

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0134.16
(212 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
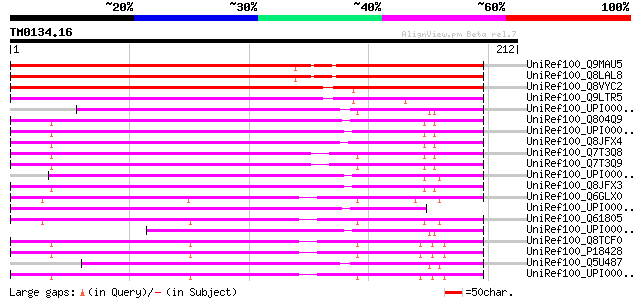
UniRef100_Q9MAU5 F13M7.4 protein [Arabidopsis thaliana] 195 6e-49
UniRef100_Q8LAL8 Hypothetical protein [Arabidopsis thaliana] 194 1e-48
UniRef100_Q8VYC2 Putative bactericidal permeability-increasing p... 170 2e-41
UniRef100_Q9LTR5 Similarity to bactericidal permeability-increas... 156 3e-37
UniRef100_UPI000027CD7C UPI000027CD7C UniRef100 entry 82 1e-14
UniRef100_Q804Q9 Bactericidal permeability-increasing protein/li... 79 8e-14
UniRef100_UPI00003373E9 UPI00003373E9 UniRef100 entry 77 2e-13
UniRef100_Q8JFX4 LBP (LPS binding protein)/BPI (Bactericidal/per... 75 1e-12
UniRef100_Q7T3Q8 Bactericidal permeability increasing protein/li... 73 4e-12
UniRef100_Q7T3Q9 Bactericidal permeability increasing protein/li... 73 6e-12
UniRef100_UPI0000365270 UPI0000365270 UniRef100 entry 72 1e-11
UniRef100_Q8JFX3 LBP (LPS binding protein)/BPI (Bactericidal/per... 71 2e-11
UniRef100_Q6GLX0 MGC84153 protein [Xenopus laevis] 67 4e-10
UniRef100_UPI0000249FDE UPI0000249FDE UniRef100 entry 66 7e-10
UniRef100_Q61805 Lipopolysaccharide-binding protein precursor [M... 58 1e-07
UniRef100_UPI0000361544 UPI0000361544 UniRef100 entry 57 2e-07
UniRef100_Q8TCF0 LBP protein [Homo sapiens] 57 2e-07
UniRef100_P18428 Lipopolysaccharide-binding protein precursor [H... 57 2e-07
UniRef100_Q5U487 Hypothetical protein [Xenopus laevis] 56 7e-07
UniRef100_UPI000036C460 UPI000036C460 UniRef100 entry 54 4e-06
>UniRef100_Q9MAU5 F13M7.4 protein [Arabidopsis thaliana]
Length = 488
Score = 195 bits (496), Expect = 6e-49
Identities = 97/200 (48%), Positives = 146/200 (72%), Gaps = 4/200 (2%)
Query: 1 VKGMQVGLTVNLKNQEGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASSV 60
V+GM++GL++ LK+ EG LKLSL + GC+V D++I+L+GG +W YQ +V+AF I SSV
Sbjct: 137 VQGMEIGLSLGLKSDEGGLKLSLSECGCHVEDITIELEGGASWFYQGMVNAFKDQIGSSV 196
Query: 61 EEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFT-- 118
E +++K+ EG++ LD FLQSLPK+IP+D + LNV+F +P+L NSSI I+GLFT
Sbjct: 197 ESTIAKKLTEGVSDLDSFLQSLPKEIPVDDNADLNVTFTSDPILRNSSITFEIDGLFTKG 256
Query: 119 ERSEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLIIDELPDQ 178
E ++V++S +KK + C G M+ +S+ E V SA+ +Y+NA +Q ++D++P+Q
Sbjct: 257 ETNQVLKS-FFKKSVSL-VICPGNSKMLGISVDEAVFNSAAALYYNADFVQWVVDKIPEQ 314
Query: 179 VILNTAECRFIVPQLYKQYP 198
+LNTA RFI+PQLYK+YP
Sbjct: 315 SLLNTARWRFIIPQLYKKYP 334
>UniRef100_Q8LAL8 Hypothetical protein [Arabidopsis thaliana]
Length = 488
Score = 194 bits (493), Expect = 1e-48
Identities = 96/200 (48%), Positives = 146/200 (73%), Gaps = 4/200 (2%)
Query: 1 VKGMQVGLTVNLKNQEGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASSV 60
V+GM++GL++ LK+ EG LKLSL + GC+V D++I+L+GG +W YQ +V+AF I SSV
Sbjct: 137 VQGMEIGLSLGLKSDEGGLKLSLSECGCHVEDITIELEGGASWFYQGMVNAFKDQIGSSV 196
Query: 61 EEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFT-- 118
E +++K+ EG++ LD FLQ+LPK+IP+D + LNV+F +P+L NSSI I+GLFT
Sbjct: 197 ESTIAKKLTEGVSDLDSFLQNLPKEIPVDDNADLNVTFTSDPILRNSSITFEIDGLFTKG 256
Query: 119 ERSEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLIIDELPDQ 178
E ++V++S +KK + C G M+ +S+ E V SA+ +Y+NA +Q ++D++P+Q
Sbjct: 257 ETNQVLKS-FFKKSVSL-VICPGNSKMLGISVDEAVFNSAAALYYNADFVQWVVDKIPEQ 314
Query: 179 VILNTAECRFIVPQLYKQYP 198
+LNTA RFI+PQLYK+YP
Sbjct: 315 SLLNTARWRFIIPQLYKKYP 334
>UniRef100_Q8VYC2 Putative bactericidal permeability-increasing protein [Arabidopsis
thaliana]
Length = 515
Score = 170 bits (431), Expect = 2e-41
Identities = 91/199 (45%), Positives = 133/199 (66%), Gaps = 5/199 (2%)
Query: 1 VKGMQVGLTVNLKNQEGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASSV 60
VKGM V +T L N G+LK++ + C V ++ I ++GG +WLYQ +VDAF I S+V
Sbjct: 173 VKGMNVRITATLVNDNGSLKIASRENDCTVKNIDIHINGGASWLYQGVVDAFQKMIISTV 232
Query: 61 EEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTER 120
E+ VS KI E + KLD FLQSLPKQ +D ++A+N++F GNPVL NSS+ + INGLF +
Sbjct: 233 EKTVSTKIVEKMKKLDSFLQSLPKQRKIDDSAAVNLTFTGNPVLGNSSVEVDINGLFMPK 292
Query: 121 SEVVESEGYKKGFKISSACGGL-PNMIKVSLHEYVIQSASLVYFNAGKMQLIIDELPDQV 179
+ ++ G + SS GG+ M+ +S+ E V SA+LVYFNA M L+++E +
Sbjct: 293 GDDIKVAGSRS----SSFFGGVNKRMVTISVEEGVFNSATLVYFNAKVMHLVMEETKNGS 348
Query: 180 ILNTAECRFIVPQLYKQYP 198
IL+T++ + I+P+LYK YP
Sbjct: 349 ILSTSDWKLILPELYKHYP 367
>UniRef100_Q9LTR5 Similarity to bactericidal permeability-increasing protein
[Arabidopsis thaliana]
Length = 1424
Score = 156 bits (395), Expect = 3e-37
Identities = 91/224 (40%), Positives = 133/224 (58%), Gaps = 30/224 (13%)
Query: 1 VKGMQVGLTVNLKNQEGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASSV 60
VKGM V +T L N G+LK++ + C V ++ I ++GG +WLYQ +VDAF I S+V
Sbjct: 259 VKGMNVRITATLVNDNGSLKIASRENDCTVKNIDIHINGGASWLYQGVVDAFQKMIISTV 318
Query: 61 EEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTER 120
E+ VS KI E + KLD FLQSLPKQ +D ++A+N++F GNPVL NSS+ + INGLF +
Sbjct: 319 EKTVSTKIVEKMKKLDSFLQSLPKQRKIDDSAAVNLTFTGNPVLGNSSVEVDINGLFMPK 378
Query: 121 SEVVESEGYKKGFKISSACGGL-PNMIKVSLHEYVIQSASLVYFN--------------- 164
+ ++ G + SS GG+ M+ +S+ E V SA+LVYFN
Sbjct: 379 GDDIKVAGSRS----SSFFGGVNKRMVTISVEEGVFNSATLVYFNVSSQILIRVWWNKTI 434
Query: 165 ----------AGKMQLIIDELPDQVILNTAECRFIVPQLYKQYP 198
A M L+++E + IL+T++ + I+P+LYK YP
Sbjct: 435 CKLIGLVLIQAKVMHLVMEETKNGSILSTSDWKLILPELYKHYP 478
>UniRef100_UPI000027CD7C UPI000027CD7C UniRef100 entry
Length = 475
Score = 81.6 bits (200), Expect = 1e-14
Identities = 49/175 (28%), Positives = 88/175 (50%), Gaps = 9/175 (5%)
Query: 29 YVGDLSIKLDGGTAWLYQLLVDAFGGNIASSVEEAVSEKINEGIAKLDKFLQSLPKQIPL 88
+VGD+ + +GG +W++Q V G + +EE + + + IAK D L ++ I +
Sbjct: 153 HVGDVHMNFEGGASWIFQPFVHQLAGRLRGEMEEKICPSLEDSIAKFDYHLLAINISIDV 212
Query: 89 DKTSALNVSFVGNPVLSNSSIAIAINGLFTERSEVVESEGYKKGFKISSACGGLPN-MIK 147
+K L++S PV+ SS+ + + G E + F ++ P+ M+
Sbjct: 213 NKDLTLDLSLTDEPVVDVSSLNVGLKGAIYSSKTHAEPPFEPQDFTMAEQ----PDFMLS 268
Query: 148 VSLHEYVIQSASLVYFNAGKMQLIIDE--LP--DQVILNTAECRFIVPQLYKQYP 198
+ + EY + SA Y++AG +Q+ I+E +P V LNT+ +PQL + YP
Sbjct: 269 LGVSEYTLNSALYAYYSAGLLQVFINESMVPSYSPVHLNTSSVGAFIPQLPEMYP 323
>UniRef100_Q804Q9 Bactericidal permeability-increasing protein/lipopolysaccharide-
binding protein precursor [Cyprinus carpio]
Length = 473
Score = 79.0 bits (193), Expect = 8e-14
Identities = 53/203 (26%), Positives = 99/203 (48%), Gaps = 8/203 (3%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
V + + TV + + + G +S+ + VG +++K GG +WLY L + ++
Sbjct: 127 VSELTISTTVAVMSDDTGHPTVSMTNCAATVGSVNVKFHGGASWLYNLFSSFINKALRNA 186
Query: 60 VEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTE 119
+++ + + + IA ++ L++L +D+ + + S VG+PV+SN+SI + + G F
Sbjct: 187 LQKQICPLVADSIADINPHLKTLNVLAKVDQYAEIEYSMVGSPVISNTSIDLGLKGEFYN 246
Query: 120 RSEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLII--DELP- 176
+ E F + S +M+ + + + I SA VY AG + L I D +P
Sbjct: 247 IGQHKEPPFSPTPFSLPSQD---TDMLYIGVSAFTINSAGFVYNRAGALSLYITDDMIPS 303
Query: 177 -DQVILNTAECRFIVPQLYKQYP 198
+ LNT +PQ+ K YP
Sbjct: 304 GSPIRLNTKTFGAFIPQIEKMYP 326
>UniRef100_UPI00003373E9 UPI00003373E9 UniRef100 entry
Length = 473
Score = 77.4 bits (189), Expect = 2e-13
Identities = 51/203 (25%), Positives = 97/203 (47%), Gaps = 8/203 (3%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
V G+ V T+ +K+ G ++S + G VG + +K GG +WLY L + S+
Sbjct: 127 VNGLSVAETLGIKSDNTGRPEVSSISCGASVGSVKVKFHGGASWLYNLFSRYINRALESA 186
Query: 60 VEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTE 119
+++ + + I+ ++ L++L +DK + + + V +P +S SSI + + G F
Sbjct: 187 LQKQICPLVTNAISDVNPRLKTLNVLANVDKYAEIEYAMVSSPDVSTSSINLKLKGQFYN 246
Query: 120 RSEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLII--DELP- 176
+ E F + S NM+ + + + + SA+ VY AG + + I D +P
Sbjct: 247 IGQHQEPPFSAPAFSLPSENS---NMLYIGMSAFTVNSAAFVYHKAGALSMYITDDMIPK 303
Query: 177 -DQVILNTAECRFIVPQLYKQYP 198
+ L T +P++ KQ+P
Sbjct: 304 SSPIRLTTNTFGTFIPEISKQFP 326
>UniRef100_Q8JFX4 LBP (LPS binding protein)/BPI (Bactericidal/permeability-increasing
protein)-1 [Oncorhynchus mykiss]
Length = 473
Score = 75.1 bits (183), Expect = 1e-12
Identities = 54/203 (26%), Positives = 98/203 (47%), Gaps = 8/203 (3%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
V G+ V ++ +K+ E G +S ++ VG SIK GG +WLY L + S+
Sbjct: 127 VDGLTVTDSITIKSDETGRPTVSSVNCVANVGSASIKFHGGASWLYNLFSAYIDKALRSA 186
Query: 60 VEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTE 119
+++ + + + I ++ L++L +DK + + S V +P +SN+SI ++ G F
Sbjct: 187 LQKQICPLVADTITDMNPHLKTLNVLAKVDKYAEVEYSMVTSPTISNASIDFSLKGEFYN 246
Query: 120 RSEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLII--DELP- 176
+ E F + + NM+ + + + SA VY NAG + L I D +P
Sbjct: 247 IGKHQEPPFSPTPFSLPPQ---VNNMLYIGMSAFTTNSAGFVYNNAGALSLYITDDMIPP 303
Query: 177 -DQVILNTAECRFIVPQLYKQYP 198
+ LNT +P++ K++P
Sbjct: 304 SSPIRLNTRTFGAFIPEIAKRFP 326
>UniRef100_Q7T3Q8 Bactericidal permeability increasing protein/lipopolysaccharide
binding protein variant b [Gadus morhua]
Length = 473
Score = 73.2 bits (178), Expect = 4e-12
Identities = 52/207 (25%), Positives = 101/207 (48%), Gaps = 16/207 (7%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
+K + + +++L++ G +++ +G +S+KL GG +WLY L + S
Sbjct: 127 IKSLSITTSISLRSDNMGLPAVAMASCTTTLGGVSVKLHGGASWLYNLFRRFIEKGLQSQ 186
Query: 60 VEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTE 119
+++ + + E ++ +++FL+++ P+D+ + + V +P +S+SSI + + G F
Sbjct: 187 LQKKLCPLVAESVSSMNQFLKTVKVMAPVDRYAEIAYPMVSSPDISSSSIGLNLKGEFYN 246
Query: 120 RSEVVESEGYKKGFKISSACGGLPN----MIKVSLHEYVIQSASLVYFNAGKMQLII--D 173
+E S A LPN M+ + L + SAS VY AG + L I D
Sbjct: 247 IGNHMEP-------PFSPAPFFLPNQEQSMLYIGLSAFTANSASFVYNKAGTLSLKITDD 299
Query: 174 ELP--DQVILNTAECRFIVPQLYKQYP 198
+P + L T ++PQ+ K +P
Sbjct: 300 MVPRSSPIRLTTNTFGVLIPQIAKLFP 326
>UniRef100_Q7T3Q9 Bactericidal permeability increasing protein/lipopolysaccharide
binding protein variant a [Gadus morhua]
Length = 473
Score = 72.8 bits (177), Expect = 6e-12
Identities = 52/207 (25%), Positives = 101/207 (48%), Gaps = 16/207 (7%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
+K + + +++L++ G +++ +G +S+KL GG +WLY L + S
Sbjct: 127 IKSLSITTSISLRSDNMGLPAVAMASCTTTLGGVSVKLHGGASWLYNLFRRFIEKGLQSQ 186
Query: 60 VEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTE 119
+++ + + E ++ +++FL+++ P+D+ + + V +P +S+SSI + + G F
Sbjct: 187 LQKKLCPLVAESVSSMNQFLKTVNVMAPVDRYAEIAYPMVSSPDISSSSIGLNLKGEFYN 246
Query: 120 RSEVVESEGYKKGFKISSACGGLPN----MIKVSLHEYVIQSASLVYFNAGKMQLII--D 173
+E S A LPN M+ + L + SAS VY AG + L I D
Sbjct: 247 IGNHMEP-------PFSPAPFFLPNQEQSMLYIGLSAFTANSASFVYNKAGTLSLKITDD 299
Query: 174 ELP--DQVILNTAECRFIVPQLYKQYP 198
+P + L T ++PQ+ K +P
Sbjct: 300 MVPRSSPIRLTTNTFGVLIPQIAKLFP 326
>UniRef100_UPI0000365270 UPI0000365270 UniRef100 entry
Length = 473
Score = 72.0 bits (175), Expect = 1e-11
Identities = 49/186 (26%), Positives = 88/186 (46%), Gaps = 7/186 (3%)
Query: 17 GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASSVEEAVSEKINEGIAKLD 76
G ++S + VG + IK GG +WLY L + S++++ + + I ++
Sbjct: 144 GRPEVSTISCAASVGRVRIKFHGGASWLYNLFSKYIEKALKSALQKQICPLVTNAIDDMN 203
Query: 77 KFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTERSEVVESEGYKKGFKIS 136
L++L +DK + + S V +P +S+SSIA+ + G F + E F +
Sbjct: 204 PRLKTLNVLANVDKYAEIEYSMVSSPAISSSSIALKLKGQFYNIGQHQEPPFSAPAFSLP 263
Query: 137 SACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLII--DELPDQ--VILNTAECRFIVPQ 192
NM+ ++L + SA+ VY AG + + I D +P + + L T +PQ
Sbjct: 264 PENS---NMLYMALSAFTANSAAFVYHKAGVLSIYITDDMIPKRSPIRLTTTTFGTFIPQ 320
Query: 193 LYKQYP 198
+ KQ+P
Sbjct: 321 ISKQFP 326
>UniRef100_Q8JFX3 LBP (LPS binding protein)/BPI (Bactericidal/permeability-increasing
protein) like-2 [Oncorhynchus mykiss]
Length = 473
Score = 70.9 bits (172), Expect = 2e-11
Identities = 50/203 (24%), Positives = 94/203 (45%), Gaps = 8/203 (3%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
V G+ + + +K+ E G +S ++ VG SIK GG +WLY L + S+
Sbjct: 127 VNGLTITADIAIKSDETGRPTVSTVNCVANVGSASIKFHGGASWLYNLFKSYIDKALRSA 186
Query: 60 VEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTE 119
+++ + + + I ++ L++ +D+ + + S V +P +S SSI ++ G F
Sbjct: 187 LQKQICPLVADVITDMNPHLKTFNVLAKVDQYAEIEYSMVTSPTISKSSIEFSLKGEFYN 246
Query: 120 RSEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLII--DELP- 176
+ E F + NM+ + + + SA VY NAG + L + D +P
Sbjct: 247 IGKHQEPPFSPTPFSLPPQDN---NMLYIGVSSFTPNSAGFVYNNAGALSLYVTDDMIPP 303
Query: 177 -DQVILNTAECRFIVPQLYKQYP 198
+ LNT +P++ K++P
Sbjct: 304 SSPIRLNTGTFGVFIPEIAKRFP 326
>UniRef100_Q6GLX0 MGC84153 protein [Xenopus laevis]
Length = 476
Score = 66.6 bits (161), Expect = 4e-10
Identities = 50/210 (23%), Positives = 99/210 (46%), Gaps = 19/210 (9%)
Query: 1 VKGMQVGLTVNL---KNQEGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIA 57
+K + + ++V L ++ G ++ D C++ ++ + + G WL L + +
Sbjct: 128 IKVLGISISVGLILGSDESGRPTIAPSDCSCHISNVEVHMSGTIGWLVDLFHNNVESELR 187
Query: 58 SSVEEAVSEKINEGIA-KLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGL 116
S+E + ++ + I+ KL LQ+LP +D+ SA++ S G P + + I + + G
Sbjct: 188 QSMENKICPEVTQSISSKLLPLLQTLPVTTKIDQISAIDYSLTGPPSVMANWIDVLLKGE 247
Query: 117 FTERSEVVESEGYKKGFKISSACGGLPN----MIKVSLHEYVIQSASLVYFNAGKM--QL 170
F + S ++ S LP M+ ++ EY+ +A VY +AG + L
Sbjct: 248 FFDIS-------HRTTPPFSPPVMSLPPEQDLMVYFAVSEYLFNTAGFVYQDAGALVFNL 300
Query: 171 IIDELPDQ--VILNTAECRFIVPQLYKQYP 198
D +P + + LNT+ ++P + K YP
Sbjct: 301 TDDMIPKESSMHLNTSSFGILIPSISKMYP 330
>UniRef100_UPI0000249FDE UPI0000249FDE UniRef100 entry
Length = 269
Score = 65.9 bits (159), Expect = 7e-10
Identities = 37/174 (21%), Positives = 80/174 (45%), Gaps = 3/174 (1%)
Query: 1 VKGMQVGLTVNLKNQEGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASSV 60
V + + + + + +++G L +S VG++ I+ GGT++ YQL D F G + +
Sbjct: 18 VYNIYIRVVLGVGDKDGHLSISSESCSNDVGNVYIQFHGGTSFFYQLFEDYFSGKASDMI 77
Query: 61 EEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFTER 120
+ + I + + ++ LQ IP+DK L+ P +++ S + + F R
Sbjct: 78 RQKICPAIQQAVTNMETILQERTVNIPVDKYVYLSAPLTSAPAVTDQSCRLEVKAEFYSR 137
Query: 121 SEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLIIDE 174
E + F + + +M+ ++ ++ + SA+ Y +G +Q I +
Sbjct: 138 RSPSEPPFSARAFDLQYSD---KHMLTLAASQFTVNSAAFAYLRSGALQTNITD 188
>UniRef100_Q61805 Lipopolysaccharide-binding protein precursor [Mus musculus]
Length = 481
Score = 58.2 bits (139), Expect = 1e-07
Identities = 50/208 (24%), Positives = 84/208 (40%), Gaps = 17/208 (8%)
Query: 1 VKGMQVGLTVNL-KNQEGTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
VKG+ + + + L + G +S + DL + + G WL L + +
Sbjct: 133 VKGVTISVDLLLGMDPSGRPTVSASGCSSRICDLDVHISGNVGWLLNLFHNQIESKLQKV 192
Query: 60 VEEAVSEKINEGIAK-LDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFT 118
+E V E I + + L +LQ+LP +D ++ S V P + + G
Sbjct: 193 LENKVCEMIQKSVTSDLQPYLQTLPVTAEIDNVLGIDYSLVAAPQAKAQVLDVMFKGEIF 252
Query: 119 ERSEVVESEGYKKGFKISSACGGLPN----MIKVSLHEYVIQSASLVYFNAGKMQLII-- 172
R+ ++ + LP M+ ++ ++ AS VY AG + I
Sbjct: 253 NRN-------HRSPVATPTPTMSLPEDSKQMVYFAISDHAFNIASRVYHQAGYLNFSITD 305
Query: 173 DELPDQ--VILNTAECRFIVPQLYKQYP 198
D LP + LNT R PQ+YK+YP
Sbjct: 306 DMLPHDSGIRLNTKAFRPFTPQIYKKYP 333
>UniRef100_UPI0000361544 UPI0000361544 UniRef100 entry
Length = 368
Score = 57.4 bits (137), Expect = 2e-07
Identities = 38/145 (26%), Positives = 71/145 (48%), Gaps = 7/145 (4%)
Query: 58 SSVEEAVSEKINEGIAKLDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLF 117
S V + ++ E I LD LQ I ++K L++ +PV+ SS+ + + G+
Sbjct: 178 SLVTPQICPQLKESIIMLDYRLQVFGVSIDVNKDLTLDLGLTDDPVIDVSSLNVGLKGVI 237
Query: 118 TERSEVVESEGYKKGFKISSACGGLPNMIKVSLHEYVIQSASLVYFNAGKMQLIIDE--L 175
E + F ++ +M+ + + E+ + SAS Y++AG +Q+++++ +
Sbjct: 238 YSIKTHAEPPSEPQPFSLAEQPD---SMLTLGVSEFTLNSASYAYYSAGLLQILLNDSMI 294
Query: 176 P--DQVILNTAECRFIVPQLYKQYP 198
P V LNT+ +PQL K YP
Sbjct: 295 PSYSPVHLNTSSFGAFIPQLPKMYP 319
>UniRef100_Q8TCF0 LBP protein [Homo sapiens]
Length = 477
Score = 57.4 bits (137), Expect = 2e-07
Identities = 49/208 (23%), Positives = 91/208 (43%), Gaps = 17/208 (8%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
VKG+ + + + L ++ G ++ + D+ + + G WL L +
Sbjct: 133 VKGISISVNLLLGSESSGRPTVTASSCSSDIADVEVDMSGDLGWLLNLFHNQIESKFQKV 192
Query: 60 VEEAVSEKINEGIAK-LDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFT 118
+E + E I + ++ L +LQ+LP +D + ++ S V P + + + G
Sbjct: 193 LESRICEMIQKSVSSDLQPYLQTLPVTTEIDSFADIDYSLVEAPRATAQMLEVMFKGEIF 252
Query: 119 ERSEVVESEGYKKGFKISSACGGLPN----MIKVSLHEYVIQSASLVYFNAGKMQL-IID 173
R+ ++ + +A LP M+ ++ +YV +ASLVY G + I D
Sbjct: 253 HRN-------HRSPVTLLAAVMSLPEEHNKMVYFAISDYVFNTASLVYHEEGYLNFSITD 305
Query: 174 EL--PDQVI-LNTAECRFIVPQLYKQYP 198
++ PD I L T R VP+L + YP
Sbjct: 306 DMIPPDSNIRLTTKSFRPFVPRLARLYP 333
>UniRef100_P18428 Lipopolysaccharide-binding protein precursor [Homo sapiens]
Length = 481
Score = 57.4 bits (137), Expect = 2e-07
Identities = 49/208 (23%), Positives = 91/208 (43%), Gaps = 17/208 (8%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
VKG+ + + + L ++ G ++ + D+ + + G WL L +
Sbjct: 133 VKGISISVNLLLGSESSGRPTVTASSCSSDIADVEVDMSGDLGWLLNLFHNQIESKFQKV 192
Query: 60 VEEAVSEKINEGIAK-LDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFT 118
+E + E I + ++ L +LQ+LP +D + ++ S V P + + + G
Sbjct: 193 LESRICEMIQKSVSSDLQPYLQTLPVTTEIDSFADIDYSLVEAPRATAQMLEVMFKGEIF 252
Query: 119 ERSEVVESEGYKKGFKISSACGGLPN----MIKVSLHEYVIQSASLVYFNAGKMQL-IID 173
R+ ++ + +A LP M+ ++ +YV +ASLVY G + I D
Sbjct: 253 HRN-------HRSPVTLLAAVMSLPEEHNKMVYFAISDYVFNTASLVYHEEGYLNFSITD 305
Query: 174 EL--PDQVI-LNTAECRFIVPQLYKQYP 198
++ PD I L T R VP+L + YP
Sbjct: 306 DMIPPDSNIRLTTKSFRPFVPRLARLYP 333
>UniRef100_Q5U487 Hypothetical protein [Xenopus laevis]
Length = 494
Score = 55.8 bits (133), Expect = 7e-07
Identities = 34/172 (19%), Positives = 82/172 (46%), Gaps = 8/172 (4%)
Query: 31 GDLSIKLDGGTAWLYQLLVDAFGGNIASSVEEAVSEKINEGIAKLDKFLQSLPKQIPLDK 90
G + + GG+ W++ + + G I + + + + ++ + +++K L SLP P+D
Sbjct: 156 GQVDLHFHGGSGWIFNMFKGSILGPIHDAFSQQLCPQFDKTVMQMEKLLSSLPVTQPVDS 215
Query: 91 TSALNVSFVGNPVLSNSSIAIAINGLFTERSEVVESEGYKKGFKISSACGGLPNMIKVSL 150
+AL V+ V P+++ ++ + + G F S+ + + A + + ++
Sbjct: 216 VAALEVALVSPPLITEQNVDLLVKGQFVGLSQHWDIPYSPVEIDLPDA----DSAVVFAV 271
Query: 151 HEYVIQSASLVYFNAGKMQLIIDE--LPDQ--VILNTAECRFIVPQLYKQYP 198
++ SA+ V++ +G ++ I + +P + LNT P+L ++P
Sbjct: 272 SQFSANSAAYVHYKSGLLRANITDNMIPKESPFRLNTKSFAAFAPELPNRFP 323
>UniRef100_UPI000036C460 UPI000036C460 UniRef100 entry
Length = 481
Score = 53.5 bits (127), Expect = 4e-06
Identities = 47/208 (22%), Positives = 89/208 (42%), Gaps = 17/208 (8%)
Query: 1 VKGMQVGLTVNLKNQE-GTLKLSLLDYGCYVGDLSIKLDGGTAWLYQLLVDAFGGNIASS 59
VKG+ + + + L ++ G ++ + + + + G WL L +
Sbjct: 133 VKGISISVNLLLGSESSGRPTVTASSCSSDIAGVEVDMSGDLGWLLNLFHNQIESKFQKV 192
Query: 60 VEEAVSEKINEGIAK-LDKFLQSLPKQIPLDKTSALNVSFVGNPVLSNSSIAIAINGLFT 118
+E + E I + ++ L +LQ+LP +D + ++ S V P + + + G
Sbjct: 193 LESRICEMIQKSVSSNLQPYLQTLPVTTEIDSFAGIDYSLVEAPRATAQMLEVMFKGEIF 252
Query: 119 ERSEVVESEGYKKGFKISSACGGLPN----MIKVSLHEYVIQSASLVYFNAGKMQL-IID 173
R+ ++ + + LP M+ ++ +YV +ASLVY G + I D
Sbjct: 253 HRN-------HRSPVTLLAPVMSLPEEHNKMVYFAISDYVFNTASLVYHEEGYLNFSITD 305
Query: 174 EL--PDQVI-LNTAECRFIVPQLYKQYP 198
++ PD I L T R VP+L + YP
Sbjct: 306 DMIPPDSNIRLTTKSFRPFVPRLARLYP 333
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.138 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 326,780,224
Number of Sequences: 2790947
Number of extensions: 12368023
Number of successful extensions: 34263
Number of sequences better than 10.0: 63
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 34206
Number of HSP's gapped (non-prelim): 65
length of query: 212
length of database: 848,049,833
effective HSP length: 122
effective length of query: 90
effective length of database: 507,554,299
effective search space: 45679886910
effective search space used: 45679886910
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 72 (32.3 bits)
Lotus: description of TM0134.16


