

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0118a.3
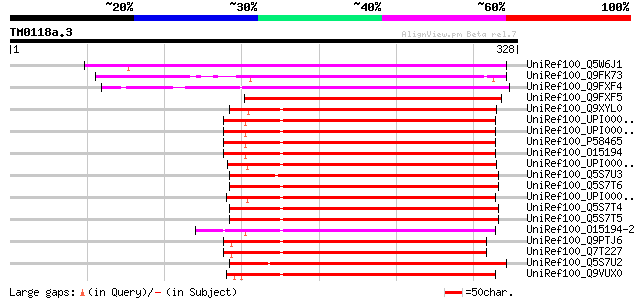
(328 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5W6J1 Hypothetical protein OSJNBb0115F21.3 [Oryza sat... 213 6e-54
UniRef100_Q9FK73 Arabidopsis thaliana genomic DNA, chromosome 5,... 199 7e-50
UniRef100_Q9FXF4 F1N18.17 protein [Arabidopsis thaliana] 185 1e-45
UniRef100_Q9FXF5 F1N18.16 protein [Arabidopsis thaliana] 173 7e-42
UniRef100_Q9XYL0 Development protein DG1148 [Dictyostelium disco... 151 3e-35
UniRef100_UPI0000435A26 UPI0000435A26 UniRef100 entry 143 6e-33
UniRef100_UPI00001D010E UPI00001D010E UniRef100 entry 143 6e-33
UniRef100_P58465 CTD small phosphatase-like protein [Mus musculus] 143 6e-33
UniRef100_O15194 CTD small phosphatase-like protein [Homo sapiens] 143 6e-33
UniRef100_UPI000042FBDF UPI000042FBDF UniRef100 entry 142 1e-32
UniRef100_Q5S7U3 Putative nuclear LIM factor interactor-interact... 142 1e-32
UniRef100_Q5S7T6 Nuclear LIM factor interactor-interacting prote... 142 1e-32
UniRef100_UPI0000364A5A UPI0000364A5A UniRef100 entry 142 2e-32
UniRef100_Q5S7T4 Nuclear LIM factor interactor-interacting prote... 142 2e-32
UniRef100_Q5S7T5 Nuclear LIM factor interactor-interacting prote... 141 2e-32
UniRef100_O15194-2 Splice isoform 2 of O15194 [Homo sapiens] 141 2e-32
UniRef100_Q9PTJ6 CTD small phosphatase-like protein [Gallus gallus] 141 3e-32
UniRef100_Q7T227 NIF [Xenopus laevis] 140 4e-32
UniRef100_Q5S7U2 Putative nuclear LIM factor interactor-interact... 140 4e-32
UniRef100_Q9VUX0 CG5830-PA [Drosophila melanogaster] 138 2e-31
>UniRef100_Q5W6J1 Hypothetical protein OSJNBb0115F21.3 [Oryza sativa]
Length = 307
Score = 213 bits (542), Expect = 6e-54
Identities = 132/280 (47%), Positives = 168/280 (59%), Gaps = 7/280 (2%)
Query: 49 SRVKRVPTKSIKDIRRNNNRRWRRKTP--IKNAVAASSAVLASIHRRITKFFSKLARL-S 105
S K P+ S R RR +P IK+ A L RR+ K F++LA L S
Sbjct: 21 SPTKPTPSPSPSPASARRRRVLRRGSPGRIKSLAATFDTSLRGCRRRLLKLFARLAVLGS 80
Query: 106 PDRKRASYKILKKTSPQDDSICRTLVFDGGDNDPSPPLPL-LPPSISHRRTVFLDLDETL 164
P ++RA+ ++ S SP LPL LPP RRT+FLDLDETL
Sbjct: 81 PTKRRAAAAGFRRLRSPPRSPSPPTPKPNQVAAVSPQLPLPLPPVSPGRRTLFLDLDETL 140
Query: 165 VHSNTAPPPES-FDFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYA 223
+HS T P + DF VRPVI G + FYV KRPGVD FL A AA FE+VVFTA L EYA
Sbjct: 141 IHSQTDPAARARHDFAVRPVIAGQAVTFYVVKRPGVDAFLAAAAAAFELVVFTAGLPEYA 200
Query: 224 SLVLDRVD-RNRLVSHRLYRDSCKQE-DGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPE 281
SLVLDR+D R L +HRLYR +C+ DG+LVKDL+ GRDL+R VIVDDNPN+++ QP+
Sbjct: 201 SLVLDRLDPRGALFAHRLYRGACRDAGDGRLVKDLAATGRDLRRAVIVDDNPNAYSLQPD 260
Query: 282 NAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAVKQF 321
NA+ + PF+DD D EL ++ DD+RDA+K++
Sbjct: 261 NAVPVAPFIDDADDHELERVMGILSIAAEFDDVRDAIKRY 300
>UniRef100_Q9FK73 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MRA19
[Arabidopsis thaliana]
Length = 272
Score = 199 bits (507), Expect = 7e-50
Identities = 127/279 (45%), Positives = 165/279 (58%), Gaps = 35/279 (12%)
Query: 56 TKSIKDIRRNNNRRWRRKTPIKNAVAASSAVLASIHRRITKFFSKLARLSPDRKRASYKI 115
TKSI +RN RR R+ + A++ V+A+I++ I + L +KI
Sbjct: 11 TKSISRHQRNYLRRHRKSQIGCSPSPATTTVIATINKSIYRCHRTFLCLFSRHATKGFKI 70
Query: 116 LKKTSPQDDSICRTLVFDGGDNDPSPPLPLLPPSISHRR----------TVFLDLDETLV 165
LK P++D F P S+ H R T+ LDLDETLV
Sbjct: 71 LK---PRED-------FQNS-----------PYSLFHERSGKSFDETKKTIVLDLDETLV 109
Query: 166 HSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASL 225
HS+ P +DFVV P IDG + F+V KRPGVDEFL+ + K+++VVFTA LREYASL
Sbjct: 110 HSSMEKPEVPYDFVVNPKIDGQILTFFVIKRPGVDEFLKKIGEKYQIVVFTAGLREYASL 169
Query: 226 VLDRVD-RNRLVSHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAI 284
VLD++D R++S YRD+C + DG+LVKDL RDL+RVVIVDDNPNS+A QPENA
Sbjct: 170 VLDKLDPERRVISRSFYRDACSEIDGRLVKDLGFVMRDLRRVVIVDDNPNSYALQPENAF 229
Query: 285 LIRPFVDDVFDRELWKLRMFFDGVDCC--DDMRDAVKQF 321
I+PF DD+ D EL KL F G DC +DMR A+K+F
Sbjct: 230 PIKPFSDDLEDVELKKLGEFLYG-DCVKFEDMRVALKEF 267
>UniRef100_Q9FXF4 F1N18.17 protein [Arabidopsis thaliana]
Length = 278
Score = 185 bits (470), Expect = 1e-45
Identities = 110/266 (41%), Positives = 159/266 (59%), Gaps = 13/266 (4%)
Query: 60 KDIRRNNNRRWRRKTPIKNAVAASSAVLASIHRRITKFFSKLARLSPDRKRASYKILKKT 119
K R+N N + R + AVAA+ ++ S++ I F ++L R + L+
Sbjct: 20 KHARKNRNHQSRC---LAAAVAAAGSIFTSLNMSIFTFHNRLLRCVS-------RFLRLA 69
Query: 120 SPQDDSICRTLVFDGGDNDPSPPLPLLPPSISHRRTVFLDLDETLVHSNTAPPPE-SFDF 178
+ + R G PL+ + +RT+FLDLDETLVHS PP + DF
Sbjct: 70 TTSSATPTRRPTMKQGYKKLHKREPLIRRN-DKKRTIFLDLDETLVHSTMEPPIRVNVDF 128
Query: 179 VVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSH 238
+VR I+G + +V KRPGV EFLE ++ + V +FTA L EYAS VLD++D+NR++S
Sbjct: 129 MVRIKIEGAVIPMFVVKRPGVTEFLERISKNYRVAIFTAGLPEYASQVLDKLDKNRVISQ 188
Query: 239 RLYRDSCKQEDGKLVKDLSL-AGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRE 297
RLYRDSC + +G+ KDLSL A DL V++VDDNP S++ QP+N + I+PF+DD+ D+E
Sbjct: 189 RLYRDSCTEVNGRYAKDLSLVAKNDLGSVLLVDDNPFSYSLQPDNGVPIKPFMDDMEDQE 248
Query: 298 LWKLRMFFDGVDCCDDMRDAVKQFFT 323
L KL FFDG +D+RDA + +
Sbjct: 249 LMKLAEFFDGCYQYEDLRDAAAELLS 274
>UniRef100_Q9FXF5 F1N18.16 protein [Arabidopsis thaliana]
Length = 247
Score = 173 bits (438), Expect = 7e-42
Identities = 90/167 (53%), Positives = 118/167 (69%), Gaps = 1/167 (0%)
Query: 153 RRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEV 212
+RT+ LDLDETLVH+ T P DF+V ++ M +V KRPGV EFLE L ++V
Sbjct: 75 KRTIILDLDETLVHATTHLPGVKHDFMVMVKMEREIMPIFVVKRPGVTEFLERLGENYKV 134
Query: 213 VVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSL-AGRDLKRVVIVDD 271
VVFTA L EYAS VLD++D+N ++S RLYRDSC + +GK VKDLSL G+DL+ +IVDD
Sbjct: 135 VVFTAGLEEYASQVLDKLDKNGVISQRLYRDSCTEVNGKYVKDLSLVVGKDLRSALIVDD 194
Query: 272 NPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAV 318
NP+S++ QPEN + I+ FVDD+ D+EL L F + +DMRDAV
Sbjct: 195 NPSSYSLQPENGVPIKAFVDDLKDQELLNLVEFLESCYAYEDMRDAV 241
>UniRef100_Q9XYL0 Development protein DG1148 [Dictyostelium discoideum]
Length = 306
Score = 151 bits (381), Expect = 3e-35
Identities = 85/176 (48%), Positives = 113/176 (63%), Gaps = 4/176 (2%)
Query: 143 LPLLPPSISHR---RTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGV 199
+PL+ P I +T+ LDLDETLVHS+ P DF+V I+G + YV KRP V
Sbjct: 123 IPLMVPMIPRHVGLKTLVLDLDETLVHSSFKPVHNP-DFIVPVEIEGTIHQVYVVKRPFV 181
Query: 200 DEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLA 259
D+FL A+A KFE+VVFTA+L +YA VLD +D R++ +RL+R+SC G VKDLS
Sbjct: 182 DDFLRAIAEKFEIVVFTASLAKYADPVLDFLDTGRVIHYRLFRESCHNHKGNYVKDLSRL 241
Query: 260 GRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMR 315
GRDLK +IVD++P+S+ PENAI I + DD DREL L D + +D+R
Sbjct: 242 GRDLKSTIIVDNSPSSYLFHPENAIPIDSWFDDKDDRELLDLLPLLDDLIKVEDVR 297
>UniRef100_UPI0000435A26 UPI0000435A26 UniRef100 entry
Length = 346
Score = 143 bits (361), Expect = 6e-33
Identities = 81/180 (45%), Positives = 113/180 (62%), Gaps = 5/180 (2%)
Query: 139 PSPPLPLLPPSIS----HRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP L P ++ ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 158 PSPPAKYLLPEVTVLDYGKKCVVIDLDETLVHSSFKPISNA-DFIVPVEIDGTIHQVYVL 216
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + RL+R+SC G VK
Sbjct: 217 KRPHVDEFLQRMGQLFECVLFTASLAKYADPVADLLDRWGVFRARLFRESCVFHRGNYVK 276
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDM 314
DLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D EL L FF+G+ DD+
Sbjct: 277 DLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDTELLDLIPFFEGLSREDDV 336
>UniRef100_UPI00001D010E UPI00001D010E UniRef100 entry
Length = 332
Score = 143 bits (361), Expect = 6e-33
Identities = 81/180 (45%), Positives = 113/180 (62%), Gaps = 5/180 (2%)
Query: 139 PSPPLPLLPPSIS----HRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP L P ++ ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 144 PSPPAKYLLPEVTVLDHGKKCVVIDLDETLVHSSFKPISNA-DFIVPVEIDGTIHQVYVL 202
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + RL+R+SC G VK
Sbjct: 203 KRPHVDEFLQRMGQLFECVLFTASLAKYADPVADLLDRWGVFRARLFRESCVFHRGNYVK 262
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDM 314
DLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D EL L FF+G+ DD+
Sbjct: 263 DLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDTELLDLIPFFEGLSREDDV 322
>UniRef100_P58465 CTD small phosphatase-like protein [Mus musculus]
Length = 276
Score = 143 bits (361), Expect = 6e-33
Identities = 81/180 (45%), Positives = 113/180 (62%), Gaps = 5/180 (2%)
Query: 139 PSPPLPLLPPSIS----HRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP L P ++ ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 88 PSPPAKYLLPEVTVLDYGKKCVVIDLDETLVHSSFKPISNA-DFIVPVEIDGTIHQVYVL 146
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + RL+R+SC G VK
Sbjct: 147 KRPHVDEFLQRMGQLFECVLFTASLAKYADPVADLLDRWGVFRARLFRESCVFHRGNYVK 206
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDM 314
DLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D EL L FF+G+ DD+
Sbjct: 207 DLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDTELLDLIPFFEGLSREDDV 266
>UniRef100_O15194 CTD small phosphatase-like protein [Homo sapiens]
Length = 276
Score = 143 bits (361), Expect = 6e-33
Identities = 81/180 (45%), Positives = 113/180 (62%), Gaps = 5/180 (2%)
Query: 139 PSPPLPLLPPSIS----HRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP L P ++ ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 88 PSPPAKYLLPEVTVLDYGKKCVVIDLDETLVHSSFKPISNA-DFIVPVEIDGTIHQVYVL 146
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + RL+R+SC G VK
Sbjct: 147 KRPHVDEFLQRMGQLFECVLFTASLAKYADPVADLLDRWGVFRARLFRESCVFHRGNYVK 206
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDM 314
DLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D EL L FF+G+ DD+
Sbjct: 207 DLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDTELLDLIPFFEGLSREDDV 266
>UniRef100_UPI000042FBDF UPI000042FBDF UniRef100 entry
Length = 613
Score = 142 bits (359), Expect = 1e-32
Identities = 78/177 (44%), Positives = 109/177 (61%), Gaps = 4/177 (2%)
Query: 142 PLPLLPPSISH---RRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPG 198
P PLLPP + R+ + LDLDETL+HS+ P + D++V I+ YV KRPG
Sbjct: 431 PAPLLPPVAAKHRGRKCLVLDLDETLLHSSFKQLPTA-DYIVPVEIESQVHNVYVIKRPG 489
Query: 199 VDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSL 258
VD FL +A +E+VVFTA+L +YA VLD +D NR+V+HRL+R+SC G VKDLS
Sbjct: 490 VDHFLTEMAKIYEIVVFTASLSKYADPVLDMLDENRVVAHRLFRESCYNHKGNYVKDLSQ 549
Query: 259 AGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMR 315
GRD++ +I+D++P S+ P NA+ + + D D EL L F + DD+R
Sbjct: 550 LGRDIEHSIIIDNSPASYIFHPNNAVPVSTWFSDPHDSELTDLCPFLADLATVDDVR 606
>UniRef100_Q5S7U3 Putative nuclear LIM factor interactor-interacting protein spore-
specific form [Phytophthora infestans]
Length = 297
Score = 142 bits (359), Expect = 1e-32
Identities = 74/174 (42%), Positives = 112/174 (63%), Gaps = 1/174 (0%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP + P+ ++ + LDLDETLVHS+ P +S+DF++ IDG YV KRPG +EF
Sbjct: 116 LPPVYPNDVDKKCLVLDLDETLVHSSFRPT-DSYDFIIPVNIDGTVHHVYVCKRPGAEEF 174
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L +A +E+V++TA+L +YA +LD++D + +RLYR+ C Q +G VKDLSL RD
Sbjct: 175 LIEMAKYYEIVIYTASLSKYADPLLDKLDPEGTIRYRLYREHCVQYEGSYVKDLSLLDRD 234
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRD 316
+ +++IVD++P ++A P NAI F+DD DREL + F +D+R+
Sbjct: 235 ITQMIIVDNSPMAYAFHPRNAIGCSSFIDDPNDRELDSIARFLTKFQDVEDVRN 288
>UniRef100_Q5S7T6 Nuclear LIM factor interactor-interacting protein [Phytophthora
sojae]
Length = 336
Score = 142 bits (359), Expect = 1e-32
Identities = 76/174 (43%), Positives = 108/174 (61%), Gaps = 1/174 (0%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP P S + + LDLDETLVHS+ P P DFV+ IDG +V KRPG +EF
Sbjct: 155 LPPRSPDDSDKMCLVLDLDETLVHSSFRPTPNP-DFVIPVEIDGTIHHVFVAKRPGAEEF 213
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L +A +E+V++TA+L +YA +LD++D ++ +RLYR C Q +G VKDLSL R+
Sbjct: 214 LVEMAKYYEIVIYTASLSKYADPLLDQLDPEGVIKYRLYRQHCVQYEGNYVKDLSLLARE 273
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRD 316
L + +IVD++P ++ P+NAI F+DD DREL + F V +D+RD
Sbjct: 274 LSQTIIVDNSPMAYIWHPKNAIGCSSFIDDPNDRELESISRFLSKVLDVEDVRD 327
>UniRef100_UPI0000364A5A UPI0000364A5A UniRef100 entry
Length = 188
Score = 142 bits (357), Expect = 2e-32
Identities = 80/177 (45%), Positives = 111/177 (62%), Gaps = 4/177 (2%)
Query: 141 PPLPLLPPSISH---RRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRP 197
P PLLPP+ SH + V +DLDETLVHS+ P ++ DF++ I+G + YV KRP
Sbjct: 5 PVKPLLPPTESHDAKKICVVIDLDETLVHSSFTPVSDA-DFIIPVEIEGTVHQVYVLKRP 63
Query: 198 GVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLS 257
VDEFL+ + FE V+FTA+L +YA V D +D +RL+R+SC G VKDLS
Sbjct: 64 HVDEFLKRMGELFECVLFTASLSKYADPVSDMLDTWGAFRNRLFRESCVFHKGNYVKDLS 123
Query: 258 LAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDM 314
GRDL +V+I+D++P S+ QPENA+ + + DD D EL L FF+ + DD+
Sbjct: 124 RLGRDLDKVIIIDNSPVSYIFQPENAVPVVSWFDDKSDTELLDLIPFFERLSQADDI 180
>UniRef100_Q5S7T4 Nuclear LIM factor interactor-interacting protein [Phytophthora
sojae]
Length = 336
Score = 142 bits (357), Expect = 2e-32
Identities = 75/174 (43%), Positives = 108/174 (61%), Gaps = 1/174 (0%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP P S + + LDLDETLVHS+ P P DFV+ IDG +V KRPG +EF
Sbjct: 155 LPPRSPDDSDKMCLVLDLDETLVHSSFRPTPNP-DFVIPVEIDGTIHHVFVAKRPGAEEF 213
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L +A +E+V++TA+L +YA +LD++D ++ +RLYR C Q +G VKDLSL R+
Sbjct: 214 LVEMAKYYEIVIYTASLSKYADPLLDQLDPEGVIKYRLYRQHCVQYEGNYVKDLSLLARE 273
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRD 316
L + +IVD++P ++ P++AI F+DD DREL + F V +D+RD
Sbjct: 274 LSQTIIVDNSPMAYIWYPKSAIGCSSFIDDPNDRELESISRFLTNVHDVEDVRD 327
>UniRef100_Q5S7T5 Nuclear LIM factor interactor-interacting protein [Phytophthora
sojae]
Length = 335
Score = 141 bits (356), Expect = 2e-32
Identities = 71/174 (40%), Positives = 111/174 (62%), Gaps = 1/174 (0%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP L P ++++ + LDLDETLVHS+ P D+++ IDG + YV KRPG +EF
Sbjct: 154 LPPLAPDDANKKCLVLDLDETLVHSSFRPTTNP-DYIIPVDIDGTIHQVYVCKRPGAEEF 212
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L ++ +E+VV+TA+L +YA +LD++D ++ +RL+R+ C Q +G VKDLSL R+
Sbjct: 213 LVEMSKYYEIVVYTASLSKYADPLLDKLDLENVIKYRLFREHCVQYEGNYVKDLSLLDRE 272
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRD 316
+ + +I+D++P S+ P NAI F+DD DREL + F + +D+RD
Sbjct: 273 IPQTIIIDNSPMSYIFHPRNAIGCSSFIDDPNDRELESISRFLTKIHDVEDVRD 326
>UniRef100_O15194-2 Splice isoform 2 of O15194 [Homo sapiens]
Length = 265
Score = 141 bits (356), Expect = 2e-32
Identities = 84/198 (42%), Positives = 118/198 (59%), Gaps = 6/198 (3%)
Query: 121 PQDDSICRTLVFDGGDNDPSPPLPLLPPSIS----HRRTVFLDLDETLVHSNTAPPPESF 176
P S+ LV + G PP L P ++ ++ V +DLDETLVHS+ P +
Sbjct: 60 PSSPSVLPPLVEENGGLQ-KPPAKYLLPEVTVLDYGKKCVVIDLDETLVHSSFKPISNA- 117
Query: 177 DFVVRPVIDGVPMEFYVKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLV 236
DF+V IDG + YV KRP VDEFL+ + FE V+FTA+L +YA V D +DR +
Sbjct: 118 DFIVPVEIDGTIHQVYVLKRPHVDEFLQRMGQLFECVLFTASLAKYADPVADLLDRWGVF 177
Query: 237 SHRLYRDSCKQEDGKLVKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDR 296
RL+R+SC G VKDLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D
Sbjct: 178 RARLFRESCVFHRGNYVKDLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDT 237
Query: 297 ELWKLRMFFDGVDCCDDM 314
EL L FF+G+ DD+
Sbjct: 238 ELLDLIPFFEGLSREDDV 255
>UniRef100_Q9PTJ6 CTD small phosphatase-like protein [Gallus gallus]
Length = 275
Score = 141 bits (355), Expect = 3e-32
Identities = 81/174 (46%), Positives = 110/174 (62%), Gaps = 5/174 (2%)
Query: 139 PSPP----LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP LP L S ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 87 PSPPAKYLLPELTASDYGKKCVVIDLDETLVHSSFKPISNA-DFIVPVEIDGTIHQVYVL 145
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + RL+R+SC G VK
Sbjct: 146 KRPHVDEFLQRMGELFECVLFTASLAKYADPVADLLDRWGVFRARLFRESCVFHRGNYVK 205
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGV 308
DLS GR+L +V+IVD++P S+ PENA+ ++ + DD+ D EL L FF+G+
Sbjct: 206 DLSRLGRELSKVIIVDNSPASYIFHPENAVPVQSWFDDMTDTELLDLIPFFEGL 259
>UniRef100_Q7T227 NIF [Xenopus laevis]
Length = 276
Score = 140 bits (354), Expect = 4e-32
Identities = 80/174 (45%), Positives = 110/174 (62%), Gaps = 5/174 (2%)
Query: 139 PSPP----LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVK 194
PSPP LP L S ++ V +DLDETLVHS+ P + DF+V IDG + YV
Sbjct: 88 PSPPTKYLLPELKVSEYGKKCVVIDLDETLVHSSFKPINNA-DFIVPVEIDGTIHQVYVL 146
Query: 195 KRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVK 254
KRP VDEFL+ + FE V+FTA+L +YA V D +DR + + RL+R+SC G VK
Sbjct: 147 KRPHVDEFLQKMGEMFECVLFTASLAKYADPVADLLDRWGVFNARLFRESCVFHRGNYVK 206
Query: 255 DLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGV 308
DLS GR+L +V+I+D++P S+ PENA+ + + DD+ D EL L FF+G+
Sbjct: 207 DLSRLGRELSKVIIIDNSPASYIFHPENAVPVMSWFDDMADTELLDLLPFFEGL 260
>UniRef100_Q5S7U2 Putative nuclear LIM factor interactor-interacting protein
cleavage- specific form [Phytophthora infestans]
Length = 339
Score = 140 bits (354), Expect = 4e-32
Identities = 76/179 (42%), Positives = 112/179 (62%), Gaps = 1/179 (0%)
Query: 143 LPLLPPSISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFYVKKRPGVDEF 202
LP P + ++ + LDLDETLVHS T P ++DF++ IDG YV KRPG +EF
Sbjct: 158 LPPASPIDADKKCLVLDLDETLVHS-TFRPTNNYDFIIPVEIDGSTHLVYVCKRPGAEEF 216
Query: 203 LEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKLVKDLSLAGRD 262
L +A +E+VV+TA+L YA +LD++D + +RLYR+ C Q +G VKDLSL RD
Sbjct: 217 LIEMAKYYEIVVYTASLSVYADPLLDKLDPEGTIRYRLYREHCVQYEGCYVKDLSLLDRD 276
Query: 263 LKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCDDMRDAVKQF 321
+ + +IVD++P ++A P NAI F DD+ DREL + F +D+R+ ++Q+
Sbjct: 277 ITQTIIVDNSPMAYAFHPRNAIGCSSFYDDLNDRELQSIGRFLTKFQDVEDVRNHMQQW 335
>UniRef100_Q9VUX0 CG5830-PA [Drosophila melanogaster]
Length = 329
Score = 138 bits (347), Expect = 2e-31
Identities = 78/182 (42%), Positives = 114/182 (61%), Gaps = 9/182 (4%)
Query: 141 PPLP-----LLPP---SISHRRTVFLDLDETLVHSNTAPPPESFDFVVRPVIDGVPMEFY 192
PPLP LLP + HR+ + +DLDETLVHS+ P P + DF+V IDG + Y
Sbjct: 69 PPLPDQQRYLLPQVRLTDMHRKCMVIDLDETLVHSSFKPIPNA-DFIVPVEIDGTVHQVY 127
Query: 193 VKKRPGVDEFLEALAAKFEVVVFTAALREYASLVLDRVDRNRLVSHRLYRDSCKQEDGKL 252
V KRP VDEFL+ + +E V+FTA+L +YA V D +D+ + RL+R+SC G
Sbjct: 128 VLKRPHVDEFLQKMGELYECVLFTASLAKYADPVADLLDKWNVFRARLFRESCVYYRGNY 187
Query: 253 VKDLSLAGRDLKRVVIVDDNPNSFANQPENAILIRPFVDDVFDRELWKLRMFFDGVDCCD 312
+KDL+ GRDL+++VIVD++P S+ P+NA+ ++ + DDV D EL +L F+ + D
Sbjct: 188 IKDLNRLGRDLQKIVIVDNSPASYIFHPDNAVPVKSWFDDVTDCELRELIPLFEKLSKVD 247
Query: 313 DM 314
+
Sbjct: 248 SV 249
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 562,213,505
Number of Sequences: 2790947
Number of extensions: 23938521
Number of successful extensions: 57605
Number of sequences better than 10.0: 314
Number of HSP's better than 10.0 without gapping: 266
Number of HSP's successfully gapped in prelim test: 48
Number of HSP's that attempted gapping in prelim test: 56971
Number of HSP's gapped (non-prelim): 348
length of query: 328
length of database: 848,049,833
effective HSP length: 127
effective length of query: 201
effective length of database: 493,599,564
effective search space: 99213512364
effective search space used: 99213512364
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Lotus: description of TM0118a.3


