

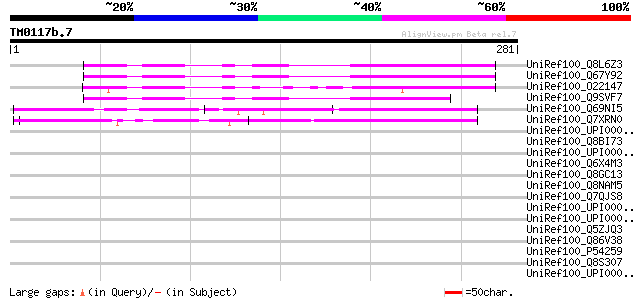
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0117b.7
(281 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8L6Z3 Hypothetical protein At4g38290 [Arabidopsis tha... 109 9e-23
UniRef100_Q67Y92 Hypothetical protein At4g38280 [Arabidopsis tha... 107 3e-22
UniRef100_O22147 Hypothetical protein At2g45250 [Arabidopsis tha... 104 2e-21
UniRef100_Q9SVF7 Hypothetical protein F22I13.50 [Arabidopsis tha... 84 3e-15
UniRef100_Q69NI5 Hypothetical protein OJ1210_A07.18 [Oryza sativa] 84 4e-15
UniRef100_Q7XRN0 OSJNBa0095E20.6 protein [Oryza sativa] 79 1e-13
UniRef100_UPI0000361BE3 UPI0000361BE3 UniRef100 entry 39 0.20
UniRef100_Q8BI73 Mus musculus 16 days embryo head cDNA, RIKEN fu... 38 0.26
UniRef100_UPI000021ACB7 UPI000021ACB7 UniRef100 entry 38 0.34
UniRef100_Q6X4M3 Rtp12.5 [Oncorhynchus mykiss] 37 0.44
UniRef100_Q8GC13 Cell envelope-associated proteinase PrtR [Lacto... 37 0.57
UniRef100_Q8NAM5 Hypothetical protein FLJ35107 [Homo sapiens] 37 0.57
UniRef100_Q7QJS8 ENSANGP00000020719 [Anopheles gambiae str. PEST] 37 0.75
UniRef100_UPI00002DAA14 UPI00002DAA14 UniRef100 entry 36 0.98
UniRef100_UPI000006F554 UPI000006F554 UniRef100 entry 36 1.3
UniRef100_Q5ZJQ3 Hypothetical protein [Gallus gallus] 36 1.3
UniRef100_Q86V38 Atrophin-1 [Homo sapiens] 36 1.3
UniRef100_P54259 Atrophin-1 [Homo sapiens] 36 1.3
UniRef100_Q8S307 BRASSINAZOLE-RESISTANT 1 protein [Arabidopsis t... 36 1.3
UniRef100_UPI000036032F UPI000036032F UniRef100 entry 35 1.7
>UniRef100_Q8L6Z3 Hypothetical protein At4g38290 [Arabidopsis thaliana]
Length = 170
Score = 109 bits (272), Expect = 9e-23
Identities = 79/228 (34%), Positives = 105/228 (45%), Gaps = 70/228 (30%)
Query: 42 DKDPDTDSNRISGTKRPLSDYPVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENP 101
+K + DS G K+P + P + A+G LVYVRR+ E + K AA
Sbjct: 12 EKANEQDSVSSIGAKKPPLESPAT--------TNAASGRLVYVRRRVEVDTSKAAA---- 59
Query: 102 TNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKS 161
T P P P AP + SS + P+
Sbjct: 60 ----------------STTNPNPP---------PTKAPLQIPSSPAQEPTPTSH------ 88
Query: 162 TMKLAPVESNCVTTSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQMLQSL 221
+ WEERY LQMLL KL+QSD+ ++VQML SL
Sbjct: 89 ---------------------------KLDWEERYLHLQMLLNKLNQSDRTDHVQMLWSL 121
Query: 222 SSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAKKFKAPAD 269
SS ELS+HAV+LEKR+IQ SLEEA+E+QRVA LN+LG+S K+ ++
Sbjct: 122 SSAELSKHAVDLEKRSIQFSLEEAREMQRVAALNMLGRSVNSLKSTSN 169
>UniRef100_Q67Y92 Hypothetical protein At4g38280 [Arabidopsis thaliana]
Length = 170
Score = 107 bits (268), Expect = 3e-22
Identities = 79/228 (34%), Positives = 104/228 (44%), Gaps = 70/228 (30%)
Query: 42 DKDPDTDSNRISGTKRPLSDYPVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENP 101
+K + DS G K+P + P + A+G LVYVRR+ E + K AA
Sbjct: 12 EKANEQDSVSSIGAKKPPLESPAT--------TNAASGRLVYVRRRVEVDTSKAAA---- 59
Query: 102 TNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKS 161
T P P P AP + SS + P+
Sbjct: 60 ----------------STTNPNPP---------PTKAPLQIPSSPAQEPTPTSH------ 88
Query: 162 TMKLAPVESNCVTTSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQMLQSL 221
+ WEERY LQMLL KL+QSD+ + VQML SL
Sbjct: 89 ---------------------------KLDWEERYLHLQMLLNKLNQSDRTDRVQMLWSL 121
Query: 222 SSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAKKFKAPAD 269
SS ELS+HAV+LEKR+IQ SLEEA+E+QRVA LN+LG+S K+ ++
Sbjct: 122 SSAELSKHAVDLEKRSIQFSLEEAREMQRVAALNMLGRSVNSLKSTSN 169
>UniRef100_O22147 Hypothetical protein At2g45250 [Arabidopsis thaliana]
Length = 211
Score = 104 bits (260), Expect = 2e-21
Identities = 84/238 (35%), Positives = 113/238 (47%), Gaps = 75/238 (31%)
Query: 41 KDKDPDTDSNRIS--GTKRPLSDYPVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAF 98
KD + + + +S G K+P D P + A+G LVYVRR+ E + K AA
Sbjct: 39 KDSEKAIEQDTVSSIGVKKPPVDSPAT--------TNAASGRLVYVRRRVEVDTSKAAA- 89
Query: 99 ENPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISC 158
T P P P AP P +P S
Sbjct: 90 -------------------STTNPNPP---------PTKAP------------PQIPSS- 108
Query: 159 GKSTMKLAPVESNCVTTSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQ-- 216
P ++ + PT + K + WEERY LQMLL KL+QSD+ ++VQ
Sbjct: 109 --------PAQAQ----AQEPTPTSHK----LDWEERYLHLQMLLNKLNQSDRTDHVQNM 152
Query: 217 -----MLQSLSSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAKKFKAPAD 269
+L SLSS ELS+HAV+LEKR+IQ SLEEA+E+QRVA LNVLG+S K+ ++
Sbjct: 153 FPLLLVLWSLSSAELSKHAVDLEKRSIQFSLEEAREMQRVAALNVLGRSVNSIKSTSN 210
>UniRef100_Q9SVF7 Hypothetical protein F22I13.50 [Arabidopsis thaliana]
Length = 159
Score = 84.3 bits (207), Expect = 3e-15
Identities = 67/203 (33%), Positives = 86/203 (42%), Gaps = 70/203 (34%)
Query: 42 DKDPDTDSNRISGTKRPLSDYPVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENP 101
+K + DS G K+P + P + A+G LVYVRR+ E + K AA
Sbjct: 12 EKANEQDSVSSIGAKKPPLESPAT--------TNAASGRLVYVRRRVEVDTSKAAA---- 59
Query: 102 TNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKS 161
T P P P AP + SS + P+
Sbjct: 60 ----------------STTNPNPP---------PTKAPLQIPSSPAQEPTPTSH------ 88
Query: 162 TMKLAPVESNCVTTSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQMLQSL 221
+ WEERY LQMLL KL+QSD+ ++VQML SL
Sbjct: 89 ---------------------------KLDWEERYLHLQMLLNKLNQSDRTDHVQMLWSL 121
Query: 222 SSVELSRHAVELEKRAIQLSLEE 244
SS ELS+HAV+LEKR+IQ SLEE
Sbjct: 122 SSAELSKHAVDLEKRSIQFSLEE 144
>UniRef100_Q69NI5 Hypothetical protein OJ1210_A07.18 [Oryza sativa]
Length = 425
Score = 84.0 bits (206), Expect = 4e-15
Identities = 56/152 (36%), Positives = 83/152 (53%), Gaps = 5/152 (3%)
Query: 109 PPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLT-SSISSSGRPSVPISCGKSTMKLAP 167
PPR L S T +P+ + S A A P +S + + S+ + S++
Sbjct: 254 PPRNLVST--TPVPRNPIAANVASSSVAAASPPRNLASTTKVSQNSIAANLASSSVSATS 311
Query: 168 VESNCVTTSSGPTIGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELS 227
S + P +P+ + W+ER+ +LQ LR +QS Q EY+ ML+SLSSV S
Sbjct: 312 TASRGAAPACYPV--DPQRSSNQDWKERFIRLQAFLRNNEQSGQEEYIHMLRSLSSVGRS 369
Query: 228 RHAVELEKRAIQLSLEEAKELQRVAVLNVLGK 259
+ A+ELE RA++L +EE KELQ++ VLNVL K
Sbjct: 370 KLAIELENRAVKLLIEEGKELQKMKVLNVLNK 401
Score = 53.5 bits (127), Expect = 6e-06
Identities = 51/182 (28%), Positives = 79/182 (43%), Gaps = 23/182 (12%)
Query: 3 LPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLSDY 62
+P D ++ KK A R+ + K + +S KDK T G KRP +
Sbjct: 1 MPAGDNPHSISEKKAALRESPKEPKNVGNQQPRTSPFPKDKAAGT-----VGIKRPQPNG 55
Query: 63 PVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETALP 122
P+N P N NGHLVYVRR+ E + K +++ + + + S ++T +
Sbjct: 56 PLN------PTNPGTNGHLVYVRRRLETDHSKVSSYASADSIS-------SLSSKKTVVD 102
Query: 123 KPQ---LKEPNVSCFPAFAP--FPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSS 177
+PQ LK N S AP TS S SG S K ++ V+ + + T++
Sbjct: 103 RPQEQGLKHQNSSLQTPLAPAAAAATSPASPSGGSLPQNSLRKQSLGKVVVQPSIIVTAN 162
Query: 178 GP 179
P
Sbjct: 163 PP 164
>UniRef100_Q7XRN0 OSJNBa0095E20.6 protein [Oryza sativa]
Length = 354
Score = 79.0 bits (193), Expect = 1e-13
Identities = 72/257 (28%), Positives = 114/257 (44%), Gaps = 8/257 (3%)
Query: 6 RDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRP--LSDYP 63
RDK + ++ D N ++ + +V K + + + T +P S
Sbjct: 91 RDKTKEQETQQNVQHDQTNKPEVSSSVAVQHDQSNKPDLSSSVAVQHDQTNKPELSSSVA 150
Query: 64 VNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETALPK 123
V H P S++ V + ++ E A ++ N P ++T K
Sbjct: 151 VQHDQSNKPDLSSSVA--VQHDQTNKPEWPSSVAVQHDQTNKPELPSSVAVQHDQTN--K 206
Query: 124 PQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSGPTIGN 183
P+L L SS++ SG P S K+ ++ + N + G T+ +
Sbjct: 207 PELPSSVAVQHDQTNKPELPSSVAESGGIVSPKSPEKTNEQIVN-KKNEPPVAPGTTVQD 265
Query: 184 PKGLIDMQ-WEERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVELEKRAIQLSL 242
Q W R+ +LQ L D+S Q Y++ML+SLS+ + S HA++LEKRAI L +
Sbjct: 266 DTHKSSNQYWNARFNRLQTYLESCDRSTQEGYMRMLRSLSAADRSMHAIDLEKRAIHLLV 325
Query: 243 EEAKELQRVAVLNVLGK 259
EE KELQR+ LNVLGK
Sbjct: 326 EEGKELQRMKALNVLGK 342
Score = 50.4 bits (119), Expect = 5e-05
Identities = 45/132 (34%), Positives = 59/132 (44%), Gaps = 20/132 (15%)
Query: 3 LPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLSDY 62
+P DKQ VKK SR+L + S G+S + KDK T+ + GTKR SD
Sbjct: 1 MPASDKQLASTVKKVTSRELPKKGADIVNMSQGTSPLPKDKGTATEPGKTVGTKR--SDA 58
Query: 63 PVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETA-- 120
P SPG H VYVRRK E + K ++ + N R E+ET
Sbjct: 59 P------SSPGY-----HNVYVRRKVENDHSKVSSSQEVKGNG-----RDKTKEQETQQN 102
Query: 121 LPKPQLKEPNVS 132
+ Q +P VS
Sbjct: 103 VQHDQTNKPEVS 114
>UniRef100_UPI0000361BE3 UPI0000361BE3 UniRef100 entry
Length = 112
Score = 38.5 bits (88), Expect = 0.20
Identities = 20/55 (36%), Positives = 33/55 (59%)
Query: 208 QSDQVEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAK 262
+S+ +E ++ L SL++ L L+ A QL LEE++E+ R LN+L +S K
Sbjct: 58 ESEDIEMLKELGSLTTANLMEKVRGLQNLAYQLGLEESREMTRGKFLNILERSKK 112
>UniRef100_Q8BI73 Mus musculus 16 days embryo head cDNA, RIKEN full-length enriched
library, clone:C130032F08 product:similar to ZINC FINGER
PROTEIN 13 [Mus musculus]
Length = 538
Score = 38.1 bits (87), Expect = 0.26
Identities = 22/55 (40%), Positives = 29/55 (52%), Gaps = 4/55 (7%)
Query: 110 PRQLCSEEETA----LPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPISCGK 160
PR L S+EET P P+ P + P AP PL+ S+S+ G V + CGK
Sbjct: 57 PRALGSQEETGGTRWAPLPEQDAPLAARVPGTAPGPLSPSLSAGGGHFVCVDCGK 111
>UniRef100_UPI000021ACB7 UPI000021ACB7 UniRef100 entry
Length = 1550
Score = 37.7 bits (86), Expect = 0.34
Identities = 33/135 (24%), Positives = 50/135 (36%), Gaps = 18/135 (13%)
Query: 46 DTDSNRISGTKRPLSDYPVNHHLQQSPGNSTANGHLV------------------YVRRK 87
D+DS R S + L+ YP+ Q G TA G ++ + +
Sbjct: 329 DSDSQRQSPIEVALAPYPITPPADQPNGPPTAVGEMLDPTHTMPKAHEHALDKYDHEAEE 388
Query: 88 SEAELGKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSIS 147
EAE + AA E PP + E +P P+ P++S + FPL +
Sbjct: 389 REAEDARKAAEELEIPIHLAMPPTRSPPSEPPRMPPPKPPAPSMSSGDGLSQFPLPPAAP 448
Query: 148 SSGRPSVPISCGKST 162
P PI +T
Sbjct: 449 PRKPPPAPIQIQTNT 463
>UniRef100_Q6X4M3 Rtp12.5 [Oncorhynchus mykiss]
Length = 112
Score = 37.4 bits (85), Expect = 0.44
Identities = 19/55 (34%), Positives = 32/55 (57%)
Query: 208 QSDQVEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAK 262
+S+ +E ++ L SL++ L L+ A QL LEE++E+ R LN+L + K
Sbjct: 58 ESEDIEMLKKLGSLTTANLMEKVKGLQNLAYQLGLEESREMTRGKFLNILERPKK 112
>UniRef100_Q8GC13 Cell envelope-associated proteinase PrtR [Lactobacillus rhamnosus]
Length = 1480
Score = 37.0 bits (84), Expect = 0.57
Identities = 48/232 (20%), Positives = 77/232 (32%), Gaps = 40/232 (17%)
Query: 8 KQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLSDYPVNHH 67
K PT D Q +S +++ KDK +T + TK+ V
Sbjct: 36 KTPTADKTTQPVNQAQTQTATSTASSQATTADAKDKTAETQPTTTTTTKQ------VTAQ 89
Query: 68 LQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLK 127
Q +P +T +++ P++ A QP + +
Sbjct: 90 SQAAPSTATKAQSQASTTNQAQPAAATKVQTGTPSSGANTQPAANTATTKSA-------- 141
Query: 128 EPNVSCFPAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSS-GP------T 180
TS+ SS+ S + +T A +S TT+ GP T
Sbjct: 142 ---------------TSTTSSAATQSAAPASNAATTNAAKTQSTAATTTDPGPANQDTLT 186
Query: 181 IGNPKGLIDMQWEERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVE 232
GN KGL W E YQ M++ +D Q L S+ +++ E
Sbjct: 187 KGNVKGL----WNEGYQGQGMVVAVIDSGVQAHDDLRLSDDSTAAITKEKAE 234
>UniRef100_Q8NAM5 Hypothetical protein FLJ35107 [Homo sapiens]
Length = 258
Score = 37.0 bits (84), Expect = 0.57
Identities = 35/150 (23%), Positives = 59/150 (39%), Gaps = 8/150 (5%)
Query: 33 SVGSSSVLKDKDPDTDSNRISGTKRPLSDYPVNHHLQQSPGNSTANGHLVYVRRKSEAEL 92
S+ SSS+L P + S+ S + S P + H SP +S++ S +
Sbjct: 44 SILSSSILSSSIPSSSSSSSSPSSSHSSSSPSSSHSSSSPSSSSSTSS---PSSSSSSSS 100
Query: 93 GKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAP-----FPLTSSIS 147
+ N ++++ P S ++ + S + +P P +SS S
Sbjct: 101 SSSPSSSNSSSSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSSSSSSPSSSSSSPSSSSSS 160
Query: 148 SSGRPSVPISCGKSTMKLAPVESNCVTTSS 177
SS S P S S+ +P SN +SS
Sbjct: 161 SSSSSSSPSSSSPSSSGSSPSSSNSSPSSS 190
>UniRef100_Q7QJS8 ENSANGP00000020719 [Anopheles gambiae str. PEST]
Length = 1405
Score = 36.6 bits (83), Expect = 0.75
Identities = 33/116 (28%), Positives = 53/116 (45%), Gaps = 11/116 (9%)
Query: 71 SPGNSTANGHLVYVRRKSEAELGKCAAFE-NPTNNAFCQPPRQLCSEEETALPKPQLKEP 129
SP S + Y+R +S + L A NP + P LC ++ L +P++K
Sbjct: 105 SPVGSPSAAAASYIRSRSVSSLKNLARSSLNPPG----RNPSSLCKLAKSQLTRPKVKPD 160
Query: 130 NVSCFPAFAPFPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSGPTIGNPK 185
N+S A +P SS SSS + + P + G + V++ S GP +GN +
Sbjct: 161 NISLPAADSP----SSSSSSSQTAGPAASGSKGGFVETVQN--PLPSPGPLLGNTR 210
>UniRef100_UPI00002DAA14 UPI00002DAA14 UniRef100 entry
Length = 977
Score = 36.2 bits (82), Expect = 0.98
Identities = 36/145 (24%), Positives = 60/145 (40%), Gaps = 30/145 (20%)
Query: 140 FPLTSSISSSGRPSVPISCGKSTMKLAPVESNCVTTSSGPTIGNPKGLIDMQW------- 192
FP+ S S P+ P+S + + V+ S P + NP G+I W
Sbjct: 421 FPVQQVASESQNPTPPVSPVPADPSSVQQD---VSISDPPQVVNPGGMIQQSWPITPSQV 477
Query: 193 --------EERYQQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVELEKRAIQLSLEE 244
E+ +Q++ L+RKL+Q ++ VE + +E+EKRA +
Sbjct: 478 PEEKDRRLHEKERQIEELMRKLEQEQKL-----------VEALKMQLEVEKRASDSVSPK 526
Query: 245 AKELQRVA-VLNVLGKSAKKFKAPA 268
+ ++ V VL S K +A A
Sbjct: 527 LSPVPAISTVPAVLNTSILKMEAAA 551
>UniRef100_UPI000006F554 UPI000006F554 UniRef100 entry
Length = 1190
Score = 35.8 bits (81), Expect = 1.3
Identities = 28/109 (25%), Positives = 42/109 (37%), Gaps = 2/109 (1%)
Query: 2 DLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLSD 61
DL + D + D + RD+ DN+ + + SV + D D+ S G RP
Sbjct: 104 DLDSLDGRSLNDDGSSDPRDIDQDNRSTSPSIYSPGSV--ENDSDSSSGLSQGPARPYHP 161
Query: 62 YPVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPP 110
P+ Q P ++ + S G A E PT+ F PP
Sbjct: 162 PPLFPPSPQPPDSTPRQPEASFEPHPSVTPTGYHAPMEPPTSRMFQAPP 210
>UniRef100_Q5ZJQ3 Hypothetical protein [Gallus gallus]
Length = 112
Score = 35.8 bits (81), Expect = 1.3
Identities = 18/55 (32%), Positives = 32/55 (57%)
Query: 208 QSDQVEYVQMLQSLSSVELSRHAVELEKRAIQLSLEEAKELQRVAVLNVLGKSAK 262
++D ++ ++ L SL++ L L+ A QL L+E++E+ R LN+L K K
Sbjct: 58 ENDDIDMLKELGSLTTANLMEKVRGLQNLAYQLGLDESREMTRGKFLNILEKPKK 112
>UniRef100_Q86V38 Atrophin-1 [Homo sapiens]
Length = 1191
Score = 35.8 bits (81), Expect = 1.3
Identities = 28/109 (25%), Positives = 42/109 (37%), Gaps = 2/109 (1%)
Query: 2 DLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLSD 61
DL + D + D + RD+ DN+ + + SV + D D+ S G RP
Sbjct: 104 DLDSLDGRSLNDDGSSDPRDIDQDNRSTSPSIYSPGSV--ENDSDSSSGLSQGPARPYHP 161
Query: 62 YPVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPP 110
P+ Q P ++ + S G A E PT+ F PP
Sbjct: 162 PPLFPPSPQPPDSTPRQPEASFEPHPSVTPTGYHAPMEPPTSRMFQAPP 210
>UniRef100_P54259 Atrophin-1 [Homo sapiens]
Length = 1185
Score = 35.8 bits (81), Expect = 1.3
Identities = 28/109 (25%), Positives = 42/109 (37%), Gaps = 2/109 (1%)
Query: 2 DLPNRDKQPTVDVKKTASRDLQNDNKIMALTSVGSSSVLKDKDPDTDSNRISGTKRPLSD 61
DL + D + D + RD+ DN+ + + SV + D D+ S G RP
Sbjct: 104 DLDSLDGRSLNDDGSSDPRDIDQDNRSTSPSIYSPGSV--ENDSDSSSGLSQGPARPYHP 161
Query: 62 YPVNHHLQQSPGNSTANGHLVYVRRKSEAELGKCAAFENPTNNAFCQPP 110
P+ Q P ++ + S G A E PT+ F PP
Sbjct: 162 PPLFPPSPQPPDSTPRQPEASFEPHPSVTPTGYHAPMEPPTSRMFQAPP 210
>UniRef100_Q8S307 BRASSINAZOLE-RESISTANT 1 protein [Arabidopsis thaliana]
Length = 336
Score = 35.8 bits (81), Expect = 1.3
Identities = 33/110 (30%), Positives = 47/110 (42%), Gaps = 18/110 (16%)
Query: 99 ENPTNNAFCQP-PRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSISSSGRPSVPIS 157
++P ++AF P P S ++ P P EPN + F PF I SS PS+ IS
Sbjct: 113 QSPLSSAFQSPIPSYQVSPSSSSFPSPSRGEPNNNMSSTFFPFLRNGGIPSS-LPSLRIS 171
Query: 158 CGKSTMKLAPVESNCVTTS--SGPTIGNPKGLIDMQWEERYQQLQMLLRK 205
++C T S PT NPK L WE +Q + ++
Sbjct: 172 ------------NSCPVTPPVSSPTSKNPKPL--PNWESIAKQSMAIAKQ 207
>UniRef100_UPI000036032F UPI000036032F UniRef100 entry
Length = 1253
Score = 35.4 bits (80), Expect = 1.7
Identities = 42/204 (20%), Positives = 85/204 (41%), Gaps = 15/204 (7%)
Query: 87 KSEAELGKCAAFENPTNNAFCQPPRQLCSEEETALPKPQLKEPNVSCFPAFAPFPLTSSI 146
K E + G+ F++ F + E+ + ++ + P + SS
Sbjct: 452 KEEEKHGRKKYFKDGELRDFAEGESAKLREDLKVSEREDKEDCSGQSSPCLSTASWASSC 511
Query: 147 SSSGRPSVPIS--CGKSTMKLAPVESNCVTTSSGPTIGNPKGLIDMQ-WEERY------- 196
S+SG ++ ++ + +KL E T+SS T + ++ W+ R
Sbjct: 512 SASGSSNIKMTSFAERKLLKLGLREGFSSTSSSQKTTPDGSEVVACPPWQLRSDSTSSWM 571
Query: 197 --QQLQMLLRKLDQSDQVEYVQMLQSLSSVELSRHAVELEKRAIQ-LSLEEAKELQRVAV 253
+ + ML + + S V ++LQ +E R A+E +K+ ++ LS + +L + A
Sbjct: 572 GKEPVSMLGKNVTLSPSVVPTELLQLHMQLEEQRRAIEYQKKKLETLSARQRLKLGKAAF 631
Query: 254 LNVL--GKSAKKFKAPADHDECSD 275
LN++ G + P H E ++
Sbjct: 632 LNIVKSGGKSDTLPLPLKHPESAE 655
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.127 0.356
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 463,355,075
Number of Sequences: 2790947
Number of extensions: 19184116
Number of successful extensions: 45624
Number of sequences better than 10.0: 88
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 65
Number of HSP's that attempted gapping in prelim test: 45555
Number of HSP's gapped (non-prelim): 118
length of query: 281
length of database: 848,049,833
effective HSP length: 126
effective length of query: 155
effective length of database: 496,390,511
effective search space: 76940529205
effective search space used: 76940529205
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0117b.7


