

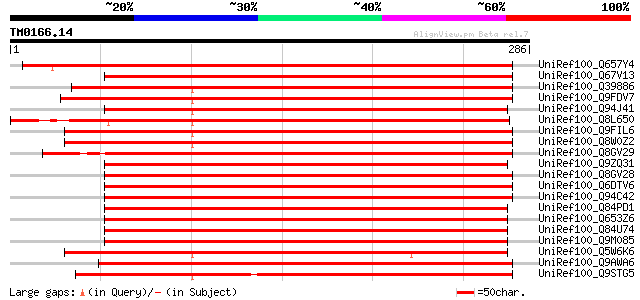
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0166.14
(286 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q657Y4 Hypothetical protein P0005A05.28 [Oryza sativa] 425 e-118
UniRef100_Q67V13 Hypothetical protein OSJNBa0019I19.51 [Oryza sa... 306 5e-82
UniRef100_Q39886 Protein kinase [Glycine max] 288 1e-76
UniRef100_Q9FDV7 Protein kinase [Fagus sylvatica] 286 4e-76
UniRef100_Q94J41 Hypothetical protein P0481E12.7 [Oryza sativa] 282 8e-75
UniRef100_Q8L650 Hypothetical protein B1015E06.24 [Oryza sativa] 272 8e-72
UniRef100_Q9FIL6 Protein-tyrosine kinase [Arabidopsis thaliana] 270 3e-71
UniRef100_Q8W0Z2 AT5g58950/k19m22_150 [Arabidopsis thaliana] 268 9e-71
UniRef100_Q8GV29 Serine/threonine protein kinase [Oryza sativa] 262 7e-69
UniRef100_Q9ZQ31 Hypothetical protein At2g24360 [Arabidopsis tha... 258 1e-67
UniRef100_Q8GV28 Serine/threonine protein kinase [Oryza sativa] 256 6e-67
UniRef100_Q6DTV6 Serine/threonine and tyrosine protein kinase [O... 256 6e-67
UniRef100_Q94C42 Serine/threonine protein kinase [Triticum aesti... 254 2e-66
UniRef100_Q84PD1 Serine/thronine protein kinase-like protein [Or... 252 7e-66
UniRef100_Q653Z6 Serine/threonine protein kinase [Oryza sativa] 252 7e-66
UniRef100_Q84U74 Serine/threonine protein kinase [Oryza sativa] 251 2e-65
UniRef100_Q9M085 Protein kinase-like protein [Arabidopsis thaliana] 249 6e-65
UniRef100_Q5W6K6 Kinase domain containing protein [Oryza sativa] 248 2e-64
UniRef100_Q9AWA6 Serine/threonine/tyrosine kinase [Arachis hypog... 241 2e-62
UniRef100_Q9STG5 Protein kinase 6-like protein [Arabidopsis thal... 215 1e-54
>UniRef100_Q657Y4 Hypothetical protein P0005A05.28 [Oryza sativa]
Length = 376
Score = 425 bits (1092), Expect = e-118
Identities = 213/275 (77%), Positives = 233/275 (84%), Gaps = 5/275 (1%)
Query: 8 KQIWSSGGGGGGKAG----RRLSLGEYKRAVSWSKYLVSP-GAAIKGEGEEEWSADMSQL 62
KQ GGG GG G RRLSLGEYK+AVSWSKYLV+P GA I+G GEE WSAD+S+L
Sbjct: 5 KQQQQQGGGNGGGQGKRLERRLSLGEYKKAVSWSKYLVAPPGAKIRGGGEELWSADLSKL 64
Query: 63 LIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVSLLLPLRHPN 122
I KFA+GRHSR+Y G Y RDVAIK+VSQPEED LA LE+QF SEV+LLL LRHPN
Sbjct: 65 EIRTKFATGRHSRVYSGRYAARDVAIKMVSQPEEDAALAAELERQFASEVALLLRLRHPN 124
Query: 123 IITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQ 182
II+FVAACKKPPVFCIITEY+AGGSLRKYLH QEPHSVP+ LVLKL+L+IARGM YLHSQ
Sbjct: 125 IISFVAACKKPPVFCIITEYMAGGSLRKYLHQQEPHSVPIELVLKLSLEIARGMSYLHSQ 184
Query: 183 GIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPEMIKEKHHTKK 242
GI+HRDLKSEN+LL DM VKV DFGISCLESQCGS KGFTGTYRWMAPEMIKEKHHT+K
Sbjct: 185 GILHRDLKSENILLDGDMSVKVADFGISCLESQCGSGKGFTGTYRWMAPEMIKEKHHTRK 244
Query: 243 VDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
VDVYSFGIVLWE+LT L PF MTPEQAA AV+ K
Sbjct: 245 VDVYSFGIVLWEILTALVPFSEMTPEQAAVAVALK 279
>UniRef100_Q67V13 Hypothetical protein OSJNBa0019I19.51 [Oryza sativa]
Length = 398
Score = 306 bits (783), Expect = 5e-82
Identities = 147/225 (65%), Positives = 178/225 (78%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW AD+S L IG KFASG +SRIY+G+YK+R VA+K+V PE DE LE QF SEV
Sbjct: 91 EEWMADLSHLFIGNKFASGANSRIYRGIYKQRAVAVKMVRIPERDEARRAVLEDQFNSEV 150
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
+ L L HPNI+ F+AACKKPPV+CIITEY++ G+LR YL+ ++P+S+ +LKLALDI
Sbjct: 151 AFLSRLYHPNIVQFIAACKKPPVYCIITEYMSQGTLRMYLNKKDPYSLSSETILKLALDI 210
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
+RGM+YLH+QG+IHRDLKS+NLLL ++M VKV DFG SCLE+ C + KG GTYRWMAPE
Sbjct: 211 SRGMEYLHAQGVIHRDLKSQNLLLNDEMRVKVADFGTSCLETACQATKGNKGTYRWMAPE 270
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
M KEK +T+KVDVYSFGIVLWEL T L PF MTP QAAYA S K
Sbjct: 271 MTKEKPYTRKVDVYSFGIVLWELTTCLLPFQGMTPVQAAYAASEK 315
>UniRef100_Q39886 Protein kinase [Glycine max]
Length = 462
Score = 288 bits (737), Expect = 1e-76
Identities = 142/246 (57%), Positives = 179/246 (72%), Gaps = 3/246 (1%)
Query: 35 SWSKYLVSPGAAIKG-EGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQ 93
+W+K L + G I E EEW+ DMSQL G+KFA G HSR+Y GVYK+ VA+K++
Sbjct: 128 AWTKLLDNGGGKITAVETAEEWNVDMSQLFFGLKFAHGAHSRLYHGVYKDEAVAVKIIMV 187
Query: 94 PEEDED--LACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKY 151
PE+D + LA LEKQF EV+LL L H N+I F AAC+KPPV+CIITEYLA GSLR Y
Sbjct: 188 PEDDGNGALASRLEKQFIREVTLLSRLHHQNVIKFSAACRKPPVYCIITEYLAEGSLRAY 247
Query: 152 LHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISC 211
LH E ++ L ++ ALDIARGM+Y+HSQG+IHRDLK EN+L+ ED +K+ DFGI+C
Sbjct: 248 LHKLEHQTISLQKLIAFALDIARGMEYIHSQGVIHRDLKPENILINEDNHLKIADFGIAC 307
Query: 212 LESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAA 271
E+ C GTYRWMAPEMIK K + KKVDVYSFG++LWE+LTG P+++M P QAA
Sbjct: 308 EEASCDLLADDPGTYRWMAPEMIKRKSYGKKVDVYSFGLILWEMLTGTIPYEDMNPIQAA 367
Query: 272 YAVSYK 277
+AV K
Sbjct: 368 FAVVNK 373
>UniRef100_Q9FDV7 Protein kinase [Fagus sylvatica]
Length = 480
Score = 286 bits (732), Expect = 4e-76
Identities = 134/252 (53%), Positives = 183/252 (72%), Gaps = 3/252 (1%)
Query: 29 EYKRAVSWSKYLVSPGAAIKG-EGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVA 87
+Y++ SW+KY G + E +EW+ D+S+L +G+KFA G HSR+Y G+Y + VA
Sbjct: 140 KYRKESSWAKYFDHGGGRVNAVETSDEWTVDLSKLFLGLKFAHGAHSRLYHGIYNDEPVA 199
Query: 88 IKLVSQPEEDED--LACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAG 145
+K++ PE+DE+ L LEKQF EV+LL L NII FVAAC+KPPV+C++TEYL+
Sbjct: 200 VKIIRVPEDDENGALGARLEKQFNREVTLLSRLHFHNIIKFVAACRKPPVYCVVTEYLSE 259
Query: 146 GSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVV 205
GSLR YLH E S+PL ++ ALDIARGM+Y+HSQG+IHRDLK EN+L+ ++ +K+
Sbjct: 260 GSLRAYLHKLERKSLPLQKLIAFALDIARGMEYIHSQGVIHRDLKPENVLIDQEFHLKIA 319
Query: 206 DFGISCLESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNM 265
DFGI+C E+ C S GTYRWMAPEMIK K + +KVDVYSFG++LWE++ G P+++M
Sbjct: 320 DFGIACEEAYCDSLADDPGTYRWMAPEMIKHKSYGRKVDVYSFGLILWEMVAGTIPYEDM 379
Query: 266 TPEQAAYAVSYK 277
P QAA+AV K
Sbjct: 380 NPVQAAFAVVNK 391
>UniRef100_Q94J41 Hypothetical protein P0481E12.7 [Oryza sativa]
Length = 637
Score = 282 bits (721), Expect = 8e-75
Identities = 125/224 (55%), Positives = 171/224 (75%), Gaps = 2/224 (0%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDED--LACFLEKQFTS 110
E+W+ D SQLLIG +FASG HSR++ G+YKE+ VA+K + QPE++ED LA LEKQF +
Sbjct: 318 EKWTVDRSQLLIGHRFASGAHSRLFHGIYKEQPVAVKFIRQPEDEEDAELAAQLEKQFNT 377
Query: 111 EVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLAL 170
EV+ L L HPN+I + AC PPVFC+ITE+L+GGSLR +LH QE S+PL ++ + L
Sbjct: 378 EVTTLSRLNHPNVIKLIGACSSPPVFCVITEFLSGGSLRTFLHKQEHKSLPLEKIISIGL 437
Query: 171 DIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMA 230
DIA G+ Y+HSQG++HRD+K EN++ + C K+VDFGISC E++C TGT+RWMA
Sbjct: 438 DIANGIGYIHSQGVVHRDVKPENIIFDSEFCAKIVDFGISCEEAECDPLANDTGTFRWMA 497
Query: 231 PEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAV 274
PEM+K K + +KVDVYSFG++LWE+ TG P++++ P QAA+AV
Sbjct: 498 PEMMKHKPYGRKVDVYSFGLILWEMFTGSVPYEDLNPFQAAFAV 541
>UniRef100_Q8L650 Hypothetical protein B1015E06.24 [Oryza sativa]
Length = 563
Score = 272 bits (695), Expect = 8e-72
Identities = 137/279 (49%), Positives = 195/279 (69%), Gaps = 16/279 (5%)
Query: 1 MKNSYWHKQIWSSGGGGGGKAGRRLSLGEYKRAVSWSKYLVSPGAAIKGEGEE--EWSAD 58
+K+S W ++ + +GG RR+S AV ++ + G ++ + +W+ D
Sbjct: 215 LKDSSWTRRYFDNGGR------RRVS------AVDATEVRRNRGVSMAQAVQTTVDWTLD 262
Query: 59 MSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDED--LACFLEKQFTSEVSLLL 116
S+LL+G KFASG +SR+YKG+Y ++ VAIK + QP++D++ +A LEKQ+ SEV+ L
Sbjct: 263 PSKLLVGHKFASGAYSRLYKGLYDDKPVAIKFIRQPDDDDNGKMAAKLEKQYNSEVNALS 322
Query: 117 PLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIARGM 176
L H N+I VAA K PPVF IITE+L GGSLR YL+ E H +PL ++ +ALD+A G+
Sbjct: 323 HLYHKNVIKLVAAYKCPPVFYIITEFLPGGSLRSYLNSTEHHPIPLEKIISIALDVACGL 382
Query: 177 QYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPEMIKE 236
+Y+HSQG++HRD+K EN+L E+ CVK+ DFGI+C ES C GTYRWMAPEMIK
Sbjct: 383 EYIHSQGVVHRDIKPENILFDENFCVKIADFGIACEESMCDVLVEDEGTYRWMAPEMIKR 442
Query: 237 KHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVS 275
K + +KVDVYSFG++LWE+++G PFD++TP QAAYAV+
Sbjct: 443 KAYNRKVDVYSFGLLLWEMISGRIPFDDLTPLQAAYAVA 481
>UniRef100_Q9FIL6 Protein-tyrosine kinase [Arabidopsis thaliana]
Length = 525
Score = 270 bits (690), Expect = 3e-71
Identities = 129/250 (51%), Positives = 177/250 (70%), Gaps = 3/250 (1%)
Query: 31 KRAVSWSKYLVSPGAAIKG-EGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIK 89
K+ WSK + G + E EE+ DMS+L G+KFA G +SR+Y G Y+++ VA+K
Sbjct: 175 KKDTGWSKLFDNTGRRVSAVEASEEFRVDMSKLFFGLKFAHGLYSRLYHGKYEDKAVAVK 234
Query: 90 LVSQPEEDED--LACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGS 147
L++ P++D++ L LEKQFT EV+LL L HPN+I FV A K PPV+C++T+YL GS
Sbjct: 235 LITVPDDDDNGCLGARLEKQFTKEVTLLSRLTHPNVIKFVGAYKDPPVYCVLTQYLPEGS 294
Query: 148 LRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDF 207
LR +LH E S+PL +++ A+DIARGM+Y+HS+ IIHRDLK EN+L+ E+ +K+ DF
Sbjct: 295 LRSFLHKPENRSLPLKKLIEFAIDIARGMEYIHSRRIIHRDLKPENVLIDEEFHLKIADF 354
Query: 208 GISCLESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTP 267
GI+C E C GTYRWMAPEMIK K H +K DVYSFG+VLWE++ G P+++M P
Sbjct: 355 GIACEEEYCDMLADDPGTYRWMAPEMIKRKPHGRKADVYSFGLVLWEMVAGAIPYEDMNP 414
Query: 268 EQAAYAVSYK 277
QAA+AV +K
Sbjct: 415 IQAAFAVVHK 424
>UniRef100_Q8W0Z2 AT5g58950/k19m22_150 [Arabidopsis thaliana]
Length = 525
Score = 268 bits (686), Expect = 9e-71
Identities = 128/250 (51%), Positives = 176/250 (70%), Gaps = 3/250 (1%)
Query: 31 KRAVSWSKYLVSPGAAIKG-EGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIK 89
K+ WSK + G + E EE+ DMS+L G+KFA G +SR+Y G Y+++ VA+K
Sbjct: 175 KKDTGWSKLFDNTGRRVSAVEASEEFRVDMSKLFFGLKFAHGLYSRLYHGKYEDKAVAVK 234
Query: 90 LVSQPEEDED--LACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGS 147
L++ P++D++ L LEKQFT EV+LL L HPN+I FV A K PPV+C++T+YL GS
Sbjct: 235 LITVPDDDDNGCLGARLEKQFTKEVTLLSRLTHPNVIKFVGAYKDPPVYCVLTQYLPEGS 294
Query: 148 LRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDF 207
LR +LH E S+PL +++ +DIARGM+Y+HS+ IIHRDLK EN+L+ E+ +K+ DF
Sbjct: 295 LRSFLHKPENRSLPLKKLIEFVIDIARGMEYIHSRRIIHRDLKPENVLIDEEFHLKIADF 354
Query: 208 GISCLESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTP 267
GI+C E C GTYRWMAPEMIK K H +K DVYSFG+VLWE++ G P+++M P
Sbjct: 355 GIACEEEYCDMLADDPGTYRWMAPEMIKRKPHGRKADVYSFGLVLWEMVAGAIPYEDMNP 414
Query: 268 EQAAYAVSYK 277
QAA+AV +K
Sbjct: 415 IQAAFAVVHK 424
>UniRef100_Q8GV29 Serine/threonine protein kinase [Oryza sativa]
Length = 422
Score = 262 bits (670), Expect = 7e-69
Identities = 131/259 (50%), Positives = 176/259 (67%), Gaps = 5/259 (1%)
Query: 19 GKAGRRLSLGEYKRAVSWSKYLVSPGAAIKGEGEEEWSADMSQLLIGMKFASGRHSRIYK 78
G+ LS RA+ +Y P +K EEW+ D+ +L +GM FA G ++YK
Sbjct: 103 GRVSHALSEDALARALMDPRY---PTETLKDY--EEWTIDLGKLHMGMPFAQGAFGKLYK 157
Query: 79 GVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCI 138
G Y DVAIKL+ +PE D + A +E+QF EV +L LRHPNI+ F+ AC+KP V+CI
Sbjct: 158 GTYNGEDVAIKLLERPEADPERAGLMEQQFVQEVMMLATLRHPNIVKFIGACRKPMVWCI 217
Query: 139 ITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGE 198
+TEY GGS+R++L ++ SVPL L +K ALD+ARGM Y+H+ G IHRDLKS+NLL+
Sbjct: 218 VTEYAKGGSVRQFLMKRQNRSVPLKLAVKQALDVARGMAYVHALGFIHRDLKSDNLLISG 277
Query: 199 DMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTG 258
D +K+ DFG++ +E + TGTYRWMAPEMI+ + + +KVDVYSFGIVLWEL+TG
Sbjct: 278 DKSIKIADFGVARIEVKTEGMTPETGTYRWMAPEMIQHRPYDQKVDVYSFGIVLWELITG 337
Query: 259 LTPFDNMTPEQAAYAVSYK 277
+ PF NMT QAA+AV K
Sbjct: 338 MLPFANMTAVQAAFAVVNK 356
>UniRef100_Q9ZQ31 Hypothetical protein At2g24360 [Arabidopsis thaliana]
Length = 411
Score = 258 bits (659), Expect = 1e-67
Identities = 119/222 (53%), Positives = 162/222 (72%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
+EW+ D+ +L +G FA G ++YKG Y DVAIK++ +PE + A F+E+QF EV
Sbjct: 121 DEWTIDLRKLNMGPAFAQGAFGKLYKGTYNGEDVAIKILERPENSPEKAQFMEQQFQQEV 180
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
S+L L+HPNI+ F+ AC+KP V+CI+TEY GGS+R++L ++ +VPL L +K ALD+
Sbjct: 181 SMLANLKHPNIVRFIGACRKPMVWCIVTEYAKGGSVRQFLTRRQNRAVPLKLAVKQALDV 240
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H + IHRDLKS+NLL+ D +K+ DFG++ +E Q TGTYRWMAPE
Sbjct: 241 ARGMAYVHGRNFIHRDLKSDNLLISADKSIKIADFGVARIEVQTEGMTPETGTYRWMAPE 300
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAV 274
MI+ + + +KVDVYSFGIVLWEL+TGL PF NMT QAA+AV
Sbjct: 301 MIQHRAYNQKVDVYSFGIVLWELITGLLPFQNMTAVQAAFAV 342
>UniRef100_Q8GV28 Serine/threonine protein kinase [Oryza sativa]
Length = 421
Score = 256 bits (653), Expect = 6e-67
Identities = 123/225 (54%), Positives = 162/225 (71%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW+ D+ +L +G+ FA G ++Y+G Y DVAIKL+ +PE D + A LE+QF EV
Sbjct: 131 EEWAIDLGRLDMGVPFAQGAFGKLYRGTYNGEDVAIKLLEKPENDPERAQALEQQFVQEV 190
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
+L LRHPNI+ F+ AC+K V+CIITEY GGS+R++L ++ SVPL L +K ALDI
Sbjct: 191 MMLSRLRHPNIVRFIGACRKSIVWCIITEYAKGGSVRQFLARRQNKSVPLRLAVKQALDI 250
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H+ G IHRDLKS+NLL+ D +K+ DFG++ +E + TGTYRWMAPE
Sbjct: 251 ARGMAYVHALGFIHRDLKSDNLLIAADKSIKIADFGVARIEVKTEGMTPETGTYRWMAPE 310
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
MI+ + + KVDVYSFGIVLWEL+TG+ PF NMT QAA+AV K
Sbjct: 311 MIQHRPYDHKVDVYSFGIVLWELITGMLPFTNMTAVQAAFAVVNK 355
>UniRef100_Q6DTV6 Serine/threonine and tyrosine protein kinase [Oryza sativa]
Length = 417
Score = 256 bits (653), Expect = 6e-67
Identities = 123/225 (54%), Positives = 161/225 (70%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW+ D+ +L IGM FA G ++Y+G Y DVAIKL+ +PE D + A LE+QF EV
Sbjct: 127 EEWTIDLGKLHIGMPFAQGAFGKLYRGTYNGGDVAIKLLERPEADPEKAQLLEQQFVQEV 186
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
+L LRH NI+ FV AC+KP V+CI+TEY GGS+R +L+ ++ SVPL L +K ALD+
Sbjct: 187 MMLATLRHSNIVKFVGACRKPMVWCIVTEYAKGGSVRNFLNRRQNRSVPLKLAVKQALDV 246
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H G IHRDLKS+NLL+ D +K+ DFG++ +E + TGTYRWMAPE
Sbjct: 247 ARGMAYVHGLGFIHRDLKSDNLLISGDKSIKIADFGVARIEVKTEGMTPETGTYRWMAPE 306
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
+I+ + + +KVDVYSFGIVLWEL+TG PF NMT QAA+AV K
Sbjct: 307 VIQHRPYDQKVDVYSFGIVLWELVTGNLPFANMTAVQAAFAVVNK 351
>UniRef100_Q94C42 Serine/threonine protein kinase [Triticum aestivum]
Length = 416
Score = 254 bits (649), Expect = 2e-66
Identities = 121/225 (53%), Positives = 160/225 (70%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW+ D+ +L +G+ FA G ++Y+G Y DVAIKL+ +PE D A LE+QF EV
Sbjct: 126 EEWTIDLGKLHMGLPFAQGAFGKLYRGTYNGMDVAIKLLERPEADPAQAQLLEQQFVQEV 185
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
+L LRHPNI+ FV AC+KP V+CI+T Y GGS+R +L+ ++ SVPL L +K ALD+
Sbjct: 186 MMLAELRHPNIVKFVGACRKPIVWCIVTGYAKGGSVRNFLNRRQNRSVPLKLAVKQALDV 245
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H G IHRDLKS+NLL+ D +K+ DFG++ +E + TGTYRWMAPE
Sbjct: 246 ARGMAYVHGLGFIHRDLKSDNLLISGDKSIKIADFGVARIEVKTEGMTPETGTYRWMAPE 305
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
MI+ + + +KVDVYSFGIVLWEL++G PF NMT QAA+AV K
Sbjct: 306 MIQHRPYNQKVDVYSFGIVLWELISGTLPFPNMTAVQAAFAVVNK 350
>UniRef100_Q84PD1 Serine/thronine protein kinase-like protein [Oryza sativa]
Length = 361
Score = 252 bits (644), Expect = 7e-66
Identities = 120/222 (54%), Positives = 159/222 (71%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW+ D+ +L +G FA G ++Y+G Y DVAIKL+ +PE D + A +E+QF EV
Sbjct: 71 EEWTIDLGKLDMGAPFAQGAFGKLYRGTYNGEDVAIKLLEKPENDPERAQLMEQQFVQEV 130
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
+L LRHPNI+ F+ AC+K V+CIITEY GGS+R++L ++ SVPL L +K ALD+
Sbjct: 131 MMLSTLRHPNIVRFIGACRKSIVWCIITEYAKGGSVRQFLARRQNKSVPLGLAVKQALDV 190
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H+ IHRDLKS+NLL+ D +K+ DFG++ +E Q TGTYRWMAPE
Sbjct: 191 ARGMAYVHALRFIHRDLKSDNLLISADKSIKIADFGVARIEVQTEGMTPETGTYRWMAPE 250
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAV 274
MI+ + + KVDVYSFGIVLWEL+TG+ PF NMT QAA+AV
Sbjct: 251 MIQHRPYDHKVDVYSFGIVLWELITGMLPFTNMTAVQAAFAV 292
>UniRef100_Q653Z6 Serine/threonine protein kinase [Oryza sativa]
Length = 428
Score = 252 bits (644), Expect = 7e-66
Identities = 120/222 (54%), Positives = 159/222 (71%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW+ D+ +L +G FA G ++Y+G Y DVAIKL+ +PE D + A +E+QF EV
Sbjct: 138 EEWTIDLGKLDMGAPFAQGAFGKLYRGTYNGEDVAIKLLEKPENDPERAQLMEQQFVQEV 197
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
+L LRHPNI+ F+ AC+K V+CIITEY GGS+R++L ++ SVPL L +K ALD+
Sbjct: 198 MMLSTLRHPNIVRFIGACRKSIVWCIITEYAKGGSVRQFLARRQNKSVPLGLAVKQALDV 257
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H+ IHRDLKS+NLL+ D +K+ DFG++ +E Q TGTYRWMAPE
Sbjct: 258 ARGMAYVHALRFIHRDLKSDNLLISADKSIKIADFGVARIEVQTEGMTPETGTYRWMAPE 317
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAV 274
MI+ + + KVDVYSFGIVLWEL+TG+ PF NMT QAA+AV
Sbjct: 318 MIQHRPYDHKVDVYSFGIVLWELITGMLPFTNMTAVQAAFAV 359
>UniRef100_Q84U74 Serine/threonine protein kinase [Oryza sativa]
Length = 428
Score = 251 bits (640), Expect = 2e-65
Identities = 119/222 (53%), Positives = 158/222 (70%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW+ D+ +L +G FA G ++Y+G Y DVAIKL+ +PE D + A +E+QF EV
Sbjct: 138 EEWTIDLGKLDMGAPFAQGAFGKLYRGTYNGEDVAIKLLEKPENDPERAQLMEQQFVQEV 197
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
+L LRHPNI+ F+ AC+K V+CIITEY GGS+R++L ++ SVPL L +K ALD+
Sbjct: 198 MMLSTLRHPNIVRFIGACRKSIVWCIITEYAKGGSVRQFLARRQNKSVPLGLAVKQALDV 257
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H+ IHRDLKS+NLL+ D +K+ DFG++ +E Q TGTYRWMAPE
Sbjct: 258 ARGMAYVHALRFIHRDLKSDNLLISADKSIKIADFGVARIEVQTEGMTPETGTYRWMAPE 317
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAV 274
MI+ + + KVDVY FGIVLWEL+TG+ PF NMT QAA+AV
Sbjct: 318 MIQHRPYDHKVDVYGFGIVLWELITGMLPFTNMTAVQAAFAV 359
>UniRef100_Q9M085 Protein kinase-like protein [Arabidopsis thaliana]
Length = 412
Score = 249 bits (636), Expect = 6e-65
Identities = 118/222 (53%), Positives = 160/222 (71%)
Query: 53 EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFTSEV 112
EEW+ D+ +L +G FA G ++Y+G Y DVAIKL+ + + + + A LE+QF EV
Sbjct: 122 EEWTIDLRKLHMGPAFAQGAFGKLYRGTYNGEDVAIKLLERSDSNPEKAQALEQQFQQEV 181
Query: 113 SLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLALDI 172
S+L L+HPNI+ F+ AC KP V+CI+TEY GGS+R++L ++ +VPL L + ALD+
Sbjct: 182 SMLAFLKHPNIVRFIGACIKPMVWCIVTEYAKGGSVRQFLTKRQNRAVPLKLAVMQALDV 241
Query: 173 ARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWMAPE 232
ARGM Y+H + IHRDLKS+NLL+ D +K+ DFG++ +E Q TGTYRWMAPE
Sbjct: 242 ARGMAYVHERNFIHRDLKSDNLLISADRSIKIADFGVARIEVQTEGMTPETGTYRWMAPE 301
Query: 233 MIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAV 274
MI+ + +T+KVDVYSFGIVLWEL+TGL PF NMT QAA+AV
Sbjct: 302 MIQHRPYTQKVDVYSFGIVLWELITGLLPFQNMTAVQAAFAV 343
>UniRef100_Q5W6K6 Kinase domain containing protein [Oryza sativa]
Length = 351
Score = 248 bits (632), Expect = 2e-64
Identities = 125/251 (49%), Positives = 172/251 (67%), Gaps = 7/251 (2%)
Query: 31 KRAVSWSKYLVSPGAAIKGEG-EEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIK 89
K+ +W K L+ P + E W+AD S+LL+G K ASG +SRI++G+Y E+ VA+K
Sbjct: 7 KKHSAWLKLLLGPKKPARSAAIVETWAADRSKLLLGPKIASGSNSRIHRGMYGEQPVAVK 66
Query: 90 LVSQPEEDED----LACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAG 145
++ P D+D + +E QF +EVSLL LRHPN++ V C++P V+ IITE +
Sbjct: 67 IMHAPVGDDDDDVQVRREMEAQFDAEVSLLSRLRHPNVVRLVGVCREPEVYWIITELMRR 126
Query: 146 GSLRKYLHHQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVV 205
G+L YLH +EP+S+P +++LALD+ARGM+YLH++G++HRDLK ENL+L VKV
Sbjct: 127 GTLSAYLHGREPYSLPPETIVRLALDVARGMEYLHARGVVHRDLKPENLMLDGGGRVKVA 186
Query: 206 DFGISCLESQCGSAK--GFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFD 263
D G SCLE+ C K GT+RWMAPEMI +K +KVDVYSFG+VLWEL T L PF
Sbjct: 187 DLGTSCLEATCRGDKCSSKAGTFRWMAPEMIHDKRCNRKVDVYSFGLVLWELTTCLVPFQ 246
Query: 264 NMTPEQAAYAV 274
N++P Q AY+V
Sbjct: 247 NLSPVQVAYSV 257
>UniRef100_Q9AWA6 Serine/threonine/tyrosine kinase [Arachis hypogaea]
Length = 411
Score = 241 bits (615), Expect = 2e-62
Identities = 116/228 (50%), Positives = 159/228 (68%)
Query: 50 EGEEEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPEEDEDLACFLEKQFT 109
E +EW+ D+ +L +G FA G ++Y+G Y DVAIK++ +PE + A +E+QF
Sbjct: 121 ENFDEWTIDLRKLNMGEAFAQGAFGKLYRGTYNGEDVAIKILERPENELSKAQLMEQQFQ 180
Query: 110 SEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLHHQEPHSVPLHLVLKLA 169
EV +L L+HPNI+ F+ AC+KP V+CI+TEY GGS+R+ L ++ SVPL L +K A
Sbjct: 181 QEVMMLATLKHPNIVRFIGACRKPMVWCIVTEYAKGGSVRQSLMKRQNRSVPLKLAVKQA 240
Query: 170 LDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLESQCGSAKGFTGTYRWM 229
LD+ARGM Y+ G+IHRDLKS+NLL+ +K+ DFG++ +E Q TGTYRWM
Sbjct: 241 LDVARGMAYVPWLGLIHRDLKSDNLLIFGAKSIKIADFGVAGIEVQTEGMTPETGTYRWM 300
Query: 230 APEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMTPEQAAYAVSYK 277
APEMI+ + +T+KVDVYSFGIVLWEL+ G+ PF NM QAA+AV K
Sbjct: 301 APEMIQHRPYTQKVDVYSFGIVLWELIPGMLPFQNMPAVQAAFAVVTK 348
>UniRef100_Q9STG5 Protein kinase 6-like protein [Arabidopsis thaliana]
Length = 475
Score = 215 bits (547), Expect = 1e-54
Identities = 113/245 (46%), Positives = 158/245 (64%), Gaps = 7/245 (2%)
Query: 37 SKYLVSPGAAIKGEGE-EEWSADMSQLLIGMKFASGRHSRIYKGVYKERDVAIKLVSQPE 95
SK + G+ + G EE D+S+L G +FA G++S+IY G Y+ + VA+K+++ PE
Sbjct: 135 SKSVDYRGSKVSSAGVLEECLIDVSKLSYGDRFAHGKYSQIYHGEYEGKAVALKIITAPE 194
Query: 96 EDED--LACFLEKQFTSEVSLLLPLRHPNIITFVAACKKPPVFCIITEYLAGGSLRKYLH 153
+ +D L LEK+F E +LL L HPN++ FV CIITEY+ GSLR YLH
Sbjct: 195 DSDDIFLGARLEKEFIVEATLLSRLSHPNVVKFVGVNTGN---CIITEYVPRGSLRSYLH 251
Query: 154 HQEPHSVPLHLVLKLALDIARGMQYLHSQGIIHRDLKSENLLLGEDMCVKVVDFGISCLE 213
E S+PL ++ LDIA+GM+Y+HS+ I+H+DLK EN+L+ D +K+ DFGI+C E
Sbjct: 252 KLEQKSLPLEQLIDFGLDIAKGMEYIHSREIVHQDLKPENVLIDNDFHLKIADFGIACEE 311
Query: 214 SQCGSAKGFTGTYRWMAPEMIKEKHHTKKVDVYSFGIVLWELLTGLTPFDNMT-PEQAAY 272
C GTYRWMAPE++K H +K DVYSFG++LWE++ G P++ M EQ AY
Sbjct: 312 EYCDVLGDNIGTYRWMAPEVLKRIPHGRKCDVYSFGLLLWEMVAGALPYEEMKFAEQIAY 371
Query: 273 AVSYK 277
AV YK
Sbjct: 372 AVIYK 376
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 527,984,049
Number of Sequences: 2790947
Number of extensions: 22600036
Number of successful extensions: 108217
Number of sequences better than 10.0: 19702
Number of HSP's better than 10.0 without gapping: 16032
Number of HSP's successfully gapped in prelim test: 3670
Number of HSP's that attempted gapping in prelim test: 61183
Number of HSP's gapped (non-prelim): 22002
length of query: 286
length of database: 848,049,833
effective HSP length: 126
effective length of query: 160
effective length of database: 496,390,511
effective search space: 79422481760
effective search space used: 79422481760
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0166.14


