

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
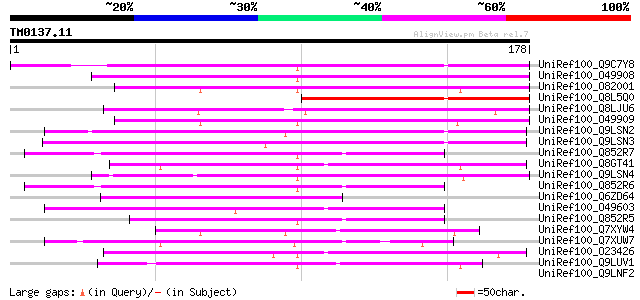
Query= TM0137.11
(178 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9C7Y8 Hypothetical protein T2J15.13 [Arabidopsis thal... 134 8e-31
UniRef100_O49908 Invertase inhibitor precursor [Nicotiana tabacum] 122 5e-27
UniRef100_O82001 Tomato invertase inhibitor precursor [Lycopersi... 110 2e-23
UniRef100_Q8L5Q0 Putative invertase inhibitor [Cicer arietinum] 100 3e-20
UniRef100_Q8LJU6 Invertase inhibitor-like protein [Ipomoea batatas] 98 1e-19
UniRef100_O49909 Vacuolar invertase inhibitor [Nicotiana tabacum] 95 7e-19
UniRef100_Q9LSN2 Invertase inhibitor-like protein [Arabidopsis t... 73 3e-12
UniRef100_Q9LSN3 Invertase inhibitor-like protein [Arabidopsis t... 70 2e-11
UniRef100_Q852R7 Pectinesterase inhibitor [Actinidia chinensis] 58 1e-07
UniRef100_Q8GT41 Putative invertase inhibitor precursor [Platanu... 58 1e-07
UniRef100_Q9LSN4 Arabidopsis thaliana genomic DNA, chromosome 3,... 57 2e-07
UniRef100_Q852R6 Pectinmethylesterase inhibitor [Actinidia delic... 56 4e-07
UniRef100_Q6ZD64 Nvertase/pectin methylesterase inhibitor-like [... 55 6e-07
UniRef100_O49603 Invertase inhibitor homologue precursor [Arabid... 54 1e-06
UniRef100_Q852R5 Pectinmethylesterase inhibitor [Actinidia delic... 54 2e-06
UniRef100_Q7XYW4 Seed specific protein Bn15D12A [Brassica napus] 51 1e-05
UniRef100_Q7XUW7 OSJNBa0027P08.11 protein [Oryza sativa] 48 1e-04
UniRef100_O23426 Hypothetical protein AT4g15750 [Arabidopsis tha... 47 3e-04
UniRef100_Q9LUV1 Gb|AAF16649.1 [Arabidopsis thaliana] 45 8e-04
UniRef100_Q9LNF2 F21D18.29 [Arabidopsis thaliana] 44 0.001
>UniRef100_Q9C7Y8 Hypothetical protein T2J15.13 [Arabidopsis thaliana]
Length = 166
Score = 134 bits (338), Expect = 8e-31
Identities = 76/179 (42%), Positives = 106/179 (58%), Gaps = 14/179 (7%)
Query: 1 MENLKSLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPR 60
M+ +K + LI + +V+ +S + ++ TCK TP + C+ L +DPR
Sbjct: 1 MKMMKVMMLIVMMMMVMVMVS------------EGSIIEPTCKETPDFNLCVSLLNSDPR 48
Query: 61 SSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGD-KPALNSCQGRYNAILKADIPQA 119
SSAD +GLALI++D I+ LN+I+ L K + K AL+ C RY IL AD+P+A
Sbjct: 49 GSSADTSGLALILIDKIKGLATKTLNEINGLYKKRPELKRALDECSRRYKTILNADVPEA 108
Query: 120 TQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNLL 178
+A+ G PKF EDGV DAGVEA+ C+ GF +G SPLTS M + V RAI+R LL
Sbjct: 109 IEAISKGVPKFGEDGVIDAGVEASVCQGGF-NGSSPLTSLTKSMQKISNVTRAIVRMLL 166
>UniRef100_O49908 Invertase inhibitor precursor [Nicotiana tabacum]
Length = 166
Score = 122 bits (305), Expect = 5e-27
Identities = 68/153 (44%), Positives = 91/153 (59%), Gaps = 3/153 (1%)
Query: 29 VLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKI 88
+LQ + LV TCKNTP CL+ L +D RS++ D+T LALIMVD I+AK N I
Sbjct: 14 LLQTNANNLVETTCKNTPNYQLCLKTLLSDKRSATGDITTLALIMVDAIKAKANQAAVTI 73
Query: 89 SQLLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTC 145
S+L K L +C Y IL A +P+A +AL G+PKFAEDG+ + +A C
Sbjct: 74 SKLRHSNPPAAWKGPLKNCAFSYKVILTASLPEAIEALTKGDPKFAEDGMVGSSGDAQEC 133
Query: 146 ESGFSSGKSPLTSENNGMHIATEVARAIIRNLL 178
E F KSP ++ N +H ++V RAI+RNLL
Sbjct: 134 EEYFKGSKSPFSALNIAVHELSDVGRAIVRNLL 166
>UniRef100_O82001 Tomato invertase inhibitor precursor [Lycopersicon esculentum]
Length = 171
Score = 110 bits (274), Expect = 2e-23
Identities = 66/149 (44%), Positives = 87/149 (58%), Gaps = 7/149 (4%)
Query: 37 LVAKTCKNTPYPSACLQFLQADPRSSSA-DVTGLALIMVDVIQAKTNGVLNKISQLLKGG 95
LV TCKNTP + C++ L D RS A D+T LALIMVD I++K N N IS+L
Sbjct: 23 LVETTCKNTPNYNLCVKTLSLDKRSEKAGDITTLALIMVDAIKSKANQAANTISKLRHSN 82
Query: 96 GD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSSG 152
K L +C Y IL A +P+A +AL G+PKFAEDG+ + +A CE F +
Sbjct: 83 PPQAWKDPLKNCAFSYKVILPASMPEALEALTKGDPKFAEDGMVGSSGDAQECEEYFKAT 142
Query: 153 K---SPLTSENNGMHIATEVARAIIRNLL 178
SPL+ N +H ++V RAI+RNLL
Sbjct: 143 TIKYSPLSKLNIDVHELSDVGRAIVRNLL 171
>UniRef100_Q8L5Q0 Putative invertase inhibitor [Cicer arietinum]
Length = 77
Score = 99.8 bits (247), Expect = 3e-20
Identities = 51/78 (65%), Positives = 58/78 (73%), Gaps = 1/78 (1%)
Query: 101 LNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSSGKSPLTSEN 160
LNSC +Y AIL ADIPQA +AL+ G+PKFAEDG DA EAN CESGF GKSPLT +N
Sbjct: 1 LNSCSDKYKAILVADIPQAIEALQKGDPKFAEDGANDAANEANYCESGF-YGKSPLTKQN 59
Query: 161 NGMHIATEVARAIIRNLL 178
N MH + VA AI+R LL
Sbjct: 60 NAMHDVSSVAAAIVRELL 77
>UniRef100_Q8LJU6 Invertase inhibitor-like protein [Ipomoea batatas]
Length = 192
Score = 97.8 bits (242), Expect = 1e-19
Identities = 62/162 (38%), Positives = 89/162 (54%), Gaps = 19/162 (11%)
Query: 33 SDAQLVAKTCKNTPYPSACLQFLQADPRSSS--ADVTGLALIMVDVIQAKTNGVLNKISQ 90
S+ L+ C NTP + C+ L +DPR+SS +V L L+MVD ++AK ++ I +
Sbjct: 34 SNGGLINTACNNTPNYALCVSVLASDPRTSSKAVNVETLGLVMVDAVKAKAEEMIETIRE 93
Query: 91 LLKGGGDKPA-----LNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTC 145
L K KP L+ C Y A++ AD+P+A ALK G PKFAEDG+ADA VEA +C
Sbjct: 94 LEKS---KPVEWRLPLSQCYIYYYAVVHADVPEAEAALKRGVPKFAEDGMADAAVEAESC 150
Query: 146 ESGFSSGKSPLTSENNGMHI---------ATEVARAIIRNLL 178
E+ F + + +G I + VA +II+ LL
Sbjct: 151 EAAFKLQNEDIILDYSGSAIDETNKDVIQLSAVATSIIKMLL 192
>UniRef100_O49909 Vacuolar invertase inhibitor [Nicotiana tabacum]
Length = 172
Score = 95.1 bits (235), Expect = 7e-19
Identities = 58/148 (39%), Positives = 85/148 (57%), Gaps = 6/148 (4%)
Query: 37 LVAKTCKNTPYPSACLQFLQADPRSSSA---DVTGLALIMVDVIQAKTNGVLNKISQLLK 93
++ TC+ T CL L +DPR+S A D+T L L+MVD ++ K+ ++ I +L K
Sbjct: 25 IINTTCRATTNYPLCLTTLHSDPRTSEAEGADLTTLGLVMVDAVKLKSIEIMKSIKKLEK 84
Query: 94 GGGD-KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSSG 152
+ + L+ C Y A+L AD+ A +ALK G PKFAE+G+ D VEA TCE F
Sbjct: 85 SNPELRLPLSQCYIVYYAVLHADVTVAVEALKRGVPKFAENGMVDVAVEAETCEFSFKYN 144
Query: 153 --KSPLTSENNGMHIATEVARAIIRNLL 178
SP++ N + + VA++IIR LL
Sbjct: 145 GLVSPVSDMNKEIIELSSVAKSIIRMLL 172
>UniRef100_Q9LSN2 Invertase inhibitor-like protein [Arabidopsis thaliana]
Length = 175
Score = 73.2 bits (178), Expect = 3e-12
Identities = 48/168 (28%), Positives = 78/168 (45%), Gaps = 5/168 (2%)
Query: 13 IFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALI 72
IF+V++ I +++ + P + LV + CK Y + C+ L DPRS ++++ GLA I
Sbjct: 10 IFVVLSQIQIALSQT-IQSPPGSSLVQRLCKRNRYQALCISTLNVDPRSKTSNLQGLASI 68
Query: 73 MVDVIQAKTNGVLNKISQLLK---GGGDKPALNSCQGRYNAILKADIPQATQALKTGNPK 129
+D K N L +LK G D +C Y A + +P LK
Sbjct: 69 SLDATTKKFNVTLTYYISVLKNVRGRVDFERYGTCIEEYGAAVDRFLPAVKADLKAKKYP 128
Query: 130 FAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNL 177
A + D + CE F +G+SP+T+ N +H ++ II+ L
Sbjct: 129 EAMSEMKDVVAKPGYCEDQF-AGESPVTARNKAVHDIADMTADIIKTL 175
>UniRef100_Q9LSN3 Invertase inhibitor-like protein [Arabidopsis thaliana]
Length = 177
Score = 70.1 bits (170), Expect = 2e-11
Identities = 47/169 (27%), Positives = 76/169 (44%), Gaps = 4/169 (2%)
Query: 12 NIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLAL 71
+IF+V I +++ + ++ + + + CK PS C+ L DPRS ++++ LA
Sbjct: 6 SIFVVFMQIQVALSSQPIRYATEPKPIQELCKFNINPSLCVSTLNLDPRSKNSNLRELAW 65
Query: 72 IMVDVIQAKTNGVLN---KISQLLKGGGDKPALNSCQGRYNAILKADIPQATQALKTGNP 128
I +D K N +LN +S+ +K D +C Y + +P A LK G
Sbjct: 66 ISIDATSNKVNKMLNYLISVSKNIKDREDLKKYKTCIDDYGTAARRFLPAALDDLKAGFF 125
Query: 129 KFAEDGVADAGVEANTCESGFSSGKSPLTSENNGMHIATEVARAIIRNL 177
A+ + + CE+ F G SPLT N H + IIR L
Sbjct: 126 SLAKSDMESVVSIPDHCEAQF-GGSSPLTGRNKATHDIANMTADIIRYL 173
>UniRef100_Q852R7 Pectinesterase inhibitor [Actinidia chinensis]
Length = 185
Score = 57.8 bits (138), Expect = 1e-07
Identities = 44/147 (29%), Positives = 67/147 (44%), Gaps = 6/147 (4%)
Query: 6 SLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSAD 65
S SL ++ +V+ IS V ++ L+++ C T PS CLQ L++DPRS+S D
Sbjct: 7 SSSLFVSLLLVILFISPLSQRPSV--KAENHLISEICPKTRNPSLCLQALESDPRSASKD 64
Query: 66 VTGLALIMVDVIQAKTNGVLNKISQLLKGGGD---KPALNSCQGRYNAILKADIPQATQA 122
+ GL +D+ QA I+ L D K +C Y + + + QA Q
Sbjct: 65 LKGLGQFSIDIAQASAKQTSKIIASLTNQATDPKLKGRYETCSENYADAIDS-LGQAKQF 123
Query: 123 LKTGNPKFAEDGVADAGVEANTCESGF 149
L +G+ + A A TCE F
Sbjct: 124 LTSGDYNSLNIYASAAFDGAGTCEDSF 150
>UniRef100_Q8GT41 Putative invertase inhibitor precursor [Platanus acerifolia]
Length = 179
Score = 57.8 bits (138), Expect = 1e-07
Identities = 48/155 (30%), Positives = 74/155 (46%), Gaps = 13/155 (8%)
Query: 35 AQLVAKTCKNTPYPSA------CLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKI 88
A +V TCK S C++ L ADP+S +AD+ GL +I ++ + + I
Sbjct: 24 ADIVQGTCKKVAQRSPNVNYDFCVKSLGADPKSHTADLQGLGVISANLAIQHGSKIQTFI 83
Query: 89 SQLLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTC 145
++LK D K LN C G Y A K+ + +A K+ + A ++ A ++ TC
Sbjct: 84 GRILKSKVDPALKKYLNDCVGLY-ADAKSSVQEAIADFKSKDYASANVKMSAALDDSVTC 142
Query: 146 ESGFSSGK---SPLTSENNGMHIATEVARAIIRNL 177
E GF K SP+T EN T ++ AI + L
Sbjct: 143 EDGFKEKKGIVSPVTKENKDYVQLTAISLAITKLL 177
>UniRef100_Q9LSN4 Arabidopsis thaliana genomic DNA, chromosome 3, TAC clone:K14A17
[Arabidopsis thaliana]
Length = 183
Score = 57.0 bits (136), Expect = 2e-07
Identities = 44/159 (27%), Positives = 76/159 (47%), Gaps = 11/159 (6%)
Query: 29 VLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKI 88
V+Q SD L+ K C+ TP+ C L+ S S D L M V+ L I
Sbjct: 24 VVQSSD-DLIDKICQATPFCDLCEASLRPFSPSPS-DPKSLGAAMASVVLGNMTDTLGYI 81
Query: 89 SQLLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTC 145
L+K D + AL C Y ++K +IPQA +A++ G FA + DA + ++C
Sbjct: 82 QSLIKHAHDPAAERALAQCAELYRPVVKFNIPQAMEAMQGGKFGFAIYVLGDAEKQTDSC 141
Query: 146 ESGFSSGKS------PLTSENNGMHIATEVARAIIRNLL 178
+ G ++ + +T+ N + +VA +++++L+
Sbjct: 142 QKGITNAGADDESSVAVTARNKLVKNLCDVAISVLKSLM 180
>UniRef100_Q852R6 Pectinmethylesterase inhibitor [Actinidia deliciosa]
Length = 185
Score = 56.2 bits (134), Expect = 4e-07
Identities = 43/147 (29%), Positives = 67/147 (45%), Gaps = 6/147 (4%)
Query: 6 SLSLICNIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSAD 65
S SL ++ +V+ IS V ++ L+++ C T PS CLQ L++DPRS+S D
Sbjct: 7 SSSLFVSLLLVILFISPLSQRPSV--KAENHLISEICPKTRNPSLCLQALESDPRSASKD 64
Query: 66 VTGLALIMVDVIQAKTNGVLNKISQLLKGGGD---KPALNSCQGRYNAILKADIPQATQA 122
+ GL +D+ QA I+ L D K +C + + + + QA Q
Sbjct: 65 LKGLGQFSIDIAQASAKQTSKIIASLTNQATDPKLKGRYETCSENFADAIDS-LGQAKQF 123
Query: 123 LKTGNPKFAEDGVADAGVEANTCESGF 149
L +G+ + A A TCE F
Sbjct: 124 LTSGDYNSLNIYASAAFDGAGTCEDSF 150
>UniRef100_Q6ZD64 Nvertase/pectin methylesterase inhibitor-like [Oryza sativa]
Length = 223
Score = 55.5 bits (132), Expect = 6e-07
Identities = 31/83 (37%), Positives = 41/83 (49%)
Query: 32 PSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQL 91
P A LV TC +T Y C+ L ADP S++ADV GL+ I V V A +G + L
Sbjct: 52 PQAAALVRATCNSTAYYDVCVSALAADPSSTTADVRGLSAIAVSVAAANASGAAQAAAAL 111
Query: 92 LKGGGDKPALNSCQGRYNAILKA 114
G A + G A+L+A
Sbjct: 112 ANGTAPLAAAAAGDGTVQALLRA 134
>UniRef100_O49603 Invertase inhibitor homologue precursor [Arabidopsis thaliana]
Length = 180
Score = 54.3 bits (129), Expect = 1e-06
Identities = 40/141 (28%), Positives = 62/141 (43%), Gaps = 5/141 (3%)
Query: 13 IFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALI 72
IF+++ T++ S + + ++ TCK T Y C+ L++DPRS +AD GLA I
Sbjct: 6 IFLLLVTLTFSASTLISAKSNTTTIIESTCKTTNYYKFCVSALKSDPRSPTADTKGLASI 65
Query: 73 MVDV----IQAKTNGVLNKISQLLKGGGDKPALNSCQGRYNAILKADIPQATQALKTGNP 128
MV V + N + +S +K K L C +Y A+ + Q L
Sbjct: 66 MVGVGMTNATSTANYIAGNLSATVKDTVLKKVLQDCSEKY-ALAADSLRLTIQDLDDEAY 124
Query: 129 KFAEDGVADAGVEANTCESGF 149
+A V A N C + F
Sbjct: 125 DYASMHVLAAQDYPNVCRNIF 145
>UniRef100_Q852R5 Pectinmethylesterase inhibitor [Actinidia deliciosa]
Length = 147
Score = 53.5 bits (127), Expect = 2e-06
Identities = 37/111 (33%), Positives = 50/111 (44%), Gaps = 4/111 (3%)
Query: 42 CKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQLLKGGGD---K 98
C T PS CLQ L++DPRS+S D+ GL +D+ QA IS L D K
Sbjct: 3 CPKTRNPSLCLQALKSDPRSASKDLKGLGQFSIDIAQASAKQTSKIISSLTNQATDPKLK 62
Query: 99 PALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGF 149
+C Y + + + QA Q L +G+ + A A TCE F
Sbjct: 63 GRYETCSENYADAIDS-LGQAKQFLTSGDYNSLNIYASAAFDGAGTCEDSF 112
>UniRef100_Q7XYW4 Seed specific protein Bn15D12A [Brassica napus]
Length = 157
Score = 51.2 bits (121), Expect = 1e-05
Identities = 38/118 (32%), Positives = 60/118 (50%), Gaps = 8/118 (6%)
Query: 51 CLQFLQADPRSSSA-DVTGLALIMVDVIQAKTNGVLNKISQLLK----GGGDKPALNSCQ 105
C+Q L+ DP+S +A ++GL L + +KT V + +LK +PAL +C
Sbjct: 25 CIQSLEQDPQSKTATSLSGLVLAATNNAASKTINVKGIVETILKSKKYAPSTEPALRTCV 84
Query: 106 GRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESGFSS--GKSPLTSENN 161
Y+ + +A +K+ + K A ++ A E TCE GF KSP+T+ENN
Sbjct: 85 KLYDDAY-GSLKEALMNVKSSDYKSANMHLSAALDEPVTCEDGFKEKHAKSPVTNENN 141
>UniRef100_Q7XUW7 OSJNBa0027P08.11 protein [Oryza sativa]
Length = 192
Score = 47.8 bits (112), Expect = 1e-04
Identities = 44/154 (28%), Positives = 65/154 (41%), Gaps = 20/154 (12%)
Query: 13 IFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSA-------CLQFLQADPRSSSAD 65
+F ++ T++ S H + + + V TC + C+ LQA P S+ AD
Sbjct: 6 VFSLLLTVACS--HAALAAAASSSAVEDTCAKATASGSRKDLAPFCVSTLQAAPGSAGAD 63
Query: 66 VTGLALIMVDVIQAKTNGVLNKISQLLKGGG----DKPALNSCQGRYNAILKADIPQATQ 121
GLA+I ++ A I L + GG ++ AL +C+ Y L + A
Sbjct: 64 ARGLAVIATNLTLANYTAAYATIKALQRRGGWSERERAALATCRQLYIEALNV-VHSAIH 122
Query: 122 ALKTGNPKFAEDGVADAGV---EANTCESGFSSG 152
AL TG + VAD GV A CE F G
Sbjct: 123 ALNTGQ---TQAYVADMGVVRRAATGCEDAFGFG 153
>UniRef100_O23426 Hypothetical protein AT4g15750 [Arabidopsis thaliana]
Length = 171
Score = 46.6 bits (109), Expect = 3e-04
Identities = 43/153 (28%), Positives = 73/153 (47%), Gaps = 9/153 (5%)
Query: 33 SDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKIS--Q 90
+D +L+ C +T Y + CL L+ADP S + D GL I++ + ++ + ++N ++ +
Sbjct: 20 ADEELIKTECNHTEYQNVCLFCLEADPISFNIDRAGLVNIIIHCLGSQLDVLINTVTSLK 79
Query: 91 LLKGGGD--KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCESG 148
L+KG G+ + L C + AI + + A L T N A + V A TCE
Sbjct: 80 LMKGEGEANENVLKDCVTGF-AIAQLRLQGANIDLITLNYDKAYELVKTALNYPRTCEEN 138
Query: 149 FSSGKSPLTSENNGMHIA----TEVARAIIRNL 177
K +S+ +A T VA+ +I L
Sbjct: 139 LQKLKFKDSSDVYDDILAYSQLTSVAKTLIHRL 171
>UniRef100_Q9LUV1 Gb|AAF16649.1 [Arabidopsis thaliana]
Length = 173
Score = 45.1 bits (105), Expect = 8e-04
Identities = 36/136 (26%), Positives = 63/136 (45%), Gaps = 8/136 (5%)
Query: 31 QPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLALIMVDVIQAKTNGVLNKISQ 90
Q +D + + KN + C +++++P++S AD+ LA I Q + KI
Sbjct: 26 QVADIKAICGKAKNQSF---CTSYMKSNPKTSGADLQTLANITFGSAQTSASEGFRKIQS 82
Query: 91 LLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNPKFAEDGVADAGVEANTCES 147
L+K + K A SC Y + + + + A Q+L +G+ K V+ A +TCE
Sbjct: 83 LVKTATNPTMKKAYTSCVQHYKSAI-SSLNDAKQSLASGDGKGLNIKVSAAMEGPSTCEQ 141
Query: 148 GFSSGK-SPLTSENNG 162
+ K P +N+G
Sbjct: 142 DMADFKVDPSAVKNSG 157
>UniRef100_Q9LNF2 F21D18.29 [Arabidopsis thaliana]
Length = 176
Score = 44.3 bits (103), Expect = 0.001
Identities = 37/137 (27%), Positives = 62/137 (45%), Gaps = 9/137 (6%)
Query: 12 NIFIVVATISMSIGHCRVLQPSDAQLVAKTCKNTPYPSACLQFLQADPRSSSADVTGLAL 71
N F+ + IG + S+ + C T PS CL+FL + +S ++ LA
Sbjct: 8 NAFLSSLMFLLLIGSSYAITSSEMSTI---CDKTLNPSFCLKFLNT--KFASPNLQALAK 62
Query: 72 IMVDVIQAKTNGVLNKISQLLKGGGD---KPALNSCQGRYNAILKADIPQATQALKTGNP 128
+D QA+ L K+ ++ GG D K A SC Y + + ++ +A + L +G+
Sbjct: 63 TTLDSTQARATQTLKKLQSIIDGGVDPRSKLAYRSCVDEYESAI-GNLEEAFEHLASGDG 121
Query: 129 KFAEDGVADAGVEANTC 145
V+ A A+TC
Sbjct: 122 MGMNMKVSAALDGADTC 138
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 265,644,093
Number of Sequences: 2790947
Number of extensions: 9656205
Number of successful extensions: 23104
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 37
Number of HSP's that attempted gapping in prelim test: 23062
Number of HSP's gapped (non-prelim): 60
length of query: 178
length of database: 848,049,833
effective HSP length: 119
effective length of query: 59
effective length of database: 515,927,140
effective search space: 30439701260
effective search space used: 30439701260
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 70 (31.6 bits)
Lotus: description of TM0137.11


