERFADLSID----CSEANQPKN 436
YN VEE+ ++FD GS++ L ++SI ++P
Sbjct: 950 YNKNKGIVEETADVQFDETNGSQEGHENLDDVGDEGLMRVMKNMSIGDVKPIEVEDKPST 1009
Query: 437 SSEDVAPEAEASEAAPTTSDQLQKKKR----------IAVSHPEELIIGNKDAPVRTRSM 486
S++D P A + + +K + ++ HP + ++G+ V+TRS
Sbjct: 1010 STQD-EPSTSAMPSQAQVEVEEEKAQEPPMPPRIHTALSKDHPIDQVLGDISKGVQTRSR 1068
Query: 487 LKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFHV 546
+ ++ VS +E K VDEAL D W+ M EEL F +N VWTL+ +P+ +V
Sbjct: 1069 VT----SICEHYSFVSCLERKHVDEALCDPDWMNAMHEELKNFARNKVWTLVERPRDHNV 1124
Query: 547 IGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVNH 606
IGTKWVFRNK +E G V RNKARLVAQG++Q EG+D+ ETF+PVARLEAI +L++F+
Sbjct: 1125 IGTKWVFRNKQDENGLVVRNKARLVAQGFTQVEGLDFGETFAPVARLEAICILLAFASCF 1184
Query: 607 NITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYERL 666
+I L QMDVKSAFLN D KYP+HVYKL K+LYGL+QAPRAWYERL
Sbjct: 1185 DIKLFQMDVKSAFLN----------------DTKYPNHVYKLSKALYGLRQAPRAWYERL 1228
Query: 667 SSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFEM 726
FLL +F GK D TLF +D + QIYVDDIIFGS N CKEF +M EFEM
Sbjct: 1229 RDFLLSKDFKIGKVDITLFTKIIGDDFFVYQIYVDDIIFGSTNEVFCKEFGDMMSREFEM 1288
Query: 727 SMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEVS 786
SM+GEL +FLG+QI Q T++ Q KY +LLK+F + D KTPM L+ +E
Sbjct: 1289 SMIGELSFFLGLQIKQLKNGTFVSQTKYIKDLLKRFGLEDAKPIKTPMATNGHLDLDEGG 1348
Query: 787 SKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTNL 846
V KLYR MIGSLLYLT SRPDI+FSV +CARFQ+ P+E HL AVKRIL+YLK ++ +
Sbjct: 1349 KPVDLKLYRSMIGSLLYLTVSRPDIMFSVCMCARFQAAPKECHLVAVKRILRYLKHSSTI 1408
Query: 847 GLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEAE 906
GL Y K +++ L G+ D D+AG +V+RKSTS SC LG +LV+WSSK+QN++ALSTAE E
Sbjct: 1409 GLWYPKGAKFKLVGYSDPDYAGCKVDRKSTSSSCQMLGRSLVSWSSKKQNSVALSTAETE 1468
Query: 907 YVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYHYI 966
YVSA +CC Q +WMK L DYG+S K P+ CDN AI ++ NP+ HSR KHI++++H++
Sbjct: 1469 YVSAGSCCAQLLWMKQTLLDYGISFTKTPLLCDNDGAIKIANNPVQHSRTKHIDIRHHFL 1528
Query: 967 RDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILENLNM 1008
RDHV K + + ++ T+ Q ADIFTKPL E RF + LN+
Sbjct: 1529 RDHVAKCDIVISHIRTEDQLADIFTKPLDETRFCKLRNELNI 1570
>UniRef100_Q850V9 Putative polyprotein [Oryza sativa]
Length = 1128
Score = 842 bits (2176), Expect = 0.0
Identities = 433/913 (47%), Positives = 599/913 (65%), Gaps = 29/913 (3%)
Query: 121 WIWLRRLGHASLRKISQLSKLNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTT 180
W+W RRL H + ++S+LSK +L+ GL +K+ + LC ACQ K K+++ST+
Sbjct: 213 WLWHRRLAHVGMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQASKQVACSHPTKSIMSTS 272
Query: 181 RPLELLHIDLFGPVKTESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQK 240
RPLELLH+DLFGP +SIGG + LVIVDDYS +TWV FL K +F F + Q
Sbjct: 273 RPLELLHMDLFGPTTYKSIGGNSHCLVIVDDYSCYTWVFFLHDKCIVAELFKKFAKRAQN 332
Query: 241 EFQTSVITVRSDHGGEFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMART 300
EF +++ +RSD+G +F+N E+ + I H S +PQQNGVVERKNRTL EMART
Sbjct: 333 EFSCTLVKIRSDNGSKFKNTNIEDYCDDLSIKHELSATYSPQQNGVVERKNRTLIEMART 392
Query: 301 MMQESSMAKHLWAEAINTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYML 360
M+ E ++ WAEAINTA + N + + +L+KT YEL GR+P+++YF FG CY+
Sbjct: 393 MLDEYGVSDSFWAEAINTACHATNRLYLHRLLKKTSYELIVGRKPNVAYFRVFGCKCYIY 452
Query: 361 NTKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIHQTVEESIQIRFDGKLGSEKSK---- 416
+L KF+S+ + + LGY+ SK +R+YN VEE+ ++FD GS++
Sbjct: 453 RKGVRLTKFESRCDEGFLLGYASNSKAYRVYNKNKGIVEETADVQFDETNGSQEGHENLD 512
Query: 417 ------LFERFADLSIDCSEANQPKNSSEDVAPEAEASEAAPTTSD-QLQKKKR------ 463
L ++SI + + ++ + ++ A+P+ + +++K+K
Sbjct: 513 DVGDEGLMRAMKNMSIGDVKPIEVEDKPSTSTQDEPSTSASPSQAQVEVEKEKAQDPPMP 572
Query: 464 ------IAVSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIEPKSVDEALEDKG 517
++ HP + ++G+ V+TRS + ++ VS +EPK VDEAL D
Sbjct: 573 PRIYTALSKDHPIDQVLGDISKGVQTRSPVA----SICEHYSFVSCLEPKHVDEALYDPD 628
Query: 518 WILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQ 577
W+ + EEL+ F +N VWTL+ +P+ +VIGTKWVFRNK +E V RNKARLVAQG++Q
Sbjct: 629 WMNAIHEELNNFARNKVWTLVERPRDHNVIGTKWVFRNKQDENRLVVRNKARLVAQGFTQ 688
Query: 578 QEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFE 637
E +D+ ETF PVARLEAIR+L++F+ +I L QMDVKSAFLNG I+E V+V+QPPGF+
Sbjct: 689 VEDLDFGETFGPVARLEAIRILLAFASCFDIKLFQMDVKSAFLNGEIAELVFVEQPPGFD 748
Query: 638 DDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQ 697
D KYP+HVYKL K+LYGLKQAPRAWYERL FLL +F GK D TLF +D + Q
Sbjct: 749 DPKYPNHVYKLSKALYGLKQAPRAWYERLRDFLLSKDFKIGKVDTTLFTKIIGDDFFVCQ 808
Query: 698 IYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLE 757
IYVDDIIFGS N CKEF +M EFEMSM+ EL +FLG+QI Q T++ Q KY +
Sbjct: 809 IYVDDIIFGSTNEVFCKEFGDMMSREFEMSMIEELSFFLGLQIKQLKDGTFVSQTKYIKD 868
Query: 758 LLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHL 817
LLK+F + D KTPM L+ +E V KLYR MIGSLLYLTASRPDI+FSV +
Sbjct: 869 LLKRFGLEDAKPIKTPMATNWHLDLDEGGKPVDLKLYRSMIGSLLYLTASRPDIMFSVCM 928
Query: 818 CARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTS 877
ARFQ+ P+E HL AVKRIL+YLK ++ + L Y K +++ L G+ D+D+AG +V+RKSTS
Sbjct: 929 YARFQAAPKECHLVAVKRILRYLKHSSTISLWYPKGAKFKLVGYSDSDYAGYKVDRKSTS 988
Query: 878 GSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSL--KKVP 935
GSC LG +LV+WSSK+QN++ALSTAEAEY+SA +CC Q +WMK L DYG+S + P
Sbjct: 989 GSCQMLGRSLVSWSSKKQNSVALSTAEAEYISAGSCCAQLLWMKQILLDYGISFTETQTP 1048
Query: 936 IYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLA 995
+ C+N + I ++ NP+ H R KHI++++H++ DHV K + + ++ T+ Q ADIFTKPL
Sbjct: 1049 LLCNNDSTIKIANNPVQHFRTKHIDIRHHFLTDHVAKCDIVISHIRTEDQLADIFTKPLD 1108
Query: 996 EDRFLFILENLNM 1008
E RF + LN+
Sbjct: 1109 ETRFCKLRNELNV 1121
>UniRef100_Q7XP45 OSJNBa0063G07.6 protein [Oryza sativa]
Length = 1539
Score = 835 bits (2157), Expect = 0.0
Identities = 453/1023 (44%), Positives = 626/1023 (60%), Gaps = 71/1023 (6%)
Query: 16 QEETLALEETRREKSLEKGSIGDGKIPVIND-----VLLVEGLFHNLLSISQIANKGYDV 70
QE+ ++ ++R+ IG GKI + ND V V+ L NLLS++QI + G
Sbjct: 551 QEKVTFVDNSKRKV------IGLGKIAISNDLSIDNVSFVKSLNFNLLSVAQICDLGLSC 604
Query: 71 IFNQTGCKAVSQTNGSVLFSGKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHA 130
F S + S +F G R +Y + + + CL++ W+W RRL H
Sbjct: 605 AFFPQEVIVSSLLDKSCVFKGFRYGNLYFVDFNSSEANLKTCLVAKTSLGWLWHRRLAHV 664
Query: 131 SLRKISQLSKLNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDL 190
+ ++S+LSK +L+ GL +K+ + LC ACQ GK K+++ST+RPLELLH+DL
Sbjct: 665 GMNQLSKLSKRDLVVGLKDVKFEKDKLCSACQAGKQVACSHPTKSIMSTSRPLELLHMDL 724
Query: 191 FGPVKTESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVR 250
FGP +SIGG + LVIVDDYSR+TWV FL K +F + Q EF +++ +R
Sbjct: 725 FGPTTYKSIGGNSHCLVIVDDYSRYTWVFFLHDKSIVAELFKKIAKRAQNEFSCTLVKIR 784
Query: 251 SDHGGEFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKH 310
SD+G EF+N E+ + GI H S +PQQNGVVERKNRTL EMARTM+ E ++
Sbjct: 785 SDNGSEFKNTNIEDYCDDLGIKHELSATYSPQQNGVVERKNRTLIEMARTMLDEYGVSDS 844
Query: 311 LWAEAINTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFD 370
WAEAINTA + N + +L+ T YEL GR+P+++YF FG CY+ +L KF+
Sbjct: 845 FWAEAINTACHATNRFYLHRLLKNTSYELIVGRKPNVAYFRVFGCKCYIYRKGVRLTKFE 904
Query: 371 SKALKCYFLGYSERSKGFRIYNIIHQTVEESIQIRFDGKLGSEKSKLFERFADLSIDCSE 430
S+ + + LGY+ SK +R+YN TVEE+ ++FD GS++ E D+ +
Sbjct: 905 SRCDEGFLLGYASNSKAYRVYNKNKGTVEETADVQFDETNGSQEG--HENLDDVGDEGLI 962
Query: 431 ANQPKNSSEDVAPEAEASEAAPTTSD-------------QLQKKK------------RIA 465
S DV P + + +T D +++++K ++
Sbjct: 963 RAMKNMSFGDVKPIEVEDKPSTSTQDEPSTFATPSQAQVEVEEEKAQDPPIPPRIHTTLS 1022
Query: 466 VSHPEELIIGNKDAPVRTRSMLKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEE 525
HP + ++G+ V+T S + ++ VS +EPK VDEAL D W+ M EE
Sbjct: 1023 KDHPIDQVLGDISKGVQTLSRV----ASICEHYSFVSCLEPKHVDEALCDPDWMNAMHEE 1078
Query: 526 LDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTE 585
L+ F +N VWTL+ +P+ +VIGTKWVFRNK +E G V RNKARLVAQG++Q EG+D+ E
Sbjct: 1079 LNNFARNKVWTLVERPRDHNVIGTKWVFRNKQDENGLVVRNKARLVAQGFTQVEGLDFGE 1138
Query: 586 TFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHV 645
TF+PVARLEAI +L++F+ +I L QMDVKSAFLNG I+E V+V+QPPGFED KYP+H
Sbjct: 1139 TFAPVARLEAICILLAFASWFDIKLFQMDVKSAFLNGEIAELVFVEQPPGFEDPKYPNHD 1198
Query: 646 YKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIF 705
+K+ GK D TLF +D + QIYVDDIIF
Sbjct: 1199 FKI-----------------------------GKVDTTLFTKIIGDDFFVCQIYVDDIIF 1229
Query: 706 GSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMS 765
GS N CKEF +M EFEMSM+GEL +FLG+QI Q T++ Q KY +LLK+F +
Sbjct: 1230 GSTNEVFCKEFGDMMSREFEMSMIGELSFFLGLQIKQLKDGTFVSQTKYIKDLLKRFGLE 1289
Query: 766 DCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDP 825
D KTPM L+ +E V KLYR MIGSLLYLTASRPDI+FSV +CA FQ+ P
Sbjct: 1290 DAKPIKTPMATNGHLDLDEGGKPVDLKLYRSMIGSLLYLTASRPDIMFSVCMCAWFQAAP 1349
Query: 826 RETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGS 885
+E HL AVKRIL+YLK ++ +GL Y K +++ L G+ D+D+AG +V+R STSGSC LG
Sbjct: 1350 KECHLVAVKRILRYLKYSSTIGLWYPKGAKFKLVGYSDSDYAGCKVDRNSTSGSCQMLGR 1409
Query: 886 NLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAIS 945
+LV+WSSK+QN++ALSTAEAEYVSA +CC Q +WMK L DYG+S K P+ CDN +AI
Sbjct: 1410 SLVSWSSKKQNSVALSTAEAEYVSAGSCCAQLLWMKQTLLDYGISFTKTPLLCDNDSAIK 1469
Query: 946 LSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILEN 1005
++ NP+ HSR KHI++++H++RDHV K + + ++ T+ Q ADIFTKPL E RF +
Sbjct: 1470 IANNPVQHSRTKHIDIRHHFLRDHVAKCDIVISHIRTEDQLADIFTKPLDETRFCKLRNE 1529
Query: 1006 LNM 1008
LN+
Sbjct: 1530 LNV 1532
>UniRef100_Q7XBD2 Retrotransposon Opie-2 [Zea mays]
Length = 1512
Score = 785 bits (2028), Expect = 0.0
Identities = 408/827 (49%), Positives = 542/827 (65%), Gaps = 58/827 (7%)
Query: 182 PLELLHIDLFGPVKTESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKE 241
PLELLH+DLFGPV SIGG KYGLVIVDD+SR+TWV FL+ K ET F+ + Q E
Sbjct: 737 PLELLHMDLFGPVAYLSIGGSKYGLVIVDDFSRFTWVFFLQDKSETQGTLKRFLRRAQNE 796
Query: 242 FQTSVITVRSDHGGEFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTM 301
F+ V +RSD+G EF+N EE +GI H FS P TPQQNGVVERKNRTL +MARTM
Sbjct: 797 FELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTPQQNGVVERKNRTLIDMARTM 856
Query: 302 MQESSMAKHLWAEAINTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLN 361
+ E + W+EA+NTA + N + + +L+KT YEL G +P++SYF FGS CY+L
Sbjct: 857 LGEFKTPECFWSEAVNTACHAINRVYLHRLLKKTSYELLTGNKPNVSYFRVFGSKCYILV 916
Query: 362 TKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIHQTVEESIQIRFDGKLGSEKSKLFERF 421
K + KF KA++ + LGY +K +R++N VE S + FD GS + ++
Sbjct: 917 KKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSSDVVFDETNGSPREQV---- 972
Query: 422 ADLSIDCSEANQPKNSSEDVAPEAEASEAAPTTSDQLQKKKRIAVSHPEELIIGNKDAPV 481
+DC + ++ EDV A + A Q Q ++ D P
Sbjct: 973 ----VDCDDVDE-----EDVPTAAIRTMAIGEVRPQEQDER---------------DQPS 1008
Query: 482 RTRSMLKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKP 541
+ ++ P+++ +P V+EAL D W+L MQEEL+ F +N+VW+L
Sbjct: 1009 SSTTVHPPTQDDE----------QPFRVEEALLDLDWVLAMQEELNNFKRNEVWSL---- 1054
Query: 542 KGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLIS 601
+E G VTRNKARLVA+GY+Q G+D+ ETF+PVARLE+IR+L++
Sbjct: 1055 ----------------DEHGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLESIRILLA 1098
Query: 602 FSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRA 661
++ +H+ L+QMDVKSAFLNG I EEVYV+QPPGFED++YPDHV KL K+LYGLKQAPRA
Sbjct: 1099 YAAHHSFRLYQMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLAKALYGLKQAPRA 1158
Query: 662 WYERLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQ 721
WYE L FL+ N F GK D TLF T D+ + QI+VDDIIFGS N C+EFS++M
Sbjct: 1159 WYECLRDFLIANAFKVGKADPTLFTKTCNGDLFVCQIFVDDIIFGSTNQKSCEEFSRVMT 1218
Query: 722 AEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILE 781
+FEMSMMG+L YFLG Q+ Q T+I Q KYT +LLK+F M D +KTPM +
Sbjct: 1219 QKFEMSMMGKLNYFLGFQVKQLKDGTFISQMKYTQDLLKRFGMKDAKPAKTPMGTDGHTD 1278
Query: 782 KEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLK 841
+ V QK YR MIGSLLYL ASRPDI+ SV +CARFQS+P+E HL AVKRIL+YL
Sbjct: 1279 LNKGGKSVDQKAYRSMIGSLLYLCASRPDIMLSVCMCARFQSEPKECHLVAVKRILRYLV 1338
Query: 842 GTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALS 901
T GL Y K S + L G+ D+D AG +V+RKSTSG+C FLG +LV+W+SK+Q ++ALS
Sbjct: 1339 ATPCFGLWYPKGSTFDLVGYSDSDNAGCKVDRKSTSGTCQFLGRSLVSWNSKKQTSVALS 1398
Query: 902 TAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEV 961
TAEAEYV+A CC Q +WM+ L D+G +L KVP+ CDN +AI +++NP+ H+R KHI++
Sbjct: 1399 TAEAEYVAAGQCCAQLLWMRQTLRDFGYNLSKVPLLCDNESAIRMAENPVEHNRTKHIDI 1458
Query: 962 KYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILENLNM 1008
++H++RDH QKG + + +V T++Q ADIFTKPL E F + LN+
Sbjct: 1459 RHHFLRDHQQKGDIEVFHVSTENQLADIFTKPLDEKTFCRLRSELNV 1505
>UniRef100_Q8W0X4 Putative pol protein [Zea mays]
Length = 1657
Score = 775 bits (2001), Expect = 0.0
Identities = 405/824 (49%), Positives = 536/824 (64%), Gaps = 42/824 (5%)
Query: 222 RHKDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNSQGISHNFSCPRTP 281
+ K ET F+ + Q EF+ V +RSD+G EF+N EE +GI H FS P TP
Sbjct: 832 KEKSETQGTLKRFLRRAQNEFELKVKKIRSDNGSEFKNLQVEEFLEEEGIKHEFSAPYTP 891
Query: 282 QQNGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISIRPILEKTPYELCK 341
QQNGVVERKNRTL +MARTM+ E + W EA+NTA + N + + +L+KT YEL
Sbjct: 892 QQNGVVERKNRTLIDMARTMLGEFKTPECFWTEAVNTACHAINRVYLHRLLKKTSYELLT 951
Query: 342 GRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIHQTVEES 401
+P++SYF FGS CY+L K + KF KA++ + LGY +K +R++N VE S
Sbjct: 952 DNKPNVSYFRVFGSKCYILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVS 1011
Query: 402 IQIRFDGKLGSEKSKLFE------------RFADLSI------DCSEANQPKNSS----- 438
+ FD GS + ++ + ++I + E +QP +S+
Sbjct: 1012 SDVVFDETNGSPREQVVDCDDVDEEDVPTAAIRTMAIGEVRPQEQDERDQPSSSTMVHPP 1071
Query: 439 ---EDVAPEAEASEAAPTTSDQL-----------QKKKRIAVSHPEELIIGNKDAPVRTR 484
++ P+ EA + DQ+ Q + I HP + I+G+ V TR
Sbjct: 1072 TEDDEQVPQVEALDQGGAQDDQVMEEEAQPAPPTQVRAMIQRDHPVDQILGDISKGVTTR 1131
Query: 485 SMLKPSEETLLSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGF 544
S L VS IEP V+EAL D W+L MQEEL+ F +N+VWTL+ +PK
Sbjct: 1132 SRL----VNFCEHYSFVSSIEPFRVEEALLDPDWVLAMQEELNNFKRNEVWTLVPRPKQ- 1186
Query: 545 HVIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSV 604
+V+GTKWVFRNK +E G VTRNKARLVA+GY+Q G+D+ ETF+PVARLE+IR+L++++
Sbjct: 1187 NVVGTKWVFRNKQDEHGVVTRNKARLVAKGYAQVAGLDFEETFAPVARLESIRILLAYAA 1246
Query: 605 NHNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYE 664
+H+ L+QMDVKSAFLNG I EEVYV+QPPGFED++YPDHV KL K+LYGLKQAPRAWYE
Sbjct: 1247 HHSFRLYQMDVKSAFLNGPIKEEVYVEQPPGFEDERYPDHVCKLSKALYGLKQAPRAWYE 1306
Query: 665 RLSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEF 724
L FLL N F GK D TLF T D+ + QIYVDDIIFGS N C+EFS++M +F
Sbjct: 1307 CLRDFLLANAFKVGKADPTLFTKTCDGDLFVCQIYVDDIIFGSTNQKSCEEFSRVMTQKF 1366
Query: 725 EMSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEE 784
EMSMMGEL YFLG Q+ Q T+I Q KYT +LLK+F M D +KTPM + +
Sbjct: 1367 EMSMMGELNYFLGFQVKQLKDGTFISQTKYTQDLLKRFGMKDAKPAKTPMGTDGHTDLNK 1426
Query: 785 VSSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTT 844
V QK YR MIGSLLYL ASRPDI+ SV +CARFQSDP+E HL AVKRIL+YL T
Sbjct: 1427 GGKSVDQKAYRSMIGSLLYLCASRPDIMLSVCMCARFQSDPKECHLVAVKRILRYLVATP 1486
Query: 845 NLGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAE 904
GL Y K S + L G+ D+D+AG +V+RKSTSG+C FLG +LV+W+SK+Q ++ALSTAE
Sbjct: 1487 CFGLWYPKGSTFDLVGYSDSDYAGCKVDRKSTSGTCQFLGRSLVSWNSKKQTSVALSTAE 1546
Query: 905 AEYVSAATCCTQTIWMKNHLEDYGLSLKKVPIYCDNTAAISLSKNPILHSRAKHIEVKYH 964
AEYV+A CC Q +WM+ L D+G +L KVP+ CDN + I +++NP+ HSR KHI++++H
Sbjct: 1547 AEYVAAGQCCAQLLWMRQTLRDFGYNLSKVPLLCDNESDIRMAENPVEHSRTKHIDIRHH 1606
Query: 965 YIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFLFILENLNM 1008
++RDH QKG + + +V T++Q ADIFTKPL E F + LN+
Sbjct: 1607 FLRDHQQKGDIEVFHVSTENQLADIFTKPLDEKTFCRLRSELNV 1650
>UniRef100_Q8S479 Putative pol protein [Zea mays]
Length = 1553
Score = 726 bits (1873), Expect = 0.0
Identities = 387/793 (48%), Positives = 514/793 (64%), Gaps = 23/793 (2%)
Query: 224 KDETHTIFTNFITQVQKEFQTSVITVRSDHGGEFENKAFEELFNSQGISHNFSCPRTPQQ 283
K ET F+ + Q EF+ V +RSD+G EF+N EE +GI H FS P TPQQ
Sbjct: 769 KSETQGTLKRFLRRAQNEFELKVKKIRSDNGSEFKNLQVEEYLEEEGIKHEFSTPYTPQQ 828
Query: 284 NGVVERKNRTLQEMARTMMQESSMAKHLWAEAINTAYYIHNIISIRPILEKTPYELCKGR 343
NGVVERKNRTL +MARTM+ E + W EA++TA + N + + +L+KT YEL G
Sbjct: 829 NGVVERKNRTLIDMARTMLGEFKTPERFWTEAVSTACHAINRVYLHRLLKKTSYELLTGN 888
Query: 344 QPDISYFHPFGSTCYMLNTKEQLGKFDSKALKCYFLGYSERSKGFRIYNIIHQTVEESIQ 403
+P++SYF FGS CY+L K + KF KA++ + LGY +K +R++N VE S
Sbjct: 889 KPNVSYFRVFGSKCYILVKKGRNSKFAPKAVEGFLLGYDSNTKAYRVFNKSSGLVEVSSD 948
Query: 404 IRFDGKLGSEKSKLFERFADLSIDCSEANQPKNSSE-----DVAPEAEASEAAPTTSDQL 458
+ FD GS + E+ DL D E + P + DV P+ + + P++S +
Sbjct: 949 VVFDETNGSPR----EQVVDLD-DVDEEDVPTAAIRTMAIGDVRPQEQKEQDQPSSSTMV 1003
Query: 459 QKKKRIAVS-HPEELII--GNKDAPVRTRSMLKPSEETLLSLKGLVSLIEPKSVDEALED 515
+ H EE G +D V +P+ T ++ + P VD+ L D
Sbjct: 1004 HPPTQDDEQVHQEEACDQGGAQDVHV-IEEEAQPAPPT--QVRATIQRDHP--VDQILGD 1058
Query: 516 KGWILGMQEELDQFTKNDVWTLMSKPKGFHVIGTKWVFRNKLNEKGEVTRNKARLVAQGY 575
+ + L F WTL+ +PK +V+GTKWVFRNK +E G VTRNKARLVA+G
Sbjct: 1059 ISKGVTTRSRLVNF----FWTLVPRPKQ-NVVGTKWVFRNKQDEHGVVTRNKARLVAKGC 1113
Query: 576 SQQEGIDYTETFSPVARLEAIRLLISFSVNHNITLHQMDVKSAFLNGYISEEVYVKQPPG 635
+Q G+D+ ETF+PVARLE+IR+L+++ +H+ L QMDVKSAFLNG I EEVYV+QPPG
Sbjct: 1114 AQVAGLDFEETFAPVARLESIRILLAYVAHHSFRLFQMDVKSAFLNGPIKEEVYVEQPPG 1173
Query: 636 FEDDKYPDHVYKLKKSLYGLKQAPRAWYERLSSFLLQNEFVRGKGDNTLFCITYKNDILI 695
FED++YPDHV KL K+LYGL QAPRAWYE L +FL+ N F GK D TLF T D+ +
Sbjct: 1174 FEDERYPDHVCKLSKALYGLMQAPRAWYECLRNFLIANAFKVGKADPTLFTKTCNGDLFV 1233
Query: 696 VQIYVDDIIFGSANPSLCKEFSKLMQAEFEMSMMGELKYFLGIQIDQRPGVTYIHQKKYT 755
QIYVDDIIFGS N C+EFS++M +FEM MMGEL YFLG Q+ Q T+I Q KYT
Sbjct: 1234 CQIYVDDIIFGSTNQKSCEEFSRVMTQKFEMPMMGELSYFLGFQVKQLKDGTFISQTKYT 1293
Query: 756 LELLKKFNMSDCNISKTPMHPTCILEKEEVSSKVCQKLYRGMIGSLLYLTASRPDILFSV 815
+L+K+F M D +KTPM ++ + V QK YR MIGSLLYL ASRPDI+ SV
Sbjct: 1294 QDLIKRFGMKDAKPAKTPMGTDGHIDLNKGGKSVDQKAYRSMIGSLLYLCASRPDIMLSV 1353
Query: 816 HLCARFQSDPRETHLTAVKRILKYLKGTTNLGLMYRKTSEYTLSGFCDADFAGDRVERKS 875
+CARFQSDPRE HL AVKRIL+YL T G+ Y K S + L G+ D+D+AG +V+RKS
Sbjct: 1354 CMCARFQSDPRECHLVAVKRILRYLVATPCFGIWYPKGSTFDLIGYSDSDYAGCKVDRKS 1413
Query: 876 TSGSCHFLGSNLVTWSSKRQNTIALSTAEAEYVSAATCCTQTIWMKNHLEDYGLSLKKVP 935
TSG+C FLG +LV+WSSK+Q ++ALSTAEAEYV+A CC Q +WM+ L D+G +L KVP
Sbjct: 1414 TSGTCQFLGRSLVSWSSKKQTSVALSTAEAEYVAAGQCCAQLLWMRQTLRDFGYNLNKVP 1473
Query: 936 IYCDNTAAISLSKNPILHSRAKHIEVKYHYIRDHVQKGTLSLEYVDTDHQWADIFTKPLA 995
+ CDN +AI ++ NP+ HSR KHI++++H++RDH QKG + + YV T++Q ADIFTKPL
Sbjct: 1474 LLCDNESAIRMADNPVEHSRTKHIDIRHHFLRDHQQKGDIEVFYVSTENQLADIFTKPLD 1533
Query: 996 EDRFLFILENLNM 1008
E F + +N+
Sbjct: 1534 ESTFCRLRSEINV 1546
Score = 44.3 bits (103), Expect = 0.019
Identities = 24/69 (34%), Positives = 38/69 (54%), Gaps = 5/69 (7%)
Query: 37 GDGKIPV-----INDVLLVEGLFHNLLSISQIANKGYDVIFNQTGCKAVSQTNGSVLFSG 91
G GKI + I++V LVE L +NLLS+SQ+ N GY+ + +++GS+ F G
Sbjct: 693 GLGKIAISSEHSISNVFLVESLGYNLLSMSQLCNMGYNCLITNVDVSVFRKSDGSLAFKG 752
Query: 92 KRKNYIYKI 100
+Y +
Sbjct: 753 VLDGKLYLV 761
>UniRef100_Q9SXB2 T28P6.8 protein [Arabidopsis thaliana]
Length = 1352
Score = 721 bits (1862), Expect = 0.0
Identities = 396/996 (39%), Positives = 569/996 (56%), Gaps = 40/996 (4%)
Query: 20 LALEETRREKSLEKGSI----GDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQT 75
+AL + + + KG+I +G I++V + + N+LS+ Q+ KGYD+
Sbjct: 363 VALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDN 422
Query: 76 GCKAVSQTNGSVLFSGKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKI 135
Q + + KN ++ + ++Q +K M +E W+W R GH + +
Sbjct: 423 NLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLK--MCYKEESWLWHLRFGHLNFGGL 480
Query: 136 SQLSKLNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVK 195
LS+ ++RGLP + + ++ +CE C GK K F ++ +PLEL+H D+ GP+K
Sbjct: 481 ELLSRKEMVRGLPCINHPNQ-VCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIK 539
Query: 196 TESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGG 255
+S+G Y L+ +DD+SR TWV FL+ K E IF F V+KE + T+RSD GG
Sbjct: 540 PKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGG 599
Query: 256 EFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEA 315
EF +K F + GI + PR+PQQNGVVERKNRT+ EMAR+M++ + K LWAEA
Sbjct: 600 EFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVVERKNRTILEMARSMLKSKRLPKELWAEA 659
Query: 316 INTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALK 375
+ A Y+ N + + KTP E GR+P +S+ FGS + E+ K D K+ K
Sbjct: 660 VACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEK 719
Query: 376 CYFLGYSERSKGFRIYN------IIHQTVEESIQIRFDGKLGSEKSKLFERFADLSIDCS 429
F+GY SKG+++YN II + + + +D E F F
Sbjct: 720 YIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHF-------- 771
Query: 430 EANQPKNSSEDVAPEAEASEAAPTTSDQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKP 489
E ++P+ + E+ E + TS Q+++ + + R RS+ +
Sbjct: 772 EEDEPEPTREEPPSEEPTTPPTSPTSSQIEES--------------SSERTPRFRSIQEL 817
Query: 490 SEETL----LSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFH 545
E T L+L L + EP +A+E K W M EE+ KND W L S P G
Sbjct: 818 YEVTENQENLTLFCLFAECEPMDFQKAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHK 877
Query: 546 VIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVN 605
IG KWV++ K N KGEV R KARLVA+GYSQ+ GIDY E F+PVARLE +RL+IS +
Sbjct: 878 AIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRVGIDYDEVFAPVARLETVRLIISLAAQ 937
Query: 606 HNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYER 665
+ +HQMDVKSAFLNG + EEVY++QP G+ D V +LKK LYGLKQAPRAW R
Sbjct: 938 NKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKVLYGLKQAPRAWNTR 997
Query: 666 LSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFE 725
+ + + +F++ ++ L+ K DILI +YVDD+IF NPS+ +EF K M EFE
Sbjct: 998 IDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPSIFEEFKKEMTKEFE 1057
Query: 726 MSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEV 785
M+ +G + Y+LGI++ Q +I Q+ Y E+LKKF M D N TPM L K+E
Sbjct: 1058 MTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEE 1117
Query: 786 SSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTN 845
V ++ ++GSL YLT +RPDIL++V + +R+ P TH A KRIL+Y+KGT N
Sbjct: 1118 GEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVN 1177
Query: 846 LGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEA 905
GL Y TS+Y L G+ D+D+ GD +RKSTSG ++G TW SK+Q + LST EA
Sbjct: 1178 FGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEA 1237
Query: 906 EYVSAATCCTQTIWMKNHLEDYGLSLKK-VPIYCDNTAAISLSKNPILHSRAKHIEVKYH 964
EYV+A +C IW++N L++ L ++ I+ DN +AI+L+KNP+ H R+KHI+ +YH
Sbjct: 1238 EYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYH 1297
Query: 965 YIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFL 1000
YIR+ V K + LEYV T Q AD FTKPL + F+
Sbjct: 1298 YIRECVSKKDVQLEYVKTHDQVADFFTKPLKRENFI 1333
>UniRef100_Q9M2D1 Copia-type polyprotein [Arabidopsis thaliana]
Length = 1352
Score = 720 bits (1858), Expect = 0.0
Identities = 395/996 (39%), Positives = 569/996 (56%), Gaps = 40/996 (4%)
Query: 20 LALEETRREKSLEKGSI----GDGKIPVINDVLLVEGLFHNLLSISQIANKGYDVIFNQT 75
+AL + + + KG+I +G I++V + + N+LS+ Q+ KGYD+
Sbjct: 363 VALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDN 422
Query: 76 GCKAVSQTNGSVLFSGKRKNYIYKIKSSELLSQKVKCLMSVNDEQWIWLRRLGHASLRKI 135
Q + + KN ++ + ++Q +K M +E W+W R GH + +
Sbjct: 423 NLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLK--MCYKEESWLWHLRFGHLNFGGL 480
Query: 136 SQLSKLNLIRGLPRLKYSSEALCEACQKGKFTKKPFKAKNVVSTTRPLELLHIDLFGPVK 195
LS+ ++RGLP + + ++ +CE C GK K F ++ +PLEL+H D+ GP+K
Sbjct: 481 ELLSRKEMVRGLPCINHPNQ-VCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIK 539
Query: 196 TESIGGKKYGLVIVDDYSRWTWVKFLRHKDETHTIFTNFITQVQKEFQTSVITVRSDHGG 255
+S+G Y L+ +DD+SR TWV FL+ K E IF F V+KE + T+RSD GG
Sbjct: 540 PKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGG 599
Query: 256 EFENKAFEELFNSQGISHNFSCPRTPQQNGVVERKNRTLQEMARTMMQESSMAKHLWAEA 315
EF +K F + GI + PR+PQQNGVVERKNRT+ EMAR+M++ + K LWAEA
Sbjct: 600 EFTSKEFLKYCEDNGIRRQLTVPRSPQQNGVVERKNRTILEMARSMLKSKRLPKELWAEA 659
Query: 316 INTAYYIHNIISIRPILEKTPYELCKGRQPDISYFHPFGSTCYMLNTKEQLGKFDSKALK 375
+ A Y+ N + + KTP E GR+P +S+ FGS + E+ K D K+ K
Sbjct: 660 VACAVYLLNRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEK 719
Query: 376 CYFLGYSERSKGFRIYN------IIHQTVEESIQIRFDGKLGSEKSKLFERFADLSIDCS 429
F+GY SKG+++YN II + + + +D E F F
Sbjct: 720 YIFIGYDNNSKGYKLYNPDTKKTIISRNIVFDEEGEWDWNSNEEDYNFFPHF-------- 771
Query: 430 EANQPKNSSEDVAPEAEASEAAPTTSDQLQKKKRIAVSHPEELIIGNKDAPVRTRSMLKP 489
E ++P+ + E+ E + TS Q+++ + + R RS+ +
Sbjct: 772 EEDEPEPTREEPPSEEPTTPPTSPTSSQIEES--------------SSERTPRFRSIQEL 817
Query: 490 SEETL----LSLKGLVSLIEPKSVDEALEDKGWILGMQEELDQFTKNDVWTLMSKPKGFH 545
E T L+L L + EP +A+E K W M EE+ KND W L S P G
Sbjct: 818 YEVTENQENLTLFCLFAECEPMDFQKAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHK 877
Query: 546 VIGTKWVFRNKLNEKGEVTRNKARLVAQGYSQQEGIDYTETFSPVARLEAIRLLISFSVN 605
IG KWV++ K N KGEV R KARLVA+GYSQ+ GIDY E F+PVARLE +RL+IS +
Sbjct: 878 AIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRVGIDYDEVFAPVARLETVRLIISLAAQ 937
Query: 606 HNITLHQMDVKSAFLNGYISEEVYVKQPPGFEDDKYPDHVYKLKKSLYGLKQAPRAWYER 665
+ +HQMDVKSAFLNG + EEVY++QP G+ D V +LKK LYGLKQAPRAW R
Sbjct: 938 NKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLKKVLYGLKQAPRAWNTR 997
Query: 666 LSSFLLQNEFVRGKGDNTLFCITYKNDILIVQIYVDDIIFGSANPSLCKEFSKLMQAEFE 725
+ + + +F++ ++ L+ K DILI +YVDD+IF NPS+ +EF K M EFE
Sbjct: 998 IDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTGNNPSIFEEFKKEMTKEFE 1057
Query: 726 MSMMGELKYFLGIQIDQRPGVTYIHQKKYTLELLKKFNMSDCNISKTPMHPTCILEKEEV 785
M+ +G + Y+LGI++ Q +I Q+ Y E+LKKF + D N TPM L K+E
Sbjct: 1058 MTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKIDDSNPVCTPMECGIKLSKKEE 1117
Query: 786 SSKVCQKLYRGMIGSLLYLTASRPDILFSVHLCARFQSDPRETHLTAVKRILKYLKGTTN 845
V ++ ++GSL YLT +RPDIL++V + +R+ P TH A KRIL+Y+KGT N
Sbjct: 1118 GEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVN 1177
Query: 846 LGLMYRKTSEYTLSGFCDADFAGDRVERKSTSGSCHFLGSNLVTWSSKRQNTIALSTAEA 905
GL Y TS+Y L G+ D+D+ GD +RKSTSG ++G TW SK+Q + LST EA
Sbjct: 1178 FGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEA 1237
Query: 906 EYVSAATCCTQTIWMKNHLEDYGLSLKK-VPIYCDNTAAISLSKNPILHSRAKHIEVKYH 964
EYV+A +C IW++N L++ L ++ I+ DN +AI+L+KNP+ H R+KHI+ +YH
Sbjct: 1238 EYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYH 1297
Query: 965 YIRDHVQKGTLSLEYVDTDHQWADIFTKPLAEDRFL 1000
YIR+ V K + LEYV T Q AD FTKPL + F+
Sbjct: 1298 YIRECVSKKDVQLEYVKTHDQVADFFTKPLKRENFI 1333
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.135 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,691,515,813
Number of Sequences: 2790947
Number of extensions: 72000822
Number of successful extensions: 183201
Number of sequences better than 10.0: 4671
Number of HSP's better than 10.0 without gapping: 2814
Number of HSP's successfully gapped in prelim test: 1857
Number of HSP's that attempted gapping in prelim test: 174995
Number of HSP's gapped (non-prelim): 6137
length of query: 1017
length of database: 848,049,833
effective HSP length: 138
effective length of query: 879
effective length of database: 462,899,147
effective search space: 406888350213
effective search space used: 406888350213
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0166.5


