

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
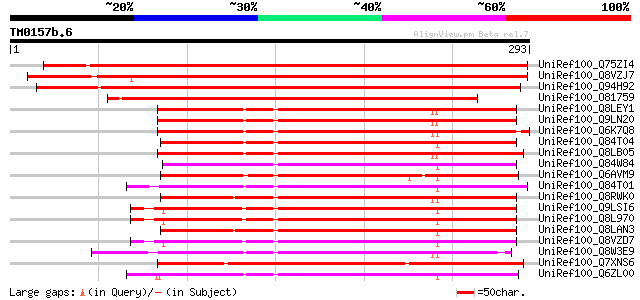
Query= TM0157b.6
(293 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q75ZI4 Prolyl 4-hydroxylase [Nicotiana tabacum] 414 e-114
UniRef100_Q8VZJ7 Hypothetical protein At4g33910 [Arabidopsis tha... 385 e-106
UniRef100_Q94H92 Putative dioxygenase [Oryza sativa] 377 e-103
UniRef100_O81759 Hypothetical protein F17I5.100 [Arabidopsis tha... 310 4e-83
UniRef100_Q8LEY1 Putative prolyl 4-hydroxylase, alpha subunit [A... 180 3e-44
UniRef100_Q9LN20 F14O10.12 protein [Arabidopsis thaliana] 179 8e-44
UniRef100_Q6K7Q8 Putative prolyl 4-hydroxylase [Oryza sativa] 178 1e-43
UniRef100_Q84T04 Putative oxidoreductase [Oryza sativa] 169 1e-40
UniRef100_Q8LB05 Putative prolyl 4-hydroxylase, alpha subunit [A... 163 6e-39
UniRef100_Q84W84 Putative prolyl 4-hydroxylase [Arabidopsis thal... 162 7e-39
UniRef100_Q6AVM9 Putative prolyl 4-hydroxylase alpha subunit [Or... 162 1e-38
UniRef100_Q84T01 Putative oxidoreductase [Oryza sativa] 162 1e-38
UniRef100_Q8RWK0 Hypothetical protein At5g18900 [Arabidopsis tha... 160 4e-38
UniRef100_Q9LSI6 Prolyl 4-hydroxylase alpha subunit-like protein... 160 5e-38
UniRef100_Q8L970 Prolyl 4-hydroxylase, putative [Arabidopsis tha... 160 5e-38
UniRef100_Q8LAN3 Prolyl 4-hydroxylase alpha subunit-like protein... 159 8e-38
UniRef100_Q8VZD7 AT3g28480/MFJ20_16 [Arabidopsis thaliana] 159 1e-37
UniRef100_Q8W3E9 Putative prolyl 4-hydroxylase, alpha subunit [O... 157 2e-37
UniRef100_Q7XNS6 OSJNBb0085H11.11 protein [Oryza sativa] 157 2e-37
UniRef100_Q6ZL00 Prolyl 4-hydroxylase alpha-1 subunit-like prote... 157 4e-37
>UniRef100_Q75ZI4 Prolyl 4-hydroxylase [Nicotiana tabacum]
Length = 286
Score = 414 bits (1064), Expect = e-114
Identities = 194/273 (71%), Positives = 234/273 (85%), Gaps = 2/273 (0%)
Query: 20 PYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGDDSIT 79
P +FL+C+FFFL GFFGS LFSH D +RPRPR LES + +++ + GE G+ S+
Sbjct: 15 PSVFLLCLFFFLLGFFGSALFSHQ--DVPSVRPRPRFLESVYQEDFDPLPIGETGEHSLI 72
Query: 80 SIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSG 139
SIPFQVLSW PRALYFPNFA+ EQC+SI+ +AKA ++PS+LALR GETE+ TKGIRTSSG
Sbjct: 73 SIPFQVLSWFPRALYFPNFASIEQCQSIIKMAKANMEPSSLALRTGETEETTKGIRTSSG 132
Query: 140 VFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQ 199
F+S+SEDKTG L +IEEKIA+ATMIP++HGEAFN+LRYE+ QRY HYD+F+PA+YGPQ
Sbjct: 133 TFISASEDKTGILDLIEEKIAKATMIPKTHGEAFNVLRYEIGQRYQSHYDAFDPAQYGPQ 192
Query: 200 KSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFP 259
KSQR ASFLLYL+DV+EGGET+FP+ENG NMD SY + CIGL+V+PR+GDGLLFYSLFP
Sbjct: 193 KSQRAASFLLYLSDVEEGGETVFPYENGQNMDASYDFSKCIGLKVKPRRGDGLLFYSLFP 252
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQD 292
NGTID TSLHGSCPVI+GEKWVATKWIR+ DQD
Sbjct: 253 NGTIDLTSLHGSCPVIRGEKWVATKWIRNQDQD 285
>UniRef100_Q8VZJ7 Hypothetical protein At4g33910 [Arabidopsis thaliana]
Length = 288
Score = 385 bits (989), Expect = e-106
Identities = 188/284 (66%), Positives = 223/284 (78%), Gaps = 5/284 (1%)
Query: 11 SSRSNKLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNL--M 68
S R KL + + C FL GF+GSTL S + ++PR R+L+ E E M
Sbjct: 7 SYRRKKLGLATVIVFCSLCFLFGFYGSTLLSQNVPR---VKPRLRMLDMVENGEEEASSM 63
Query: 69 TAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETE 128
G G++SI SIPFQVLSW+PRA+YFPNFATAEQC++I+ AK LKPSALALR+GET
Sbjct: 64 PHGVTGEESIGSIPFQVLSWRPRAIYFPNFATAEQCQAIIERAKVNLKPSALALRKGETA 123
Query: 129 QNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHY 188
+NTKG RTSSG F+S+SE+ TG L +E KIARATMIPRSHGE+FNILRYE+ Q+Y+ HY
Sbjct: 124 ENTKGTRTSSGTFISASEESTGALDFVERKIARATMIPRSHGESFNILRYELGQKYDSHY 183
Query: 189 DSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQ 248
D FNP EYGPQ SQR+ASFLLYL+DV+EGGETMFPFENG NM + Y Y+ CIGL+V+PR+
Sbjct: 184 DVFNPTEYGPQSSQRIASFLLYLSDVEEGGETMFPFENGSNMGIGYDYKQCIGLKVKPRK 243
Query: 249 GDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQD 292
GDGLLFYS+FPNGTID TSLHGSCPV KGEKWVATKWIRD DQ+
Sbjct: 244 GDGLLFYSVFPNGTIDQTSLHGSCPVTKGEKWVATKWIRDQDQE 287
>UniRef100_Q94H92 Putative dioxygenase [Oryza sativa]
Length = 310
Score = 377 bits (969), Expect = e-103
Identities = 184/273 (67%), Positives = 216/273 (78%), Gaps = 1/273 (0%)
Query: 16 KLTFPYIFLICIFFFLAGFFGSTLFSHSQDDGRGLRPRPRLLESSEKAEYNLMTAGEFGD 75
+L P + L C FFLAGFFGS LF+ L P E +A + M GE G+
Sbjct: 34 RLRLPVVLLSCSLFFLAGFFGSILFTQDPQGEEEL-DTPMRRERLMEAAWPGMAYGESGE 92
Query: 76 DSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIR 135
+ IP+Q+LSW+PRALYFP FAT++QC++IV AK L PS LALR+GETE++TKGIR
Sbjct: 93 PEPSLIPYQILSWQPRALYFPQFATSQQCENIVKTAKQRLMPSTLALRKGETEESTKGIR 152
Query: 136 TSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAE 195
TSSG F+SS ED TGTLA +E+KIA+ATMIPR HGE FNILRYE+ QRY HYD+F+PA+
Sbjct: 153 TSSGTFLSSDEDPTGTLAEVEKKIAKATMIPRHHGEPFNILRYEIGQRYASHYDAFDPAQ 212
Query: 196 YGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFY 255
YGPQKSQR+ASFLLYLTDV+EGGETMFP+ENG NMD+ Y YE CIGL+V+PR+GDGLLFY
Sbjct: 213 YGPQKSQRVASFLLYLTDVEEGGETMFPYENGENMDIGYDYEKCIGLKVKPRKGDGLLFY 272
Query: 256 SLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRD 288
SL NGTIDPTSLHGSCPVIKGEKWVATKWIRD
Sbjct: 273 SLMVNGTIDPTSLHGSCPVIKGEKWVATKWIRD 305
>UniRef100_O81759 Hypothetical protein F17I5.100 [Arabidopsis thaliana]
Length = 257
Score = 310 bits (793), Expect = 4e-83
Identities = 147/209 (70%), Positives = 178/209 (84%), Gaps = 1/209 (0%)
Query: 56 LLESSEKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGL 115
++E+ E+ E + M G G++SI SIPFQVLSW+PRA+YFPNFATAEQC++I+ AK L
Sbjct: 1 MVENGEE-EASSMPHGVTGEESIGSIPFQVLSWRPRAIYFPNFATAEQCQAIIERAKVNL 59
Query: 116 KPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNI 175
KPSALALR+GET +NTKG RTSSG F+S+SE+ TG L +E KIARATMIPRSHGE+FNI
Sbjct: 60 KPSALALRKGETAENTKGTRTSSGTFISASEESTGALDFVERKIARATMIPRSHGESFNI 119
Query: 176 LRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSYR 235
LRYE+ Q+Y+ HYD FNP EYGPQ SQR+ASFLLYL+DV+EGGETMFPFENG NM + Y
Sbjct: 120 LRYELGQKYDSHYDVFNPTEYGPQSSQRIASFLLYLSDVEEGGETMFPFENGSNMGIGYD 179
Query: 236 YEDCIGLRVRPRQGDGLLFYSLFPNGTID 264
Y+ CIGL+V+PR+GDGLLFYS+FPNGTID
Sbjct: 180 YKQCIGLKVKPRKGDGLLFYSVFPNGTID 208
>UniRef100_Q8LEY1 Putative prolyl 4-hydroxylase, alpha subunit [Arabidopsis thaliana]
Length = 287
Score = 180 bits (457), Expect = 3e-44
Identities = 94/207 (45%), Positives = 130/207 (62%), Gaps = 7/207 (3%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVS 143
+VLSW+PRA + NF + E+C+ ++ +AK + S + E ++++ +RTSSG F+
Sbjct: 77 EVLSWEPRAFVYHNFLSKEECEYLISLAKPHMVKSTVVDSETGKSKDSR-VRTSSGTFLR 135
Query: 144 SSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQR 203
DK + IE++IA T IP HGE +L YE Q+Y PHYD F QR
Sbjct: 136 RGRDKI--IKTIEKRIADYTFIPADHGEGLQVLHYEAGQKYEPHYDYFVDEFNTKNGGQR 193
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IGLRVRPRQGDGLLFYSLFP 259
MA+ L+YL+DV+EGGET+FP N V + E +C GL V+PR GD LLF+S+ P
Sbjct: 194 MATMLMYLSDVEEGGETVFPAANMNFSSVPWYNELSECGKKGLSVKPRMGDALLFWSMRP 253
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWI 286
+ T+DPTSLHG CPVI+G KW +TKWI
Sbjct: 254 DATLDPTSLHGGCPVIRGNKWSSTKWI 280
>UniRef100_Q9LN20 F14O10.12 protein [Arabidopsis thaliana]
Length = 287
Score = 179 bits (454), Expect = 8e-44
Identities = 93/207 (44%), Positives = 130/207 (61%), Gaps = 7/207 (3%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVS 143
+VLSW+PRA + NF + E+C+ ++ +AK + S + E ++++ +RTSSG F+
Sbjct: 77 EVLSWEPRAFVYHNFLSKEECEYLISLAKPHMVKSTVVDSETGKSKDSR-VRTSSGTFLR 135
Query: 144 SSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQR 203
DK + IE++IA T IP HGE +L YE Q+Y PHYD F QR
Sbjct: 136 RGRDKI--IKTIEKRIADYTFIPADHGEGLQVLHYEAGQKYEPHYDYFVDEFNTKNGGQR 193
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IGLRVRPRQGDGLLFYSLFP 259
MA+ L+YL+DV+EGGET+FP N V + E +C GL V+PR GD LLF+S+ P
Sbjct: 194 MATMLMYLSDVEEGGETVFPAANMNFSSVPWYNELSECGKKGLSVKPRMGDALLFWSMRP 253
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWI 286
+ T+DPTSLHG CPVI+G KW +TKW+
Sbjct: 254 DATLDPTSLHGGCPVIRGNKWSSTKWM 280
>UniRef100_Q6K7Q8 Putative prolyl 4-hydroxylase [Oryza sativa]
Length = 310
Score = 178 bits (452), Expect = 1e-43
Identities = 92/214 (42%), Positives = 135/214 (62%), Gaps = 9/214 (4%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVS 143
+V+SW+PRA + NF + E+C ++G+AK + S + ++++ +RTSSG+F+
Sbjct: 100 EVISWEPRAFVYHNFLSKEECDYLIGLAKPHMVKSTVVDSTTGKSKDSR-VRTSSGMFLQ 158
Query: 144 SSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQR 203
DK + IE++IA T IP HGE +L YEV Q+Y PH+D F QR
Sbjct: 159 RGRDKV--IRAIEKRIADYTFIPMEHGEGLQVLHYEVGQKYEPHFDYFLDEYNTKNGGQR 216
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DCI--GLRVRPRQGDGLLFYSLFP 259
MA+ L+YL+DV+EGGET+FP N + + + E +C GL V+P+ GD LLF+S+ P
Sbjct: 217 MATLLMYLSDVEEGGETIFPDANVNSSSLPWYNELSECARKGLAVKPKMGDALLFWSMKP 276
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQDY 293
+ T+DP SLHG CPVIKG KW +TKW+ H ++Y
Sbjct: 277 DATLDPLSLHGGCPVIKGNKWSSTKWM--HVREY 308
>UniRef100_Q84T04 Putative oxidoreductase [Oryza sativa]
Length = 299
Score = 169 bits (427), Expect = 1e-40
Identities = 87/204 (42%), Positives = 131/204 (63%), Gaps = 6/204 (2%)
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
LSW+PRA + F + ++C +V +AK ++ S +A + ++ +RTSSG F+S
Sbjct: 40 LSWRPRAFLYSGFLSHDECDHLVNLAKGRMEKSMVADNDSGKSIMSQ-VRTSSGTFLSKH 98
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
ED ++ IE+++A T +P + E+ IL YE+ Q+Y+ H+D F+ + R+A
Sbjct: 99 EDDI--VSGIEKRVAAWTFLPEENAESIQILHYELGQKYDAHFDYFHDKNNLKRGGHRVA 156
Query: 206 SFLLYLTDVQEGGETMFPFENGLNMDVSYR-YEDCI--GLRVRPRQGDGLLFYSLFPNGT 262
+ L+YLTDV++GGET+FP G ++ + + DC GL V+P++GD LLF+SL N T
Sbjct: 157 TVLMYLTDVKKGGETVFPNAAGRHLQLKDETWSDCARSGLAVKPKKGDALLFFSLHVNAT 216
Query: 263 IDPTSLHGSCPVIKGEKWVATKWI 286
DP SLHGSCPVI+GEKW ATKWI
Sbjct: 217 TDPASLHGSCPVIEGEKWSATKWI 240
>UniRef100_Q8LB05 Putative prolyl 4-hydroxylase, alpha subunit [Arabidopsis thaliana]
Length = 291
Score = 163 bits (412), Expect = 6e-39
Identities = 83/211 (39%), Positives = 130/211 (61%), Gaps = 7/211 (3%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVS 143
+V+SW+PRA+ + NF T E+C+ ++ +AK + S + + ++++ +RTSSG F+
Sbjct: 81 EVISWEPRAVVYHNFLTNEECEHLISLAKPSMVKSTVVDEKTGGSKDSR-VRTSSGTFLR 139
Query: 144 SSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQR 203
D+ + +IE++I+ T IP +GE +L Y+V Q+Y PHYD F QR
Sbjct: 140 RGHDEV--VEVIEKRISDFTFIPVENGEGLQVLHYQVGQKYEPHYDYFLDEFNTKNGGQR 197
Query: 204 MASFLLYLTDVQEGGETMFPFENGLNMDVSYRYE--DC--IGLRVRPRQGDGLLFYSLFP 259
+A+ L+YL+DV +GGET+FP G V + E C GL V P+ D LLF+++ P
Sbjct: 198 IATVLMYLSDVDDGGETVFPAARGNISAVPWWNELSKCGKEGLSVLPKXRDALLFWNMRP 257
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWIRDHD 290
+ ++DP+SLHG CPV+KG KW +TKW H+
Sbjct: 258 DASLDPSSLHGGCPVVKGNKWSSTKWFHVHE 288
>UniRef100_Q84W84 Putative prolyl 4-hydroxylase [Arabidopsis thaliana]
Length = 253
Score = 162 bits (411), Expect = 7e-39
Identities = 85/203 (41%), Positives = 122/203 (59%), Gaps = 5/203 (2%)
Query: 87 SWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSE 146
SW PRA + F + E+C ++ +AK L+ S + E +RTSSG+F++ +
Sbjct: 1 SWTPRAFLYKGFLSDEECDHLIKLAKGKLEKSMVVADVDSGESEDSEVRTSSGMFLTKRQ 60
Query: 147 DKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMAS 206
D +A +E K+A T +P +GEA IL YE Q+Y+PH+D F + R+A+
Sbjct: 61 DDI--VANVEAKLAAWTFLPEENGEALQILHYENGQKYDPHFDYFYDKKALELGGHRIAT 118
Query: 207 FLLYLTDVQEGGETMFPFENGLNMDVSY-RYEDCI--GLRVRPRQGDGLLFYSLFPNGTI 263
L+YL++V +GGET+FP G + + C G V+PR+GD LLF++L NGT
Sbjct: 119 VLMYLSNVTKGGETVFPNWKGKTPQLKDDSWSKCAKQGYAVKPRKGDALLFFNLHLNGTT 178
Query: 264 DPTSLHGSCPVIKGEKWVATKWI 286
DP SLHGSCPVI+GEKW AT+WI
Sbjct: 179 DPNSLHGSCPVIEGEKWSATRWI 201
>UniRef100_Q6AVM9 Putative prolyl 4-hydroxylase alpha subunit [Oryza sativa]
Length = 319
Score = 162 bits (410), Expect = 1e-38
Identities = 92/210 (43%), Positives = 131/210 (61%), Gaps = 13/210 (6%)
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALR-EGETEQNTKGIRTSSGVFVSS 144
+SWKPR + +F + ++ +V +A+ LK SA+A G++E + RTSSG F+
Sbjct: 61 ISWKPRVFLYQHFLSDDEANHLVSLARTELKRSAVADNLSGKSELSDA--RTSSGTFIRK 118
Query: 145 SEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRM 204
S+D +A IEEKIA T +P+ +GE +LRY+ ++Y HYD F+ + R+
Sbjct: 119 SQDPI--VAGIEEKIAAWTFLPKENGEDIQVLRYKHGEKYERHYDYFSDNVNTLRGGHRI 176
Query: 205 ASFLLYLTDVQEGGETMFPF-----ENGLNMDVSYRYEDCI--GLRVRPRQGDGLLFYSL 257
A+ L+YLTDV EGGET+FP E+G N + S +C G+ V+PR+GD LLF++L
Sbjct: 177 ATVLMYLTDVAEGGETVFPLAEEFTESGTNNEDS-TLSECAKKGVAVKPRKGDALLFFNL 235
Query: 258 FPNGTIDPTSLHGSCPVIKGEKWVATKWIR 287
P+ + D SLH CPVIKGEKW ATKWIR
Sbjct: 236 SPDASKDSLSLHAGCPVIKGEKWSATKWIR 265
>UniRef100_Q84T01 Putative oxidoreductase [Oryza sativa]
Length = 310
Score = 162 bits (409), Expect = 1e-38
Identities = 91/229 (39%), Positives = 134/229 (57%), Gaps = 11/229 (4%)
Query: 67 LMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGE 126
++ A F S+T ++SWKPR ++ F + ++C +V + K LK S +A E
Sbjct: 37 VVAAPPFNASSVT-----IISWKPRIFFYKGFLSDDECDHLVKLGKEKLKRSMVADNESG 91
Query: 127 TEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNP 186
++ +RTSSG+F+ +D ++ IEE+IA T++P+ + E ILRYE Q+Y+P
Sbjct: 92 KSVMSE-VRTSSGMFLDKQQDPV--VSGIEERIAAWTLLPQENAENIQILRYENGQKYDP 148
Query: 187 HYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGL-NMDVSYRYEDCI--GLR 243
H+D F Q R A+ L YL+ V++GGET+FP G + + DC GL
Sbjct: 149 HFDYFQDKVNQLQGGHRYATVLTYLSTVEKGGETVFPNAEGWESQPKDDSFSDCAKKGLA 208
Query: 244 VRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIRDHDQD 292
V+ +GD +LF++L P+GT DP SLHGSCPVI+GEKW A KWI D
Sbjct: 209 VKAVKGDSVLFFNLQPDGTPDPLSLHGSCPVIEGEKWSAPKWIHVRSYD 257
>UniRef100_Q8RWK0 Hypothetical protein At5g18900 [Arabidopsis thaliana]
Length = 298
Score = 160 bits (405), Expect = 4e-38
Identities = 90/207 (43%), Positives = 127/207 (60%), Gaps = 9/207 (4%)
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
+S KPRA + F T +C +V +AKA LK SA+A + E +RTSSG F+S
Sbjct: 40 VSSKPRAFVYEGFLTELECDHMVSLAKASLKRSAVADNDSG-ESKFSEVRTSSGTFISKG 98
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
+D ++ IE+KI+ T +P+ +GE +LRYE Q+Y+ H+D F+ + RMA
Sbjct: 99 KDPI--VSGIEDKISTWTFLPKENGEDIQVLRYEHGQKYDAHFDYFHDKVNIVRGGHRMA 156
Query: 206 SFLLYLTDVQEGGETMFPFENGLNMDVSYRYE----DCI--GLRVRPRQGDGLLFYSLFP 259
+ L+YL++V +GGET+FP + V E DC G+ V+PR+GD LLF++L P
Sbjct: 157 TILMYLSNVTKGGETVFPDAEIPSRRVLSENEEDLSDCAKRGIAVKPRKGDALLFFNLHP 216
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWI 286
+ DP SLHG CPVI+GEKW ATKWI
Sbjct: 217 DAIPDPLSLHGGCPVIEGEKWSATKWI 243
>UniRef100_Q9LSI6 Prolyl 4-hydroxylase alpha subunit-like protein [Arabidopsis
thaliana]
Length = 332
Score = 160 bits (404), Expect = 5e-38
Identities = 90/224 (40%), Positives = 135/224 (60%), Gaps = 15/224 (6%)
Query: 69 TAGEFGDDSITSIPFQV--LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALRE-G 125
+A FG D P +V LSW PR + F + E+C + +AK L+ S +A + G
Sbjct: 61 SASSFGFD-----PTRVTQLSWTPRVFLYEGFLSDEECDHFIKLAKGKLEKSMVADNDSG 115
Query: 126 ETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYN 185
E+ ++ +RTSSG+F+S +D ++ +E K+A T +P +GE+ IL YE Q+Y
Sbjct: 116 ESVESE--VRTSSGMFLSKRQDDI--VSNVEAKLAAWTFLPEENGESMQILHYENGQKYE 171
Query: 186 PHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSY-RYEDCI--GL 242
PH+D F+ R+A+ L+YL++V++GGET+FP G + + +C G
Sbjct: 172 PHFDYFHDQANLELGGHRIATVLMYLSNVEKGGETVFPMWKGKATQLKDDSWTECAKQGY 231
Query: 243 RVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWI 286
V+PR+GD LLF++L PN T D SLHGSCPV++GEKW AT+WI
Sbjct: 232 AVKPRKGDALLFFNLHPNATTDSNSLHGSCPVVEGEKWSATRWI 275
>UniRef100_Q8L970 Prolyl 4-hydroxylase, putative [Arabidopsis thaliana]
Length = 316
Score = 160 bits (404), Expect = 5e-38
Identities = 90/224 (40%), Positives = 135/224 (60%), Gaps = 15/224 (6%)
Query: 69 TAGEFGDDSITSIPFQV--LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALRE-G 125
+A FG D P +V LSW PR + F + E+C + +AK L+ S +A + G
Sbjct: 45 SASSFGFD-----PTRVTQLSWTPRVFLYEGFLSDEECDHFIKLAKGKLEKSMVADNDSG 99
Query: 126 ETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYN 185
E+ ++ +RTSSG+F+S +D ++ +E K+A T +P +GE+ IL YE Q+Y
Sbjct: 100 ESVESE--VRTSSGMFLSKRQDDI--VSNVEAKLAAWTFLPEENGESMQILHYENGQKYE 155
Query: 186 PHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSY-RYEDCI--GL 242
PH+D F+ R+A+ L+YL++V++GGET+FP G + + +C G
Sbjct: 156 PHFDYFHDQANLELGGHRIATVLMYLSNVEKGGETVFPMWKGKATQLKDDSWTECAKQGY 215
Query: 243 RVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWI 286
V+PR+GD LLF++L PN T D SLHGSCPV++GEKW AT+WI
Sbjct: 216 AVKPRKGDALLFFNLHPNATTDSNSLHGSCPVVEGEKWSATRWI 259
>UniRef100_Q8LAN3 Prolyl 4-hydroxylase alpha subunit-like protein [Arabidopsis
thaliana]
Length = 298
Score = 159 bits (402), Expect = 8e-38
Identities = 90/207 (43%), Positives = 127/207 (60%), Gaps = 9/207 (4%)
Query: 86 LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSS 145
+S KPRA + F T +C +V +AKA LK SA+A + E +RTSSG F+S
Sbjct: 40 VSSKPRAFVYEGFLTELECDHMVSLAKASLKRSAVADNDSG-ESKFSEVRTSSGTFISKG 98
Query: 146 EDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMA 205
+D ++ IE+KI+ T +P+ +GE +LRYE Q+Y+ H+D F+ + RMA
Sbjct: 99 KDPI--VSGIEDKISTWTFLPKENGEDIQVLRYEHGQKYDAHFDYFHDKVNIVRGGHRMA 156
Query: 206 SFLLYLTDVQEGGETMFP-FENGLNMDVSYRYEDCI-----GLRVRPRQGDGLLFYSLFP 259
+ L+YL++V +GGET+FP E +S ED G+ V+PR+GD LLF++L P
Sbjct: 157 TILMYLSNVTKGGETVFPDAEIPSRRVLSENKEDLSDCAKRGIAVKPRKGDALLFFNLHP 216
Query: 260 NGTIDPTSLHGSCPVIKGEKWVATKWI 286
+ DP SLHG CPVI+GEKW ATKWI
Sbjct: 217 DAIPDPLSLHGGCPVIEGEKWSATKWI 243
>UniRef100_Q8VZD7 AT3g28480/MFJ20_16 [Arabidopsis thaliana]
Length = 316
Score = 159 bits (401), Expect = 1e-37
Identities = 90/224 (40%), Positives = 134/224 (59%), Gaps = 15/224 (6%)
Query: 69 TAGEFGDDSITSIPFQV--LSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALRE-G 125
+A FG D P +V LSW PR + F + E+C + +AK L+ S +A + G
Sbjct: 45 SASSFGFD-----PTRVTQLSWTPRVFLYEGFLSDEECDHFIKLAKGKLEKSMVADNDSG 99
Query: 126 ETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYN 185
E+ ++ +RTSSG+F+S +D + +E K+A T +P +GE+ IL YE Q+Y
Sbjct: 100 ESVESE--VRTSSGMFLSKRQDDI--VNNVEAKLAAWTFLPEENGESMQILHYENGQKYE 155
Query: 186 PHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGLNMDVSY-RYEDCI--GL 242
PH+D F+ R+A+ L+YL++V++GGET+FP G + + +C G
Sbjct: 156 PHFDYFHDQANLELGGHRIATVLMYLSNVEKGGETVFPMWKGKATQLKDDSWTECAKQGY 215
Query: 243 RVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWI 286
V+PR+GD LLF++L PN T D SLHGSCPV++GEKW AT+WI
Sbjct: 216 AVKPRKGDALLFFNLHPNATTDSNSLHGSCPVVEGEKWSATRWI 259
>UniRef100_Q8W3E9 Putative prolyl 4-hydroxylase, alpha subunit [Oryza sativa]
Length = 343
Score = 157 bits (398), Expect = 2e-37
Identities = 92/242 (38%), Positives = 141/242 (58%), Gaps = 17/242 (7%)
Query: 47 GRGL-RPRPRLLESSEKAEYNLMTAGEFGDDSITSIPFQVLSWKPRALYFPNFATAEQCK 105
G GL RPRPR S+ M GE G+ +VLSW+PRA + NF + E+C+
Sbjct: 78 GIGLPRPRPRFRRSAAFESGLEMRGGEKGEPWT-----EVLSWEPRAFLYHNFLSKEECE 132
Query: 106 SIVGVAKAGLKPSALALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMI 165
++ +AK +K S + ++++ +RTSSG+F+ +DK + IE++I+ T I
Sbjct: 133 YLISLAKPHMKKSTVVDASTGGSKDSR-VRTSSGMFLGRGQDKI--IRTIEKRISDYTFI 189
Query: 166 PRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFE 225
P +GE +L YEV Q+Y PH+D F+ QR+A+ L+YL+DV+EGGET+FP
Sbjct: 190 PVENGEGLQVLHYEVGQKYEPHFDYFHDEFNTKNGGQRIATLLMYLSDVEEGGETIFPSS 249
Query: 226 NGLNMDVSYRYE--DCI--GLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWV 281
+ + E +C GL V+P+ GD LLF+S+ P+G++D TSLHG P++ W+
Sbjct: 250 KANSSSSPFYNELSECAKKGLAVKPKMGDALLFWSMRPDGSLDATSLHGEIPIL----WL 305
Query: 282 AT 283
T
Sbjct: 306 LT 307
>UniRef100_Q7XNS6 OSJNBb0085H11.11 protein [Oryza sativa]
Length = 267
Score = 157 bits (398), Expect = 2e-37
Identities = 85/208 (40%), Positives = 127/208 (60%), Gaps = 5/208 (2%)
Query: 84 QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKPSALALREGETEQNTKG-IRTSSGVFV 142
+V+SW PR + F NF ++E+C + +A+ L+ S + + T + K +RTSSG+FV
Sbjct: 62 EVISWSPRIIVFHNFLSSEECDYLRSIARPRLQISTVV--DVATGKGVKSNVRTSSGMFV 119
Query: 143 SSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILRYEVDQRYNPHYDSFNPAEYGPQKSQ 202
SS E K + IE++I+ + IP +GE +LRYE Q Y PH+D F+ + Q
Sbjct: 120 SSEERKLPVIQSIEKRISVYSQIPEENGELIQVLRYEPSQYYRPHHDYFSDTFNIKRGGQ 179
Query: 203 RMASFLLYLTDVQEGGETMFPFENGLNMDVSYRYEDCIGLRVRPRQGDGLLFYSLFPNGT 262
R+A+ L+YLTD EGGET FP + + S + GL V+P +GD +LF+S+ +G
Sbjct: 180 RVATMLMYLTDGVEGGETHFP--QAGDGECSCGGKMVKGLCVKPNKGDAVLFWSMGLDGE 237
Query: 263 IDPTSLHGSCPVIKGEKWVATKWIRDHD 290
D S+HG CPV++GEKW ATKW+R +
Sbjct: 238 TDSNSIHGGCPVLEGEKWSATKWMRQKE 265
>UniRef100_Q6ZL00 Prolyl 4-hydroxylase alpha-1 subunit-like protein [Oryza sativa]
Length = 313
Score = 157 bits (396), Expect = 4e-37
Identities = 89/233 (38%), Positives = 135/233 (57%), Gaps = 15/233 (6%)
Query: 67 LMTAGEFGDDSITSI----PF-----QVLSWKPRALYFPNFATAEQCKSIVGVAKAGLKP 117
L+ GE GDD + ++ PF + +SW+PR + F + ++C +V + K ++
Sbjct: 26 LLLLGEAGDDGVGAVAAAPPFNASRVRAVSWRPRVFVYKGFLSDDECDHLVKLGKRKMQR 85
Query: 118 SALALREGETEQNTKGIRTSSGVFVSSSEDKTGTLAIIEEKIARATMIPRSHGEAFNILR 177
S +A + ++ +RTSSG+F+ +D ++ IE++IA T +P + E ILR
Sbjct: 86 SMVADNKSGKSVMSE-VRTSSGMFLDKRQDPV--VSRIEKRIAAWTFLPEENAENIQILR 142
Query: 178 YEVDQRYNPHYDSFNPAEYGPQKSQRMASFLLYLTDVQEGGETMFPFENGL-NMDVSYRY 236
YE Q+Y PH+D F+ R A+ L+YL+ V++GGET+FP G N +
Sbjct: 143 YEHGQKYEPHFDYFHDKVNQALGGHRYATVLMYLSTVEKGGETVFPNAEGWENQPKDDTF 202
Query: 237 EDCI--GLRVRPRQGDGLLFYSLFPNGTIDPTSLHGSCPVIKGEKWVATKWIR 287
+C GL V+P +GD +LF+SL +G DP SLHGSCPVI+GEKW A KWIR
Sbjct: 203 SECAQKGLAVKPVKGDTVLFFSLHIDGVPDPLSLHGSCPVIEGEKWSAPKWIR 255
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 503,608,857
Number of Sequences: 2790947
Number of extensions: 21080915
Number of successful extensions: 42224
Number of sequences better than 10.0: 292
Number of HSP's better than 10.0 without gapping: 199
Number of HSP's successfully gapped in prelim test: 93
Number of HSP's that attempted gapping in prelim test: 41559
Number of HSP's gapped (non-prelim): 319
length of query: 293
length of database: 848,049,833
effective HSP length: 126
effective length of query: 167
effective length of database: 496,390,511
effective search space: 82897215337
effective search space used: 82897215337
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0157b.6


