

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0162a.11
(401 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
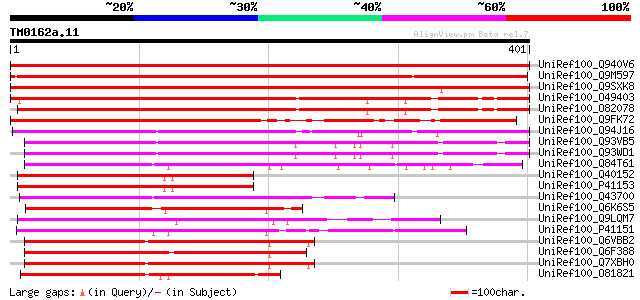
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q940V6 Heat shock transcription factor [Phaseolus acut... 638 0.0
UniRef100_Q9M597 Heat shock transcription factor [Medicago sativa] 554 e-156
UniRef100_Q9SXK8 Heat shock factor [Nicotiana tabacum] 464 e-129
UniRef100_O49403 Heat shock transcription factor-like protein [A... 391 e-107
UniRef100_O82078 Heat shock transcription factor 21 [Arabidopsis... 390 e-107
UniRef100_Q9FK72 Heat shock transcription factor [Arabidopsis th... 355 1e-96
UniRef100_Q94J16 Heat shock factor RHSF9 [Oryza sativa] 308 2e-82
UniRef100_Q93VB5 Heat stress transcription factor Spl7 [Oryza sa... 251 2e-65
UniRef100_Q93WD1 Spl7 protein [Oryza sativa] 246 6e-64
UniRef100_Q84T61 Putative heat shock transcription factor [Oryza... 218 2e-55
UniRef100_Q40152 Heat shock factor protein HSF8 [Lycopersicon es... 214 3e-54
UniRef100_P41153 Heat shock factor protein HSF8 [Lycopersicon pe... 214 4e-54
UniRef100_Q43700 Heat shock factor [Zea mays] 208 2e-52
UniRef100_Q6K6S5 Putative heat stress transcription factor Spl7 ... 205 2e-51
UniRef100_Q9LQM7 F5D14.8 protein [Arabidopsis thaliana] 201 4e-50
UniRef100_P41151 Heat shock factor protein 1 [Arabidopsis thaliana] 201 4e-50
UniRef100_Q6VBB2 Heat shock factor RHSF5 [Oryza sativa] 200 5e-50
UniRef100_Q6F388 Putative HSF-type DNA-binding protein [Oryza sa... 200 5e-50
UniRef100_Q7XBH0 Heat stress protein [Oryza sativa] 199 9e-50
UniRef100_O81821 Heat shock factor protein 3 [Arabidopsis thaliana] 197 4e-49
>UniRef100_Q940V6 Heat shock transcription factor [Phaseolus acutifolius]
Length = 402
Score = 638 bits (1645), Expect = 0.0
Identities = 306/402 (76%), Positives = 352/402 (87%), Gaps = 1/402 (0%)
Query: 1 MDEAQNSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHN 60
MDEA SS SLPPFLAKTYEMVDD S++ IVSWS +++SF+VWNPP+FAR+LLPRFFKHN
Sbjct: 1 MDEALGSSCSLPPFLAKTYEMVDDLSTNSIVSWSVSSKSFIVWNPPEFARDLLPRFFKHN 60
Query: 61 NFSSFIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPL 120
NFSSFIRQLNTYGF+K+DPEQWEFAND FVRG+PHLMKNIHRRKPVHSHS+QNLQA PL
Sbjct: 61 NFSSFIRQLNTYGFKKIDPEQWEFANDDFVRGQPHLMKNIHRRKPVHSHSLQNLQAQGPL 120
Query: 121 GESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSS 180
G+SER+ D IEKLK +KERLL+ELQ+ QHEWQ YEIQ++C ++LEKLEQKQ KMVSS
Sbjct: 121 GDSERQGFTDGIEKLKRDKERLLVELQKFQHEWQTYEIQIHCSNDRLEKLEQKQHKMVSS 180
Query: 181 VSQVLHKPVIALNLLPPTETMDRKRRLPGGGYFNDEASIEDAVDTSQMLPRENAEGASVL 240
+S VL KPV+A+N+LP TETMDRKRRLP G++ DE+SIEDA++TSQMLPRENAE +VL
Sbjct: 181 ISHVLQKPVLAVNILPLTETMDRKRRLPRSGHYYDESSIEDAIETSQMLPRENAENTTVL 240
Query: 241 TLNMERLDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCADSLSISCPQLEVEVRS 300
TLN+ERLDQLESS+ FWE IAH+ G+ F I +NMD DESTSCADS SISC QL+V+VR
Sbjct: 241 TLNVERLDQLESSVAFWEAIAHDIGDNFAQIQSNMDFDESTSCADSPSISCAQLDVDVRP 300
Query: 301 KSSGIDMNSEPTAAAVLEPVASKEQPAG-KAAATGVNDVFWEQFLTEDPGASEAQEVQSE 359
KSSGIDMNSEPTAAAV +P+ASK+QPAG AATGVNDVFWEQFLTEDPGASE QEVQSE
Sbjct: 301 KSSGIDMNSEPTAAAVPDPLASKDQPAGITVAATGVNDVFWEQFLTEDPGASETQEVQSE 360
Query: 360 RKEYNGRKSEGKPTDHGRFWWNARNANNLPEQVGHVSQAEKT 401
RK+ +GRK+EGKP DH +FWWN RNANNL E +GHV QAEKT
Sbjct: 361 RKDCDGRKNEGKPNDHSKFWWNIRNANNLSEPMGHVGQAEKT 402
>UniRef100_Q9M597 Heat shock transcription factor [Medicago sativa]
Length = 402
Score = 554 bits (1428), Expect = e-156
Identities = 281/403 (69%), Positives = 326/403 (80%), Gaps = 5/403 (1%)
Query: 1 MDEAQNSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHN 60
MDEA SSSSLPPFLAKTYEMVDD SSD IVSWSA+N+SFVVWNPP+FAR LL KHN
Sbjct: 1 MDEA-GSSSSLPPFLAKTYEMVDDRSSDPIVSWSASNKSFVVWNPPEFARVLLSEILKHN 59
Query: 61 NFSSFIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPL 120
NFSSFIRQLNTYGFRKVDPEQWEFAND F+RG+PHLMKNIHRRKPVHSHS+ NLQA A L
Sbjct: 60 NFSSFIRQLNTYGFRKVDPEQWEFANDDFIRGQPHLMKNIHRRKPVHSHSLHNLQAQASL 119
Query: 121 GESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSS 180
ESER+S+ DEIEKLK ++E+LL+E +R+QH+W+ +EIQ++C K+QLEKLE KQQKM+SS
Sbjct: 120 TESERQSMIDEIEKLKQDREQLLVETKRYQHDWERHEIQMHCSKDQLEKLEHKQQKMLSS 179
Query: 181 VSQVLHKPVIALNLLPPTETMDRKRRLPGGGY-FNDEASIEDAVDTSQMLPRENAEGASV 239
VS+ L KP+IA+NLLP E M+RKRRLP FN+EAS+EDA++TS LPREN+E
Sbjct: 180 VSEALQKPMIAVNLLPLAEAMERKRRLPARSVCFNNEASVEDAMETSVALPRENSEDNFY 239
Query: 240 LTLNMERLDQL-ESSMLFWETIAHESGETFIHIHTNMDVDESTSCADSLSISCPQLEVEV 298
+ + F +T+AHE G+ F+H H+NMD+DEST CAD LSISC QL+ EV
Sbjct: 240 FDVKHREIGSTSRHPWHFGKTLAHEVGDNFVHTHSNMDLDESTCCADRLSISCQQLDGEV 299
Query: 299 RSKSSGIDMNSEPTAAAVLEPVASKEQPAG-KAAATGVNDVFWEQFLTEDPGASEAQEVQ 357
R KS IDMN EP AAA LE VA KEQPA AATGVNDVFWEQFLTEDPGASEAQEVQ
Sbjct: 300 RPKSPEIDMNVEP-AAAALEAVAVKEQPARITTAATGVNDVFWEQFLTEDPGASEAQEVQ 358
Query: 358 SERKEYNGRKSEGKPTDHGRFWWNARNANNLPEQVGHVSQAEK 400
SERK+ + RK+EGKP+DHGRFWWN R +NN PEQ+GHVSQ EK
Sbjct: 359 SERKDNSSRKNEGKPSDHGRFWWNMRKSNNHPEQMGHVSQVEK 401
>UniRef100_Q9SXK8 Heat shock factor [Nicotiana tabacum]
Length = 408
Score = 464 bits (1193), Expect = e-129
Identities = 229/408 (56%), Positives = 303/408 (74%), Gaps = 7/408 (1%)
Query: 1 MDEAQNSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHN 60
MDEA S+++LPPFL KTYEMVDDPSSD IVSWS++N+SFVVWNPPDFAR+LLPR+FKHN
Sbjct: 1 MDEATCSTNALPPFLTKTYEMVDDPSSDAIVSWSSSNKSFVVWNPPDFARDLLPRYFKHN 60
Query: 61 NFSSFIRQLNTYGFRKVDPEQWEFAN-DGFVRGEPHLMKNIHRRKPVHSHSMQNLQA-HA 118
NFSSFIRQLNTYGFRKVDPE+WEFAN D F RG+PHL+KNIHRRKPVHSHS QNL +
Sbjct: 61 NFSSFIRQLNTYGFRKVDPEKWEFANEDNFFRGQPHLLKNIHRRKPVHSHSAQNLHGLSS 120
Query: 119 PLGESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMV 178
PL ESER+ ++I+KLKH E L ++LQRHQ + Q E+Q+ E+++ +E +Q+ M+
Sbjct: 121 PLTESERQGYKEDIQKLKHENESLHLDLQRHQQDRQGLELQMQVFTERVQHVEHRQKTML 180
Query: 179 SSVSQVLHKPVIALNLLPPTETMDRKRRLPGGGYFNDEASIEDA-VDTSQMLPRENAEGA 237
S+++++L KPV L+ +P + DRKRRLPG +E +ED +S+ L EN +
Sbjct: 181 SALARMLDKPVTDLSRMPQLQVNDRKRRLPGNSCLYNETDLEDTRAISSRALTWENMNPS 240
Query: 238 SVLTLNMERLDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCADSLSISCPQLEVE 297
S+LT+N E L+QL+SS+ FWE + + + +I + ++++DESTSCADS +IS QL V+
Sbjct: 241 SLLTINAELLNQLDSSLTFWENVLQDVDQAWIQQNCSLELDESTSCADSPAISYTQLNVD 300
Query: 298 VRSKSSGIDMNSEPTAAAVLEPVASKEQPAGKAAA----TGVNDVFWEQFLTEDPGASEA 353
V K+S IDMNSEP A E A ++Q A TGVND+FWEQFLTE+PG+ +A
Sbjct: 301 VGPKASDIDMNSEPNANTNPEVAAPEDQAAVAGTTTNVPTGVNDIFWEQFLTENPGSVDA 360
Query: 354 QEVQSERKEYNGRKSEGKPTDHGRFWWNARNANNLPEQVGHVSQAEKT 401
EVQSERK+ +K+E KP D G+FWWN ++ N+L EQ+GH++ AEKT
Sbjct: 361 SEVQSERKDIGNKKNESKPVDSGKFWWNMKSVNSLAEQLGHLTPAEKT 408
>UniRef100_O49403 Heat shock transcription factor-like protein [Arabidopsis thaliana]
Length = 401
Score = 391 bits (1005), Expect = e-107
Identities = 213/411 (51%), Positives = 282/411 (67%), Gaps = 20/411 (4%)
Query: 1 MDEAQN--SSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFK 58
MDE + SSSSLPPFL KTYEMVDD SSD IVSWS +N+SF+VWNPP+F+R+LLPRFFK
Sbjct: 1 MDENNHGVSSSSLPPFLTKTYEMVDDSSSDSIVSWSQSNKSFIVWNPPEFSRDLLPRFFK 60
Query: 59 HNNFSSFIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHA 118
HNNFSSFIRQLNTYGFRK DPEQWEFAND FVRG+PHLMKNIHRRKPVHSHS+ NLQA
Sbjct: 61 HNNFSSFIRQLNTYGFRKADPEQWEFANDDFVRGQPHLMKNIHRRKPVHSHSLPNLQAQL 120
Query: 119 -PLGESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKM 177
PL +SER +N++IE+L KE LL EL + E + +E+Q+ LKE+L+ +E++Q+ M
Sbjct: 121 NPLTDSERVRMNNQIERLTKEKEGLLEELHKQDEEREVFEMQVKELKERLQHMEKRQKTM 180
Query: 178 VSSVSQVLHKPVIALNLLP-PTETMDRKRRLPGGGYFNDEASIEDAVDTSQMLPRENAEG 236
VS VSQVL KP +ALNL P ET +RKRR P +F DE +E+ + + ++ RE
Sbjct: 181 VSFVSQVLEKPGLALNLSPCVPETNERKRRFPRIEFFPDEPMLEE--NKTCVVVREEGST 238
Query: 237 ASVLTLNMERLDQLESSMLFWETIAHESGETFIHIHTNM--DVDESTSCADSLSISCPQL 294
+ +++QLESS+ WE + +S E+ + + M DVDES++ +S +SC QL
Sbjct: 239 SPSSHTREHQVEQLESSIAIWENLVSDSCESMLQSRSMMTLDVDESSTFPESPPLSCIQL 298
Query: 295 EVEVRSKSSG----IDMNSEPTAAAVLEPVASKEQPAGKAAATGVNDVFWEQFLTEDPGA 350
V+ R KS IDMN EP + VA+ P G ND FW+QF +E+PG+
Sbjct: 299 SVDSRLKSPPSPRIIDMNCEPDGSKEQNTVAAPPPP----PVAGANDGFWQQFFSENPGS 354
Query: 351 SEAQEVQSERKEYNGRKSEGKPTDHGRFWWNARNANNLPEQVGHVSQAEKT 401
+E +EVQ ERK+ + G T+ + WWN+RN N + EQ+GH++ +E++
Sbjct: 355 TEQREVQLERKD--DKDKAGVRTE--KCWWNSRNVNAITEQLGHLTSSERS 401
>UniRef100_O82078 Heat shock transcription factor 21 [Arabidopsis thaliana]
Length = 401
Score = 390 bits (1003), Expect = e-107
Identities = 210/403 (52%), Positives = 278/403 (68%), Gaps = 18/403 (4%)
Query: 7 SSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFI 66
SSSSLPPFL KTYEMVDD SSD IVSWS +N+SF+VWNPP+F+R+LLPRFFKHNNFSSFI
Sbjct: 9 SSSSLPPFLTKTYEMVDDSSSDSIVSWSQSNKSFIVWNPPEFSRDLLPRFFKHNNFSSFI 68
Query: 67 RQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHA-PLGESER 125
RQLNTYGFRK DPEQWEFAND FVRG+PHLMKNIHRRKPVHSHS+ NLQA PL +SER
Sbjct: 69 RQLNTYGFRKADPEQWEFANDDFVRGQPHLMKNIHRRKPVHSHSLPNLQAQLNPLTDSER 128
Query: 126 KSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVL 185
+N++IE+L KE LL EL + E + +E+Q+ LKE+L+ +E++Q+ MVS VSQVL
Sbjct: 129 VRMNNQIERLTKEKEGLLEELHKQDEEREVFEMQVKELKERLQHMEKRQKTMVSFVSQVL 188
Query: 186 HKPVIALNLLP-PTETMDRKRRLPGGGYFNDEASIEDAVDTSQMLPRENAEGASVLTLNM 244
KP +ALNL P ET +RKRR P +F DE +E+ + + ++ RE +
Sbjct: 189 EKPGLALNLSPCVPETNERKRRFPRIEFFPDEPMLEE--NKTCVVVREEGSTSPSSHTRE 246
Query: 245 ERLDQLESSMLFWETIAHESGETFIHIHTNM--DVDESTSCADSLSISCPQLEVEVRSKS 302
+++QLESS+ WE + +S E+ + + M DVDES++ +S +SC QL V+ R KS
Sbjct: 247 HQVEQLESSIAIWENLVSDSCESMLQSRSMMTLDVDESSTFPESPPLSCIQLSVDSRLKS 306
Query: 303 SG----IDMNSEPTAAAVLEPVASKEQPAGKAAATGVNDVFWEQFLTEDPGASEAQEVQS 358
IDMN EP + VA+ P G ND FW+QF +E+PG++E +EVQ
Sbjct: 307 PPSPRIIDMNCEPDGSKEQNTVAAPPPP----PVAGANDGFWQQFFSENPGSTEQREVQL 362
Query: 359 ERKEYNGRKSEGKPTDHGRFWWNARNANNLPEQVGHVSQAEKT 401
ERK+ + G T+ + WWN+RN N + EQ+GH++ +E++
Sbjct: 363 ERKD--DKDKAGVRTE--KCWWNSRNVNAITEQLGHLTSSERS 401
>UniRef100_Q9FK72 Heat shock transcription factor [Arabidopsis thaliana]
Length = 345
Score = 355 bits (911), Expect = 1e-96
Identities = 198/391 (50%), Positives = 257/391 (65%), Gaps = 48/391 (12%)
Query: 1 MDEAQNSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHN 60
MDE SSSLPPFL KTYEMVDD SSD +V+WS N+SF+V NP +F+R+LLPRFFKH
Sbjct: 1 MDENNGGSSSLPPFLTKTYEMVDDSSSDSVVAWSENNKSFIVKNPAEFSRDLLPRFFKHK 60
Query: 61 NFSSFIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPL 120
NFSSFIRQLNTYGFRKVDPE+WEF ND FVRG P+LMKNIHRRKPVHSHS+ NLQA PL
Sbjct: 61 NFSSFIRQLNTYGFRKVDPEKWEFLNDDFVRGRPYLMKNIHRRKPVHSHSLVNLQAQNPL 120
Query: 121 GESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSS 180
ESER+S+ D+IE+LK+ KE LL ELQ + E + +E+Q+ LK++L+ +EQ Q+ +V+
Sbjct: 121 TESERRSMEDQIERLKNEKEGLLAELQNQEQERKEFELQVTTLKDRLQHMEQHQKSIVAY 180
Query: 181 VSQVLHKPVIALNLLPPTETMDRKRRLPGGGYFNDEASIEDAVDTSQMLPRENAEGASVL 240
VSQVL KP ++LNL E +R++R F + + LP ++
Sbjct: 181 VSQVLGKPGLSLNL----ENHERRKRR-----FQENS-----------LPPSSS------ 214
Query: 241 TLNMERLDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCADSLSISCPQLEVEVRS 300
++E++++LESS+ FWE + ES E ++MD D + S SLSI + R
Sbjct: 215 --HIEQVEKLESSLTFWENLVSESCEKSGLQSSSMDHDAAES---SLSIG------DTRP 263
Query: 301 KSSGIDMNSEPTAAAVLEPVASKEQPAGKAAATGVNDVFWEQFLTEDPGASEAQEVQSER 360
KSS IDMNSEP PA K TGVND FWEQ LTE+PG++E QEVQSER
Sbjct: 264 KSSKIDMNSEPPVTVTA--------PAPK---TGVNDDFWEQCLTENPGSTEQQEVQSER 312
Query: 361 KEYNGRKSEGKPTDHGRFWWNARNANNLPEQ 391
++ + K + +WWN+ N NN+ E+
Sbjct: 313 RDVGNDNNGNKIGNQRTYWWNSGNVNNITEK 343
>UniRef100_Q94J16 Heat shock factor RHSF9 [Oryza sativa]
Length = 440
Score = 308 bits (789), Expect = 2e-82
Identities = 179/448 (39%), Positives = 254/448 (55%), Gaps = 58/448 (12%)
Query: 3 EAQNSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNF 62
E SLPPFL+KTYEMVDDPS+D +V W+ SFVV N P+F R+LLP++FKHNNF
Sbjct: 2 EGGGGGGSLPPFLSKTYEMVDDPSTDAVVGWTPAGTSFVVANQPEFCRDLLPKYFKHNNF 61
Query: 63 SSFIRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGE 122
SSF+RQLNTYGFRKVDPEQWEFAN+ F++G+ H +KNIHRRKP+ SHS + Q PL +
Sbjct: 62 SSFVRQLNTYGFRKVDPEQWEFANEDFIKGQRHRLKNIHRRKPIFSHSSHS-QGAGPLTD 120
Query: 123 SERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVS 182
+ERK +EIE+LK + L ELQ + + E ++ L+E+L +E +Q+ ++S V
Sbjct: 121 NERKDYEEEIERLKSDNAALSSELQNNTLKKLNMEKRMQALEEKLFVVEDQQRSLISYVR 180
Query: 183 QVLHKPVIALNLLPPTETMDRKRRLPGGGYFNDEASIEDAVDTSQMLPRENAEGASVLTL 242
+++ P + + + +KRRLP F+++A+ ++ +Q++P + + T
Sbjct: 181 EIVKAPGFLSSFVQQQDHHRKKRRLPIPISFHEDANTQE----NQIMPCD-LTNSPAQTF 235
Query: 243 NMERLDQLESSMLFWETIAHESGETF---------------------IH----------- 270
E D++ESS+ E E+ E F +H
Sbjct: 236 YRESFDKMESSLNSLENFLREASEEFGNDISYDDGVPGPSSTVVLTELHSPGESDPRVSS 295
Query: 271 --------------IHTNMDVDESTSCADSLSISCPQLEVEVRSKSSGIDMNSEPTAAAV 316
H++ DV ESTSCA+S I V+ R+K S ID+NSEP AV
Sbjct: 296 PPTRMRTSSAGAGDSHSSRDVAESTSCAESPPIPQMHSRVDTRAKVSEIDVNSEP---AV 352
Query: 317 LEPVASKEQPAGK--AAATGVNDVFWEQFLTEDPGASEA-QEVQSERKEYNGRKSEGKPT 373
E S++QPA + A G ND FW+QFLTE PG+S+A QE QSER++ + E K
Sbjct: 353 TETGPSRDQPAEEPPAVTPGANDGFWQQFLTEQPGSSDAHQEAQSERRDGGNKVDEMKSG 412
Query: 374 DHGRFWWNARNANNLPEQVGHVSQAEKT 401
D WW RN + E++G ++ EKT
Sbjct: 413 DRQHLWWGKRNVEQITEKLGLLTSTEKT 440
>UniRef100_Q93VB5 Heat stress transcription factor Spl7 [Oryza sativa]
Length = 459
Score = 251 bits (642), Expect = 2e-65
Identities = 157/447 (35%), Positives = 237/447 (52%), Gaps = 66/447 (14%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KTYEMV+D +++ +VSW SFVVWNP DF+R+LLP++FKHNNFSSFIRQLNT
Sbjct: 19 PPFLIKTYEMVEDAATNHVVSWGPGGASFVVWNPLDFSRDLLPKYFKHNNFSSFIRQLNT 78
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLNDE 131
YGFRK+DPE+WEFAN+ F+RG HL+KNIHRRKPVHSHS+QN Q + PL ESER+ L +E
Sbjct: 79 YGFRKIDPERWEFANEDFIRGHTHLLKNIHRRKPVHSHSLQN-QINGPLAESERRELEEE 137
Query: 132 IEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPVIA 191
I +LK+ K L+ +LQR + Q+ ++ +L +EQ+Q+ +V+S+ ++L + A
Sbjct: 138 INRLKYEKSILVADLQRQNQQQYVINWQMQAMEGRLVAMEQRQKNIVASLCEMLQRRGGA 197
Query: 192 L-NLLPPTETMDRKRRLPGGGYFNDEASI-------------EDAVDTSQMLPRENAEGA 237
+ + L ++ +KRR+P F D+ + DA +LP N E
Sbjct: 198 VSSSLLESDHFSKKRRVPKMDLFVDDCAAGEEQKVFQFQGIGTDAPAMPPVLPVTNGEAF 257
Query: 238 SVLTLNMERLDQL-----ESSMLFWETIAHESG------------------ETFI----- 269
+ L++ L++L ++ E +H G ET I
Sbjct: 258 DRVELSLVSLEKLFQRANDACTAAEEMYSHGHGGTEPSTAICPEEMNTAPMETGIDLQLP 317
Query: 270 -------------HIHTNMDVDESTSCADSLSISCPQL--EVEVRSKSSGIDMNSEPTAA 314
H+H + ++ ES S + ++ ++ V + D+NSE ++
Sbjct: 318 ASLHPSSPNTGNAHLHLSTELTESPGFVQSPELPMAEIREDIHVTRYPTQADVNSEIASS 377
Query: 315 AVLEPVASKEQPAGKAAATGVNDVFWEQFLTEDPGASEAQEVQSERKEYNGRKSEGKPTD 374
+ E A NDVFWE+FLTE P + + + E + + + G
Sbjct: 378 TDTSQDGTSETEASHGP---TNDVFWERFLTETPRSCLDESERQESPKDDVKAELGCNGF 434
Query: 375 HGRFWWNARNANNLPEQVGHVSQAEKT 401
H R + + EQ+GH++ AE+T
Sbjct: 435 HHR-----EKVDQITEQMGHLASAEQT 456
>UniRef100_Q93WD1 Spl7 protein [Oryza sativa]
Length = 459
Score = 246 bits (629), Expect = 6e-64
Identities = 156/447 (34%), Positives = 236/447 (51%), Gaps = 66/447 (14%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KTYEMV+D +++ +VS SFVVWNP DF+R+LLP++FKHNNFSSFIRQLNT
Sbjct: 19 PPFLIKTYEMVEDAATNHVVSCGPGGASFVVWNPLDFSRDLLPKYFKHNNFSSFIRQLNT 78
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLNDE 131
YGFRK+DPE+WEFAN+ F+RG HL+KNIHRRKPVHSHS+QN Q + PL ESER+ L +E
Sbjct: 79 YGFRKIDPERWEFANEDFIRGHTHLLKNIHRRKPVHSHSLQN-QINGPLAESERRELEEE 137
Query: 132 IEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPVIA 191
I +LK+ K L+ +LQR + Q+ ++ +L +EQ+Q+ +V+S+ ++L + A
Sbjct: 138 INRLKYEKSILVADLQRQNQQQYVINWQMQAMEGRLVAMEQRQKNIVASLCEMLQRRGGA 197
Query: 192 L-NLLPPTETMDRKRRLPGGGYFNDEASI-------------EDAVDTSQMLPRENAEGA 237
+ + L ++ +KRR+P F D+ + DA +LP N E
Sbjct: 198 VSSSLLESDHFSKKRRVPKMDLFVDDCAAGEEQKVFQFQGIGTDAPAMPPVLPVTNGEAF 257
Query: 238 SVLTLNMERLDQL-----ESSMLFWETIAHESG------------------ETFI----- 269
+ L++ L++L ++ E +H G ET I
Sbjct: 258 DRVELSLVSLEKLFQRANDACTAAEEMYSHGHGGTEPSTAICPEEMNTAPMETGIDLQLP 317
Query: 270 -------------HIHTNMDVDESTSCADSLSISCPQL--EVEVRSKSSGIDMNSEPTAA 314
H+H + ++ ES S + ++ ++ V + D+NSE ++
Sbjct: 318 ASLHPSSPNTGNAHLHLSTELTESPGFVQSPELPMAEIREDIHVTRYPTQADVNSEIASS 377
Query: 315 AVLEPVASKEQPAGKAAATGVNDVFWEQFLTEDPGASEAQEVQSERKEYNGRKSEGKPTD 374
+ E A NDVFWE+FLTE P + + + E + + + G
Sbjct: 378 TDTSQDGTSETEASHGP---TNDVFWERFLTETPRSCLDESERQESPKDDVKAELGCNGF 434
Query: 375 HGRFWWNARNANNLPEQVGHVSQAEKT 401
H R + + EQ+GH++ AE+T
Sbjct: 435 HHR-----EKVDQITEQMGHLASAEQT 456
>UniRef100_Q84T61 Putative heat shock transcription factor [Oryza sativa]
Length = 506
Score = 218 bits (555), Expect = 2e-55
Identities = 156/475 (32%), Positives = 226/475 (46%), Gaps = 99/475 (20%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KTYEMVDDP++D +VSW N SFVVWN P+FAR+LLP++FKH+NFSSF+RQLNT
Sbjct: 36 PPFLMKTYEMVDDPATDAVVSWGPGNNSFVVWNTPEFARDLLPKYFKHSNFSSFVRQLNT 95
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLG------ESER 125
YGFRKVDP++WEFAN+GF+RG+ HL+K I+RRKP H ++ Q Q P E +
Sbjct: 96 YGFRKVDPDRWEFANEGFLRGQKHLLKTINRRKPTHGNN-QVQQPQLPAAPVPACVEVGK 154
Query: 126 KSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVL 185
+ +EIE LK +K L+ EL R + + Q + QL L ++L+ +EQ+QQ+M+S +++ +
Sbjct: 155 FGMEEEIEMLKRDKNVLMQELVRLRQQQQTTDHQLQTLGKRLQGMEQRQQQMMSFLAKAM 214
Query: 186 HKPVIALNLLPPTE-------TMDRKRRLP--GGGYFNDEASIEDAVDTSQMLPRENAEG 236
H P + E ++KRRLP G ++ AS++ + Q + E A+
Sbjct: 215 HSPGFLAQFVQQNENSRRRIVASNKKRRLPKQDGSLDSESASLDGQIVKYQPMINEAAKA 274
Query: 237 ASVLTLNMERLDQLES----------------SMLFWETIAHESGETFIHIHTNMD---V 277
L ++ + ES L + SG T + N V
Sbjct: 275 MLRKILKLDSSHRFESMGNSDNFLLENYMPNGQGLDSSSSTRNSGVTLAEVPANSGLPYV 334
Query: 278 DESTSCADSLSISCPQLEVEVRSKSSGI-----DMNSEPTAAAVLEP------------- 319
S+ + S S PQ++ V +GI +M++ P+ + P
Sbjct: 335 ATSSGLSAICSTSTPQIQCPV-VLDNGIPKEVPNMSAVPSVPKAVAPGPTDINILEFPDL 393
Query: 320 --VASKEQ---PAGKAAATGVNDVF---------------------------------WE 341
+ ++E P G G VF WE
Sbjct: 394 QDIVAEENVDIPGGGFEMPGPEGVFSLPEEGDDSVPIETDEILYNDDTQKLPAIIDSFWE 453
Query: 342 QFLTEDPGASEAQEVQSERKEYNGRKSEGKPTDHGRFWWNARNANNLPEQVGHVS 396
QFL P + + EV S + K T G W A N NL EQ+G +S
Sbjct: 454 QFLVASPLSVDNDEVDS-------GVLDQKETQQGNGWTKAENMANLTEQMGLLS 501
>UniRef100_Q40152 Heat shock factor protein HSF8 [Lycopersicon esculentum]
Length = 527
Score = 214 bits (546), Expect = 3e-54
Identities = 101/198 (51%), Positives = 145/198 (73%), Gaps = 16/198 (8%)
Query: 7 SSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFI 66
S+++ PPFL KTY+MVDDPS+D+IVSWS TN SFVVW+PP+FA++LLP++FKHNNFSSF+
Sbjct: 33 SANAPPPFLVKTYDMVDDPSTDKIVSWSPTNNSFVVWDPPEFAKDLLPKYFKHNNFSSFV 92
Query: 67 RQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHA-------P 119
RQLNTYGFRKVDP++WEFAN+GF+RG+ HL+K+I RRKP H H+ Q Q H P
Sbjct: 93 RQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKSISRRKPAHGHAQQQQQPHGNAQQQMQP 152
Query: 120 LGESE---------RKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKL 170
G S + L +E+E+LK +K L+ EL R + + Q + QL + ++L+ +
Sbjct: 153 PGHSASVGACVEVGKFGLEEEVERLKRDKNVLMQELVRLRQQQQATDNQLQGMVQRLQGM 212
Query: 171 EQKQQKMVSSVSQVLHKP 188
E +QQ+M+S +++ +++P
Sbjct: 213 ELRQQQMMSFLAKAVNRP 230
>UniRef100_P41153 Heat shock factor protein HSF8 [Lycopersicon peruvianum]
Length = 527
Score = 214 bits (544), Expect = 4e-54
Identities = 101/198 (51%), Positives = 144/198 (72%), Gaps = 16/198 (8%)
Query: 7 SSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFI 66
S+++ PPFL KTY+MVDDPS+D+IVSWS TN SFVVW+PP+FA++LLP++FKHNNFSSF+
Sbjct: 35 SANAPPPFLVKTYDMVDDPSTDKIVSWSPTNNSFVVWDPPEFAKDLLPKYFKHNNFSSFV 94
Query: 67 RQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHA-------P 119
RQLNTYGFRKVDP++WEFAN+GF+RG+ HL+K+I RRKP H H+ Q Q H P
Sbjct: 95 RQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKSISRRKPAHGHAQQQQQPHGHAQQQMQP 154
Query: 120 LGESE---------RKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKL 170
G S + L +E+E+LK +K L+ EL R + + Q + QL + ++L+ +
Sbjct: 155 PGHSASVGACVEVGKFGLEEEVERLKRDKNVLMQELVRLRQQQQSTDNQLQGMVQRLQGM 214
Query: 171 EQKQQKMVSSVSQVLHKP 188
E +QQ+M+S +++ ++ P
Sbjct: 215 ELRQQQMMSFLAKAVNSP 232
>UniRef100_Q43700 Heat shock factor [Zea mays]
Length = 308
Score = 208 bits (529), Expect = 2e-52
Identities = 116/291 (39%), Positives = 172/291 (58%), Gaps = 15/291 (5%)
Query: 8 SSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIR 67
+SSLPPFL+KTYEMVDDP++D +V+W+ SFVV N +F R+LLP++FKHNNFSSF+R
Sbjct: 4 ASSLPPFLSKTYEMVDDPATDAVVAWTPLGTSFVVANQAEFWRDLLPKYFKHNNFSSFVR 63
Query: 68 QLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKS 127
QLNTYGF+K+DPEQWEFAND F+RG+ H +KNIHRRKP+ SHS + Q PL ++ER+
Sbjct: 64 QLNTYGFKKIDPEQWEFANDDFIRGQQHRLKNIHRRKPIFSHS-SHTQGSGPLPDTERRD 122
Query: 128 LNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHK 187
+EIE+LK + L EL+++ + E ++ L+++L LE +Q+ +++ V ++
Sbjct: 123 YEEEIERLKCDNAALTSELEKNAQKKLVTEKRMQDLEDKLIFLEDRQKNLMAYVRDIVQA 182
Query: 188 PVIALNLLPPTETMDRKRRLPGG-GYFNDEASIEDAVDTSQMLPRENAEGASVLTLNMER 246
P + + + +KRRLP + D + + V + A E
Sbjct: 183 PGSFSSFVQQPDHHGKKRRLPVPISLYQDSNAKGNQVVHGSFITNPPA--------CRES 234
Query: 247 LDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCADSLSISCPQLEVE 297
D+ ESS+ E E+ E F N+ D+ LS+S + E
Sbjct: 235 FDKTESSLNSLENFLREASEAF-----NISYDDGLPGLHLLSLSQSSIRPE 280
>UniRef100_Q6K6S5 Putative heat stress transcription factor Spl7 [Oryza sativa]
Length = 475
Score = 205 bits (522), Expect = 2e-51
Identities = 110/224 (49%), Positives = 146/224 (65%), Gaps = 21/224 (9%)
Query: 13 PFLAKTYEMVDDPSSDRIVSWS-ATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PFL KTYEMVDDPS+D +VSWS A++ SFVVWN P+FA LLP +FKH+NFSSFIRQLNT
Sbjct: 21 PFLLKTYEMVDDPSTDAVVSWSDASDASFVVWNHPEFAARLLPAYFKHSNFSSFIRQLNT 80
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAP--LGESERKSLN 129
YGFRK+DPE+WEFAN+ F++G+ HL+KNIHRRKP+HSHS H P L ++ER
Sbjct: 81 YGFRKIDPERWEFANEYFIKGQKHLLKNIHRRKPIHSHS------HPPGALPDNERAIFE 134
Query: 130 DEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPV 189
DEIE+L K L +L + + + Q+ L+ ++ +EQ+Q KM++ + Q P
Sbjct: 135 DEIERLSREKSNLQADLWKSKQQQSGTMNQIEDLERRVLGMEQRQTKMIAFLQQASKNPQ 194
Query: 190 IALNLLPP-------TETMDRKRRLPGGGYFNDEASIEDAVDTS 226
L+ T+ ++KRRLPG Y SIE+ TS
Sbjct: 195 FVNKLVKMAEASSIFTDAFNKKRRLPGLDY-----SIENTETTS 233
Score = 45.4 bits (106), Expect = 0.003
Identities = 32/79 (40%), Positives = 38/79 (47%), Gaps = 5/79 (6%)
Query: 301 KSSGIDMNSEPTAAAVLEPVASKEQPAGKAAATGVNDVFWEQFLTEDPGASEAQEVQSER 360
K S ++ T VAS E PA AA VND FWEQFLTE PG SE +E S
Sbjct: 389 KKSADSRTADATTPRADARVAS-EAPAAPAAV--VNDKFWEQFLTERPGCSETEEASSGL 445
Query: 361 KEYNGRK--SEGKPTDHGR 377
+ R+ + DH R
Sbjct: 446 RTDTSREQMENRQAYDHSR 464
>UniRef100_Q9LQM7 F5D14.8 protein [Arabidopsis thaliana]
Length = 482
Score = 201 bits (510), Expect = 4e-50
Identities = 121/349 (34%), Positives = 185/349 (52%), Gaps = 50/349 (14%)
Query: 7 SSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFI 66
SS++ PPFL+KTY+MVDD ++D IVSWSA N SF+VW PP+FAR+LLP+ FKHNNFSSF+
Sbjct: 31 SSNAPPPFLSKTYDMVDDHNTDSIVSWSANNNSFIVWKPPEFARDLLPKNFKHNNFSSFV 90
Query: 67 RQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERK 126
RQLNTYGFRKVDP++WEFAN+GF+RG+ HL+++I RRKP H + ++ G++
Sbjct: 91 RQLNTYGFRKVDPDRWEFANEGFLRGQKHLLQSITRRKPAHGQGQGHQRSQHSNGQNSSV 150
Query: 127 S---------LNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKM 177
S L +E+E+LK +K L+ EL R + + Q + QL + ++L+ +E +QQ++
Sbjct: 151 SACVEVGKFGLEEEVERLKRDKNVLMQELVRLRQQQQSTDNQLQTMVQRLQGMENRQQQL 210
Query: 178 VSSVSQVLHKPVIALNLLPPTETMD----------RKRRLPGGGYF--NDEASIEDAVDT 225
+S +++ + P L + +KRR G ND A+ + +
Sbjct: 211 MSFLAKAVQSPHFLSQFLQQQNQQNESNRRISDTSKKRRFKRDGIVRNNDSATPDGQIVK 270
Query: 226 SQMLPRENAEGASVLTLNMERLDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCAD 285
Q E A+ + ME +++G+ + E T
Sbjct: 271 YQPPMHEQAKAMFKQLMKME---------------PYKTGDDGFLLGNGTSTTEGT---- 311
Query: 286 SLSISCPQLEVEVRSKS-SGIDMNSEPTAAAVLEPVASKEQPAGKAAAT 333
E+E S SGI + PTA+ + + P +AA+
Sbjct: 312 ---------EMETSSNQVSGITLKEMPTASEIQSSSPIETTPENVSAAS 351
>UniRef100_P41151 Heat shock factor protein 1 [Arabidopsis thaliana]
Length = 495
Score = 201 bits (510), Expect = 4e-50
Identities = 122/370 (32%), Positives = 199/370 (52%), Gaps = 36/370 (9%)
Query: 6 NSSSSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSF 65
N++S PPFL+KTY+MV+DP++D IVSWS TN SF+VW+PP+F+R+LLP++FKHNNFSSF
Sbjct: 45 NANSLPPPFLSKTYDMVEDPATDAIVSWSPTNNSFIVWDPPEFSRDLLPKYFKHNNFSSF 104
Query: 66 IRQLNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSH--------SMQNLQAH 117
+RQLNTYGFRKVDP++WEFAN+GF+RG+ HL+K I RRK V H S Q Q
Sbjct: 105 VRQLNTYGFRKVDPDRWEFANEGFLRGQKHLLKKISRRKSVQGHGSSSSNPQSQQLSQGQ 164
Query: 118 APLG------ESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLE 171
+ E + L +E+E+LK +K L+ EL + + + Q + +L L + L+ +E
Sbjct: 165 GSMAALSSCVEVGKFGLEEEVEQLKRDKNVLMQELVKLRQQQQTTDNKLQVLVKHLQVME 224
Query: 172 QKQQKMVSSVSQVLHKPVIALNLLPP-------TETMDRKRRLPGGGYFNDEASIEDAVD 224
Q+QQ+++S +++ + P + ++KRRL D + ++
Sbjct: 225 QRQQQIMSFLAKAVQNPTFLSQFIQKQTDSNMHVTEANKKRRLR-----EDSTAATESNS 279
Query: 225 TSQMLPRENAEGASVLTLNMERLDQLESSMLFWETIAHESGETFIHIHTNMDVDESTSCA 284
S L E ++G V + L + + W + + F+ ++ + +
Sbjct: 280 HSHSL--EASDGQIV------KYQPLRNDSMMWNMMKTDDKYPFLDGFSSPNQVSGVTLQ 331
Query: 285 DSLSISCPQLEVEVRSKSSGIDMNSEP-TAAAVLEPVASKEQPAGKAAATGVNDVFWEQF 343
+ L I+ Q + S SG ++ P T+ ++ + + + + +ND E F
Sbjct: 332 EVLPITSGQSQA-YASVPSGQPLSYLPSTSTSLPDTIMPETSQIPQLTRESINDFPTENF 390
Query: 344 LTEDPGASEA 353
+ + EA
Sbjct: 391 MDTEKNVPEA 400
>UniRef100_Q6VBB2 Heat shock factor RHSF5 [Oryza sativa]
Length = 372
Score = 200 bits (509), Expect = 5e-50
Identities = 106/235 (45%), Positives = 150/235 (63%), Gaps = 12/235 (5%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KTY+MVDD +D VSWSAT+ SFVVW+P FA LLPRFFKHNNFSSF+RQLNT
Sbjct: 51 PPFLTKTYDMVDDAGTDAAVSWSATSNSFVVWDPHAFATVLLPRFFKHNNFSSFVRQLNT 110
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLNDE 131
YGFRKVDP++WEFAN+ F+RG+ HL+KNI RRKP SH+ N Q+ P E + E
Sbjct: 111 YGFRKVDPDRWEFANENFLRGQRHLLKNIKRRKP-PSHTASNQQSLGPYLEVGHFGYDAE 169
Query: 132 IEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPVIA 191
I++LK +K+ L+ E+ + + E Q + L ++++L+ EQ+QQ+M++ +++V+ P
Sbjct: 170 IDRLKRDKQLLMAEVVKLRQEQQNTKANLKAMEDRLQGTEQRQQQMMAFLARVMKNPEFL 229
Query: 192 LNLLPPTE---------TMDRKRRLPGGGYFNDEASIEDAVDTSQML--PRENAE 235
L+ E + R+RR+ G +D + S L P+E+ E
Sbjct: 230 KQLMSQNEMRKELQDAISKKRRRRIDQGPEVDDVGTSSSIEQESPALFDPQESVE 284
>UniRef100_Q6F388 Putative HSF-type DNA-binding protein [Oryza sativa]
Length = 357
Score = 200 bits (509), Expect = 5e-50
Identities = 104/228 (45%), Positives = 148/228 (64%), Gaps = 13/228 (5%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KTY+MVDDP++D +VSWSATN SFVVW+P F LLPR+FKHNNFSSF+RQLNT
Sbjct: 37 PPFLTKTYDMVDDPTTDAVVSWSATNNSFVVWDPHLFGNVLLPRYFKHNNFSSFVRQLNT 96
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSH-SMQNLQAHAPLGESERKSLND 130
YGFRKVDP++WEFAN+GF+RG+ HL+K+I RRKP +S S Q+L + +G
Sbjct: 97 YGFRKVDPDKWEFANEGFLRGQKHLLKSIKRRKPPNSSPSQQSLGSFLEVGHF---GYEG 153
Query: 131 EIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPVI 190
EI++LK +K L+ E+ + + E Q + L ++++L+ EQKQQ M++ +S+V+H P
Sbjct: 154 EIDQLKRDKHLLMAEVVKLRQEQQNTKSDLQAMEQKLQGTEQKQQHMMAFLSRVMHNPEF 213
Query: 191 ALNLLPPTE---------TMDRKRRLPGGGYFNDEASIEDAVDTSQML 229
L +E + R+RR+ G + + SQ++
Sbjct: 214 IRQLFSQSEMRKELEEFVSKKRRRRIDQGPELDSMGTGSSPEQVSQVM 261
>UniRef100_Q7XBH0 Heat stress protein [Oryza sativa]
Length = 372
Score = 199 bits (507), Expect = 9e-50
Identities = 105/235 (44%), Positives = 149/235 (62%), Gaps = 12/235 (5%)
Query: 12 PPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQLNT 71
PPFL KTY+MVDD +D VSWSAT+ SFVVW+P FA LLPRFFKHNNFSSF+RQLNT
Sbjct: 51 PPFLTKTYDMVDDAGTDAAVSWSATSNSFVVWDPHAFATVLLPRFFKHNNFSSFVRQLNT 110
Query: 72 YGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQAHAPLGESERKSLNDE 131
YGFRKVDP++WEFAN+ F+RG+ HL KNI RRKP SH+ N Q+ P E + E
Sbjct: 111 YGFRKVDPDRWEFANENFLRGQRHLFKNIKRRKP-PSHTASNQQSFGPYLEVGHFGYDAE 169
Query: 132 IEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSVSQVLHKPVIA 191
I++LK +K+ L+ E+ + + E Q + L ++++L+ EQ+Q++M++ +++V+ P
Sbjct: 170 IDRLKRDKQLLMAEVVKLRQEQQNTKANLKAMEDRLQGTEQRQKQMIAFLARVMKNPEFL 229
Query: 192 LNLLPPTE---------TMDRKRRLPGGGYFNDEASIEDAVDTSQML--PRENAE 235
L+ E + R+RR+ G +D + S L P+E+ E
Sbjct: 230 KQLMSQNEMRKELQDAISKKRRRRIDQGPEVDDVGTSSSIEQESPALFDPQESVE 284
>UniRef100_O81821 Heat shock factor protein 3 [Arabidopsis thaliana]
Length = 481
Score = 197 bits (501), Expect = 4e-49
Identities = 100/208 (48%), Positives = 147/208 (70%), Gaps = 10/208 (4%)
Query: 9 SSLPPFLAKTYEMVDDPSSDRIVSWSATNRSFVVWNPPDFARELLPRFFKHNNFSSFIRQ 68
+S+PPFL+KTY+MVDDP ++ +VSWS+ N SFVVW+ P+F++ LLP++FKHNNFSSF+RQ
Sbjct: 23 NSVPPFLSKTYDMVDDPLTNEVVSWSSGNNSFVVWSAPEFSKVLLPKYFKHNNFSSFVRQ 82
Query: 69 LNTYGFRKVDPEQWEFANDGFVRGEPHLMKNIHRRKPVHSHSMQNLQ----AHAPLG--- 121
LNTYGFRKVDP++WEFAN+GF+RG L+K+I RRKP SH QN Q + +G
Sbjct: 83 LNTYGFRKVDPDRWEFANEGFLRGRKQLLKSIVRRKP--SHVQQNQQQTQVQSSSVGACV 140
Query: 122 ESERKSLNDEIEKLKHNKERLLMELQRHQHEWQPYEIQLNCLKEQLEKLEQKQQKMVSSV 181
E + + +E+E+LK +K L+ EL R + + Q E QL + ++++ +EQ+QQ+M+S +
Sbjct: 141 EVGKFGIEEEVERLKRDKNVLMQELVRLRQQQQATENQLQNVGQKVQVMEQRQQQMMSFL 200
Query: 182 SQVLHKPVIALNLLPPTETMDRKRRLPG 209
++ + P LN L D R++PG
Sbjct: 201 AKAVQSPGF-LNQLVQQNNNDGNRQIPG 227
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.312 0.128 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 686,646,589
Number of Sequences: 2790947
Number of extensions: 29027176
Number of successful extensions: 91712
Number of sequences better than 10.0: 626
Number of HSP's better than 10.0 without gapping: 297
Number of HSP's successfully gapped in prelim test: 340
Number of HSP's that attempted gapping in prelim test: 90011
Number of HSP's gapped (non-prelim): 1636
length of query: 401
length of database: 848,049,833
effective HSP length: 130
effective length of query: 271
effective length of database: 485,226,723
effective search space: 131496441933
effective search space used: 131496441933
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0162a.11


