

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
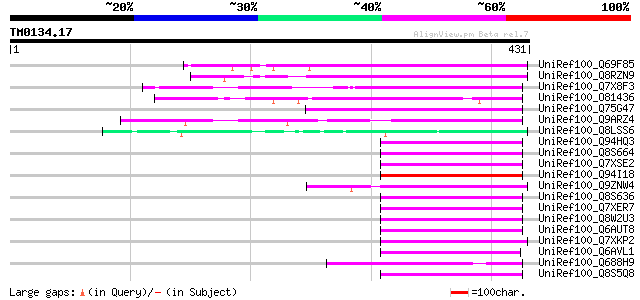
Query= TM0134.17
(431 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q69F85 Gag-pol polyprotein [Phaseolus vulgaris] 191 3e-47
UniRef100_Q8RZN9 Putative polyprotein [Oryza sativa] 97 9e-19
UniRef100_Q7X8F3 OSJNBa0093P23.5 protein [Oryza sativa] 94 8e-18
UniRef100_O81436 T24H24.13 protein [Arabidopsis thaliana] 90 1e-16
UniRef100_Q75G47 Putative polyprotein [Oryza sativa] 89 2e-16
UniRef100_Q9ARZ4 Putative polyprotein [Oryza sativa] 89 3e-16
UniRef100_Q8LSS6 Putative gag-pol [Oryza sativa] 89 3e-16
UniRef100_Q94HQ3 Putative retroelement [Oryza sativa] 88 4e-16
UniRef100_Q8S664 Putative retroelement [Oryza sativa] 88 4e-16
UniRef100_Q7XSE2 OSJNBb0033G08.12 protein [Oryza sativa] 88 5e-16
UniRef100_Q94I18 Putative retroelement [Oryza sativa] 88 5e-16
UniRef100_Q9ZNW4 Polyprotein [Sorghum bicolor] 87 7e-16
UniRef100_Q8S636 Putative polyprotein [Oryza sativa] 87 7e-16
UniRef100_Q7XER7 Contains similarity to gag-pol polyprotein [Ory... 87 7e-16
UniRef100_Q8W2U3 Putative polyprotein [Oryza sativa] 87 1e-15
UniRef100_Q6AUT8 Putative gag-pol [Oryza sativa] 87 1e-15
UniRef100_Q7XKP2 OSJNBa0032N05.7 protein [Oryza sativa] 86 2e-15
UniRef100_Q6AVL1 Putative retrotransposon protein [Oryza sativa] 86 2e-15
UniRef100_Q688H9 Putative polyprotein [Oryza sativa] 86 2e-15
UniRef100_Q8S5Q8 Putative retroelement [Oryza sativa] 86 2e-15
>UniRef100_Q69F85 Gag-pol polyprotein [Phaseolus vulgaris]
Length = 1859
Score = 191 bits (486), Expect = 3e-47
Identities = 112/311 (36%), Positives = 172/311 (55%), Gaps = 31/311 (9%)
Query: 145 PPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLIRAG-------------RSQLNDRG 191
PP D +K C+YHR+ G+ T C LK++IE L++AG RS D
Sbjct: 455 PPNA--DTSKRCQYHRNYGHTTEGCQALKDKIEELVQAGHLRKFVKTTITAPRSPQRDHD 512
Query: 192 DRWRG*RQ----QGNRYKGDR*QDDRRRDD---RRRTPTSDKKETLATRKKGSEETFNKD 244
R R R+ + N Y+ R R+R + RR P S+ E + K+ N
Sbjct: 513 PRERSGRRDDRTRDNHYRSSR----RKRSESPIRRTRPKSESPERRSRTKQKVRTVINTI 568
Query: 245 LEP-----PVGTINTIAGGFGGGGDIASTRRRNVRAVTSVREYEAPFGFHHPDIVISSAD 299
P P IN IAGGF GGG S R++++RA+ SV H P I + D
Sbjct: 569 AGPVSLGQPPQEINYIAGGFAGGGCSNSARKKHLRAIQSVHSTPTQRRPHIPPITFTDED 628
Query: 300 F*RIKTHKDDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLV 359
F I +DDP+V+ + I+ F + +VL++QGSS DI+Y + F ++ + + ++ PY +V
Sbjct: 629 FTAIDPSQDDPMVITVEIDKFAIAKVLVDQGSSVDILYWETFKKMKIPEAEIQPYNEQIV 688
Query: 360 GFSGEQVWVRGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTD 419
GFS E+V +G++DL+T FG D +K + +RYL++ SYN+++GR ++NRL A++ST
Sbjct: 689 GFSRERVDTKGFIDLYTTFGDDYLSKTINIRYLLVNANTSYNILLGRPSINRLKAIVSTP 748
Query: 420 HLAVKYPLICG 430
HLA+K+P + G
Sbjct: 749 HLAMKFPSVNG 759
>UniRef100_Q8RZN9 Putative polyprotein [Oryza sativa]
Length = 2001
Score = 97.1 bits (240), Expect = 9e-19
Identities = 82/289 (28%), Positives = 129/289 (44%), Gaps = 32/289 (11%)
Query: 151 DKTKWCEYHRSAGYDTGDCFTLKNEIE-------RLIRAGRSQLNDRGDRWRG*RQQGNR 203
D+TK+C+ H G+ T +C++ I R L +R+ N+
Sbjct: 630 DQTKFCQTHGPGGHSTEECYSKFCHIHGPGGHSTEECRQMTHLLEKHVNRYE------NK 683
Query: 204 YKGDR*QDDRRRDDRRRTPTSDKKETLATRKKGSEETFNKDLEPPVGTINTIAGGFGGGG 263
Y+G R D+R + P K E + E P IN I GG G
Sbjct: 684 YEGAR---DQRGQNAIEAPQVMKIEAIE--------------EVPKRVINAITGGSSLGV 726
Query: 264 DIASTRRRNVRAVTSV-REYEA-PFGFHHPDIVISSADF*RIKTHKDDPVVVMIRINSFN 321
+ R+ VR V V Y++ P + I D I DP+VV + I
Sbjct: 727 ESKRQRKAYVRQVNHVGTSYQSNPPVYSKTVISFGPEDAEGILFPHQDPLVVSVEIAQCE 786
Query: 322 VRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWVRGYLDLHTVFGVD 381
V+RVL++ GSSAD+++ DAF ++ + + L L GF G+QV G + L VFG
Sbjct: 787 VQRVLIDGGSSADVLFYDAFKKMQIPEDRLTNAGVPLQGFGGQQVHAIGKISLQVVFGKG 846
Query: 382 ENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYPLICG 430
N + + + V+ + YN I+GR+T+N A+I +++ +K P + G
Sbjct: 847 TNVRKEEIVFDVVDMPYQYNAILGRSTINIFEAIIHHNYICMKLPGLRG 895
>UniRef100_Q7X8F3 OSJNBa0093P23.5 protein [Oryza sativa]
Length = 2074
Score = 94.0 bits (232), Expect = 8e-18
Identities = 83/316 (26%), Positives = 132/316 (41%), Gaps = 57/316 (18%)
Query: 111 RQEESGKSLNAHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCF 170
R + +GK++ A E ++ ++ + P R V K +C +H + + T C
Sbjct: 1325 RTDRAGKAVMA--VEEVQALRKEFDTQQASNHQQPARKKVRKDLYCAFHGRSSHTTEQC- 1381
Query: 171 TLKNEIERLIRAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDRRRDDRRRTPTSDKKETL 230
R R RG Q +G + R + TP +++T
Sbjct: 1382 -------------------RNIRQRGNVQDPRPQQGTTVEAPREAVQEQTTPAKQRQDTQ 1422
Query: 231 ATRKKGSEETFNKDLEPPVGTINTIAGGFGGGGDIASTRRRNVRAVTSVREYEAPFGFHH 290
RK + R V ++TS E AP + +
Sbjct: 1423 RRRKM---------------------------------QIRMVHSITSAGE-GAP-QYVN 1447
Query: 291 PDIVISSADF*RIKTHKDDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKD 350
I D + DP+V+ I F VRR+L++ GSSAD+IY +A+ ++GL +
Sbjct: 1448 QLISFGPEDAEGVLFPHQDPLVISAEIAGFEVRRILVDGGSSADVIYAEAYAKMGLPTQA 1507
Query: 351 LMPYTGTLVGFSGEQVWVRGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLN 410
L P +L GFSGE V V G L FG EN + ++ + V+ + +YN I GR TLN
Sbjct: 1508 LTPALTSLRGFSGEAVQVLGQALLLIAFGSGENRREEQILFDVVNIPYNYNAIFGRATLN 1567
Query: 411 RLCAVISTDHLAVKYP 426
+L A+ ++L +K P
Sbjct: 1568 KLEAISHHNYLKLKMP 1583
>UniRef100_O81436 T24H24.13 protein [Arabidopsis thaliana]
Length = 681
Score = 90.1 bits (222), Expect = 1e-16
Identities = 77/324 (23%), Positives = 142/324 (43%), Gaps = 42/324 (12%)
Query: 121 AHLTELLRKVKATHVVEAGEKETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLI 180
AHL + ++ + + + +D K+C+YH+ GY T +C +K +LI
Sbjct: 351 AHLRSFRFAITRAYLTDEENEAAENESPELDLGKYCKYHKKRGYSTEECRAVK----KLI 406
Query: 181 RAGRSQLNDRGDRWRG*RQQGNRYKGDR*QDDRRRDD-------RRRTPTSDKKETLATR 233
AG G ++G+ K + D + ++ R RTP A
Sbjct: 407 AAG------------GKTKKGSNPKVETPPPDEQEEEQTPKQKKRERTPEGITTPPPAPH 454
Query: 234 KKGSE---------ETFNKDLEPPVGTINTIAGGFGGGGDIASTRRRNVRAVTSVREYEA 284
KK + + L P I+ I GG D ++ + + R S + ++
Sbjct: 455 KKNMRFPLRLLKFCRSATEHLSPK--RIDFIMGGSQLCNDSINSIKTHQRKADSYTKGKS 512
Query: 285 PFGFHHPDIVISSADF*RIKTHKDDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQL 344
P + I ++ + DD +V+ I + ++ + ++++ GSS D+++ DAF ++
Sbjct: 513 PMMGPNHQITFWESETTDLNKPHDDALVIRIDVGNYELSHIMIDTGSSVDVLFYDAFKRM 572
Query: 345 GLTDKDLMPYTGTLVGFSGEQVWVRGYLDLHTVFGVDENAKLLR--VRYLVLQVVASYNV 402
G D +L L GF+G+ + G + L T+ A+ +R +LV+ A +N
Sbjct: 573 GHLDSELQGRKTPLTGFAGDTTFSLGTIQLPTI------ARGVRRLTSFLVVNKKAPFNA 626
Query: 403 IIGRNTLNRLCAVISTDHLAVKYP 426
I+GR L+ + AV ST H +K+P
Sbjct: 627 ILGRPWLHAMKAVPSTYHQCIKFP 650
>UniRef100_Q75G47 Putative polyprotein [Oryza sativa]
Length = 1459
Score = 89.0 bits (219), Expect = 2e-16
Identities = 59/183 (32%), Positives = 93/183 (50%), Gaps = 2/183 (1%)
Query: 246 EPPVGTINTIAGGFGGGGDIASTRRRNVRAVTSV-REYEA-PFGFHHPDIVISSADF*RI 303
E P IN I GG G + R+ VR + V Y++ P + + I D I
Sbjct: 205 EAPKRVINAITGGSSLGVESKRQRKAYVRQIHHVGTSYQSVPPAYSNTVISFGPEDAEGI 264
Query: 304 KTHKDDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSG 363
DP+V+ + I V+RVL++ GSSAD+++ DAF ++ + + L L GF G
Sbjct: 265 LFPHQDPLVISVEIAQCEVQRVLVDGGSSADVLFYDAFKKMQIPEDRLTHAGIPLQGFGG 324
Query: 364 EQVWVRGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAV 423
QV G + L VFG EN + V + V+ + YN ++GR+T+N A+I +++ +
Sbjct: 325 HQVHTIGKISLQVVFGGGENKRREEVVFDVVDMPYQYNAVLGRSTINIFEAIIHHNYICM 384
Query: 424 KYP 426
K P
Sbjct: 385 KLP 387
>UniRef100_Q9ARZ4 Putative polyprotein [Oryza sativa]
Length = 1579
Score = 88.6 bits (218), Expect = 3e-16
Identities = 85/350 (24%), Positives = 143/350 (40%), Gaps = 60/350 (17%)
Query: 93 PRKESFERRRPWHQTDSRRQEESGKSLNAHLTELLRK-VKATHVVEAGEKETD------- 144
P S + +P Q RQ ++ + T+ K V A V+A KE D
Sbjct: 253 PSPLSVRQSQPAIQGQPPRQGQAPITWRKFRTDCAGKAVMAVEEVQALRKEFDAQQASNH 312
Query: 145 --PPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLIRAGRSQLNDRGDRWRG*RQQGN 202
P R V K +C +H + + T C R R RG Q
Sbjct: 313 QQPARKKVRKDLYCAFHGRSSHTTEQC--------------------RNIRQRGNAQDPR 352
Query: 203 RYKGDR*QDDRRRDDRRRTPTSDKKET------LATRKKGSEETFNKDLEPPVGTINTIA 256
KG + R + +P ++ + TR + + + + +++I
Sbjct: 353 PQKGTTFEASREAIQEQTSPAEQCQDVQRRVIQVITRADPPSQLSKRQKKMQIRMVHSIT 412
Query: 257 GGFGGGGDIASTRRRNVRAVTSVREYEAPFGFHHPDIVISSADF*RIKTHKDDPVVVMIR 316
+ + + + S +A F H D+++ SA+
Sbjct: 413 -------SVGEGAPQYLNQLISFGPEDAGVMFPHQDLLVISAE----------------- 448
Query: 317 INSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWVRGYLDLHT 376
I F VRR+L++ GSSAD+I+ +A+ ++GL+ + L P +L GF GE V V G L
Sbjct: 449 IAGFEVRRILVDGGSSADVIFAEAYAKMGLSTQALTPAPASLRGFGGEAVQVLGQALLLI 508
Query: 377 VFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
FG EN + ++ + V+ + +YN I GR +LN+L A+ ++L +K P
Sbjct: 509 AFGSGENRREEQILFDVIDIPYNYNAIFGRASLNKLEAISHHNYLKLKMP 558
>UniRef100_Q8LSS6 Putative gag-pol [Oryza sativa]
Length = 1112
Score = 88.6 bits (218), Expect = 3e-16
Identities = 96/366 (26%), Positives = 147/366 (39%), Gaps = 47/366 (12%)
Query: 78 KKIKSGERTAKEPLYPRKESFERRRPWHQTDSRRQEESGKSLNAHLTELL-RKVKATHVV 136
+KI S E L+ + R+ + R + G L A L RK KAT
Sbjct: 330 EKIASKEPKTTAELFELADKVARKE---EAKPARSNDEGHVLAADGPALAPRKGKAT--- 383
Query: 137 EAGEK--ETDPPRVVVDKTKWCEYHRSAGYDTGDCFTLKNEIERLIRAGRSQLNDRGDRW 194
G+K T P KWC H S + DC ++KN ER +A ++ R +
Sbjct: 384 --GDKPSSTAPSGEGRSADKWCSVHNSYRHSLADCRSVKNLAERFRKADENKRQGRRESK 441
Query: 195 RG*RQQGNRYKGDR*QDDRRRDDRRRTPTSDKKETLATRKKGSEETFNKDLEPPVGTINT 254
G DRR + + + P D + SE+ D + P GT+ T
Sbjct: 442 APATSTG----------DRREEAKNKAPADDGGD--------SEDL---DFQIPQGTVAT 480
Query: 255 IAGGFGGGGDIASTRRRNVRAVTSVREYEAPFGFHHPDIVISSADF*RIKTHKDDP---- 310
+ GG GGG A T RR + + RE A H + D P
Sbjct: 481 LDGG--GGGACAHTSRRGFKVMR--RELLAAVPTHEAARKARWLEVKLTFDQSDHPTVLS 536
Query: 311 ------VVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGE 364
+VV I++ ++RVL++ G S II +FD L L P +++G +
Sbjct: 537 RGGKLALVVSPTIHNVKMKRVLVDGGDSLSIISPASFDALKAPGMKLQPSL-SIIGVTPG 595
Query: 365 QVWVRGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVK 424
+W G+++L FG N + R+ + V + YNV++GR L + A +L +K
Sbjct: 596 HMWPLGHVELPVTFGDSTNFRTERIDFDVADLNLPYNVVLGRPALVKFMAATHYAYLQMK 655
Query: 425 YPLICG 430
P + G
Sbjct: 656 MPGLAG 661
>UniRef100_Q94HQ3 Putative retroelement [Oryza sativa]
Length = 1705
Score = 88.2 bits (217), Expect = 4e-16
Identities = 46/118 (38%), Positives = 70/118 (58%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ SSAD+I+ +A+ ++GLT + + P L GF GE V V
Sbjct: 656 DPLVISAEIAGFEVRRILVDGESSADVIFAEAYAKMGLTTQAITPAPALLRGFGGEAVQV 715
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG EN + +V + V+ + +YN I GR TLN+ A+ ++L +K P
Sbjct: 716 LGQAQLMVAFGTGENRREEQVLFDVVDIPYNYNAIFGRATLNKFEAISHHNYLKLKMP 773
>UniRef100_Q8S664 Putative retroelement [Oryza sativa]
Length = 1367
Score = 88.2 bits (217), Expect = 4e-16
Identities = 46/118 (38%), Positives = 70/118 (58%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ SSAD+I+ +A+ ++GLT + + P L GF GE V V
Sbjct: 514 DPLVISAEIAGFEVRRILVDGESSADVIFAEAYAKMGLTTQAITPAPALLRGFGGEAVQV 573
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG EN + +V + V+ + +YN I GR TLN+ A+ ++L +K P
Sbjct: 574 LGQAQLMVAFGTGENRREEQVLFDVVDIPYNYNAIFGRATLNKFEAISHHNYLKLKMP 631
>UniRef100_Q7XSE2 OSJNBb0033G08.12 protein [Oryza sativa]
Length = 1847
Score = 87.8 bits (216), Expect = 5e-16
Identities = 46/118 (38%), Positives = 71/118 (59%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ GSSAD+I+ +A+ ++GL + L P +L GF GE V V
Sbjct: 782 DPLVISAEIAGFEVRRILVDGGSSADVIFAEAYAKIGLPTQALTPAPASLRGFGGEAVQV 841
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG EN + ++ + V+ + +YN I GR TLN+ A+ ++L +K P
Sbjct: 842 LGQALLLIAFGSGENRREEQILFDVVDIPYNYNAIFGRATLNKFEAIFHHNYLKLKMP 899
>UniRef100_Q94I18 Putative retroelement [Oryza sativa]
Length = 1497
Score = 87.8 bits (216), Expect = 5e-16
Identities = 46/118 (38%), Positives = 72/118 (60%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L+++GSSADII+ +A+ ++GL + L P +L GF GE V V
Sbjct: 441 DPLVISAEIAGFEVRRILVDEGSSADIIFAEAYAKMGLPTQALTPAPASLRGFGGEPVQV 500
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG +N + ++ + V+ + +YN I GR TLN+ A+ ++L +K P
Sbjct: 501 LGQALLLIAFGSGDNRREEQILFDVVDIPYNYNAIFGRATLNKFEAISHHNYLKLKMP 558
>UniRef100_Q9ZNW4 Polyprotein [Sorghum bicolor]
Length = 1877
Score = 87.4 bits (215), Expect = 7e-16
Identities = 58/193 (30%), Positives = 88/193 (45%), Gaps = 16/193 (8%)
Query: 247 PPVGTINTIAGGFGGGGDIASTRRRNVRAVTSVREY---------EAPFGFHHPDIVISS 297
P +G I IAGG ++ ++R V +V P F D+ + S
Sbjct: 572 PTIGMILPIAGGSSMEFQTKKQKKDHLRLVNNVAVQGPVRCTDWSRTPITFTEEDLRLES 631
Query: 298 ADF*RIKTHKDDPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGT 357
D +V+ + + V R+L++ GSSADII+ FDQ+ L+ L P
Sbjct: 632 YPH-------TDALVITTNVAGWGVSRILVDSGSSADIIFAGTFDQMKLSRSQLQPSESP 684
Query: 358 LVGFSGEQVWVRGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVIS 417
L+GF G+Q+ G + L FG ENA+ V + V+ + YN I GR LN+ A
Sbjct: 685 LIGFGGKQIHALGKVALPVSFGTTENARTEYVTFDVVDLHYPYNAIFGRGFLNKFNAAAH 744
Query: 418 TDHLAVKYPLICG 430
+L +K P + G
Sbjct: 745 MGYLCMKIPALHG 757
>UniRef100_Q8S636 Putative polyprotein [Oryza sativa]
Length = 1722
Score = 87.4 bits (215), Expect = 7e-16
Identities = 47/118 (39%), Positives = 70/118 (58%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ GSSAD+I+ +A+ ++GLT + L P +L GF GE V V
Sbjct: 534 DPLVISAEIAGFEVRRILVDGGSSADVIFAEAYAKMGLTSQALTPAPASLRGFGGEAVQV 593
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG EN + +V + V+ + +YN I GR TLN + ++L +K P
Sbjct: 594 LGQALLLIAFGSGENRREEQVLFDVVDIPYNYNTIFGRATLNMFEVISHHNYLELKMP 651
>UniRef100_Q7XER7 Contains similarity to gag-pol polyprotein [Oryza sativa]
Length = 1260
Score = 87.4 bits (215), Expect = 7e-16
Identities = 46/118 (38%), Positives = 70/118 (58%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ GSSAD+I+ +A+ ++GL L P +L GF G+ V V
Sbjct: 404 DPLVISAEIAGFEVRRILVDGGSSADVIFAEAYAKMGLPTLALTPAPASLRGFGGKAVQV 463
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG EN + +V + V+ + +YN I GR TLN+ A+ ++L +K P
Sbjct: 464 LGQAQLLVAFGTGENRREEQVLFDVVDIPYNYNAIFGRATLNKFEAISHHNYLKLKMP 521
>UniRef100_Q8W2U3 Putative polyprotein [Oryza sativa]
Length = 1931
Score = 86.7 bits (213), Expect = 1e-15
Identities = 46/118 (38%), Positives = 70/118 (58%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ GSSAD+I+ +A+ +GL + L P +L GF GE V V
Sbjct: 788 DPLVISAEIAGFEVRRILVDGGSSADVIFAEAYATMGLPTQSLTPAPTSLHGFGGEAVQV 847
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG EN + ++ + V+ + +YN I GR TLN+ A+ ++L +K P
Sbjct: 848 LGQALLLIAFGSGENRREEQILFDVVDIPYNYNAIFGRATLNKFEAICHHNYLKLKMP 905
>UniRef100_Q6AUT8 Putative gag-pol [Oryza sativa]
Length = 1631
Score = 86.7 bits (213), Expect = 1e-15
Identities = 46/118 (38%), Positives = 71/118 (59%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ GSSAD+I+ +A+ ++GL + L P +L GF GE V V
Sbjct: 550 DPLVISAEIAGFEVRRILVDGGSSADVIFAEAYAKMGLPTQALTPAPASLRGFGGEAVRV 609
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG EN + ++ + V+ + +YN I GR TLN+ A+ ++L +K P
Sbjct: 610 LGQALLLIAFGSGENRREEQILFDVVDIPYNYNAIFGRATLNKFEAISHHNYLKLKMP 667
>UniRef100_Q7XKP2 OSJNBa0032N05.7 protein [Oryza sativa]
Length = 598
Score = 86.3 bits (212), Expect = 2e-15
Identities = 44/122 (36%), Positives = 72/122 (58%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP++++ I F VRR+L++ GSSAD+I +A+ ++GL L +L GF GE V V
Sbjct: 85 DPLIILAEIVGFEVRRILVDGGSSADVIIAEAYARMGLPTLALTQAPTSLQGFGGEAVQV 144
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYPLI 428
G + L FG +N + ++ + V+ + +YN I GR TLN+ A+ ++L +K P +
Sbjct: 145 LGQVQLAVAFGTSDNRREEQILFDVVDIPYNYNTIFGRATLNKFKAISHHNYLKLKMPGL 204
Query: 429 CG 430
G
Sbjct: 205 AG 206
>UniRef100_Q6AVL1 Putative retrotransposon protein [Oryza sativa]
Length = 1168
Score = 86.3 bits (212), Expect = 2e-15
Identities = 45/116 (38%), Positives = 69/116 (58%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ GSS D+I+ +A+ ++GLT + L P +L GF GE V V
Sbjct: 369 DPLVISAEIAGFEVRRILVDGGSSTDVIFAEAYAKMGLTTQALTPAPTSLRGFGGEAVQV 428
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVK 424
G L FG EN + +V + V+ + +YN I GR TLN+ + ++L +K
Sbjct: 429 LGQAQLIVAFGTGENRREEQVLFDVVDIPYNYNAIFGRATLNKFEVISHHNYLKLK 484
>UniRef100_Q688H9 Putative polyprotein [Oryza sativa]
Length = 1528
Score = 86.3 bits (212), Expect = 2e-15
Identities = 55/163 (33%), Positives = 78/163 (47%), Gaps = 10/163 (6%)
Query: 264 DIASTRRRNVRAVTSVREYEAPFGFHHPDIVISSADF*RIKTHKDDPVVVMIRINSFNVR 323
D T R N +V E + + I D + DP+V+ I F VR
Sbjct: 457 DALRTPRSNAETFGNVATREGAPQYLNQQIFFGPEDAEGVMFPHQDPLVISAEIAGFEVR 516
Query: 324 RVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWVRGYLDLHTVFGVDEN 383
R+L+N GSSAD+I+ +A+ ++GLT + L P +L GF GE V V G L FG EN
Sbjct: 517 RILVNGGSSADVIFAEAYAKMGLTTQALTPALTSLRGFGGEAVQVLGQAHLIVAFGTGEN 576
Query: 384 AKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
+ +YN I GR TLN+ A+ ++L +K P
Sbjct: 577 RR----------EEQNYNTIFGRATLNKFEAISHHNYLKLKMP 609
>UniRef100_Q8S5Q8 Putative retroelement [Oryza sativa]
Length = 1292
Score = 85.9 bits (211), Expect = 2e-15
Identities = 46/118 (38%), Positives = 71/118 (59%)
Query: 309 DPVVVMIRINSFNVRRVLLNQGSSADIIYGDAFDQLGLTDKDLMPYTGTLVGFSGEQVWV 368
DP+V+ I F VRR+L++ GSSAD+I+ +A+ ++GL + L P +L GF GE V V
Sbjct: 361 DPLVISAEIAGFEVRRILVDGGSSADVIFAEAYAKMGLPTQALTPAPVSLRGFGGEAVQV 420
Query: 369 RGYLDLHTVFGVDENAKLLRVRYLVLQVVASYNVIIGRNTLNRLCAVISTDHLAVKYP 426
G L FG EN + ++ + V+ + +YN I GR TLN+ A+ ++L +K P
Sbjct: 421 LGQTLLLIAFGSGENRREEQILFDVVDIPYNYNAIFGRATLNKFEAISHHNYLKLKMP 478
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.327 0.143 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 708,340,368
Number of Sequences: 2790947
Number of extensions: 30123990
Number of successful extensions: 104309
Number of sequences better than 10.0: 561
Number of HSP's better than 10.0 without gapping: 296
Number of HSP's successfully gapped in prelim test: 272
Number of HSP's that attempted gapping in prelim test: 102876
Number of HSP's gapped (non-prelim): 1019
length of query: 431
length of database: 848,049,833
effective HSP length: 130
effective length of query: 301
effective length of database: 485,226,723
effective search space: 146053243623
effective search space used: 146053243623
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0134.17


