

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
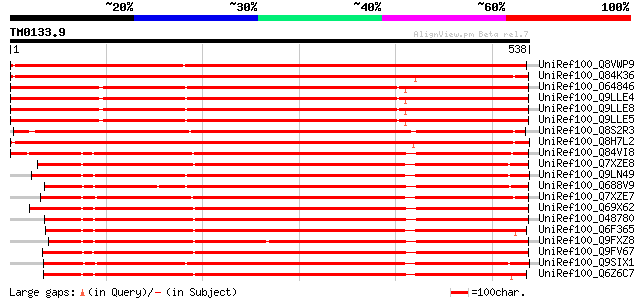
Query= TM0133.9
(538 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8VWP9 Fiddlehead-like protein [Gossypium hirsutum] 924 0.0
UniRef100_Q84K36 Fiddlehead-like protein [Tropaeolum majus] 880 0.0
UniRef100_O64846 Putative beta-ketoacyl-CoA synthase FIDDLEHEAD ... 853 0.0
UniRef100_Q9LLE4 Fiddlehead protein [Arabidopsis thaliana] 852 0.0
UniRef100_Q9LLE8 Fiddlehead protein [Arabidopsis thaliana] 851 0.0
UniRef100_Q9LLE5 Fiddlehead protein [Arabidopsis thaliana] 850 0.0
UniRef100_Q8S2R3 Putative beta-ketoacyl-CoA synthase [Antirrhinu... 836 0.0
UniRef100_Q8H7L2 Putative fiddlehead-like protein [Oryza sativa] 793 0.0
UniRef100_Q84VI8 Beta-ketoacyl-CoA-synthase [Marchantia polymorpha] 636 0.0
UniRef100_Q7XZE8 FAE3 [Marchantia polymorpha] 630 e-179
UniRef100_Q9LN49 F18O14.21 [Arabidopsis thaliana] 629 e-179
UniRef100_Q688V9 Putative beta-ketoacyl-CoA synthase [Oryza sativa] 628 e-178
UniRef100_Q7XZE7 FAE1 [Marchantia polymorpha] 626 e-178
UniRef100_Q69X62 Putative beta-ketoacyl-CoA synthase [Oryza sativa] 622 e-177
UniRef100_O48780 Putative beta-ketoacyl-CoA synthase [Arabidopsi... 618 e-175
UniRef100_Q6F365 Putative beta-ketoacyl synthase [Oryza sativa] 605 e-171
UniRef100_Q9FXZ8 Putative fatty acid elongase [Zea mays] 603 e-171
UniRef100_Q9FV67 Fatty acid elongase 1-like protein [Limnanthes ... 597 e-169
UniRef100_Q9SIX1 Putative beta-ketoacyl-CoA synthase [Arabidopsi... 593 e-168
UniRef100_Q6Z6C7 Putative beta-ketoacyl-CoA-synthase [Oryza sativa] 590 e-167
>UniRef100_Q8VWP9 Fiddlehead-like protein [Gossypium hirsutum]
Length = 535
Score = 924 bits (2389), Expect = 0.0
Identities = 449/535 (83%), Positives = 492/535 (91%), Gaps = 2/535 (0%)
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
MARN E+D+ S EIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN
Sbjct: 1 MARN-EQDLLSTEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 59
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSK 120
HGIYL TIP+L++VFSAEVGSLS+E+LWKKLWEDA YDLA+VL+ FAVFVFT+SVYFMS+
Sbjct: 60 HGIYLATIPVLVLVFSAEVGSLSREELWKKLWEDARYDLATVLSFFAVFVFTVSVYFMSR 119
Query: 121 PRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPK 180
PR IYLIDFACYRP D+LKV+++Q+IELA SGKFDE SL FQK+IL SSGIGDETY+PK
Sbjct: 120 PRSIYLIDFACYRPHDDLKVTKDQFIELARASGKFDEASLEFQKKILKSSGIGDETYVPK 179
Query: 181 AVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAM 240
V+ S N ATMKEGR EASTVMFGALDELFEKT IRPKD+G+LVVNCSIFNPTPSLSAM
Sbjct: 180 -VIMSKENCATMKEGRLEASTVMFGALDELFEKTRIRPKDVGVLVVNCSIFNPTPSLSAM 238
Query: 241 IINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKE 300
IINHYKMRGNILSYNLGGMGCSAGII VDLARD+LQ+NPNNYAVVVSTEMVG+NWY G++
Sbjct: 239 IINHYKMRGNILSYNLGGMGCSAGIIAVDLARDMLQANPNNYAVVVSTEMVGYNWYPGRD 298
Query: 301 RSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKF 360
RSML+PNCFFRMGCSA+LLSNRR DY RAKYRLEH+VRTHKGADDRSFR +YQEED+Q F
Sbjct: 299 RSMLVPNCFFRMGCSAVLLSNRRRDYRRAKYRLEHLVRTHKGADDRSFRSIYQEEDEQGF 358
Query: 361 KGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMK 420
KGLK+SKDL EIGGDALKTNITTLGPLVLPFSEQL FFATL+WRH FGG S
Sbjct: 359 KGLKVSKDLTEIGGDALKTNITTLGPLVLPFSEQLFFFATLIWRHFFGGDKSKTSLSPSS 418
Query: 421 KPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYEL 480
KPYIP+YKLAFEHFCVHAASK +LDELQ+NLELSE NMEASRMTLHRFGNTSSSSIWYEL
Sbjct: 419 KPYIPDYKLAFEHFCVHAASKTVLDELQKNLELSENNMEASRMTLHRFGNTSSSSIWYEL 478
Query: 481 AYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPV 535
AY+EAKERV+RGDR+WQ+AFGSGFKCNSVVW SM+RV KPSR+NPWLDCI+RYPV
Sbjct: 479 AYLEAKERVKRGDRIWQIAFGSGFKCNSVVWRSMRRVRKPSRDNPWLDCIDRYPV 533
>UniRef100_Q84K36 Fiddlehead-like protein [Tropaeolum majus]
Length = 538
Score = 880 bits (2275), Expect = 0.0
Identities = 425/540 (78%), Positives = 489/540 (89%), Gaps = 5/540 (0%)
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
MA N E+D+ S EIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN
Sbjct: 1 MATN-EQDLLSTEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 59
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSK 120
HGIYL TIP+L+++FSAEVG+L+KE++W KLW + YDL +VLA F VFVFTL+VYFMS+
Sbjct: 60 HGIYLVTIPILVLLFSAEVGTLTKEEMWNKLWHETRYDLVTVLAFFVVFVFTLAVYFMSR 119
Query: 121 PRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPK 180
PR IYLIDFACYR D+LKV++ ++IE A KSGKFDE SL FQ+RIL SSGIGDETYIPK
Sbjct: 120 PRSIYLIDFACYRAHDDLKVTKAEFIEQARKSGKFDEASLDFQQRILESSGIGDETYIPK 179
Query: 181 AVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAM 240
+V+S+ N++TM EGR EA+TVMF ALDELFEKT +RPKD+G+LVVNCSIFNPTPSLSAM
Sbjct: 180 SVMSTEVNSSTMSEGRLEATTVMFDALDELFEKTKVRPKDVGVLVVNCSIFNPTPSLSAM 239
Query: 241 IINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKE 300
IINHYKMRGNILSYNLGGMGCSAGIIGVDLARD+L++NPNNYAVVVSTEMVG+NWY G++
Sbjct: 240 IINHYKMRGNILSYNLGGMGCSAGIIGVDLARDMLEANPNNYAVVVSTEMVGYNWYPGQD 299
Query: 301 RSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKF 360
RSMLIPNC+FRMGCSA+LLSNRR DY RAKY L+H+VRTHKGADDRSFRC+YQEED++ +
Sbjct: 300 RSMLIPNCYFRMGCSAVLLSNRRGDYRRAKYSLQHLVRTHKGADDRSFRCIYQEEDEEGY 359
Query: 361 KGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSM- 419
KGLK+SK+L+EIGGDALKTNITTLGPLVLPFSEQL+FFATL+WR+L G + N +
Sbjct: 360 KGLKVSKELMEIGGDALKTNITTLGPLVLPFSEQLIFFATLIWRNLLGTDPNKNKTSKQS 419
Query: 420 --KKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIW 477
KPYIP+YKLAFEHFCVHAASK +LDELQ+NLELS+ NMEASRMTLHRFGNTSSSSIW
Sbjct: 420 LSSKPYIPDYKLAFEHFCVHAASKIVLDELQKNLELSDDNMEASRMTLHRFGNTSSSSIW 479
Query: 478 YELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSL 537
YELAYMEAKERV+RGDR+WQ++FGSGFKCNS+VW SM+RV KP+R NPWLD I+RYPV+L
Sbjct: 480 YELAYMEAKERVKRGDRIWQISFGSGFKCNSLVWRSMRRVRKPNR-NPWLDWIDRYPVTL 538
>UniRef100_O64846 Putative beta-ketoacyl-CoA synthase FIDDLEHEAD [Arabidopsis
thaliana]
Length = 550
Score = 853 bits (2205), Expect = 0.0
Identities = 417/555 (75%), Positives = 483/555 (86%), Gaps = 23/555 (4%)
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
M R++E+D+ S EIVNRGIE SGPNAGS TFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN
Sbjct: 1 MGRSNEQDLLSTEIVNRGIEPSGPNAGSPTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSK 120
H +YL TIP+L++VFSAEVGSLS+E++WKKLW+ YDLA+V+ F VFV T VYFMS+
Sbjct: 61 HAVYLATIPVLVLVFSAEVGSLSREEIWKKLWD---YDLATVIGFFGVFVLTACVYFMSR 117
Query: 121 PRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPK 180
PR +YLIDFACY+P DE KV++E++IELA KSGKFDEE+L F+KRIL +SGIGDETY+P+
Sbjct: 118 PRSVYLIDFACYKPSDEHKVTKEEFIELARKSGKFDEETLGFKKRILQASGIGDETYVPR 177
Query: 181 AVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAM 240
++ SS+ N TMKEGR EASTV+FGALDELFEKT ++PKD+G+LVVNCSIFNPTPSLSAM
Sbjct: 178 SI-SSSENITTMKEGREEASTVIFGALDELFEKTRVKPKDVGVLVVNCSIFNPTPSLSAM 236
Query: 241 IINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKE 300
+INHYKMRGNILSYNLGGMGCSAGII +DLARD+LQSNPN+YAVVVSTEMVG+NWY G +
Sbjct: 237 VINHYKMRGNILSYNLGGMGCSAGIIAIDLARDMLQSNPNSYAVVVSTEMVGYNWYVGSD 296
Query: 301 RSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKF 360
+SM+IPNCFFRMGCSA++LSNRR D+ AKYRLEHIVRTHK ADDRSFR VYQEED+Q F
Sbjct: 297 KSMVIPNCFFRMGCSAVMLSNRRRDFRHAKYRLEHIVRTHKAADDRSFRSVYQEEDEQGF 356
Query: 361 KGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFG------------ 408
KGLKIS+DL+E+GG+ALKTNITTLGPLVLPFSEQLLFFA L+ R F
Sbjct: 357 KGLKISRDLMEVGGEALKTNITTLGPLVLPFSEQLLFFAALL-RRTFSPAAKTSTTTSFS 415
Query: 409 ----GKSDG--NSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASR 462
K++G +SS + KPYIP+YKLAFEHFC HAASK +L+ELQ+NL LSE+NMEASR
Sbjct: 416 TSATAKTNGIKSSSSDLSKPYIPDYKLAFEHFCFHAASKVVLEELQKNLGLSEENMEASR 475
Query: 463 MTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSR 522
MTLHRFGNTSSS IWYELAYMEAKE VRRGDRVWQ+AFGSGFKCNSVVW +M++V KP+R
Sbjct: 476 MTLHRFGNTSSSGIWYELAYMEAKESVRRGDRVWQIAFGSGFKCNSVVWKAMRKVKKPTR 535
Query: 523 NNPWLDCINRYPVSL 537
NNPW+DCINRYPV L
Sbjct: 536 NNPWVDCINRYPVPL 550
>UniRef100_Q9LLE4 Fiddlehead protein [Arabidopsis thaliana]
Length = 550
Score = 852 bits (2201), Expect = 0.0
Identities = 416/555 (74%), Positives = 483/555 (86%), Gaps = 23/555 (4%)
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
M R++E+D+ S EIVNRGIE SGPNAGS TFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN
Sbjct: 1 MGRSNEQDLLSTEIVNRGIEPSGPNAGSPTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSK 120
H +YL TIP+L++VFSAEVGSLS+E++WKKLW+ YDLA+V+ F VFV T VYFMS+
Sbjct: 61 HAVYLATIPVLVLVFSAEVGSLSREEIWKKLWD---YDLATVIGFFGVFVLTACVYFMSR 117
Query: 121 PRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPK 180
PR +YLIDFACY+P DE KV++E++IELA KSGKFDEE+L F+KRIL +SGIGDETY+P+
Sbjct: 118 PRSVYLIDFACYKPSDEHKVTKEEFIELARKSGKFDEETLGFKKRILQASGIGDETYVPR 177
Query: 181 AVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAM 240
++ SS+ N TMKEGR EASTV+FGALDELFEKT ++PKD+G+LVVNCSIFNPTPSLSAM
Sbjct: 178 SI-SSSENITTMKEGREEASTVIFGALDELFEKTRVKPKDVGVLVVNCSIFNPTPSLSAM 236
Query: 241 IINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKE 300
+INHYKMRGNILSYNLGGMGCSAGII +DLARD+LQSNPN+YAVVVSTEMVG+NWY G +
Sbjct: 237 VINHYKMRGNILSYNLGGMGCSAGIIAIDLARDMLQSNPNSYAVVVSTEMVGYNWYVGSD 296
Query: 301 RSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKF 360
+SM+IPNCFFRMGCSA++LSNRR D+ AKYRLEHIVRTHK ADDRSFR VYQEED+Q F
Sbjct: 297 KSMVIPNCFFRMGCSAVMLSNRRRDFRHAKYRLEHIVRTHKAADDRSFRSVYQEEDEQGF 356
Query: 361 KGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFG------------ 408
KGLKIS+DL+E+GG+ALKTNITTLGPLVLPFSEQLLFFA L+ R F
Sbjct: 357 KGLKISRDLMEVGGEALKTNITTLGPLVLPFSEQLLFFAALL-RRTFSPAAKTSTTTSFS 415
Query: 409 ----GKSDG--NSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASR 462
K++G +SS + KPYIP+YKLAFEHFC HAASK +L+ELQ+NL LSE+NMEASR
Sbjct: 416 TSATAKTNGIKSSSSDLSKPYIPDYKLAFEHFCFHAASKVVLEELQKNLGLSEENMEASR 475
Query: 463 MTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSR 522
MTLHRFGNTSSS IWYELAYM+AKE VRRGDRVWQ+AFGSGFKCNSVVW +M++V KP+R
Sbjct: 476 MTLHRFGNTSSSGIWYELAYMKAKESVRRGDRVWQIAFGSGFKCNSVVWKAMRKVKKPTR 535
Query: 523 NNPWLDCINRYPVSL 537
NNPW+DCINRYPV L
Sbjct: 536 NNPWVDCINRYPVPL 550
>UniRef100_Q9LLE8 Fiddlehead protein [Arabidopsis thaliana]
Length = 550
Score = 851 bits (2198), Expect = 0.0
Identities = 416/555 (74%), Positives = 482/555 (85%), Gaps = 23/555 (4%)
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
M R++E+D+ S EIVNRGIE SGPNAGS TFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN
Sbjct: 1 MGRSNEQDLLSTEIVNRGIEPSGPNAGSPTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSK 120
H +YL TIP+L++VFSAEVGSLS+E++WKKLW+ YDLA+V+ F VFV T VYFMS+
Sbjct: 61 HAVYLATIPVLVLVFSAEVGSLSREEIWKKLWD---YDLATVIGFFGVFVLTACVYFMSR 117
Query: 121 PRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPK 180
PR +YLIDFACY+P DE KV++E++IELA KSGKFDEE+L F+KRIL +SGIGDETY+P+
Sbjct: 118 PRSVYLIDFACYKPSDEHKVTKEEFIELARKSGKFDEETLGFKKRILQASGIGDETYVPR 177
Query: 181 AVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAM 240
++ SS+ N TMKEGR EASTV+FGALDELFEKT ++PKD+G+LVVNCSIFNPTPSLSAM
Sbjct: 178 SI-SSSENITTMKEGREEASTVIFGALDELFEKTRVKPKDVGVLVVNCSIFNPTPSLSAM 236
Query: 241 IINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKE 300
+INHYKMRGNILSYNLGGMGCSAGII +DLARD+LQSNPN+YAVVVSTEMVG+NWY G +
Sbjct: 237 VINHYKMRGNILSYNLGGMGCSAGIIAIDLARDMLQSNPNSYAVVVSTEMVGYNWYVGSD 296
Query: 301 RSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKF 360
+SM+IPNCFFRMGCSA++LSNRR D+ AKYRLEHIVRTHK ADDRSFR VYQEED+Q F
Sbjct: 297 KSMVIPNCFFRMGCSAVMLSNRRRDFRHAKYRLEHIVRTHKAADDRSFRSVYQEEDEQGF 356
Query: 361 KGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFG------------ 408
KGLKIS+DL+E+GG+ALKTNITTLGPLVLPFSEQLLFFA L+ R F
Sbjct: 357 KGLKISRDLMEVGGEALKTNITTLGPLVLPFSEQLLFFAALL-RRTFSPAAKTSTTTSFS 415
Query: 409 ----GKSDG--NSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASR 462
K++G +SS + KPYIP+YKLAFEHFC HAASK +L+ELQ+NL LSE+NMEASR
Sbjct: 416 TSATAKTNGIKSSSSDLSKPYIPDYKLAFEHFCFHAASKVVLEELQKNLGLSEENMEASR 475
Query: 463 MTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSR 522
MTLHRFGNTSSS IWYELAYMEAKE VRRGDRVWQ+AF SGFKCNSVVW +M++V KP+R
Sbjct: 476 MTLHRFGNTSSSGIWYELAYMEAKESVRRGDRVWQIAFDSGFKCNSVVWKAMRKVKKPTR 535
Query: 523 NNPWLDCINRYPVSL 537
NNPW+DCINRYPV L
Sbjct: 536 NNPWVDCINRYPVPL 550
>UniRef100_Q9LLE5 Fiddlehead protein [Arabidopsis thaliana]
Length = 550
Score = 850 bits (2197), Expect = 0.0
Identities = 416/555 (74%), Positives = 482/555 (85%), Gaps = 23/555 (4%)
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
M R++E+D+ S EIVNRGIE SGPNAGS TFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN
Sbjct: 1 MGRSNEQDLLSTEIVNRGIEPSGPNAGSPTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSK 120
H +YL TIP+L++VFSAEVGSLS+E++WKKLW+ YDLA+V+ F VFV T VYFMS+
Sbjct: 61 HAVYLATIPVLVLVFSAEVGSLSREEIWKKLWD---YDLATVIGFFGVFVLTACVYFMSR 117
Query: 121 PRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPK 180
PR +YLIDFACY+P DE KV++E++IELA KSGKFDEE+L F+KRIL +SGIGDETY+P+
Sbjct: 118 PRSVYLIDFACYKPSDEHKVTKEEFIELARKSGKFDEETLGFKKRILQASGIGDETYVPR 177
Query: 181 AVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAM 240
++ SS+ N TMKEGR EASTV+FGALDELFEKT ++PKD+G+LVVNCSIFNPTPSLSAM
Sbjct: 178 SI-SSSENITTMKEGREEASTVIFGALDELFEKTRVKPKDVGVLVVNCSIFNPTPSLSAM 236
Query: 241 IINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKE 300
+INHYKMRGNILSYNLGGM CSAGII +DLARD+LQSNPN+YAVVVSTEMVG+NWY G +
Sbjct: 237 VINHYKMRGNILSYNLGGMRCSAGIIAIDLARDMLQSNPNSYAVVVSTEMVGYNWYVGSD 296
Query: 301 RSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKF 360
+SM+IPNCFFRMGCSA++LSNRR D+ AKYRLEHIVRTHK ADDRSFR VYQEED+Q F
Sbjct: 297 KSMVIPNCFFRMGCSAVMLSNRRRDFRHAKYRLEHIVRTHKAADDRSFRSVYQEEDEQGF 356
Query: 361 KGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFG------------ 408
KGLKIS+DL+E+GG+ALKTNITTLGPLVLPFSEQLLFFA L+ R F
Sbjct: 357 KGLKISRDLMEVGGEALKTNITTLGPLVLPFSEQLLFFAALL-RRTFSPAAKTSTTTSFS 415
Query: 409 ----GKSDG--NSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASR 462
K++G +SS + KPYIP+YKLAFEHFC HAASK +L+ELQ+NL LSE+NMEASR
Sbjct: 416 TSATAKTNGIKSSSSDLSKPYIPDYKLAFEHFCFHAASKVVLEELQKNLGLSEENMEASR 475
Query: 463 MTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSR 522
MTLHRFGNTSSS IWYELAYMEAKE VRRGDRVWQ+AFGSGFKCNSVVW +M++V KP+R
Sbjct: 476 MTLHRFGNTSSSGIWYELAYMEAKESVRRGDRVWQIAFGSGFKCNSVVWKAMRKVKKPTR 535
Query: 523 NNPWLDCINRYPVSL 537
NNPW+DCINRYPV L
Sbjct: 536 NNPWVDCINRYPVPL 550
>UniRef100_Q8S2R3 Putative beta-ketoacyl-CoA synthase [Antirrhinum majus]
Length = 526
Score = 836 bits (2159), Expect = 0.0
Identities = 404/531 (76%), Positives = 472/531 (88%), Gaps = 12/531 (2%)
Query: 5 SERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLINHGIY 64
SE+ M S EIVNRGIE AG++TFSVRVRRRLPDFL SVNLKYVKLGYHYLINHGIY
Sbjct: 3 SEQHMLSTEIVNRGIE-----AGAMTFSVRVRRRLPDFLNSVNLKYVKLGYHYLINHGIY 57
Query: 65 LFTIPLLLVVFSAEVGSLSKEDLWKKLWED-ASYDLASVLASFAVFVFTLSVYFMSKPRP 123
L TIP++++VF AEVGSLSKE++WK++W+ A YDLA+VL AVFVFT+SVYFMS+PR
Sbjct: 58 LATIPVIILVFGAEVGSLSKEEMWKRIWDSTAGYDLATVLVFLAVFVFTISVYFMSRPRS 117
Query: 124 IYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKAVV 183
IYL+DFACY+P D+LKV++E++IELA KSGKF E SL F+KRIL SSGIGDETY+PK++
Sbjct: 118 IYLLDFACYKPSDDLKVTKEEFIELARKSGKFTESSLEFKKRILQSSGIGDETYVPKSIA 177
Query: 184 SSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIIN 243
SS NTATMKEGR EAS V+FGALDELFEKT IRPKD+G+LVVNCSIFNPTPSLSAM+IN
Sbjct: 178 SSE-NTATMKEGRTEASAVIFGALDELFEKTHIRPKDVGVLVVNCSIFNPTPSLSAMVIN 236
Query: 244 HYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKERSM 303
HYKMRGNILS+NLGGMGCSAGI+ VDLARD+L++NPN+YAVVVSTE+VG+NWY G ERSM
Sbjct: 237 HYKMRGNILSFNLGGMGCSAGIVAVDLARDMLEANPNSYAVVVSTEIVGYNWYPGNERSM 296
Query: 304 LIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGL 363
LIPNC+FRMGCSA+LLSNRR DY RAKYRLEHIVRTHKGADDRSFR VYQEED Q FKG+
Sbjct: 297 LIPNCYFRMGCSAVLLSNRRRDYGRAKYRLEHIVRTHKGADDRSFRSVYQEEDSQTFKGI 356
Query: 364 KISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPY 423
K+SKDL+EIGG+A+KTNITTLGPLVLPFSEQLLFF+TLVW+ + G ++ SS PY
Sbjct: 357 KVSKDLVEIGGEAIKTNITTLGPLVLPFSEQLLFFSTLVWKLMSGSGANSMSS----NPY 412
Query: 424 IPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYM 483
IP+YKLAFEHFC+HAASK +LDELQRNLELS+KN+EASR LHRFGNTSSSSIWYELAY+
Sbjct: 413 IPDYKLAFEHFCMHAASKTVLDELQRNLELSDKNLEASRAVLHRFGNTSSSSIWYELAYL 472
Query: 484 EAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYP 534
EAK +++RGDRVWQL+FGSGFKCN+ VW +++++ KP R NPW+DCI+ YP
Sbjct: 473 EAKGQIKRGDRVWQLSFGSGFKCNTPVWKAVRKIRKPER-NPWVDCIDGYP 522
>UniRef100_Q8H7L2 Putative fiddlehead-like protein [Oryza sativa]
Length = 595
Score = 793 bits (2047), Expect = 0.0
Identities = 383/544 (70%), Positives = 459/544 (83%), Gaps = 9/544 (1%)
Query: 1 MARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLIN 60
MAR E+ + EIVNRG+E SGP+AGS TFSVRVRRRLPDFLQSVNLKYV+LGYHYLI+
Sbjct: 54 MAR--EQALLLTEIVNRGVEPSGPDAGSPTFSVRVRRRLPDFLQSVNLKYVRLGYHYLIS 111
Query: 61 HGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSK 120
HG+YL TIP++++V AEVGSLS+++LW+K+W +A+YDLA+VLA AV FT+SVY MS+
Sbjct: 112 HGVYLATIPVIVLVCGAEVGSLSRDELWRKVWGEATYDLATVLAFLAVLAFTISVYIMSR 171
Query: 121 PRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPK 180
PRP+YLIDFACY+P DELKVS+ ++I+LA KSGKFDE+SLAFQ R+L SGIGDE+Y+P+
Sbjct: 172 PRPVYLIDFACYKPADELKVSKAEFIDLARKSGKFDEDSLAFQSRLLAKSGIGDESYMPR 231
Query: 181 AVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAM 240
V TN ATMKEGRAEAS MF ALDELF+K +RPKD+G+LVVNCS+FNPTPSLSAM
Sbjct: 232 CVFEPGTNCATMKEGRAEASAAMFAALDELFDKCRVRPKDVGVLVVNCSLFNPTPSLSAM 291
Query: 241 IINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKE 300
I+NHYKMRGNILSYNLGGMGCSAG+I VDLARD+LQ++ AVVVSTE V F WY GK
Sbjct: 292 IVNHYKMRGNILSYNLGGMGCSAGVIAVDLARDMLQASGAGLAVVVSTEAVSFTWYAGKR 351
Query: 301 RSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKF 360
RSMLIPN FFR GC+A+LLSNRR D++RAKY+LEHIVRTHKGADDRSFR VYQEED+Q+
Sbjct: 352 RSMLIPNAFFRAGCAAVLLSNRRRDFHRAKYQLEHIVRTHKGADDRSFRSVYQEEDEQRI 411
Query: 361 KGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSP--- 417
KGL IS+DL+E+GG ALKTNITTLGPLVLPFSEQ+LFFA +++RHLF K+ P
Sbjct: 412 KGLSISRDLVEVGGHALKTNITTLGPLVLPFSEQILFFAGVLFRHLFPSKTSAPPPPSAD 471
Query: 418 ---SMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSS 474
S PYIP++K AFEHFC+HAAS+ +L+ LQ NL L + ++EASR LHRFGNTSSS
Sbjct: 472 GDASAAAPYIPDFKRAFEHFCMHAASRDVLEHLQGNLGLRDGDLEASRAALHRFGNTSSS 531
Query: 475 SIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYP 534
SIWYELAY+EAK RVRRGDRVWQLAFGSGFKCNS VW +++RV +P+R +PWLDC+ +YP
Sbjct: 532 SIWYELAYLEAKGRVRRGDRVWQLAFGSGFKCNSAVWRAVRRVRRPAR-SPWLDCVEQYP 590
Query: 535 VSLN 538
+N
Sbjct: 591 ARMN 594
>UniRef100_Q84VI8 Beta-ketoacyl-CoA-synthase [Marchantia polymorpha]
Length = 537
Score = 636 bits (1640), Expect = 0.0
Identities = 317/536 (59%), Positives = 403/536 (75%), Gaps = 14/536 (2%)
Query: 2 ARNSERDMFSAEIVNRGIESSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLINH 61
A+ SE ++ + +R E+ P G+ S+++R RLPDFL SVNLKYVKLGYHY+I H
Sbjct: 8 AKGSEEEVPLKQSSSRK-EAVNPADGNAAVSIKIRTRLPDFLNSVNLKYVKLGYHYVITH 66
Query: 62 GIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSKP 121
+YL +PLL+VV +AE G L ED + +LWE ++L SVL VF ++YFMS+P
Sbjct: 67 ALYLLMVPLLMVV-AAEFGRLGHED-FGQLWEQLQFNLVSVLVCSGTLVFGGTLYFMSRP 124
Query: 122 RPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKA 181
P+YL+DFACY+P DE KV+RE ++E + +G F+E+SL FQ++IL SG+G +TY+P A
Sbjct: 125 HPVYLVDFACYKPIDERKVTREIFMECSRITGNFNEKSLEFQRKILERSGLGQDTYLPPA 184
Query: 182 VVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMI 241
V+ N M E R EA TVMFGALDELFEKTG++PK++GILVVNCS+FNPTPSLSAMI
Sbjct: 185 VLRVPPNP-NMHEARLEAETVMFGALDELFEKTGVKPKEVGILVVNCSLFNPTPSLSAMI 243
Query: 242 INHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKER 301
+NHYKMRGNI S NLGGMGCSAG+I +DLARD+LQ + + YAVVVS E + NWY G +R
Sbjct: 244 VNHYKMRGNIKSLNLGGMGCSAGVISIDLARDLLQVHNSTYAVVVSMENITLNWYMGNDR 303
Query: 302 SMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFK 361
S ++ NC FRMG +A+LLSN+ + RAKY L H VRTHKGAD + F CV QEEDD+
Sbjct: 304 SKMLSNCIFRMGGAAILLSNKMKERRRAKYELVHTVRTHKGADPKCFSCVVQEEDDEGKI 363
Query: 362 GLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKK 421
G+ +S+DL+ + GDALK NITTLGPLVLP SEQ+LFFATLV R LF K K
Sbjct: 364 GVSLSRDLMGVAGDALKANITTLGPLVLPLSEQILFFATLVGRKLFKMKI---------K 414
Query: 422 PYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELA 481
PYIP++KLAFEHFC+HA + +LDEL++NL L+E +ME SRMTL+RFGNTSSSS+WYELA
Sbjct: 415 PYIPDFKLAFEHFCIHAGGRAVLDELEKNLNLTEWHMEPSRMTLYRFGNTSSSSLWYELA 474
Query: 482 YMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSL 537
Y EAK RVRRGDR+WQ+AFGSGFKCNS VW +++ V P + +PW D I+ +PV +
Sbjct: 475 YTEAKGRVRRGDRLWQIAFGSGFKCNSAVWRALRTVKSP-KQSPWADEIDSFPVEV 529
>UniRef100_Q7XZE8 FAE3 [Marchantia polymorpha]
Length = 529
Score = 630 bits (1624), Expect = e-179
Identities = 300/508 (59%), Positives = 395/508 (77%), Gaps = 12/508 (2%)
Query: 30 TFSVRVRRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWK 89
T ++ +R+RLPDFLQSVN+KYVKLGYHYLI H ++L T+P +V +AE+G L E +++
Sbjct: 31 TVAIGIRQRLPDFLQSVNMKYVKLGYHYLITHAMFLLTLPAFFLV-AAEIGRLGHERIYR 89
Query: 90 KLWEDASYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELA 149
+LW +L S++A + V ++YFMS+PRP+YL++FACYRP++ LKVS++ +++++
Sbjct: 90 ELWTHLHLNLVSIMACSSALVAGATLYFMSRPRPVYLVEFACYRPDERLKVSKDFFLDMS 149
Query: 150 SKSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDE 209
++G F S+ FQ +I SG+GDETY+P A+++S N M+E R EA+ VMFGALDE
Sbjct: 150 RRTGLFSSSSMDFQTKITQRSGLGDETYLPPAILASPPNPC-MREAREEAAMVMFGALDE 208
Query: 210 LFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVD 269
LFE+TG++PK+IG+LVVNCS+FNPTPS+SAMI+NHY MRGNI S NLGGMGCSAG+I +D
Sbjct: 209 LFEQTGVKPKEIGVLVVNCSLFNPTPSMSAMIVNHYHMRGNIKSLNLGGMGCSAGLISID 268
Query: 270 LARDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRA 329
LARD+LQ + N YAVVVSTE + NWY G +RS L+ NC FRMG +A+LLSN+R + RA
Sbjct: 269 LARDLLQVHGNTYAVVVSTENITLNWYFGDDRSKLMSNCIFRMGGAAVLLSNKRRERRRA 328
Query: 330 KYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVL 389
KY L H VRTHKGADD+ FRCVYQEED G+ +S++L+ + G+ALK NITTLGPLVL
Sbjct: 329 KYELLHTVRTHKGADDKCFRCVYQEEDSTGSLGVSLSRELMAVAGNALKANITTLGPLVL 388
Query: 390 PFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQR 449
P SEQ+LFFA+LV R K KPYIP++KLAFEHFC+HA + +LDEL++
Sbjct: 389 PLSEQILFFASLVARKFLNMK---------MKPYIPDFKLAFEHFCIHAGGRAVLDELEK 439
Query: 450 NLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSV 509
NL+L+E +ME SRMTL+RFGNTSSSS+WYELAY EA+ RV+RGDR+WQ+AFGSGFKCNS
Sbjct: 440 NLDLTEWHMEPSRMTLYRFGNTSSSSLWYELAYTEAQGRVKRGDRLWQIAFGSGFKCNSA 499
Query: 510 VWLSMKRVNKPSRNNPWLDCINRYPVSL 537
VW +++ V KP NN W D I+R+PV L
Sbjct: 500 VWRALRTV-KPPVNNAWSDVIDRFPVKL 526
>UniRef100_Q9LN49 F18O14.21 [Arabidopsis thaliana]
Length = 516
Score = 629 bits (1621), Expect = e-179
Identities = 308/516 (59%), Positives = 396/516 (76%), Gaps = 13/516 (2%)
Query: 23 GPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSL 82
G GS+ +R R LPDFLQSVNLKYVKLGYHYLI++ + L PL +V+ S E +
Sbjct: 13 GGGDGSVGVQIRQTRMLPDFLQSVNLKYVKLGYHYLISNLLTLCLFPLAVVI-SVEASQM 71
Query: 83 SKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSR 142
+ +DL K+LW Y+L S++ A+ VF L+VY M++PRP+YL+DF+CY P D LK
Sbjct: 72 NPDDL-KQLWIHLQYNLVSIIICSAILVFGLTVYVMTRPRPVYLVDFSCYLPPDHLKAPY 130
Query: 143 EQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTV 202
+++E + +G FD+ +L FQ++IL SG+G++TY+P+A+ +M R EA V
Sbjct: 131 ARFMEHSRLTGDFDDSALEFQRKILERSGLGEDTYVPEAM-HYVPPRISMAAAREEAEQV 189
Query: 203 MFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCS 262
MFGALD LF T ++PKDIGILVVNCS+FNPTPSLSAMI+N YK+RGNI SYNLGGMGCS
Sbjct: 190 MFGALDNLFANTNVKPKDIGILVVNCSLFNPTPSLSAMIVNKYKLRGNIRSYNLGGMGCS 249
Query: 263 AGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNR 322
AG+I VDLA+D+L + N YAVVVSTE + NWY G ++SMLIPNC FR+G SA+LLSN+
Sbjct: 250 AGVIAVDLAKDMLLVHRNTYAVVVSTENITQNWYFGNKKSMLIPNCLFRVGGSAVLLSNK 309
Query: 323 RFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNIT 382
D R+KYRL H+VRTH+GADD++FRCVYQE+DD G+ +SKDL+ I G+ LKTNIT
Sbjct: 310 SRDKRRSKYRLVHVVRTHRGADDKAFRCVYQEQDDTGRTGVSLSKDLMAIAGETLKTNIT 369
Query: 383 TLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKP 442
TLGPLVLP SEQ+LFF TLV + LF GK KPYIP++KLAFEHFC+HA +
Sbjct: 370 TLGPLVLPISEQILFFMTLVVKKLFNGK---------VKPYIPDFKLAFEHFCIHAGGRA 420
Query: 443 ILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGS 502
++DEL++NL+LS ++EASRMTLHRFGNTSSSSIWYELAY+EAK R+RRG+RVWQ+AFGS
Sbjct: 421 VIDELEKNLQLSPVHVEASRMTLHRFGNTSSSSIWYELAYIEAKGRMRRGNRVWQIAFGS 480
Query: 503 GFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSLN 538
GFKCNS +W +++ V KPS N+PW DCI++YPV+L+
Sbjct: 481 GFKCNSAIWEALRHV-KPSNNSPWEDCIDKYPVTLS 515
>UniRef100_Q688V9 Putative beta-ketoacyl-CoA synthase [Oryza sativa]
Length = 514
Score = 628 bits (1620), Expect = e-178
Identities = 311/501 (62%), Positives = 389/501 (77%), Gaps = 14/501 (2%)
Query: 37 RRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDAS 96
RRLPDFLQSVNLKYVKLGYHYLI H + L +PL+ V+ E G +DL ++LW
Sbjct: 16 RRLPDFLQSVNLKYVKLGYHYLITHLLTLLLLPLMAVIV-LEAGRTDPDDL-RQLWLHLQ 73
Query: 97 YDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFD 156
Y+L SVL AV VF +VY +++PRP+YL+DFACY+P D+LKV ++++ + G F
Sbjct: 74 YNLVSVLVLSAVLVFGATVYVLTRPRPVYLVDFACYKPPDKLKVRFDEFLHHSKLCG-FS 132
Query: 157 EESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGI 216
++ L FQ++IL SG+ +ETY+P+A+ TM RAEA +VMFGALD+LF+ TG+
Sbjct: 133 DDCLEFQRKILERSGLSEETYVPEAM-HLIPPEPTMANARAEAESVMFGALDKLFKFTGV 191
Query: 217 RPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQ 276
+PKD+G+LVVNCS+FNPTPSLSAMI+N YK+RGNI S+NLGGMGCSAG+I VDLARD+LQ
Sbjct: 192 KPKDVGVLVVNCSLFNPTPSLSAMIVNKYKLRGNIKSFNLGGMGCSAGVIAVDLARDMLQ 251
Query: 277 SNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHI 336
+ N YAVVVSTE + NWY G +SMLIPNC FR+G +A+LLSNR D RAKY L+H+
Sbjct: 252 VHRNTYAVVVSTENITQNWYFGNRKSMLIPNCLFRVGGAAVLLSNRGADRRRAKYALKHV 311
Query: 337 VRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLL 396
VRTHKGAD+++F CVYQE+DD+ G+ +SKDL+ I G ALKTNITTLGPLVLPFSEQLL
Sbjct: 312 VRTHKGADNKAFNCVYQEQDDEGKTGVSLSKDLMAIAGGALKTNITTLGPLVLPFSEQLL 371
Query: 397 FFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEK 456
FFATLV + LF K KPYIP++KLAFEHFC+HA + ++DEL++NL+L
Sbjct: 372 FFATLVAKKLFNAKI---------KPYIPDFKLAFEHFCIHAGGRAVIDELEKNLQLQPV 422
Query: 457 NMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKR 516
++EASRMTLHRFGNTSSSSIWYELAYMEAK RVRRG R+WQ+AFGSGFKCNS VW +++
Sbjct: 423 HVEASRMTLHRFGNTSSSSIWYELAYMEAKGRVRRGHRIWQIAFGSGFKCNSAVWHALRN 482
Query: 517 VNKPSRNNPWLDCINRYPVSL 537
VN PS +PW DCI+RYPV L
Sbjct: 483 VN-PSPESPWEDCIDRYPVEL 502
>UniRef100_Q7XZE7 FAE1 [Marchantia polymorpha]
Length = 527
Score = 626 bits (1615), Expect = e-178
Identities = 305/505 (60%), Positives = 390/505 (76%), Gaps = 13/505 (2%)
Query: 33 VRVRRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLW 92
+ RRRLPDFLQSVNLKYVKLGYHYLI H + L IPLLL V + EVG + E++W+ LW
Sbjct: 30 INSRRRLPDFLQSVNLKYVKLGYHYLITHLVILLFIPLLLAV-TLEVGRMGPEEMWQ-LW 87
Query: 93 EDASYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKS 152
+ ++L SVL A+ VF ++VY MS+PRP+YL+D+AC+ P + +V +++ + KS
Sbjct: 88 VNLQFNLVSVLICSALLVFGVTVYVMSRPRPVYLVDYACHLPAENTRVKFSLFMDHSEKS 147
Query: 153 GKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFE 212
G FDE +L FQ++IL SG+G+ET++P ++ N A M E R EA VMFGALDELFE
Sbjct: 148 GFFDERALEFQRKILERSGLGEETHLPVSLHRLPAN-ANMAEARNEAEEVMFGALDELFE 206
Query: 213 KTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLAR 272
KT ++PKDIGILVVNCS+FNPTPSLSAMI+N Y MRGNI +YNLGGMGCSAG+I +DLA+
Sbjct: 207 KTKVKPKDIGILVVNCSLFNPTPSLSAMIVNKYHMRGNIRTYNLGGMGCSAGVISIDLAK 266
Query: 273 DILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYR 332
D+LQ + YA+VVSTE + NWY G RSMLIPNC FR+G +A+LLSN+R + RAKY+
Sbjct: 267 DMLQVHGGTYAIVVSTENITQNWYYGNRRSMLIPNCLFRVGGAAILLSNKRSEKRRAKYQ 326
Query: 333 LEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFS 392
L H VRTHKGADD+ FRCVYQEED++ F G+ +SKDL+ I GDALKTNITTLGPLVLP S
Sbjct: 327 LVHTVRTHKGADDKCFRCVYQEEDEKNFMGVSLSKDLMAIAGDALKTNITTLGPLVLPLS 386
Query: 393 EQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLE 452
EQLLFF LV R +F K KPYIP++KLAF+HFC+HA + ++DEL++NL+
Sbjct: 387 EQLLFFGILVARKVFNMK---------VKPYIPDFKLAFDHFCIHAGGRAVIDELEKNLQ 437
Query: 453 LSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWL 512
L++ ++E SRMTLHRFGNTSSSSIWYEL Y+E+K R+++GDRVWQ+AFGSGFKCNS VW
Sbjct: 438 LTKDHVEPSRMTLHRFGNTSSSSIWYELNYIESKGRMKKGDRVWQIAFGSGFKCNSAVWQ 497
Query: 513 SMKRVNKPSRNNPWLDCINRYPVSL 537
+++ + P+R PW CI+ YPV +
Sbjct: 498 AVRTLKPPAR-GPWTHCIDSYPVKI 521
>UniRef100_Q69X62 Putative beta-ketoacyl-CoA synthase [Oryza sativa]
Length = 519
Score = 622 bits (1604), Expect = e-177
Identities = 297/517 (57%), Positives = 395/517 (75%), Gaps = 12/517 (2%)
Query: 21 SSGPNAGSLTFSVRVRRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVG 80
++ PN G+ + RRRLPDF QSV LKYVKLGYHYLI+HG+YL PL+ +V + ++
Sbjct: 6 AAAPNGGAAAAEQQQRRRLPDFQQSVRLKYVKLGYHYLISHGMYLLLSPLMALV-AVQLS 64
Query: 81 SLSKEDLWKKLWEDASYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKV 140
++S D+ LWE ++L SV+A + VF ++YF+++PRP+YL+DFACY+P+ + K
Sbjct: 65 TVSPGDI-ADLWEQLRFNLLSVVACSTLLVFLSTLYFLTRPRPVYLLDFACYKPDPQRKC 123
Query: 141 SREQYIELASKSGKFDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEAS 200
+RE ++ +S +G F + +L FQ++IL SG+G++TY+P AV+ N M E R EA
Sbjct: 124 TRETFMRCSSLTGSFTDANLDFQRKILERSGLGEDTYLPPAVLRVPPNPC-MDEARKEAR 182
Query: 201 TVMFGALDELFEKTGIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMG 260
TVMFGA+D+L EKTG++PKDIG++VVNCS+FNPTPSLSAM++NHYK+RGN++SYNLGGMG
Sbjct: 183 TVMFGAIDQLLEKTGVKPKDIGVVVVNCSLFNPTPSLSAMVVNHYKLRGNVISYNLGGMG 242
Query: 261 CSAGIIGVDLARDILQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLS 320
CSAG++ VDLA+D+LQ +PN+YA+VVS E + NWY G RSML+ NC FRMG +A+LLS
Sbjct: 243 CSAGLLSVDLAKDLLQVHPNSYALVVSMENITLNWYFGNNRSMLVSNCLFRMGGAAILLS 302
Query: 321 NRRFDYNRAKYRLEHIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTN 380
NRR D R+KY L H VRTHKGA+D+ F CV QEED+ G+ +SKDL+ + GDALKTN
Sbjct: 303 NRRSDRRRSKYELVHTVRTHKGANDKCFGCVTQEEDEIGKIGVSLSKDLMAVAGDALKTN 362
Query: 381 ITTLGPLVLPFSEQLLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAAS 440
ITTLGPLVLP SEQLLF ATLV + + K KPYIP++KLAFEHFC+HA
Sbjct: 363 ITTLGPLVLPLSEQLLFMATLVAKKVLKMKI---------KPYIPDFKLAFEHFCIHAGG 413
Query: 441 KPILDELQRNLELSEKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAF 500
+ +LDEL++NLEL++ +ME SRMTL+RFGNTSSSS+WYELAY EAK R+R+ DR+WQ+AF
Sbjct: 414 RAVLDELEKNLELTDWHMEPSRMTLYRFGNTSSSSLWYELAYTEAKGRIRKRDRIWQIAF 473
Query: 501 GSGFKCNSVVWLSMKRVNKPSRNNPWLDCINRYPVSL 537
GSGFKCNS VW +++ VN NPW+D I+ +PV +
Sbjct: 474 GSGFKCNSAVWKALQTVNPAKEKNPWMDEIDNFPVEV 510
>UniRef100_O48780 Putative beta-ketoacyl-CoA synthase [Arabidopsis thaliana]
Length = 509
Score = 618 bits (1594), Expect = e-175
Identities = 293/501 (58%), Positives = 387/501 (76%), Gaps = 12/501 (2%)
Query: 37 RRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDAS 96
R LPDF +SV LKYVKLGYHYLI HG+YLF PL+LV+ +A++ + S DL + LWE
Sbjct: 15 RNLPDFKKSVKLKYVKLGYHYLITHGMYLFLSPLVLVI-AAQISTFSVTDL-RSLWEHLQ 72
Query: 97 YDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFD 156
Y+L SV+ + VF +++YFM++PRP+YL++F+C++P++ K +++ +++ + +G F
Sbjct: 73 YNLISVVVCSMLLVFLMTIYFMTRPRPVYLVNFSCFKPDESRKCTKKIFMDRSKLTGSFT 132
Query: 157 EESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGI 216
EE+L FQ++IL SG+G+ TY+P+AV++ N MKE R EA TVMFGA+DEL KT +
Sbjct: 133 EENLEFQRKILQRSGLGESTYLPEAVLNVPPNPC-MKEARKEAETVMFGAIDELLAKTNV 191
Query: 217 RPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQ 276
PKDIGIL+VNCS+FNPTPSLSAM++NHYK+RGNILSYNLGGMGCSAG+I +DLA+ +L
Sbjct: 192 NPKDIGILIVNCSLFNPTPSLSAMVVNHYKLRGNILSYNLGGMGCSAGLISIDLAKHLLH 251
Query: 277 SNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHI 336
S PN YA+V+S E + NWY G +RS L+ NC FRMG +A+LLSN+R+D R+KY L
Sbjct: 252 SIPNTYAMVISMENITLNWYFGNDRSKLVSNCLFRMGGAAILLSNKRWDRRRSKYELVDT 311
Query: 337 VRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLL 396
VRTHKGADD+ F C+ QEED G+ +SK+L+ + GDALKTNITTLGPLVLP SEQLL
Sbjct: 312 VRTHKGADDKCFGCITQEEDSASKIGVTLSKELMAVAGDALKTNITTLGPLVLPTSEQLL 371
Query: 397 FFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEK 456
FFATLV R LF K KPYIP++KLAFEHFC+HA + +LDEL++NL+L+E
Sbjct: 372 FFATLVGRKLFKMKI---------KPYIPDFKLAFEHFCIHAGGRAVLDELEKNLKLTEW 422
Query: 457 NMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKR 516
+ME SRMTL+RFGNTSSSS+WYELAY EAK R+++GDR+WQ+AFGSGFKCNS VW +++
Sbjct: 423 HMEPSRMTLYRFGNTSSSSLWYELAYSEAKGRIKKGDRIWQIAFGSGFKCNSSVWRAVRS 482
Query: 517 VNKPSRNNPWLDCINRYPVSL 537
VN NPW+D I+ +PV +
Sbjct: 483 VNPKKEKNPWMDEIHEFPVEV 503
>UniRef100_Q6F365 Putative beta-ketoacyl synthase [Oryza sativa]
Length = 520
Score = 605 bits (1559), Expect = e-171
Identities = 294/501 (58%), Positives = 376/501 (74%), Gaps = 15/501 (2%)
Query: 38 RLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASY 97
RLPDF QSV LKYVKLGYHYLI HG YL PL +V +A + + + DL LW++ Y
Sbjct: 24 RLPDFKQSVKLKYVKLGYHYLITHGAYLLLAPLPGLV-AAHLSTFTLGDL-ADLWQNLQY 81
Query: 98 DLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDE 157
+L SVL + V + YF+++PRP+YL+DFACY+P+DE K SR +++ + G F
Sbjct: 82 NLVSVLLCSTLLVLVSTAYFLTRPRPVYLVDFACYKPDDERKCSRARFMNCTERLGTFTP 141
Query: 158 ESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIR 217
E++ FQ++I+ SG+G++TY+P+AV++ N + M R EA VM+GALDEL KTG+
Sbjct: 142 ENVEFQRKIIERSGLGEDTYLPEAVLNIPPNPS-MANARKEAEMVMYGALDELLAKTGVN 200
Query: 218 PKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQS 277
PKDIGILVVNCS+FNPTPSLSAM++NHYK+RGN++SYNLGGMGCSAG+I +DLA+D+LQ
Sbjct: 201 PKDIGILVVNCSLFNPTPSLSAMVVNHYKLRGNVVSYNLGGMGCSAGLISIDLAKDLLQV 260
Query: 278 NPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIV 337
PN YAVV+S E + NWY G +RSML+ NC FRMG +A+LLSNR R+KY+L H V
Sbjct: 261 YPNTYAVVISMENITLNWYFGNDRSMLVSNCLFRMGGAAILLSNRGSARRRSKYQLVHTV 320
Query: 338 RTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLF 397
RTH+GADDR F CV Q ED G+ +SKDL+ + G+ALKTNITTLGPLVLP SEQLLF
Sbjct: 321 RTHRGADDRCFGCVTQREDADGKTGVSLSKDLMAVAGEALKTNITTLGPLVLPMSEQLLF 380
Query: 398 FATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKN 457
FATLV R + K KPYIP++KLAFEHFC+HA + +LDEL++NL+LS+ +
Sbjct: 381 FATLVTRKVLKRK---------VKPYIPDFKLAFEHFCIHAGGRAVLDELEKNLQLSDWH 431
Query: 458 MEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRV 517
ME SRMTLHRFGNTSSSS+WYELAY EAK R+++GDR WQ+AFGSGFKCNS VW +++ V
Sbjct: 432 MEPSRMTLHRFGNTSSSSLWYELAYAEAKGRIKKGDRTWQIAFGSGFKCNSAVWRALRSV 491
Query: 518 NKPSRN---NPWLDCINRYPV 535
N N NPW+D I+R+PV
Sbjct: 492 NPAKENNFTNPWIDEIHRFPV 512
>UniRef100_Q9FXZ8 Putative fatty acid elongase [Zea mays]
Length = 513
Score = 603 bits (1555), Expect = e-171
Identities = 291/497 (58%), Positives = 380/497 (75%), Gaps = 14/497 (2%)
Query: 41 DFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDASYDLA 100
DF QSV LKYVKLGYHYLI+HG+YL PL+ +V + ++ ++S DL LWE ++L
Sbjct: 22 DFQQSVRLKYVKLGYHYLISHGMYLLLSPLMALV-AVQLSTVSPHDL-ADLWEQLRFNLL 79
Query: 101 SVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKFDEESL 160
SV+A + VF +VYF+++PRP+YL+DFACY+PE E K +RE ++ + +G F +E+L
Sbjct: 80 SVVACSTLLVFLSTVYFLTRPRPVYLLDFACYKPESERKCTRETFMHCSKLTGSFTDENL 139
Query: 161 AFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTGIRPKD 220
FQ++IL SG+G++TY+P AV+ N M E R EA VMFGA+D+L EKTG+RPKD
Sbjct: 140 EFQRKILERSGLGEDTYLPPAVLRVPPNPC-MDEARKEARAVMFGAIDQLLEKTGVRPKD 198
Query: 221 IGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDILQSNPN 280
IG+LVVNCS+FNPTPSLSAM++NHYK+RGNI+SYNLGGMGCSAG+I DLA+D+LQ +PN
Sbjct: 199 IGVLVVNCSLFNPTPSLSAMVVNHYKLRGNIVSYNLGGMGCSAGLI--DLAKDLLQVHPN 256
Query: 281 NYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEHIVRTH 340
+YA+V+S E + NWY G RSML+ NC FRMG +A+LLSNRR D R+KY L H VRTH
Sbjct: 257 SYALVISMENITLNWYFGNNRSMLVSNCLFRMGGAAILLSNRRSDRRRSKYELVHTVRTH 316
Query: 341 KGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQLLFFAT 400
KGA+D+ F CV QEED+ G+ +SKDL+ + GDALKTNITTLGPLVLP SEQLLF T
Sbjct: 317 KGANDKCFGCVTQEEDEIGKIGVSLSKDLMAVAGDALKTNITTLGPLVLPLSEQLLFMGT 376
Query: 401 LVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSEKNMEA 460
L+ + + K KPYIP++KLAFEHFC+HA + +LDEL++NLEL++ +ME
Sbjct: 377 LIAKKVLKMKI---------KPYIPDFKLAFEHFCIHAGGRAVLDELEKNLELTDWHMEP 427
Query: 461 SRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMKRVNKP 520
SRMTL+RFGNTSSSS+WYELAY EAK R+R+ DR+WQ+AFGSGFKCNS VW +++ VN
Sbjct: 428 SRMTLYRFGNTSSSSLWYELAYTEAKGRIRKRDRIWQIAFGSGFKCNSAVWKALRTVNPA 487
Query: 521 SRNNPWLDCINRYPVSL 537
+PW+D I+ +PV +
Sbjct: 488 KEKSPWMDEIDNFPVDV 504
>UniRef100_Q9FV67 Fatty acid elongase 1-like protein [Limnanthes douglasii]
Length = 505
Score = 597 bits (1538), Expect = e-169
Identities = 283/503 (56%), Positives = 384/503 (76%), Gaps = 12/503 (2%)
Query: 35 VRRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWED 94
V+ LPD S+NLK+VKLGYHYLI HG+YL PL LV+F A++ +LS +D + +WE
Sbjct: 14 VKNTLPDLKLSINLKHVKLGYHYLITHGMYLCLPPLALVLF-AQISTLSLKD-FNDIWEQ 71
Query: 95 ASYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGK 154
++L SV+ S + V L +YFM++PRP+YL+DFACY+P++ K +RE +++ G
Sbjct: 72 LQFNLISVVVSSTLLVSLLILYFMTRPRPVYLMDFACYKPDETRKSTREHFMKCGESLGS 131
Query: 155 FDEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKT 214
F E+++ FQ++++ SG+GD TY+P+A+ + + + MK R EA VMFGA+D+L EKT
Sbjct: 132 FTEDNIDFQRKLVARSGLGDATYLPEAIGTIPAHPS-MKAARREAELVMFGAIDQLLEKT 190
Query: 215 GIRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDI 274
+ PKDIGILVVNCS+F+PTPSLS+MI+NHYK+RGNI+SYNLGGMGCSAG+I VDLA+ +
Sbjct: 191 KVNPKDIGILVVNCSLFSPTPSLSSMIVNHYKLRGNIISYNLGGMGCSAGLISVDLAKRL 250
Query: 275 LQSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLE 334
L++NPN YA+V+STE + NWY G +RS L+ NC FRMG +A+LLSN+ D R+KY+L
Sbjct: 251 LETNPNTYALVMSTENITLNWYMGNDRSKLVSNCLFRMGGAAVLLSNKTSDKKRSKYQLV 310
Query: 335 HIVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQ 394
VR+HKGADD + C++QEED G+ +SK+L+ + GDALKTNITTLGPLVLP SEQ
Sbjct: 311 TTVRSHKGADDNCYGCIFQEEDSNGKIGVSLSKNLMAVAGDALKTNITTLGPLVLPMSEQ 370
Query: 395 LLFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELS 454
LLFFATLV R +F K KPYIP++KLAF+HFC+HA + +LDEL++NL+LS
Sbjct: 371 LLFFATLVARKVFKKKI---------KPYIPDFKLAFDHFCIHAGGRAVLDELEKNLQLS 421
Query: 455 EKNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSM 514
++E SRMTL+RFGNTSSSS+WYELAY EAK R+R+G+RVWQ+ FGSGFKCNS VW ++
Sbjct: 422 SWHLEPSRMTLYRFGNTSSSSLWYELAYSEAKGRIRKGERVWQIGFGSGFKCNSAVWKAL 481
Query: 515 KRVNKPSRNNPWLDCINRYPVSL 537
K V+ NPW+D I+++PV++
Sbjct: 482 KSVDPKKEKNPWMDEIHQFPVAV 504
>UniRef100_Q9SIX1 Putative beta-ketoacyl-CoA synthase [Arabidopsis thaliana]
Length = 512
Score = 593 bits (1528), Expect = e-168
Identities = 287/503 (57%), Positives = 386/503 (76%), Gaps = 13/503 (2%)
Query: 36 RRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDA 95
RR+LP+FLQSVN+KYVKLGYHYLI H L +PL+ V+ + E+ L+ +DL++ +W
Sbjct: 22 RRKLPNFLQSVNMKYVKLGYHYLITHLFKLCLVPLMAVLVT-EISRLTTDDLYQ-IWLHL 79
Query: 96 SYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKF 155
Y+L + + A+ +F +VY MS+PR +YL+D++CY P + L+V +++++ + F
Sbjct: 80 QYNLVAFIFLSALAIFGSTVYIMSRPRSVYLVDYSCYLPPESLQVKYQKFMDHSKLIEDF 139
Query: 156 DEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTG 215
+E SL FQ++IL SG+G+ETY+P+A+ TM R E+ VMFGALD+LFE T
Sbjct: 140 NESSLEFQRKILERSGLGEETYLPEAL-HCIPPRPTMMAAREESEQVMFGALDKLFENTK 198
Query: 216 IRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDIL 275
I P+DIG+LVVNCS+FNPTPSLSAMI+N YK+RGN+ S+NLGGMGCSAG+I +DLA+D+L
Sbjct: 199 INPRDIGVLVVNCSLFNPTPSLSAMIVNKYKLRGNVKSFNLGGMGCSAGVISIDLAKDML 258
Query: 276 QSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEH 335
Q + N YAVVVSTE + NWY G +++MLIPNC FR+G SA+LLSN+ D R+KY+L H
Sbjct: 259 QVHRNTYAVVVSTENITQNWYFGNKKAMLIPNCLFRVGGSAILLSNKGKDRRRSKYKLVH 318
Query: 336 IVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQL 395
VRTHKGA +++F CVYQE+DD G+ +SKDL+ I G+ALK NITTLGPLVLP SEQ+
Sbjct: 319 TVRTHKGAVEKAFNCVYQEQDDNGKTGVSLSKDLMAIAGEALKANITTLGPLVLPISEQI 378
Query: 396 LFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSE 455
LFF TLV + LF K KPYIP++KLAF+HFC+HA + ++DEL++NL+LS+
Sbjct: 379 LFFMTLVTKKLFNSK---------LKPYIPDFKLAFDHFCIHAGGRAVIDELEKNLQLSQ 429
Query: 456 KNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMK 515
++EASRMTLHRFGNTSSSSIWYELAY+EAK R+++G+RVWQ+AFGSGFKCNS VW+++
Sbjct: 430 THVEASRMTLHRFGNTSSSSIWYELAYIEAKGRMKKGNRVWQIAFGSGFKCNSAVWVALN 489
Query: 516 RVNKPSRNNPWLDCINRYPVSLN 538
V KPS ++PW CI+RYPV L+
Sbjct: 490 NV-KPSVSSPWEHCIDRYPVKLD 511
>UniRef100_Q6Z6C7 Putative beta-ketoacyl-CoA-synthase [Oryza sativa]
Length = 519
Score = 590 bits (1520), Expect = e-167
Identities = 290/503 (57%), Positives = 376/503 (74%), Gaps = 14/503 (2%)
Query: 36 RRRLPDFLQSVNLKYVKLGYHYLINHGIYLFTIPLLLVVFSAEVGSLSKEDLWKKLWEDA 95
RRRLPDF QSV LKYVKLGYHYLI +G+YL PL+ +V + + +L+ D+ LW
Sbjct: 17 RRRLPDFQQSVRLKYVKLGYHYLITNGVYLLLTPLIALV-AVHLSTLTAGDV-AGLWSHL 74
Query: 96 SYDLASVLASFAVFVFTLSVYFMSKPRPIYLIDFACYRPEDELKVSREQYIELASKSGKF 155
++L SV+A + VF +V F+++PRP+YL+DFACY+P E + SR+ ++ + +G F
Sbjct: 75 RFNLVSVVACTTLLVFLSTVRFLTRPRPVYLVDFACYKPPPERRCSRDAFMRCSRLAGCF 134
Query: 156 DEESLAFQKRILMSSGIGDETYIPKAVVSSTTNTATMKEGRAEASTVMFGALDELFEKTG 215
SL FQ+RI+ SG+GD+TY+P AV+ N + M E R EA VMFGA+D+L KTG
Sbjct: 135 TAASLDFQRRIVERSGLGDDTYLPAAVLREPPNPS-MAEARREAEAVMFGAVDDLLAKTG 193
Query: 216 IRPKDIGILVVNCSIFNPTPSLSAMIINHYKMRGNILSYNLGGMGCSAGIIGVDLARDIL 275
+ K+IG+LVVNCS+FNPTPSLSAM++NHYK+RGNI+SYNLGGMGCSAG++ +DLA+D+L
Sbjct: 194 VSAKEIGVLVVNCSLFNPTPSLSAMVVNHYKLRGNIVSYNLGGMGCSAGLLAIDLAKDLL 253
Query: 276 QSNPNNYAVVVSTEMVGFNWYQGKERSMLIPNCFFRMGCSALLLSNRRFDYNRAKYRLEH 335
Q + N+YA+V+S E + NWY G +RSML+ NC FRMG +A+LLSNR + R+KY L H
Sbjct: 254 QVHRNSYALVISMENITLNWYSGNDRSMLVSNCLFRMGGAAILLSNRWSERRRSKYELVH 313
Query: 336 IVRTHKGADDRSFRCVYQEEDDQKFKGLKISKDLIEIGGDALKTNITTLGPLVLPFSEQL 395
VRTHKG DD+ F CV QEED + G+ +SKDL+ + GDALKTNITTLGPLVLP SEQL
Sbjct: 314 TVRTHKGGDDKCFGCVTQEEDGEGNVGVALSKDLMAVAGDALKTNITTLGPLVLPLSEQL 373
Query: 396 LFFATLVWRHLFGGKSDGNSSPSMKKPYIPNYKLAFEHFCVHAASKPILDELQRNLELSE 455
LF ATLV + L K+ KPYIP++KLAFEHFCVHA + +LDE+++NL L E
Sbjct: 374 LFMATLVAKKLLKMKN--------VKPYIPDFKLAFEHFCVHAGGRAVLDEIEKNLSLGE 425
Query: 456 KNMEASRMTLHRFGNTSSSSIWYELAYMEAKERVRRGDRVWQLAFGSGFKCNSVVWLSMK 515
ME SRMTL+RFGNTSSSS+WYELAY EAK RVRR DRVWQ+AFGSGFKCNS VW +++
Sbjct: 426 WQMEPSRMTLYRFGNTSSSSLWYELAYSEAKGRVRRRDRVWQIAFGSGFKCNSAVWRALR 485
Query: 516 RVN---KPSRNNPWLDCINRYPV 535
V+ + + NPW+D I+R+PV
Sbjct: 486 SVDPEEEALKKNPWMDEIDRFPV 508
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 888,462,962
Number of Sequences: 2790947
Number of extensions: 36766591
Number of successful extensions: 91760
Number of sequences better than 10.0: 718
Number of HSP's better than 10.0 without gapping: 509
Number of HSP's successfully gapped in prelim test: 209
Number of HSP's that attempted gapping in prelim test: 90528
Number of HSP's gapped (non-prelim): 1058
length of query: 538
length of database: 848,049,833
effective HSP length: 132
effective length of query: 406
effective length of database: 479,644,829
effective search space: 194735800574
effective search space used: 194735800574
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Lotus: description of TM0133.9


