

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0135.9
(645 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
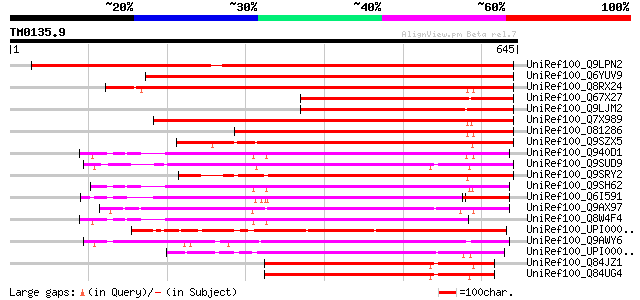
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LPN2 F2J10.1 protein [Arabidopsis thaliana] 845 0.0
UniRef100_Q6YUV9 Transitional endoplasmic reticulum ATPase-like ... 710 0.0
UniRef100_Q8RX24 Hypothetical protein At4g02480 [Arabidopsis tha... 477 e-133
UniRef100_Q67X27 Spastin-like [Oryza sativa] 467 e-130
UniRef100_Q9LJM2 Similarity to unknown protein [Arabidopsis thal... 460 e-128
UniRef100_Q7X989 Putative MSP1(Mitochondrial sorting of proteins... 433 e-120
UniRef100_O81286 T14P8.7 protein [Arabidopsis thaliana] 407 e-112
UniRef100_Q9SZX5 Hypothetical protein F6I7.60 [Arabidopsis thali... 397 e-109
UniRef100_Q940D1 At1g64110/F22C12_22 [Arabidopsis thaliana] 396 e-108
UniRef100_Q9SUD9 Hypothetical protein T13J8.110 [Arabidopsis tha... 395 e-108
UniRef100_Q9SRY2 F22D16.11 protein [Arabidopsis thaliana] 394 e-108
UniRef100_Q9SH62 F22C12.12 [Arabidopsis thaliana] 388 e-106
UniRef100_Q6I591 Hypothetical protein OSJNBa0009C07.5 [Oryza sat... 365 2e-99
UniRef100_Q9AX97 Cell division cycle gene CDC48-like [Oryza sativa] 361 4e-98
UniRef100_Q8W4F4 Similar to homeobox protein [Arabidopsis thaliana] 357 8e-97
UniRef100_UPI000049A4BB UPI000049A4BB UniRef100 entry 355 2e-96
UniRef100_Q9AWY6 P0492F05.26 protein [Oryza sativa] 336 1e-90
UniRef100_UPI0000499829 UPI0000499829 UniRef100 entry 322 3e-86
UniRef100_Q84JZ1 Hypothetical protein [Arabidopsis lyrata] 315 3e-84
UniRef100_Q84UG4 Hypothetical protein [Arabidopsis thaliana] 313 1e-83
>UniRef100_Q9LPN2 F2J10.1 protein [Arabidopsis thaliana]
Length = 627
Score = 845 bits (2182), Expect = 0.0
Identities = 426/615 (69%), Positives = 512/615 (82%), Gaps = 15/615 (2%)
Query: 28 RPLANGQRGEVYEVNGDRVAVILDISEDRENEVELESVNNDGRKPPVYWINVKNIEHDLD 87
RPL++GQRGEVYEV G+RVAVI + +D+ +E + + P++W++VK++++DLD
Sbjct: 26 RPLSSGQRGEVYEVIGNRVAVIFEYGDDKTSEGSEKKPAEQPQMLPIHWLDVKDLKYDLD 85
Query: 88 AQSQDCYLAMEALREVLNSKQPLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSGP 147
Q+ D Y+AMEAL EVL S QPLIVYFPD +QWL ++VPK+ R EF KV+EMFD+LSGP
Sbjct: 86 MQAVDGYIAMEALNEVLQSIQPLIVYFPDSTQWLSRAVPKTRRKEFVDKVKEMFDKLSGP 145
Query: 148 IVLICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTD--KGQKTSDDDEINKLFS 205
IV+ICGQNK ++G+KE+ +FTM+LPN +R+ KLPL LK LT+ G+ S+++EI KLF+
Sbjct: 146 IVMICGQNKIETGSKEREKFTMVLPNLSRVVKLPLPLKGLTEGFTGRGKSEENEIYKLFT 205
Query: 206 NVVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVI 265
NV+ + PPK+E+ L FKKQL EDR+IVISRSN+NEL K + ++ V
Sbjct: 206 NVMRLHPPKEEDTLRLFKKQLGEDRRIVISRSNINELLKKRPQKLVAFVFA--------- 256
Query: 266 LTKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLK 325
+AEK +GWAKNHYL+SC +P VKG +L LPRESLEI+I+RL+ +E S +PSQ+LK
Sbjct: 257 ----EAEKAIGWAKNHYLASCPVPLVKGGRLSLPRESLEISIARLRKLEDNSLKPSQNLK 312
Query: 326 NLAKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRP 385
N+AKDE+E NF+SAVV PGEIGVKF+DIGALEDVKKALNELVILPMRRPELF GNLLRP
Sbjct: 313 NIAKDEYERNFVSAVVAPGEIGVKFEDIGALEDVKKALNELVILPMRRPELFARGNLLRP 372
Query: 386 VKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLA 445
KGILLFGPPGTGKTL+AKALATEAGANFIS++GSTLTSKWFGDAEKLTKALFSFA+KLA
Sbjct: 373 CKGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSFATKLA 432
Query: 446 PVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLD 505
PVIIFVDE+DSLLGARGG+ EHEATRRMRNEFMAAWDGLRSK++QRILILGATNRPFDLD
Sbjct: 433 PVIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNRPFDLD 492
Query: 506 DAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCI 565
DAVIRRLPRRIYVDLPDA+NR KIL+I L ENL+ DFQF+KLA T+GYSGSDLKNLCI
Sbjct: 493 DAVIRRLPRRIYVDLPDAENRLKILKIFLTPENLESDFQFEKLAKETEGYSGSDLKNLCI 552
Query: 566 TAAYRPVQELLEEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNE 625
AAYRPVQELL+EE+ G + LR L+LDDF+QSK+KV PSVAYDA ++NELRKWNE
Sbjct: 553 AAAYRPVQELLQEEQKGARAEASPGLRSLSLDDFIQSKAKVSPSVAYDATTMNELRKWNE 612
Query: 626 MYGEGGTRTKSPFGF 640
YGEGG+RTKSPFGF
Sbjct: 613 QYGEGGSRTKSPFGF 627
>UniRef100_Q6YUV9 Transitional endoplasmic reticulum ATPase-like [Oryza sativa]
Length = 473
Score = 710 bits (1832), Expect = 0.0
Identities = 352/469 (75%), Positives = 406/469 (86%)
Query: 173 NFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSNVVSIQPPKDENLLATFKKQLEEDRKI 232
N +RL+ LP SLKRL K S I+KLF+N + + P+++ F Q+EEDRKI
Sbjct: 4 NLSRLSSLPSSLKRLVGGRPKYSRSSGISKLFTNSLIVPLPEEDEQRRIFNNQIEEDRKI 63
Query: 233 VISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVK 292
+ISR NL EL KVL+EH+LSCV+LLH+ SD V+LT+QKAEKVVGWA++HYLSS +LP +K
Sbjct: 64 IISRHNLVELHKVLQEHELSCVELLHVKSDGVVLTRQKAEKVVGWARSHYLSSAVLPNIK 123
Query: 293 GDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLAKDEFESNFISAVVPPGEIGVKFDD 352
GD+L +PRESL++AI RLK +++PSQ++KNLAKDE+E NFISAVVPP EIGVKFDD
Sbjct: 124 GDRLIIPRESLDVAIERLKEQGIKTKRPSQNIKNLAKDEYERNFISAVVPPDEIGVKFDD 183
Query: 353 IGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGA 412
IGALEDVK+ L+ELV LPMRRPELF+HGNLLRP KG+LLFGPPGTGKTL+AKALATEAGA
Sbjct: 184 IGALEDVKRTLDELVTLPMRRPELFSHGNLLRPCKGVLLFGPPGTGKTLLAKALATEAGA 243
Query: 413 NFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRR 472
NFIS++GSTLTSKWFGDAEKLTKALFSFAS+LAPVIIFVDEVDSLLGARGGAFEHEATRR
Sbjct: 244 NFISITGSTLTSKWFGDAEKLTKALFSFASRLAPVIIFVDEVDSLLGARGGAFEHEATRR 303
Query: 473 MRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRI 532
MRNEFMAAWDGLRSKE+QRILILGATNRPFDLDDAVIRRLPRRIYVDLPDA NR KIL+I
Sbjct: 304 MRNEFMAAWDGLRSKESQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAQNRMKILKI 363
Query: 533 ILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALR 592
+L ENL+ DF+FD+LAN T+GYSGSDLKNLCI +AYRPV ELLEEEK G N LR
Sbjct: 364 LLAKENLESDFRFDELANSTEGYSGSDLKNLCIASAYRPVHELLEEEKKGGPCSQNTGLR 423
Query: 593 PLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFGFG 641
PL LDDF+Q+K+KV PSV+YDA S+NELRKWNE YGEGG+RT+SPFGFG
Sbjct: 424 PLRLDDFIQAKAKVSPSVSYDATSMNELRKWNEQYGEGGSRTRSPFGFG 472
>UniRef100_Q8RX24 Hypothetical protein At4g02480 [Arabidopsis thaliana]
Length = 1265
Score = 477 bits (1228), Expect = e-133
Identities = 256/540 (47%), Positives = 363/540 (66%), Gaps = 23/540 (4%)
Query: 123 KSVPKSI--RSEFFHKVEEMFDQLSGPIVLICGQNKAQSGTKEKGQ-----FTMILPNFA 175
K + KS+ S+ + ++ + L IV+I Q + S KEK FT N
Sbjct: 725 KDIEKSLVGNSDVYATLKSKLETLPENIVVIASQTQLDS-RKEKSHPGGFLFTKFGGNQT 783
Query: 176 RLAKL--PLSLKRLTDKGQKTSDD-DEINKLFSNVVSIQPPKDENLLATFKKQLEEDRKI 232
L L P + +L D+ ++T +I +LF N ++IQ P++E LL+ +K++L+ D +I
Sbjct: 784 ALLDLAFPDNFGKLHDRSKETPKSMKQITRLFPNKIAIQLPQEEALLSDWKEKLDRDTEI 843
Query: 233 VISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVK 292
+ ++N+ + VL +++L C DL L D L + EKVVGWA H+L C P VK
Sbjct: 844 LKVQANITSILAVLAKNKLDCPDLGTLCIKDQTLPSESVEKVVGWAFGHHLMICTEPIVK 903
Query: 293 GDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLA-KDEFESNFISAVVPPGEIGVKFD 351
+KL + ES+ + L ++ ++ +SLK++ ++EFE +S V+PP +IGV FD
Sbjct: 904 DNKLVISAESISYGLQTLHDIQNENKSLKKSLKDVVTENEFEKKLLSDVIPPSDIGVSFD 963
Query: 352 DIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAG 411
DIGALE+VK+ L ELV+LP++RPELF G L +P KGILLFGPPGTGKT++AKA+ATEAG
Sbjct: 964 DIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAG 1023
Query: 412 ANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATR 471
ANFI++S S++TSKWFG+ EK KA+FS ASK+AP +IFVDEVDS+LG R EHEA R
Sbjct: 1024 ANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMR 1083
Query: 472 RMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILR 531
+M+NEFM WDGLR+K+ +R+L+L ATNRPFDLD+AVIRRLPRR+ V+LPDA NR KIL
Sbjct: 1084 KMKNEFMVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDATNRSKILS 1143
Query: 532 IILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK------AGDNN 585
+IL E + PD + +AN+TDGYSGSDLKNLC+TAA+ P++E+LE+EK +N
Sbjct: 1144 VILAKEEIAPDVDLEAIANMTDGYSGSDLKNLCVTAAHFPIREILEKEKKEKTAAQAENR 1203
Query: 586 ITN-----VALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFGF 640
T +R L ++DF + +V SV+ D+ ++NEL++WNE+YGEGG+R K+ +
Sbjct: 1204 PTPPLYSCTDVRSLTMNDFKAAHDQVCASVSSDSSNMNELQQWNELYGEGGSRKKTSLSY 1263
>UniRef100_Q67X27 Spastin-like [Oryza sativa]
Length = 271
Score = 467 bits (1201), Expect = e-130
Identities = 230/271 (84%), Positives = 252/271 (92%), Gaps = 1/271 (0%)
Query: 371 MRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDA 430
MRRPELF+HGNLLRP KGILLFGPPGTGKTL+AKALATEAGANFIS++GS LTSKWFGDA
Sbjct: 1 MRRPELFSHGNLLRPCKGILLFGPPGTGKTLLAKALATEAGANFISITGSNLTSKWFGDA 60
Query: 431 EKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQ 490
EKLTKALFSFAS+LAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQ
Sbjct: 61 EKLTKALFSFASRLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQ 120
Query: 491 RILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLAN 550
RILILGATNRPFDLDDAVIRRLPRRIYVDLPD+ NR KIL+I+L ENL+ DF+FD+LAN
Sbjct: 121 RILILGATNRPFDLDDAVIRRLPRRIYVDLPDSQNRMKILKILLAKENLESDFRFDELAN 180
Query: 551 LTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSV 610
T+GYSGSDLKNLCI AAYRPV ELLEEEK G + T ++LRPL L+DFVQ+K+KV PSV
Sbjct: 181 ATEGYSGSDLKNLCIAAAYRPVHELLEEEKGGVSG-TKISLRPLKLEDFVQAKAKVSPSV 239
Query: 611 AYDAVSVNELRKWNEMYGEGGTRTKSPFGFG 641
A+DA S+NELRKWNE YGEGG+R+KSPFGFG
Sbjct: 240 AFDATSMNELRKWNEQYGEGGSRSKSPFGFG 270
>UniRef100_Q9LJM2 Similarity to unknown protein [Arabidopsis thaliana]
Length = 270
Score = 460 bits (1184), Expect = e-128
Identities = 231/272 (84%), Positives = 251/272 (91%), Gaps = 4/272 (1%)
Query: 371 MRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDA 430
MRRPELFT GNLLRP KGILLFGPPGTGKTL+AKALATEAGANFIS++GSTLTSKWFGDA
Sbjct: 1 MRRPELFTRGNLLRPCKGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDA 60
Query: 431 EKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQ 490
EKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSK++Q
Sbjct: 61 EKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKDSQ 120
Query: 491 RILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLAN 550
RILILGATNRPFDLDDAVIRRLPRRIYVDLPDA+NR KIL+I L ENL+ F+FDKLA
Sbjct: 121 RILILGATNRPFDLDDAVIRRLPRRIYVDLPDAENRLKILKIFLTPENLETGFEFDKLAK 180
Query: 551 LTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVA--LRPLNLDDFVQSKSKVGP 608
T+GYSGSDLKNLCI AAYRPVQELL+EE +++TN + LRPL+LDDF+QSK+KV P
Sbjct: 181 ETEGYSGSDLKNLCIAAAYRPVQELLQEE--NKDSVTNASPDLRPLSLDDFIQSKAKVSP 238
Query: 609 SVAYDAVSVNELRKWNEMYGEGGTRTKSPFGF 640
SVAYDA ++NELRKWNE YGEGGTRTKSPFGF
Sbjct: 239 SVAYDATTMNELRKWNEQYGEGGTRTKSPFGF 270
>UniRef100_Q7X989 Putative MSP1(Mitochondrial sorting of proteins) protein [Oryza
sativa]
Length = 1081
Score = 433 bits (1113), Expect = e-120
Identities = 229/475 (48%), Positives = 329/475 (69%), Gaps = 17/475 (3%)
Query: 183 SLKRLTDKGQKTSD-DDEINKLFSNVVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNE 241
S R+ DK ++ + KLF N V+IQ P+DE L+ +K+ L+ D +I+ +++N ++
Sbjct: 605 SFGRVNDKNKEALKIAKHLTKLFPNKVTIQTPQDELELSQWKQLLDRDVEILKAKANTSK 664
Query: 242 LRKVLEEHQLSCVDL-LHLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKGDKL-CLP 299
++ L + L C D+ D ILT + +KVVG+A +H +P + D L L
Sbjct: 665 IQSFLTRNGLECADIETSACVKDRILTNECVDKVVGYALSHQFKHSTIPTRENDGLLALS 724
Query: 300 RESLEIAISRLKGVETMSRQPS--QSLKNLA-KDEFESNFISAVVPPGEIGVKFDDIGAL 356
ESL+ + L +++ ++ S +SLK++ ++EFE + V+PP EIGV F+DIGAL
Sbjct: 725 GESLKHGVELLDSMQSDPKKKSTKKSLKDVTTENEFEKRLLGDVIPPDEIGVTFEDIGAL 784
Query: 357 EDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFIS 416
E+VK+ L ELV+LP++RPELF+ G L++P KGILLFGPPGTGKT++AKA+ATEAGANFI+
Sbjct: 785 ENVKETLKELVMLPLQRPELFSKGQLMKPCKGILLFGPPGTGKTMLAKAVATEAGANFIN 844
Query: 417 VSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNE 476
+S S++ SKWFG+ EK KA+FS ASK+AP +IFVDEVD +LG R EHEA R+M+NE
Sbjct: 845 ISMSSIASKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDGMLGRRENPGEHEAMRKMKNE 904
Query: 477 FMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKH 536
FM WDGLR+K+ +R+L+L ATNRPFDLD+AV+RRLPRR+ V+LPDA NRKKIL +IL
Sbjct: 905 FMVNWDGLRTKDKERVLVLAATNRPFDLDEAVVRRLPRRLMVNLPDASNRKKILSVILAK 964
Query: 537 ENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK------AGDNN----- 585
E+L D + LANLTDGYSGSD+KNLC+TAA+ P++E+LE EK +N
Sbjct: 965 EDLADDVDLEALANLTDGYSGSDMKNLCVTAAHCPIREILEREKKERASAEAENKPLPPP 1024
Query: 586 ITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFGF 640
++ +R L ++DF + +V S+ D+ ++ EL +WN++YGEGG+R K+ +
Sbjct: 1025 RSSSDVRSLRMNDFKHAHEQVCASITSDSRNMTELIQWNDLYGEGGSRKKTSLSY 1079
>UniRef100_O81286 T14P8.7 protein [Arabidopsis thaliana]
Length = 371
Score = 407 bits (1045), Expect = e-112
Identities = 201/367 (54%), Positives = 272/367 (73%), Gaps = 12/367 (3%)
Query: 286 CLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLA-KDEFESNFISAVVPPG 344
C P VK +KL + ES+ + L ++ ++ +SLK++ ++EFE +S V+PP
Sbjct: 3 CTEPIVKDNKLVISAESISYGLQTLHDIQNENKSLKKSLKDVVTENEFEKKLLSDVIPPS 62
Query: 345 EIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAK 404
+IGV FDDIGALE+VK+ L ELV+LP++RPELF G L +P KGILLFGPPGTGKT++AK
Sbjct: 63 DIGVSFDDIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAK 122
Query: 405 ALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGA 464
A+ATEAGANFI++S S++TSKWFG+ EK KA+FS ASK+AP +IFVDEVDS+LG R
Sbjct: 123 AVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENP 182
Query: 465 FEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAD 524
EHEA R+M+NEFM WDGLR+K+ +R+L+L ATNRPFDLD+AVIRRLPRR+ V+LPDA
Sbjct: 183 GEHEAMRKMKNEFMVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDAT 242
Query: 525 NRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK---- 580
NR KIL +IL E + PD + +AN+TDGYSGSDLKNLC+TAA+ P++E+LE+EK
Sbjct: 243 NRSKILSVILAKEEIAPDVDLEAIANMTDGYSGSDLKNLCVTAAHFPIREILEKEKKEKT 302
Query: 581 --AGDNNITN-----VALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTR 633
+N T +R L ++DF + +V SV+ D+ ++NEL++WNE+YGEGG+R
Sbjct: 303 AAQAENRPTPPLYSCTDVRSLTMNDFKAAHDQVCASVSSDSSNMNELQQWNELYGEGGSR 362
Query: 634 TKSPFGF 640
K+ +
Sbjct: 363 KKTSLSY 369
>UniRef100_Q9SZX5 Hypothetical protein F6I7.60 [Arabidopsis thaliana]
Length = 442
Score = 397 bits (1021), Expect = e-109
Identities = 209/443 (47%), Positives = 292/443 (65%), Gaps = 19/443 (4%)
Query: 213 PKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDL----LHLDSDDVILTK 268
P+DE L +K Q++ D + +SN N LR VL L C L + D+ L +
Sbjct: 2 PQDEKRLTLWKHQMDRDAETSKVKSNFNHLRMVLRRRGLGCEGLETTWSRMCLKDLTLQR 61
Query: 269 QKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLA 328
EK++GWA +++S P K+ L RES+E I L+ + S +
Sbjct: 62 DSVEKIIGWAFGNHISKN--PDTDPAKVTLSRESIEFGIGLLQN--DLKGSTSSKKDIVV 117
Query: 329 KDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKG 388
++ FE +S V+ P +I V FDDIGALE VK L ELV+LP++RPELF G L +P KG
Sbjct: 118 ENVFEKRLLSDVILPSDIDVTFDDIGALEKVKDILKELVMLPLQRPELFCKGELTKPCKG 177
Query: 389 ILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVI 448
ILLFGPPGTGKT++AKA+A EA ANFI++S S++TSKWFG+ EK KA+FS ASK++P +
Sbjct: 178 ILLFGPPGTGKTMLAKAVAKEADANFINISMSSITSKWFGEGEKYVKAVFSLASKMSPSV 237
Query: 449 IFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAV 508
IFVDEVDS+LG R EHEA+R+++NEFM WDGL ++E +R+L+L ATNRPFDLD+AV
Sbjct: 238 IFVDEVDSMLGRREHPREHEASRKIKNEFMMHWDGLTTQERERVLVLAATNRPFDLDEAV 297
Query: 509 IRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAA 568
IRRLPRR+ V LPD NR IL++IL E+L PD ++A++T+GYSGSDLKNLC+TAA
Sbjct: 298 IRRLPRRLMVGLPDTSNRAFILKVILAKEDLSPDLDIGEIASMTNGYSGSDLKNLCVTAA 357
Query: 569 YRPVQELLEEEKAGDNNIT-----------NVALRPLNLDDFVQSKSKVGPSVAYDAVSV 617
+RP++E+LE+EK + + LR LN++DF + V SV+ ++ ++
Sbjct: 358 HRPIKEILEKEKRERDAALAQGKVPPPLSGSSDLRALNVEDFRDAHKWVSASVSSESATM 417
Query: 618 NELRKWNEMYGEGGTRTKSPFGF 640
L++WN+++GEGG+ + F F
Sbjct: 418 TALQQWNKLHGEGGSGKQQSFSF 440
>UniRef100_Q940D1 At1g64110/F22C12_22 [Arabidopsis thaliana]
Length = 824
Score = 396 bits (1017), Expect = e-108
Identities = 239/613 (38%), Positives = 352/613 (56%), Gaps = 104/613 (16%)
Query: 90 SQDCYLAMEALREVL---NSKQPLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSG 146
S D L +++L +VL + P+++Y D +L +S ++ +++ +LSG
Sbjct: 242 SFDEKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRS------QRTYNLFQKLLQKLSG 295
Query: 147 PIVLICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSN 206
P VLI G +++ + D++++ +F
Sbjct: 296 P-VLILGSRIVDLSSEDAQEI-----------------------------DEKLSAVFPY 325
Query: 207 VVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVIL 266
+ I+PP+DE L ++K QLE D ++ ++ N N + +VL E+ L C DL + +D +
Sbjct: 326 NIDIRPPEDETHLVSWKSQLERDMNMIQTQDNRNHIMEVLSENDLICDDLESISFEDTKV 385
Query: 267 TKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAIS------------------ 308
E++V A +++L + P + KL + SL S
Sbjct: 386 LSNYIEEIVVSALSYHLMNNKDPEYRNGKLVISSISLSHGFSLFREGKAGGREKLKQKTK 445
Query: 309 ----------------RLKGVETMS--RQPSQSLK---------NLAKD-EFESNFISAV 340
+ + V T+S +P + K +A D EFE V
Sbjct: 446 EESSKEVKAESIKPETKTESVTTVSSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEV 505
Query: 341 VPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKT 400
+P EI V F DIGAL+++K++L ELV+LP+RRP+LFT G LL+P +GILLFGPPGTGKT
Sbjct: 506 IPAEEINVTFKDIGALDEIKESLQELVMLPLRRPDLFT-GGLLKPCRGILLFGPPGTGKT 564
Query: 401 LMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGA 460
++AKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ ASK++P IIFVDEVDS+LG
Sbjct: 565 MLAKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQ 624
Query: 461 RGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDL 520
R EHEA R+++NEFM+ WDGL +K +RIL+L ATNRPFDLD+A+IRR RRI V L
Sbjct: 625 RTRVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGL 684
Query: 521 PDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEE- 579
P +NR+KILR +L E +D + + +LA +T+GY+GSDLKNLC TAAYRPV+EL+++E
Sbjct: 685 PAVENREKILRTLLAKEKVDENLDYKELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQER 744
Query: 580 -------------KAGDNNITN----VALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRK 622
KAG+ + + LRPLN DF ++K++V S A + + EL++
Sbjct: 745 IKDTEKKKQREPTKAGEEDEGKEERVITLRPLNRQDFKEAKNQVAASFAAEGAGMGELKQ 804
Query: 623 WNEMYGEGGTRTK 635
WNE+YGEGG+R K
Sbjct: 805 WNELYGEGGSRKK 817
>UniRef100_Q9SUD9 Hypothetical protein T13J8.110 [Arabidopsis thaliana]
Length = 726
Score = 395 bits (1016), Expect = e-108
Identities = 238/582 (40%), Positives = 343/582 (58%), Gaps = 80/582 (13%)
Query: 95 LAMEALREVLNS---KQPLIVYFPDGSQWLHKSVPKSIRSEFFHKV-EEMFDQLSGPIVL 150
L +++L +VL S P+I+Y D V K +SE F+K+ + + +LSGP+++
Sbjct: 187 LFLQSLYKVLVSISETNPIIIYLRD--------VEKLCQSERFYKLFQRLLTKLSGPVLV 238
Query: 151 ICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSNVVSI 210
+ + L D Q+ + I+ LF + I
Sbjct: 239 LGSR-----------------------------LLEPEDDCQEVGEG--ISALFPYNIEI 267
Query: 211 QPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQK 270
+PP+DEN L ++K + E+D K++ + N N + +VL + L C DL + D +
Sbjct: 268 RPPEDENQLMSWKTRFEDDMKVIQFQDNKNHIAEVLAANDLECDDLGSICHADTMFLSSH 327
Query: 271 AEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKG-----VETMSRQPSQSLK 325
E++V A +++L + P K +L + SL ++ L+ +++ + K
Sbjct: 328 IEEIVVSAISYHLMNNKEPEYKNGRLVISSNSLSHGLNILQEGQGCFEDSLKLDTNIDSK 387
Query: 326 NLAKD-EFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLR 384
+A D EFE V+P EIGV F DIG+L++ K++L ELV+LP+RRP+LF G LL+
Sbjct: 388 EVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLFK-GGLLK 446
Query: 385 PVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKL 444
P +GILLFGPPGTGKT+MAKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ A+K+
Sbjct: 447 PCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKV 506
Query: 445 APVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDL 504
+P IIFVDEVDS+LG R EHEA R+++NEFM WDGL S RIL+L ATNRPFDL
Sbjct: 507 SPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFDL 566
Query: 505 DDAVIRRLPRRIYVDLPDADNRKKILRIIL---KHENLDPDFQFDKLANLTDGYSGSDLK 561
D+A+IRR RRI V LP ++R+KILR +L K ENLD F +LA +TDGYSGSDLK
Sbjct: 567 DEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTENLD----FQELAQMTDGYSGSDLK 622
Query: 562 NLCITAAYRPVQELLEEEKAGD-----------------------NNITNVALRPLNLDD 598
N C TAAYRPV+EL+++E D + + LRPL+++D
Sbjct: 623 NFCTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGITLRPLSMED 682
Query: 599 FVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTKSPFGF 640
+KS+V S A + +NEL++WN++YGEGG+R K +
Sbjct: 683 MKVAKSQVAASFAAEGAGMNELKQWNDLYGEGGSRKKEQLSY 724
>UniRef100_Q9SRY2 F22D16.11 protein [Arabidopsis thaliana]
Length = 1217
Score = 394 bits (1012), Expect = e-108
Identities = 208/437 (47%), Positives = 287/437 (65%), Gaps = 45/437 (10%)
Query: 215 DENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKV 274
DE L +K +LE D +I+ +++N+ +R A V
Sbjct: 813 DEASLVDWKDKLERDTEILKAQANITSIR---------------------------AHLV 845
Query: 275 VGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLAKDEFES 334
+ +NH ++ C + LC S E+ + +G + + ++EFE
Sbjct: 846 ICLIENHMINRC----GESGWLCFQSPSYELLRTYSQGQQAYHLGRKDVV---TENEFEK 898
Query: 335 NFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGP 394
+S V+PP +IGV F DIGALE+VK L ELV+LP++RPELF G L +P KGILLFGP
Sbjct: 899 KLLSDVIPPSDIGVSFSDIGALENVKDTLKELVMLPLQRPELFGKGQLTKPTKGILLFGP 958
Query: 395 PGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEV 454
PGTGKT++AKA+ATEAGANFI++S S++TSKWFG+ EK KA+FS ASK+AP +IFVDEV
Sbjct: 959 PGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEV 1018
Query: 455 DSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPR 514
DS+LG R EHEA R+M+NEFM WDGLR+K+ +R+L+L ATNRPFDLD+AVIRRLPR
Sbjct: 1019 DSMLGRRENPGEHEAMRKMKNEFMINWDGLRTKDKERVLVLAATNRPFDLDEAVIRRLPR 1078
Query: 515 RIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQE 574
R+ V+LPD+ NR KIL +IL E + D + +AN+TDGYSGSDLKNLC+TAA+ P++E
Sbjct: 1079 RLMVNLPDSANRSKILSVILAKEEMAEDVDLEAIANMTDGYSGSDLKNLCVTAAHLPIRE 1138
Query: 575 LLEEEK-----------AGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKW 623
+LE+EK A ++ +RPLN++DF + +V SVA D+ ++NEL++W
Sbjct: 1139 ILEKEKKERSVAQAENRAMPQLYSSTDVRPLNMNDFKTAHDQVCASVASDSSNMNELQQW 1198
Query: 624 NEMYGEGGTRTKSPFGF 640
NE+YGEGG+R K+ +
Sbjct: 1199 NELYGEGGSRKKTSLSY 1215
>UniRef100_Q9SH62 F22C12.12 [Arabidopsis thaliana]
Length = 825
Score = 388 bits (996), Expect = e-106
Identities = 234/619 (37%), Positives = 344/619 (54%), Gaps = 124/619 (20%)
Query: 104 LNSKQPLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSGPIVLICGQNKAQSGTKE 163
++ P+++Y D +L +S ++ +++ +LSGP VLI G +++
Sbjct: 237 VSKANPIVLYLRDVENFLFRS------QRTYNLFQKLLQKLSGP-VLILGSRIVDLSSED 289
Query: 164 KGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSNVVSIQPPKDENLLATFK 223
+ D++++ +F + I+PP+DE L ++K
Sbjct: 290 AQEI-----------------------------DEKLSAVFPYNIDIRPPEDETHLVSWK 320
Query: 224 KQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGWAKNHYL 283
QLE D ++ ++ N N + +VL E+ L C DL + +D + E++V A +++L
Sbjct: 321 SQLERDMNMIQTQDNRNHIMEVLSENDLICDDLESISFEDTKVLSNYIEEIVVSALSYHL 380
Query: 284 SSCLLPCVKGDKLCLPRESLEIAIS----------------------------------R 309
+ P + KL + SL S +
Sbjct: 381 MNNKDPEYRNGKLVISSISLSHGFSLFREGKAGGREKLKQKTKEESSKEVKAESIKPETK 440
Query: 310 LKGVETMS--RQPSQSLK---------NLAKD-EFESNFISAVVPPGEIGVKFDDIGALE 357
+ V T+S +P + K +A D EFE V+P EI V F DIGAL+
Sbjct: 441 TESVTTVSSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIPAEEINVTFKDIGALD 500
Query: 358 DVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISV 417
++K++L ELV+LP+RRP+LFT G LL+P +GILLFGPPGTGKT++AKA+A EAGA+FI+V
Sbjct: 501 EIKESLQELVMLPLRRPDLFT-GGLLKPCRGILLFGPPGTGKTMLAKAIAKEAGASFINV 559
Query: 418 SGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEF 477
S ST+TSKWFG+ EK +ALF+ ASK++P IIFVDEVDS+LG R EHEA R+++NEF
Sbjct: 560 SMSTITSKWFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEF 619
Query: 478 MAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHE 537
M+ WDGL +K +RIL+L ATNRPFDLD+A+IRR RRI V LP +NR+KILR +L E
Sbjct: 620 MSHWDGLMTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPAVENREKILRTLLAKE 679
Query: 538 NLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGD---NNIT------- 587
+D + + +LA +T+GY+GSDLKNLC TAAYRPV+EL+++E+ D NNI+
Sbjct: 680 KVDENLDYKELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQERIKDTVRNNISLRLFLYT 739
Query: 588 -------------------------------NVALRPLNLDDFVQSKSKVGPSVAYDAVS 616
+ LRPLN DF ++K++V S A +
Sbjct: 740 SIFILVLTDCEKKKQREPTKAGEEDEGKEERVITLRPLNRQDFKEAKNQVAASFAAEGAG 799
Query: 617 VNELRKWNEMYGEGGTRTK 635
+ EL++WNE+YGEGG+R K
Sbjct: 800 MGELKQWNELYGEGGSRKK 818
>UniRef100_Q6I591 Hypothetical protein OSJNBa0009C07.5 [Oryza sativa]
Length = 855
Score = 365 bits (937), Expect = 2e-99
Identities = 217/523 (41%), Positives = 305/523 (57%), Gaps = 75/523 (14%)
Query: 91 QDCYLAMEALREVLNSKQPLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSGPIVL 150
Q Y M ++ E P+I+Y D Q LH+S + ++M +L+G +++
Sbjct: 249 QSLYKVMVSVAE----NNPVILYIRDVDQLLHRS------QRTYSLFQKMLAKLTGQVLI 298
Query: 151 ICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSNVVSI 210
L + L T D+ ++ LF V I
Sbjct: 299 -------------------------------LGSRLLDSDSDHTDVDERVSSLFPFHVDI 327
Query: 211 QPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVILTKQK 270
+PP++E L ++K Q+EED K + + N N + +VL + L C DL + D ++
Sbjct: 328 KPPEEETHLDSWKTQMEEDTKKIQIQDNRNHIIEVLSANDLDCDDLSSICQADTMVLSNY 387
Query: 271 AEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLK-----GVETMSRQ------ 319
E+++ A ++++ P K KL L +SL +S + G ET+ +
Sbjct: 388 IEEIIVSAVSYHMIHNKDPEYKNGKLVLSSKSLSHGLSIFQESGFGGKETLKLEDDLKGA 447
Query: 320 --PSQS---------LKN-----------LAKDEFESNFISAVVPPGEIGVKFDDIGALE 357
P +S LK+ + +EFE V+P EIGV FDDIGAL
Sbjct: 448 TGPKKSETEKSATVPLKDGDGPLPPPKPEIPDNEFEKRIRPEVIPASEIGVTFDDIGALA 507
Query: 358 DVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISV 417
D+K++L ELV+LP+RRP+LF G LL+P +GILLFGPPGTGKT++AKA+A +AGA+FI+V
Sbjct: 508 DIKESLQELVMLPLRRPDLFK-GGLLKPCRGILLFGPPGTGKTMLAKAIANDAGASFINV 566
Query: 418 SGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEF 477
S ST+TSKWFG+ EK +ALFS A+K+AP IIFVDEVDS+LG R EHEA R+++NEF
Sbjct: 567 SMSTITSKWFGEDEKNVRALFSLAAKVAPTIIFVDEVDSMLGQRARCGEHEAMRKIKNEF 626
Query: 478 MAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHE 537
M+ WDGL SK +RIL+L ATNRPFDLD+A+IRR RRI V LP D+R+ ILR +L E
Sbjct: 627 MSHWDGLLSKSGERILVLAATNRPFDLDEAIIRRFERRIMVGLPTLDSRELILRTLLSKE 686
Query: 538 NLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK 580
+ D + +LA +T+GYSGSDLKNLC+TAAYRPV+ELL+ E+
Sbjct: 687 KVAEDIDYKELATMTEGYSGSDLKNLCVTAAYRPVRELLKRER 729
Score = 55.5 bits (132), Expect = 5e-06
Identities = 24/59 (40%), Positives = 38/59 (63%)
Query: 577 EEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTK 635
+ KA + LRPL ++D Q+K++V S A + +NEL++WN++YGEGG+R K
Sbjct: 790 DNSKAEGGTEGTIDLRPLTMEDLRQAKNQVAASFATEGAVMNELKQWNDLYGEGGSRKK 848
>UniRef100_Q9AX97 Cell division cycle gene CDC48-like [Oryza sativa]
Length = 812
Score = 361 bits (926), Expect = 4e-98
Identities = 217/588 (36%), Positives = 337/588 (56%), Gaps = 72/588 (12%)
Query: 115 PDGSQWLHKSVP-----KSIRSEFFHKVEEMFDQLSGPIVLICGQNKAQSGTKEKGQFTM 169
P+ S++L P + + + +K+ ++S P++L G+ EK + M
Sbjct: 223 PESSKYLASVKPCWSLNEKVLIQSLYKIIVSASEIS-PVILYIRDVDDLLGSSEKA-YCM 280
Query: 170 ILPNFARLAK--LPLSLKRLTDKGQKTSDDDEINKLFSNVVSIQPPKDENLLATFKKQLE 227
+L+ + + + L D + ++ + LF ++ +PPKD+ LL +K Q+E
Sbjct: 281 FQKMLKKLSGRVIVIGSQFLDDDEDREDIEESVCALFPCILETKPPKDKVLLEKWKTQME 340
Query: 228 EDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVI-LTKQKAEKVVGWAKNHYLSSC 286
ED ++ N + +VL E+ L C DL +++DD + E+++ + +++L +
Sbjct: 341 EDSNNNNNQVVQNYIAEVLAENNLECEDLSSINADDDCKIIVAYLEEIITPSVSYHLMNN 400
Query: 287 LLPCVKGDKLCLPRESLEIAI-----SRLKGVETMSRQPSQSLKNLAKDEFESNFISAVV 341
P + L + ESL + S G +T+ + + + +E+E V+
Sbjct: 401 KNPKYRNGNLVISSESLSHGLRIFQESNDLGKDTVEAKDETEMV-VPDNEYEKKIRPTVI 459
Query: 342 PPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTL 401
P EIGV FDDIGAL D+K+ L+ELV+LP++RP+ F G LL+P KG+LLFGPPGTGKT+
Sbjct: 460 PANEIGVTFDDIGALADIKECLHELVMLPLQRPDFFK-GGLLKPCKGVLLFGPPGTGKTM 518
Query: 402 MAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGAR 461
+AKALA AGA+F+++S +++TSKW+G++EK +ALFS A+KLAP IIF+DEVDS+LG R
Sbjct: 519 LAKALANAAGASFLNISMASMTSKWYGESEKCIQALFSLAAKLAPAIIFIDEVDSMLGKR 578
Query: 462 GGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLP 521
E+EA+RR++NEFMA WDGL SK N+RIL+L ATNRPFDLDDAVIRR RI V LP
Sbjct: 579 DNHSENEASRRVKNEFMAHWDGLLSKSNERILVLAATNRPFDLDDAVIRRFEHRIMVGLP 638
Query: 522 DADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPV--------- 572
++R+ IL+ +L E ++ + F +LA +T+GY+ SDLKN+C+TAAY PV
Sbjct: 639 TLESRELILKTLLSKETVE-NIDFKELAKMTEGYTSSDLKNICVTAAYHPVRELLQKEKN 697
Query: 573 -----------QELLEEEKAGDNNITN--------------------------------- 588
QE E+ K +N +
Sbjct: 698 KVKKETAPETKQEPKEKTKIQENGTKSSDSKTEKDKLDNKEGKKDKPADKKDKSDKGDAG 757
Query: 589 -VALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTK 635
LRPLN++D ++K +V S A + V +N++++WNE+YG+GG+R +
Sbjct: 758 ETTLRPLNMEDLRKAKDEVAASFASEGVVMNQIKEWNELYGKGGSRKR 805
>UniRef100_Q8W4F4 Similar to homeobox protein [Arabidopsis thaliana]
Length = 752
Score = 357 bits (915), Expect = 8e-97
Identities = 214/543 (39%), Positives = 315/543 (57%), Gaps = 86/543 (15%)
Query: 90 SQDCYLAMEALREVL---NSKQPLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSG 146
S D L +++L +VL + P+++Y D +L +S ++ +++ +LSG
Sbjct: 247 SFDEKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRS------QRTYNLFQKLLQKLSG 300
Query: 147 PIVLICGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSN 206
P VLI G +++ + D++++ +F
Sbjct: 301 P-VLILGSRIVDLSSEDAQEI-----------------------------DEKLSAVFPY 330
Query: 207 VVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVIL 266
+ I+PP+DE L ++K QLE D ++ ++ N N + +VL E+ L C DL + +D +
Sbjct: 331 NIDIRPPEDETHLVSWKSQLERDMNMIQTQDNRNHIMEVLSENDLICDDLESISFEDTKV 390
Query: 267 TKQKAEKVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAIS------------------ 308
E++V A +++L + P + KL + SL S
Sbjct: 391 LSNYIEEIVVSALSYHLMNNKDPEYRNGKLVISSISLSHGFSLFREGKAGGREKLKQKTK 450
Query: 309 ----------------RLKGVETMS--RQPSQSLK---------NLAKD-EFESNFISAV 340
+ + V T+S +P + K +A D EFE V
Sbjct: 451 EESSKEVKAESIKPETKTESVTTVSSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEV 510
Query: 341 VPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKT 400
+P EI V F DIGAL+++K++L ELV+LP+RRP+LFT G LL+P +GILLFGPPGTGKT
Sbjct: 511 IPAEEINVTFKDIGALDEIKESLQELVMLPLRRPDLFT-GGLLKPCRGILLFGPPGTGKT 569
Query: 401 LMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGA 460
++AKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ ASK++P IIFVDEVDS+LG
Sbjct: 570 MLAKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQ 629
Query: 461 RGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDL 520
R EHEA R+++NEFM+ WDGL +K +RIL+L ATNRPFDLD+A+IRR RRI V L
Sbjct: 630 RTRVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGL 689
Query: 521 PDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEK 580
P +NR+KILR +L E +D + + +LA +T+GY+GSDLKNLC TAAYRPV+EL+++E+
Sbjct: 690 PAVENREKILRTLLAKEKVDENLDYKELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQER 749
Query: 581 AGD 583
D
Sbjct: 750 IKD 752
>UniRef100_UPI000049A4BB UPI000049A4BB UniRef100 entry
Length = 936
Score = 355 bits (912), Expect = 2e-96
Identities = 208/481 (43%), Positives = 297/481 (61%), Gaps = 25/481 (5%)
Query: 155 NKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSNVVSIQPPK 214
+K+QSG + M+ R +P S + T QK D + I+KLF NV+ PP
Sbjct: 468 SKSQSGPQMDQSLCMLGVKEFRFETIPFS--KATKDQQK--DLNNISKLFGNVLKFSPPT 523
Query: 215 DENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSCVDLLHLDSDDVI--LTKQKAE 272
E L +K + ED K+ N ++ + + LL +++ T ++ E
Sbjct: 524 GE-LERPWKALVNEDAKMEKIAKNRE---MIIAQTTALRIKLLQYPGEEMTETYTPEQIE 579
Query: 273 KVVGWAKNHYLSSCLLPCVKGDKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLAKD-E 331
K +G A ++ G L +E + G+ T+ + + ++ + D E
Sbjct: 580 KAIGIAIEEARNTT------GIANELSKEQIA------HGLNTVREKKNIDIEEMETDNE 627
Query: 332 FESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILL 391
FE +S V+ +I V FDDIGAL+DVK+ LNE + LP++R ELF L + KG+LL
Sbjct: 628 FEKKLLSDVIRADDINVSFDDIGALDDVKEVLNETITLPLKRSELF-FSKLTQGAKGVLL 686
Query: 392 FGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFV 451
FGPPGTGKT++AKA+ATE+ +NFI+VS S+L SKWFG+AEK KALF+ ASKL+P +IFV
Sbjct: 687 FGPPGTGKTMLAKAVATESKSNFINVSMSSLGSKWFGEAEKYVKALFTLASKLSPCVIFV 746
Query: 452 DEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRR 511
DEVD+LLG R + EHEA R+M+NEFM+ WDGL+SKE +R++++ ATNRPFDLDDAV+RR
Sbjct: 747 DEVDALLGKRSSS-EHEAVRKMKNEFMSLWDGLKSKEMERVIVMAATNRPFDLDDAVLRR 805
Query: 512 LPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRP 571
L RRI VDLP+ NR IL+ IL+ E+++ D + +A T+G+SGSDL L A RP
Sbjct: 806 LSRRILVDLPNETNRVLILKKILRREDVEKDLNYSIIAQQTEGFSGSDLFALGQMVAMRP 865
Query: 572 VQELLEEEKAGDNNITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGG 631
++E L +E G N LRPL+ DF++ K+ PSV+ D+ S+NELR+WN +YGEG
Sbjct: 866 IKEYLAKEVKGQKKEMNPVLRPLSTQDFLEEVKKINPSVSKDSSSLNELRRWNSLYGEGA 925
Query: 632 T 632
+
Sbjct: 926 S 926
>UniRef100_Q9AWY6 P0492F05.26 protein [Oryza sativa]
Length = 810
Score = 336 bits (862), Expect = 1e-90
Identities = 214/583 (36%), Positives = 324/583 (54%), Gaps = 62/583 (10%)
Query: 95 LAMEALREVLNS---KQPLIVYFPDGSQWLHKSVPKSIRSEFFHKVEEMFDQLSGPIVLI 151
+ ++A+ +VL+S K P+++Y D ++LHKS + + E++ ++L GP++++
Sbjct: 241 ILVQAVYKVLHSVSKKNPIVLYIRDVEKFLHKS------KKMYVMFEKLLNKLEGPVLVL 294
Query: 152 CGQNKAQSGTKEKGQFTMILPNFARLAKLPLSLKRLTDKGQKTSDDDEINKLFSNVVSIQ 211
+ +E + L + K P + L + +D ++ + N I
Sbjct: 295 GSRIVDMDFDEELDERLTALFPYNIEIKPPENENHLVSWNSQLEEDMKMIQFQDNRNHIT 354
Query: 212 PPKDENLLA-------------TFKKQLEE-------------------DRKIVISRSNL 239
EN L + +EE + K+++S +L
Sbjct: 355 EVLAENDLECDDLGSICLSDTMVLGRYIEEIVVSAVSYHLMNKKDPEYRNGKLLLSAKSL 414
Query: 240 NELRKVLEEHQLSCVDLLHLDSDDVILTKQKAEKVVGW------AKNHYLSSCLLPCVKG 293
+ ++ +E+++ D + L++ K+ A KV AK+ + LLP V
Sbjct: 415 SHALEIFQENKMYDKDSMKLEA------KRDASKVADRGIAPFAAKSETKPATLLPPVPP 468
Query: 294 DKLCLPRESLEIAISRLKGVETMSRQPSQSLKNLAKD-EFESNFISAVVPPGEIGVKFDD 352
P + + + + S P+ + D EFE V+P EIGV FDD
Sbjct: 469 TAAAAPPVESKAEPEKFEKKDNPS--PAAKAPEMPPDNEFEKRIRPEVIPANEIGVTFDD 526
Query: 353 IGALEDVKKALNELVILPMRRPELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGA 412
IGAL D+K++L ELV+LP+RRP+LF G LL+P +GILLFGPPGTGKT++AKA+A EA A
Sbjct: 527 IGALSDIKESLQELVMLPLRRPDLFK-GGLLKPCRGILLFGPPGTGKTMLAKAIANEAQA 585
Query: 413 NFISVSGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRR 472
+FI+VS ST+TSKWFG+ EK +ALF+ A+K++P IIFVDEVDS+LG R A EHEA R+
Sbjct: 586 SFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRNRAGEHEAMRK 645
Query: 473 MRNEFMAAWDGLRSKENQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRI 532
++NEFM WDGL S+ +Q+IL+L ATNRPFDLD+A+IRR RRI V LP ++R+ ILR
Sbjct: 646 IKNEFMTHWDGLLSRPDQKILVLAATNRPFDLDEAIIRRFERRIMVGLPSLESRELILRS 705
Query: 533 ILKHENLDPDFQFDKLANLTDGYSGSDLKNLCITAAYRPVQELLEEEKAGDNNITNVALR 592
+L E +D + +LA +T+GYSGSDLK +EK + LR
Sbjct: 706 LLSKEKVDGGLDYKELATMTEGYSGSDLKEKKREQGGNASDASKMKEKD-----ETIILR 760
Query: 593 PLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGEGGTRTK 635
PLN+ D ++K++V S A + + EL++WNE+YGEGG+R K
Sbjct: 761 PLNMKDLKEAKNQVAASFAAEGTIMGELKQWNELYGEGGSRKK 803
>UniRef100_UPI0000499829 UPI0000499829 UniRef100 entry
Length = 912
Score = 322 bits (824), Expect = 3e-86
Identities = 192/469 (40%), Positives = 274/469 (57%), Gaps = 56/469 (11%)
Query: 200 INKLFSNVVSIQPPKDENLLATFKKQLEEDRKIVISRSNLNELRKVLEEHQLSC--VDLL 257
+ K+F N ++IQ P E + + ++ED K + + + ++ L +H L +D
Sbjct: 448 LQKMFGNSITIQTPTGEEARSWWI-MMQEDAKQMSASISKRSIKNELLKHGLEMEKIDDS 506
Query: 258 HLDSDDVILTKQKAEKVVGWAKNHYLSSCLLPCVKGDK--LCLPRESLEIAISRLKGVET 315
L D L ++ EK+VGWA H + + DK + +ES+ AI+
Sbjct: 507 ELQLD---LKEEDVEKIVGWAFAHEIEK------RPDKNIRTISKESIMHAIAM-----Q 552
Query: 316 MSRQPSQSLKNL--AKDEFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRR 373
M P + + + A++EFE ++ V+ G++ V F DIGALE VK+ L E + LP+ R
Sbjct: 553 MQLNPVKDVVDTLEAENEFEKKLMNDVIRAGDVDVSFSDIGALEKVKETLYESITLPLLR 612
Query: 374 PELFTHGNLLRPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKL 433
PELF G+L + KGIL FGPPGTGKT++AKA+A E+ ANFI+ S S+L SKWFG+AEK
Sbjct: 613 PELFKKGSLTKRSKGILFFGPPGTGKTMLAKAVAKESKANFINASLSSLESKWFGEAEKF 672
Query: 434 TKALFSFASKLAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRIL 493
KALFS A+KL+P +IF+DEVD+LLG R E+E R+M+NEFM WDGL+S+ ++I+
Sbjct: 673 VKALFSLAAKLSPCVIFIDEVDALLGKRTSQNENETLRKMKNEFMTLWDGLKSQNLEQII 732
Query: 494 ILGATNRPFDLDDAVIRRLPRRIYVDLPDADNRKKILRIILKHENLDPDFQFDKLANLTD 553
+LGATNRPFDLDDA++RR RRI VDLP ++R+ IL+IILK E +D D K+A T
Sbjct: 733 VLGATNRPFDLDDAILRRFSRRILVDLPTKEDRENILKIILKGEKIDCD--ISKIAEKTP 790
Query: 554 GYSGSDLKNLCITAAYRPVQELL------------------EEEKAGDN----------- 584
GYSG DL NLC AA RP+++ + E E G N
Sbjct: 791 GYSGCDLFNLCCAAAMRPIRDYIAKENKEKERIEQLKKEQKEMESKGINPSPFVKVEEFV 850
Query: 585 ----NITNVALRPLNLDDFVQSKSKVGPSVAYDAVSVNELRKWNEMYGE 629
+ +R +N +DF + S + PS D+ + E+R WNE +GE
Sbjct: 851 NPTIEVAKEQIRAMNDNDFFEVLSTMNPSTNKDSPFLTEIRNWNEQFGE 899
>UniRef100_Q84JZ1 Hypothetical protein [Arabidopsis lyrata]
Length = 316
Score = 315 bits (806), Expect = 3e-84
Identities = 171/320 (53%), Positives = 219/320 (68%), Gaps = 32/320 (10%)
Query: 325 KNLAKD-EFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLL 383
K +A D EFE V+P EIGV F DIG+L++ K++L ELV+LP+RRP+LF G LL
Sbjct: 2 KEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLFK-GGLL 60
Query: 384 RPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASK 443
+P +GILLFGPPGTGKT+MAKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ A+K
Sbjct: 61 KPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 120
Query: 444 LAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFD 503
++P IIFVDEVDS+LG R EHEA R+++NEFM WDGL S RIL+L ATNRPFD
Sbjct: 121 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFD 180
Query: 504 LDDAVIRRLPRRIYVDLPDADNRKKILRIIL---KHENLDPDFQFDKLANLTDGYSGSDL 560
LD+A+IRR RRI V LP ++R+KILR +L K ENLD F +LA +TDGYSGSDL
Sbjct: 181 LDEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTENLD----FHELAQMTDGYSGSDL 236
Query: 561 KNLCITAAYRPVQELLEEEKAGDNNITN-----------------------VALRPLNLD 597
KN C TAAYRPV+EL+++E D + LRPL+++
Sbjct: 237 KNFCTTAAYRPVRELIKQECLKDQERKKKEEAEKSSEEGSETKEEVSEERVITLRPLSME 296
Query: 598 DFVQSKSKVGPSVAYDAVSV 617
D +KS+V S A + +
Sbjct: 297 DMKVAKSQVAASFAAEGAGM 316
>UniRef100_Q84UG4 Hypothetical protein [Arabidopsis thaliana]
Length = 316
Score = 313 bits (802), Expect = 1e-83
Identities = 170/320 (53%), Positives = 219/320 (68%), Gaps = 32/320 (10%)
Query: 325 KNLAKD-EFESNFISAVVPPGEIGVKFDDIGALEDVKKALNELVILPMRRPELFTHGNLL 383
K +A D EFE V+P EIGV F DIG+L++ K++L ELV+LP+RRP+LF G LL
Sbjct: 2 KEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLFK-GGLL 60
Query: 384 RPVKGILLFGPPGTGKTLMAKALATEAGANFISVSGSTLTSKWFGDAEKLTKALFSFASK 443
+P +GILLFGPPGTGKT+MAKA+A EAGA+FI+VS ST+TSKWFG+ EK +ALF+ A+K
Sbjct: 61 KPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 120
Query: 444 LAPVIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKENQRILILGATNRPFD 503
++P IIFVDEVDS+LG R EHEA R+++NEFM WDGL S RIL+L ATNRPFD
Sbjct: 121 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFD 180
Query: 504 LDDAVIRRLPRRIYVDLPDADNRKKILRIIL---KHENLDPDFQFDKLANLTDGYSGSDL 560
LD+A+IRR RRI V LP ++R+KILR +L K ENLD F +LA +TDGYSGSDL
Sbjct: 181 LDEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTENLD----FQELAQMTDGYSGSDL 236
Query: 561 KNLCITAAYRPVQELLEEEKAGD-----------------------NNITNVALRPLNLD 597
KN C TAAYRPV+EL+++E D + + LRPL+++
Sbjct: 237 KNFCTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGITLRPLSME 296
Query: 598 DFVQSKSKVGPSVAYDAVSV 617
D +K +V S A + +
Sbjct: 297 DMKVAKIQVAASFAAEGAGM 316
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,102,542,702
Number of Sequences: 2790947
Number of extensions: 47950435
Number of successful extensions: 162594
Number of sequences better than 10.0: 5156
Number of HSP's better than 10.0 without gapping: 3123
Number of HSP's successfully gapped in prelim test: 2034
Number of HSP's that attempted gapping in prelim test: 152507
Number of HSP's gapped (non-prelim): 6765
length of query: 645
length of database: 848,049,833
effective HSP length: 134
effective length of query: 511
effective length of database: 474,062,935
effective search space: 242246159785
effective search space used: 242246159785
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0135.9


