

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
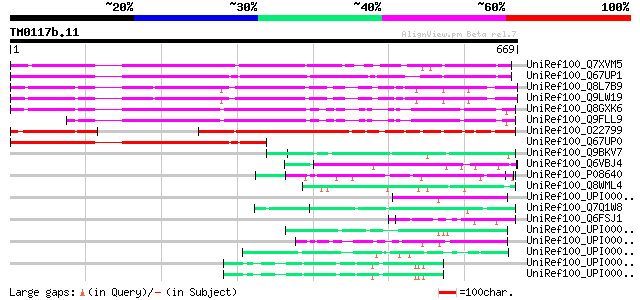
Query= TM0117b.11
(669 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XVM5 OSJNBa0072K14.9 protein [Oryza sativa] 478 e-133
UniRef100_Q67UP1 Hydroxyproline-rich glycoprotein-like [Oryza sa... 444 e-123
UniRef100_Q8L7B9 Hypothetical protein At3g26910 [Arabidopsis tha... 431 e-119
UniRef100_Q9LW19 Gb|AAB80666.1 [Arabidopsis thaliana] 431 e-119
UniRef100_Q8GXK6 Hypothetical protein At5g41100/MEE6_17 [Arabido... 405 e-111
UniRef100_Q9FLL9 Gb|AAB80666.1 [Arabidopsis thaliana] 349 1e-94
UniRef100_O22799 Hypothetical protein At2g33490 [Arabidopsis tha... 310 9e-83
UniRef100_Q67UP0 Hydroxyproline-rich glycoprotein-like [Oryza sa... 300 1e-79
UniRef100_Q9BKV7 Ppg3 [Leishmania major] 68 8e-10
UniRef100_Q6VBJ4 Epa5p [Candida glabrata] 67 2e-09
UniRef100_P08640 Glucoamylase S1/S2 precursor [Saccharomyces cer... 65 5e-09
UniRef100_Q8WML4 MUC1 protein precursor [Bos taurus] 60 2e-07
UniRef100_UPI0000337A4E UPI0000337A4E UniRef100 entry 57 2e-06
UniRef100_Q7Q1W8 ENSANGP00000020885 [Anopheles gambiae str. PEST] 57 2e-06
UniRef100_Q6FSJ1 Similarities with sp|P47179 Saccharomyces cerev... 57 2e-06
UniRef100_UPI000035F9F4 UPI000035F9F4 UniRef100 entry 55 7e-06
UniRef100_UPI000030431E UPI000030431E UniRef100 entry 55 7e-06
UniRef100_UPI0000364562 UPI0000364562 UniRef100 entry 54 1e-05
UniRef100_UPI00003A9848 UPI00003A9848 UniRef100 entry 54 1e-05
UniRef100_UPI00003A9847 UPI00003A9847 UniRef100 entry 54 1e-05
>UniRef100_Q7XVM5 OSJNBa0072K14.9 protein [Oryza sativa]
Length = 624
Score = 478 bits (1231), Expect = e-133
Identities = 295/678 (43%), Positives = 397/678 (58%), Gaps = 90/678 (13%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MKS L+K RG GLH+H H P +L+ELA A ++M++MR+CYD+LLSAAAA
Sbjct: 1 MKSPLRKFRGFGLHSHRERKDHRP-----PPAKLDELADAAQEMEEMRNCYDSLLSAAAA 55
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLI 120
T +S YEFAE++ +MG+CLLEKTALN ++++G+VL+MLGK QF+LQK +D Y
Sbjct: 56 TMNSVYEFAEAMEEMGTCLLEKTALNYDDDDSGRVLMMLGKAQFELQKFVDNY------- 108
Query: 121 PSLYFIFCFCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKR 180
R++II TIT PSESLL EL++VEEMK CD KR
Sbjct: 109 ---------------------------RTNIINTITNPSESLLKELQVVEEMKELCDHKR 141
Query: 181 DVYEYMATRFRERGRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLLT 240
YE M +RE+GRS+ KTET S +QLQ +Y E+A LF+FRLKSLKQGQ RS+LT
Sbjct: 142 QEYEAMRAAYREKGRSRHSKTETLSSEQLQAYFLDYQEDAALFIFRLKSLKQGQFRSILT 201
Query: 241 QAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHIDYHFNGLEEEDGDEGDEGYEDDDG 300
QAARHH+AQL FF++ +K LE +EPHVK+V E+QHI+Y NGL+++ ++ Y+
Sbjct: 202 QAARHHSAQLSFFRRGLKYLEALEPHVKAVAEKQHIEYPLNGLDDDTDNDEYSSYQG--- 258
Query: 301 YDENDDGELSFDYGQIEQEQDVSTSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFS 360
+++DD ELSFDY ++++D SR+SM+LDQ E +E +++Q + +
Sbjct: 259 -NQSDDSELSFDYEINDRDKDFPASRSSMDLDQSNQACSPEPLKEHKQEYTEQIQADFAA 317
Query: 361 FRVR--TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPK 418
RV+ G+QSAP+ ADN D S + R+M S +R SY LPTP D K+S S+ +N
Sbjct: 318 PRVKLEIGTQSAPISADNVFDPSTRFRKMNTS-NRTNYSYKLPTPDDDKNSTSAHTNRSP 376
Query: 419 PSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPP 478
S + + NLWHSSPL K FS P ++T + P
Sbjct: 377 HSDQPESKSHVAENLWHSSPL--VKGFKPNSMFSGPV-----------KMPSSTEGISAP 423
Query: 479 LVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPP 538
LV + Y ++ KK+KR AFSGP+ S +P +FS L P+ P
Sbjct: 424 LV-------YPYATSDFKKMKREAFSGPIPSKAGLNKP-------LFSATDLRAPMNYPR 469
Query: 539 SSSPK---------VSPSASPTIVS----SPKISELHELPRPPTNFPSNSRLLGLVGYSG 585
+ S K V+P +P I S SP+ISELHELPRPP N + GLVGYSG
Sbjct: 470 AMSTKSYGPGWQSSVAPKFTPRITSLPTTSPRISELHELPRPPANV--GAARPGLVGYSG 527
Query: 586 PLVPRGQKVSAPNNL-VISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHIS 644
PLV R Q + P S ASPLP PP AM RS+SIPS+ R L + LE+ H S
Sbjct: 528 PLVSRRQVPNVPTRASPPSQTASPLPRPPAAMTRSYSIPSNSQRTPILTVNKLLEARH-S 586
Query: 645 SISEDIASPPLTPIALSN 662
S +++SPPLTPI+L++
Sbjct: 587 RESSEVSSPPLTPISLAD 604
>UniRef100_Q67UP1 Hydroxyproline-rich glycoprotein-like [Oryza sativa]
Length = 623
Score = 444 bits (1141), Expect = e-123
Identities = 276/666 (41%), Positives = 380/666 (56%), Gaps = 68/666 (10%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MKS L +L+G G H + K + +L+ELA A +D+++MR+CYD +SAAAA
Sbjct: 1 MKSPLLRLKGFGHHQQHRERKSRQPQPQPTPAKLDELADAAQDVEEMRNCYDGFISAAAA 60
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLI 120
T + YEFAE+L ++GSCLL K LND ++++G+VL+MLGK Q++LQK D+Y
Sbjct: 61 TTNGVYEFAEALEELGSCLLAKPVLNDDDDDSGRVLMMLGKAQYELQKSADRY------- 113
Query: 121 PSLYFIFCFCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKR 180
R++II TIT PSESLL EL+ +EEMK+QCD KR
Sbjct: 114 ---------------------------RTNIIHTITTPSESLLKELQTLEEMKQQCDMKR 146
Query: 181 DVYEYMATRFRERGRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLLT 240
D YE M + ++G S+ KTE+FS +QL + EY E++ LF FRLKSLKQGQ +SLLT
Sbjct: 147 DAYETMRASYSDKGGSRHSKTESFSTEQLDASFLEYQEDSALFTFRLKSLKQGQFQSLLT 206
Query: 241 QAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHIDYHFNGLEEEDGDEGDEGYEDDDG 300
QAARHHAAQL FF+K +K LE +EP VK+++E+ HIDY+F+GLE++ D ++GY D
Sbjct: 207 QAARHHAAQLSFFRKGLKCLEALEPRVKAISEKHHIDYNFSGLEDDGSD--NDGYSTYDS 264
Query: 301 YDENDDGELSFDYGQIEQEQDVSTSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLF- 359
+DDGELSFDY +++QD TSR SM+ D+ + T E +E + + +
Sbjct: 265 C--SDDGELSFDYEINDRDQDFLTSRGSMDFDKSDQTTSPKPIKENKQEEAKQAEAEIVF 322
Query: 360 -SFRVRTGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPK 418
+ + SAPLFA N D +++LRQMRPS ++ SY LPTPV A + + SGS+
Sbjct: 323 PQLKPEFATHSAPLFAGNLLDETDRLRQMRPSSTK--HSYRLPTPVGADNPVPSGSHRLH 380
Query: 419 PS-KMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPP 477
S + A TNLWHSSPL + + + + + P+ ++ LK+ + + P
Sbjct: 381 HSAQFFETKPHAPTNLWHSSPLTKDYNGAMHNAATKPSSSSSTDDLKKLKRESWSG--PI 438
Query: 478 PLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQP 537
P+ G S + A + A+ G + + R S SV
Sbjct: 439 PIKAG--SGGKPFSQADHRPSPTMAYPGAMPAAKPHVRHASSSSV--------------- 481
Query: 538 PSSSPKVSPSASPT-IVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSA 596
SPKVSP SP SS KISELH LP PP N R GLVGYSGPLV + A
Sbjct: 482 ---SPKVSPKMSPVPPASSLKISELHLLPLPPANV-DPVRPSGLVGYSGPLVSKRAPTPA 537
Query: 597 PNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLT 656
+ S ASPLP PP A+ARS+SIPS+ R + + LE+ H S D +SPPLT
Sbjct: 538 RASPKASRTASPLPRPPAALARSYSIPSNSQRTPIITVNKLLEAKH-SREGSDASSPPLT 596
Query: 657 PIALSN 662
P++LS+
Sbjct: 597 PLSLSD 602
>UniRef100_Q8L7B9 Hypothetical protein At3g26910 [Arabidopsis thaliana]
Length = 608
Score = 431 bits (1108), Expect = e-119
Identities = 289/690 (41%), Positives = 386/690 (55%), Gaps = 113/690 (16%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MK+S++KLR L H+H + K + ++ Q++EL +A +DMQDMR+CYD LL+AAAA
Sbjct: 1 MKASIEKLRRLTSHSHKVDVK--EKGDVMATTQIDELDRAGKDMQDMRECYDRLLAAAAA 58
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLI 120
TA+SAYEF+ESL +MGSCL + ND EE+ ++L MLGK+Q +LQ+L+D Y
Sbjct: 59 TANSAYEFSESLGEMGSCLEQIAPHND--EESSRILFMLGKVQSELQRLLDTY------- 109
Query: 121 PSLYFIFCFCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKR 180
RSHI +TIT PSE+LL +LR VE+MK+QCD KR
Sbjct: 110 ---------------------------RSHIFETITSPSEALLKDLRYVEDMKQQCDGKR 142
Query: 181 DVYEYMATRFRERGRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLLT 240
+VYE + E+GR K K E + + A E+ +EAT+ +FRLKSLK+GQ+RSLL
Sbjct: 143 NVYEMSLVK--EKGRPKSSKGERHIPPESRPAYSEFHDEATMCIFRLKSLKEGQARSLLI 200
Query: 241 QAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHID----YHFNGLEEEDGDEGDEGYE 296
QA RHH AQ+ F +KSLE VE HVK E+QHID H N +E + D
Sbjct: 201 QAVRHHTAQMRLFHTGLKSLEAVERHVKVAVEKQHIDCDLSVHGNEMEASEDD------- 253
Query: 297 DDDGYDENDDGELSFDYGQIEQEQDVSTSRN--SMELDQVELTLPRGSTAEAAKENLDKL 354
DDDG N +GELSFDY EQ+ + S+ + ++D +L+ PR ST A N D
Sbjct: 254 DDDGRYMNREGELSFDYRTNEQKVEASSLSTPWATKMDDTDLSFPRPSTTRPAAVNADHR 313
Query: 355 QRNLFSFRVR-TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAK-SSISS 412
+ S R + S SAPLF + KPD SE+LRQ PS F++YVLPTP D++ S S
Sbjct: 314 EEYPVSTRDKYLSSHSAPLFPEKKPDVSERLRQANPS----FNAYVLPTPNDSRYSKPVS 369
Query: 413 GSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTAT 472
+ NP+P +N + N+WHSSPLE K + K++ SN+
Sbjct: 370 QALNPRP------TNHSAGNIWHSSPLEPIK---------------SGKDGKDAESNSFY 408
Query: 473 TRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPT 532
RLP P S D + RHAFSGPL P T+P+++ +SG P
Sbjct: 409 GRLPRP-------STTDTHHHQQQAAGRHAFSGPL--RPSSTKPITM--ADSYSGAFCPL 457
Query: 533 PIP------QPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNF---PSNSRLLGLVGY 583
P P SSSP+VSP+ASP SSP+++ELHELPRPP +F P ++ GLVG+
Sbjct: 458 PTPPVLQSHPHSSSSPRVSPTASPPPASSPRLNELHELPRPPGHFAPPPRRAKSPGLVGH 517
Query: 584 SGPLVPRGQKVSAPNNLVISS---AASPLPIPPQAMARSFSIPSSGARVTALHSSSALES 640
S PL Q+ S V S+ ASPLP+PP + RS+SIPS RV S
Sbjct: 518 SAPLTAWNQERSTVTVAVPSATNIVASPLPVPPLVVPRSYSIPSRNQRVV---------S 568
Query: 641 SHISSISEDI-ASPPLTPIALSNSRPSSDG 669
+ +DI ASPPLTP++LS P + G
Sbjct: 569 QRLVERRDDIVASPPLTPMSLSRPLPQATG 598
>UniRef100_Q9LW19 Gb|AAB80666.1 [Arabidopsis thaliana]
Length = 621
Score = 431 bits (1108), Expect = e-119
Identities = 289/690 (41%), Positives = 386/690 (55%), Gaps = 113/690 (16%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MK+S++KLR L H+H + K + ++ Q++EL +A +DMQDMR+CYD LL+AAAA
Sbjct: 1 MKASIEKLRRLTSHSHKVDVK--EKGDVMATTQIDELDRAGKDMQDMRECYDRLLAAAAA 58
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLI 120
TA+SAYEF+ESL +MGSCL + ND EE+ ++L MLGK+Q +LQ+L+D Y
Sbjct: 59 TANSAYEFSESLGEMGSCLEQIAPHND--EESSRILFMLGKVQSELQRLLDTY------- 109
Query: 121 PSLYFIFCFCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKR 180
RSHI +TIT PSE+LL +LR VE+MK+QCD KR
Sbjct: 110 ---------------------------RSHIFETITSPSEALLKDLRYVEDMKQQCDGKR 142
Query: 181 DVYEYMATRFRERGRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLLT 240
+VYE + E+GR K K E + + A E+ +EAT+ +FRLKSLK+GQ+RSLL
Sbjct: 143 NVYEMSLVK--EKGRPKSSKGERHIPPESRPAYSEFHDEATMCIFRLKSLKEGQARSLLI 200
Query: 241 QAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHID----YHFNGLEEEDGDEGDEGYE 296
QA RHH AQ+ F +KSLE VE HVK E+QHID H N +E + D
Sbjct: 201 QAVRHHTAQMRLFHTGLKSLEAVERHVKVAVEKQHIDCDLSVHGNEMEASEDD------- 253
Query: 297 DDDGYDENDDGELSFDYGQIEQEQDVSTSRN--SMELDQVELTLPRGSTAEAAKENLDKL 354
DDDG N +GELSFDY EQ+ + S+ + ++D +L+ PR ST A N D
Sbjct: 254 DDDGRYMNREGELSFDYRTNEQKVEASSLSTPWATKMDDTDLSFPRPSTTRPAAVNADHR 313
Query: 355 QRNLFSFRVR-TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAK-SSISS 412
+ S R + S SAPLF + KPD SE+LRQ PS F++YVLPTP D++ S S
Sbjct: 314 EEYPVSTRDKYLSSHSAPLFPEKKPDVSERLRQANPS----FNAYVLPTPNDSRYSKPVS 369
Query: 413 GSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTAT 472
+ NP+P +N + N+WHSSPLE K + K++ SN+
Sbjct: 370 QALNPRP------TNHSAGNIWHSSPLEPIK---------------SGKDGKDAESNSFY 408
Query: 473 TRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPT 532
RLP P S D + RHAFSGPL P T+P+++ +SG P
Sbjct: 409 GRLPRP-------STTDTHHHQQQAAGRHAFSGPL--RPSSTKPITM--ADSYSGAFCPL 457
Query: 533 PIP------QPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNF---PSNSRLLGLVGY 583
P P SSSP+VSP+ASP SSP+++ELHELPRPP +F P ++ GLVG+
Sbjct: 458 PTPPVLQSHPHSSSSPRVSPTASPPPASSPRLNELHELPRPPGHFAPPPRRAKSPGLVGH 517
Query: 584 SGPLVPRGQKVSAPNNLVISS---AASPLPIPPQAMARSFSIPSSGARVTALHSSSALES 640
S PL Q+ S V S+ ASPLP+PP + RS+SIPS RV S
Sbjct: 518 SAPLTAWNQERSTVTVAVPSATNIVASPLPVPPLVVPRSYSIPSRNQRVV---------S 568
Query: 641 SHISSISEDI-ASPPLTPIALSNSRPSSDG 669
+ +DI ASPPLTP++LS P + G
Sbjct: 569 QRLVERRDDIVASPPLTPMSLSRPLPQATG 598
>UniRef100_Q8GXK6 Hypothetical protein At5g41100/MEE6_17 [Arabidopsis thaliana]
Length = 582
Score = 405 bits (1040), Expect = e-111
Identities = 289/680 (42%), Positives = 377/680 (54%), Gaps = 133/680 (19%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MK+S +LR L + D + P Q+E LA+A +DMQDMR+ YD LL AAA
Sbjct: 2 MKASFGRLRRFALPKADAI----DIGELFPTAQIEGLARAAKDMQDMREGYDRLLEVAAA 57
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLI 120
A+SAYEF+ESL +MGSCL + ND +E+G +LLMLGK+QF+L+KL+D Y
Sbjct: 58 MANSAYEFSESLGEMGSCLEQIAPHND--QESGGILLMLGKVQFELKKLVDTY------- 108
Query: 121 PSLYFIFCFCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKR 180
RS I +TIT PSESLL++LR VE+MK+QC+EKR
Sbjct: 109 ---------------------------RSQIFKTITRPSESLLSDLRTVEDMKQQCEEKR 141
Query: 181 DVYEYMATRF-RERGRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLL 239
DV ++M +++ + KG K E +QL+TARDE +EATL +FRLKSLK+GQ+RSLL
Sbjct: 142 DVVKHMLMEHVKDKVQVKGTKGERLIRRQLETARDELQDEATLCIFRLKSLKEGQARSLL 201
Query: 240 TQAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHIDYHFNGLEEEDGDEGDEGYE-DD 298
TQAARHH AQ+ F +KSLE VE HV+ ++QHID + + G+E D + DD
Sbjct: 202 TQAARHHTAQMHMFFAGLKSLEAVEQHVRIAADRQHIDC----VLSDPGNEMDCSEDNDD 257
Query: 299 DGYDENDDGELSFDYGQIEQEQDV-STSRNSMELDQVELTLPRGSTAEAAKENLDKLQRN 357
D N DGELSFDY EQ +V ST SM++D +L+ R S A +A N D + +
Sbjct: 258 DDRLVNRDGELSFDYITSEQRVEVISTPHGSMKMDDTDLSFQRPSPAGSATVNADPREEH 317
Query: 358 LFSFR-VRTGSQSAPLFADNKPDSSEK-LRQMRPSLSRKFSSYVLPTPVDAKSSISSGSN 415
S R RT S SAPLF D K D +++ +RQM PS ++Y+LPTPVD+KSS
Sbjct: 318 SVSNRDRRTSSHSAPLFPDKKADLADRSMRQMTPSA----NAYILPTPVDSKSS------ 367
Query: 416 NPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRL 475
P +K T +N + NLWHSSPLE K + K++ SN +RL
Sbjct: 368 -PIFTKPVTQTNH-SANLWHSSPLEPIK-----------------TAHKDAESN-LYSRL 407
Query: 476 PPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIP 535
P P HAFSGPL P TR LP P+
Sbjct: 408 PRP--------------------SEHAFSGPL--KPSSTR--------------LPVPVA 431
Query: 536 -QPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNF--PSNSRLLGLVGYSGPLVPRGQ 592
Q SSSP++SP+ASP + SSP+I+ELHELPRPP F P S+ GLVG+S PL Q
Sbjct: 432 VQAQSSSPRISPTASPPLASSPRINELHELPRPPGQFAPPRRSKSPGLVGHSAPLTAWNQ 491
Query: 593 KVSAPNNLVISS--AASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDI 650
+ S N+V+S+ ASPLP+PP + RS+SIPS R A + +
Sbjct: 492 ERS---NVVVSTNIVASPLPVPPLVVPRSYSIPSRNQRAMAQQPLPERNQNR-------V 541
Query: 651 ASP---PLTPIALSNSRPSS 667
ASP PLTP +L N R S
Sbjct: 542 ASPPPLPLTPASLMNLRSLS 561
>UniRef100_Q9FLL9 Gb|AAB80666.1 [Arabidopsis thaliana]
Length = 534
Score = 349 bits (896), Expect = 1e-94
Identities = 254/606 (41%), Positives = 331/606 (53%), Gaps = 129/606 (21%)
Query: 75 MGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLIPSLYFIFCFCFNFI 134
MGSCL + ND +E+G +LLMLGK+QF+L+KL+D Y
Sbjct: 1 MGSCLEQIAPHND--QESGGILLMLGKVQFELKKLVDTY--------------------- 37
Query: 135 TNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKRDVYEYMATRF-RER 193
RS I +TIT PSESLL++LR VE+MK+QC+EKRDV ++M +++
Sbjct: 38 -------------RSQIFKTITRPSESLLSDLRTVEDMKQQCEEKRDVVKHMLMEHVKDK 84
Query: 194 GRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLLTQAARHHAAQLCFF 253
+ KG K E +QL+TARDE +EATL +FRLKSLK+GQ+RSLLTQAARHH AQ+ F
Sbjct: 85 VQVKGTKGERLIRRQLETARDELQDEATLCIFRLKSLKEGQARSLLTQAARHHTAQMHMF 144
Query: 254 KKAVKSLETVEPHVKSVTEQQHIDYHFNGLEEEDGDEGDEGYE-DDDGYDENDDGELSFD 312
+KSLE VE HV+ ++QHID + + G+E D + DDD N DGELSFD
Sbjct: 145 FAGLKSLEAVEQHVRIAADRQHIDC----VLSDPGNEMDCSEDNDDDDRLVNRDGELSFD 200
Query: 313 YGQIEQEQDV-STSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFSFR-VRTGSQSA 370
Y EQ +V ST SM++D +L+ R S A +A N D + + S R RT S SA
Sbjct: 201 YITSEQRVEVISTPHGSMKMDDTDLSFQRPSPAGSATVNADPREEHSVSNRDRRTSSHSA 260
Query: 371 PLFADNKPDSSEK-LRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEA 429
PLF D K D +++ +RQM PS ++Y+LPTPVD+KSS P +K T +N
Sbjct: 261 PLFPDKKADLADRSMRQMTPSA----NAYILPTPVDSKSS-------PIFTKPVTQTNH- 308
Query: 430 TTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHD 489
+ NLWHSSPLE K + K++ SN +RLP P
Sbjct: 309 SANLWHSSPLEPIK-----------------TAHKDAESN-LYSRLPRP----------- 339
Query: 490 YVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIP-QPPSSSPKVSPSA 548
HAFSGPL P TR LP P+ Q SSSP++SP+A
Sbjct: 340 ---------SEHAFSGPL--KPSSTR--------------LPVPVAVQAQSSSPRISPTA 374
Query: 549 SPTIVSSPKISELHELPRPPTNF--PSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISS-- 604
SP + SSP+I+ELHELPRPP F P S+ GLVG+S PL Q+ S N+V+S+
Sbjct: 375 SPPLASSPRINELHELPRPPGQFAPPRRSKSPGLVGHSAPLTAWNQERS---NVVVSTNI 431
Query: 605 AASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASP---PLTPIALS 661
ASPLP+PP + RS+SIPS R A + +ASP PLTP +L
Sbjct: 432 VASPLPVPPLVVPRSYSIPSRNQRAMAQQPLPERNQNR-------VASPPPLPLTPASLM 484
Query: 662 NSRPSS 667
N R S
Sbjct: 485 NLRSLS 490
>UniRef100_O22799 Hypothetical protein At2g33490 [Arabidopsis thaliana]
Length = 521
Score = 310 bits (794), Expect = 9e-83
Identities = 195/423 (46%), Positives = 267/423 (63%), Gaps = 31/423 (7%)
Query: 250 LCFFKKAVKSLETVEPHVKSVTEQQHIDYHFNGLEEEDGDEGDEGYEDDDGYDENDDGEL 309
LCFFKKA+ SLE V+PHV+ VTE QHIDYHF+GLE++DGD+ E E +DG + +DDGEL
Sbjct: 108 LCFFKKALSSLEEVDPHVQMVTESQHIDYHFSGLEDDDGDDEIENNE-NDGSEVHDDGEL 166
Query: 310 SFDYGQIEQEQDV-STSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFSFR--VRTG 366
SF+Y +++QD S++ S EL ++T P+ A+EN + R SFR VR
Sbjct: 167 SFEYRVNDKDQDADSSAGGSSELGNSDITFPQIGGPYTAQENEEGNYRKSHSFRRDVRAV 226
Query: 367 SQSAPLFADNK-PDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTN 425
SQSAPLF +N+ SEKL +MR +L+RKF++Y LPTPV+ S SS ++ + +N
Sbjct: 227 SQSAPLFPENRTTPPSEKLLRMRSTLTRKFNTYALPTPVETTRSPSSTTSPGHKNVGSSN 286
Query: 426 SNEA-TTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLL 484
+A T +W+SSPLE + + S V + VL+ESN N T+RLPPPL DGLL
Sbjct: 287 PTKAITKQIWYSSPLETRGPAKVS---SRSMVALKEQVLRESNKN--TSRLPPPLADGLL 341
Query: 485 SSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKV 544
S +KR +FSGPLTS P+P +P+S S ++SGP+ P+ + P S
Sbjct: 342 FSR-------LGTLKRRSFSGPLTSKPLPNKPLSTTS-HLYSGPIPRNPVSKLPKVSS-- 391
Query: 545 SPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISS 604
SP+ASPT VS+PKISELHELPRPP S+++ +GYS PLV R Q +S P +I++
Sbjct: 392 SPTASPTFVSTPKISELHELPRPPPR--SSTKSSRELGYSAPLVSRSQLLSKP---LITN 446
Query: 605 AASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSR 664
+ASPLPIPP A+ RSFSIP+S R + L S + + + SPPLTP++L +
Sbjct: 447 SASPLPIPP-AITRSFSIPTSNLRASDLDMS----KTSLGTKKLGTPSPPLTPMSLIHPP 501
Query: 665 PSS 667
P +
Sbjct: 502 PQA 504
Score = 146 bits (369), Expect = 2e-33
Identities = 72/115 (62%), Positives = 99/115 (85%), Gaps = 6/115 (5%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MK+SL++LRG+ H H+ D + + L Q +ELAQA++D++DMRDCYD+LL+AAAA
Sbjct: 1 MKTSLRRLRGV-----LHKHESKDRRDLRALVQKDELAQASQDVEDMRDCYDSLLNAAAA 55
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVC 115
TA+SAYEF+ESLR++G+CLLEKTALND +EE+G+VL+MLGK+QF+LQKL+DKY+C
Sbjct: 56 TANSAYEFSESLRELGACLLEKTALND-DEESGRVLIMLGKLQFELQKLVDKYLC 109
>UniRef100_Q67UP0 Hydroxyproline-rich glycoprotein-like [Oryza sativa]
Length = 320
Score = 300 bits (767), Expect = 1e-79
Identities = 159/338 (47%), Positives = 220/338 (65%), Gaps = 38/338 (11%)
Query: 1 MKSSLKKLRGLGLHNHNHNHKHHDSKTILPLGQLEELAQATRDMQDMRDCYDTLLSAAAA 60
MKS L +L+G G H + K + +L+ELA A +D+++MR+CYD +SAAAA
Sbjct: 1 MKSPLLRLKGFGHHQQHRERKSRQPQPQPTPAKLDELADAAQDVEEMRNCYDGFISAAAA 60
Query: 61 TASSAYEFAESLRDMGSCLLEKTALNDHEEETGKVLLMLGKIQFKLQKLIDKYVCISLLI 120
T + YEFAE+L ++GSCLL K LND ++++G+VL+MLGK Q++LQK D+Y
Sbjct: 61 TTNGVYEFAEALEELGSCLLAKPVLNDDDDDSGRVLMMLGKAQYELQKSADRY------- 113
Query: 121 PSLYFIFCFCFNFITNFSVFLFFQFAQRSHIIQTITIPSESLLNELRIVEEMKRQCDEKR 180
R++II TIT PSESLL EL+ +EEMK+QCD KR
Sbjct: 114 ---------------------------RTNIIHTITTPSESLLKELQTLEEMKQQCDMKR 146
Query: 181 DVYEYMATRFRERGRSKGGKTETFSLQQLQTARDEYDEEATLFVFRLKSLKQGQSRSLLT 240
D YE M + ++G S+ KTE+FS +QL + EY E++ LF FRLKSLKQGQ +SLLT
Sbjct: 147 DAYETMRASYSDKGGSRHSKTESFSTEQLDASFLEYQEDSALFTFRLKSLKQGQFQSLLT 206
Query: 241 QAARHHAAQLCFFKKAVKSLETVEPHVKSVTEQQHIDYHFNGLEEEDGDEGDEGYEDDDG 300
QAARHHAAQL FF+K +K LE +EP VK+++E+ HIDY+F+GLE++ D ++GY D
Sbjct: 207 QAARHHAAQLSFFRKGLKCLEALEPRVKAISEKHHIDYNFSGLEDDGSD--NDGYSTYDS 264
Query: 301 YDENDDGELSFDYGQIEQEQDVSTSRNSMELDQVELTL 338
+DDGELSFDY +++QD TSR SM++ + + L
Sbjct: 265 C--SDDGELSFDYEINDRDQDFLTSRGSMDVSSIYIWL 300
>UniRef100_Q9BKV7 Ppg3 [Leishmania major]
Length = 1325
Score = 68.2 bits (165), Expect = 8e-10
Identities = 78/306 (25%), Positives = 120/306 (38%), Gaps = 13/306 (4%)
Query: 367 SQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNS 426
S SAP + + P +S S + SS P+ + S SS S S + S
Sbjct: 690 SSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSAS 749
Query: 427 NEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSS 486
+ + + S+P S+P+ ++ + S++ +A++ P SS
Sbjct: 750 SSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPS------SS 803
Query: 487 NHDYVSAYSKKIKRHAFSGP-LTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVS 545
+ SA S + S P +S+ P+ S S S P + P SSS S
Sbjct: 804 SSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSS 863
Query: 546 PSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSA 605
S++P+ SS S P ++ +S S P SAP+ S+
Sbjct: 864 SSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAP 923
Query: 606 ASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLT----PIALS 661
+S P A S S PSS + SSS+ SS S+ S +S P + P A S
Sbjct: 924 SSSSSSAPS--ASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASS 981
Query: 662 NSRPSS 667
+S PSS
Sbjct: 982 SSAPSS 987
Score = 58.2 bits (139), Expect = 8e-07
Identities = 78/305 (25%), Positives = 120/305 (38%), Gaps = 26/305 (8%)
Query: 367 SQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNS 426
S SAP + + P +S S + SS P+ + SS S S++ PS +
Sbjct: 812 SSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPS---SSSSAPSASSSSAPSSSSSAP 868
Query: 427 NEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSS 486
+ ++++ SS + SS + +A S S+S+++ SS
Sbjct: 869 SASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSS 928
Query: 487 NHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSP 546
+ S+ S + + +S+ P+ S S S P + P SSS S
Sbjct: 929 SAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSS 988
Query: 547 SASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAA 606
S+S SS P ++ PS S S P SAP+ SS++
Sbjct: 989 SSSAPSASS------SSAPSSSSSAPSASS--SSAPSSSSSAPSASSSSAPS----SSSS 1036
Query: 607 SPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLT----PIALSN 662
+P A S S PSS + + SSSA SS S+ S +S P + P A S+
Sbjct: 1037 AP-------SASSSSAPSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSS 1089
Query: 663 SRPSS 667
S PSS
Sbjct: 1090 SAPSS 1094
Score = 54.7 bits (130), Expect = 9e-06
Identities = 85/337 (25%), Positives = 130/337 (38%), Gaps = 19/337 (5%)
Query: 339 PRGSTAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSY 398
P ++ A EN K + S S SA + + P +S S + SS
Sbjct: 647 PELLASDCATENACKPETESSSSAPSASSSSASSSSSSAPSASSSSAPSSSSSAPSASSS 706
Query: 399 VLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRN 458
P+ + SS S S++ PS +S+ A + S+P + + + +
Sbjct: 707 SAPS---SSSSAPSASSSSAPS----SSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSS 759
Query: 459 AQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVS 518
A S S +++++ P SS+ SA S + S S + P S
Sbjct: 760 APSASSSSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSS-SSSAPSASSSSAPSS 818
Query: 519 VDSVQMFSGPLLPTPIPQPPSSSPKVSPSAS---PTIVSSPKISELHELP-RPPTNFPSN 574
S S P+ PS+S +PS+S P+ SS S P ++ PS+
Sbjct: 819 SSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSSSSSAPSASSSSAPSS 878
Query: 575 SRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHS 634
S S P SAP+ S+ +S P A + S SS + +A S
Sbjct: 879 SS--SAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSASSSSAPSSSSSSAPSA-SS 935
Query: 635 SSALESSHISSISEDIASPPLT----PIALSNSRPSS 667
SSA SS S+ S +S P + P A S+S PSS
Sbjct: 936 SSAPSSSSSSAPSASSSSAPSSSSSAPSASSSSAPSS 972
>UniRef100_Q6VBJ4 Epa5p [Candida glabrata]
Length = 1218
Score = 66.6 bits (161), Expect = 2e-09
Identities = 76/324 (23%), Positives = 131/324 (39%), Gaps = 30/324 (9%)
Query: 363 VRTGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKM 422
+ + S S+P + + SS S S SS + SS SS S++P PS
Sbjct: 329 INSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSS--------SSSSSSSSSSSPSPSSS 380
Query: 423 QTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDG 482
++S+ ++++ SSP + SS + ++ S S+S+++++ P P
Sbjct: 381 SSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSRSS 440
Query: 483 LLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSP 542
SS+ S+ S + S + +P + S S S +P P SSS
Sbjct: 441 SSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSS 500
Query: 543 KVSPSASPTIVSSPKISELHELPRPPTNFPSN--------SRLLGLVGYSGPLVPRGQKV 594
S S+SP+I P + P P+N PS+ S + P V + V
Sbjct: 501 SSSSSSSPSI--QPSSKPVDPSPADPSNNPSSVNPSSVNPSSINSSPSIISPSVST-KTV 557
Query: 595 SAPNNLVISSAASPLPIPP-----QAMARSFSIPSSGARVTALHSSSALESSHI----SS 645
+ N I+ + P P + + SFS+P T + S+ E+ + S+
Sbjct: 558 TLSNGSTITVTVTVTPTSPNGGNSNSDSDSFSLPP--PYTTTVSKSTTTETDIVSFFPST 615
Query: 646 ISEDIASPPLTPIALSNSRPSSDG 669
S+ T I L+ S+P ++G
Sbjct: 616 DSDGHTRTGTTTITLTGSKPGNNG 639
Score = 55.5 bits (132), Expect = 5e-06
Identities = 65/275 (23%), Positives = 114/275 (40%), Gaps = 16/275 (5%)
Query: 401 PTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQ 460
P P ++ SS S++P PS ++S+ ++++ SS + SS ++
Sbjct: 321 PLPTPSQDINSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSS 380
Query: 461 SVLKESNSNTATTRLPPP---LVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPV 517
S S+S+++++ P P SS+ S+ S + S +S+P P+R
Sbjct: 381 SSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSRSS 440
Query: 518 SVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRL 577
S S S + SSS SPS+S + SS S P+ S+S
Sbjct: 441 SSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSS 500
Query: 578 LGLVGYSGPLVPRGQKV----SAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALH 633
S + P + V + P+N +P + P ++ S SI SS + ++
Sbjct: 501 SSSSSSSPSIQPSSKPVDPSPADPSN-------NPSSVNPSSVNPS-SINSSPSIISPSV 552
Query: 634 SSSALESSHISSISEDIASPPLTPIALSNSRPSSD 668
S+ + S+ S+I+ + P +P NS SD
Sbjct: 553 STKTVTLSNGSTITVTVTVTPTSPNG-GNSNSDSD 586
>UniRef100_P08640 Glucoamylase S1/S2 precursor [Saccharomyces cerevisiae]
Length = 1367
Score = 65.5 bits (158), Expect = 5e-09
Identities = 81/308 (26%), Positives = 115/308 (37%), Gaps = 34/308 (11%)
Query: 365 TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNN-PKPSKMQ 423
T S SAP+ P SS P S S P P + S+ S S P PS
Sbjct: 547 TESSSAPV---PTPSSSTTESSSTPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSST 603
Query: 424 TNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGL 483
T S+ A SS E SS T ++ V S+S T ++ P P
Sbjct: 604 TESSSAPAPTPSSSTTESSSAPVT----SSTTESSSAPVPTPSSSTTESSSAPVPTPSSS 659
Query: 484 LSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPK 543
+ + + S P+TS+ + V S S P+P P SS+ +
Sbjct: 660 TTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSS---APVPTPSSSTTE 716
Query: 544 VSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAP-NNLVI 602
S + PT SS S +P P ++ +S SAP +
Sbjct: 717 SSSAPVPTPSSSTTESSSAPVPTPSSSTTESS-------------------SAPVTSSTT 757
Query: 603 SSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSN 662
S+++P+P P + S S P + SSSA + SS +E +P TP + SN
Sbjct: 758 ESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSVAPVPTPSSSSN 817
Query: 663 ---SRPSS 667
S PSS
Sbjct: 818 ITSSAPSS 825
Score = 65.1 bits (157), Expect = 6e-09
Identities = 90/359 (25%), Positives = 139/359 (38%), Gaps = 38/359 (10%)
Query: 325 SRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPDSSEKL 384
S ++ E P ST E++ + S V T S S + SS
Sbjct: 516 SSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSTPVTSSTTE 575
Query: 385 RQMRP-----SLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPL 439
P S + + SS +PTP + ++ SS + P PS T S+ A +S
Sbjct: 576 SSSAPVPTPSSSTTESSSAPVPTP-SSSTTESSSAPAPTPSSSTTESSSAPV----TSST 630
Query: 440 EQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIK 499
+ + SS T ++ V S+S T ++ P P SS + SA
Sbjct: 631 TESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPS---SSTTESSSAPVTSST 687
Query: 500 RHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSS------PKVSPSASPTIV 553
+ S P+TS+ + V + + P+P P SS+ P +PS+S T
Sbjct: 688 TESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTES 747
Query: 554 SS-PKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIP 612
SS P S E P PS+S S P V P++ S+++P+P P
Sbjct: 748 SSAPVTSSTTESSSAPVPTPSSSTT---ESSSAP-------VPTPSSSTTESSSAPVPTP 797
Query: 613 PQAMARS----FSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
+ S PSS + +T SSA S+ SS +E + P TP + + S+
Sbjct: 798 SSSTTESSVAPVPTPSSSSNIT----SSAPSSTPFSSSTESSSVPVPTPSSSTTESSSA 852
Score = 58.2 bits (139), Expect = 8e-07
Identities = 68/291 (23%), Positives = 111/291 (37%), Gaps = 10/291 (3%)
Query: 365 TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQT 424
T S SAP+ + SS + S + SS +PTP + ++ SS + P PS T
Sbjct: 619 TESSSAPVTSSTTESSSAPVPTPSSSTTES-SSAPVPTP-SSSTTESSSAPVPTPSSSTT 676
Query: 425 NSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLL 484
S+ A SS E SS T ++ V S+S T ++ P P
Sbjct: 677 ESSSAPVT---SSTTESSSAPVT----SSTTESSSAPVPTPSSSTTESSSAPVPTPSSST 729
Query: 485 SSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKV 544
+ + + S P+TS+ + V + + P+P P SS+ +
Sbjct: 730 TESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTES 789
Query: 545 SPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISS 604
S + PT SS S + +P P ++ S +S V P++ S
Sbjct: 790 SSAPVPTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTES 849
Query: 605 AASPL-PIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPP 654
+++P+ ++ PSS + +T+ SS SS S S P
Sbjct: 850 SSAPVSSSTTESSVAPVPTPSSSSNITSSAPSSIPFSSTTESFSTGTTVTP 900
Score = 57.4 bits (137), Expect = 1e-06
Identities = 74/313 (23%), Positives = 130/313 (40%), Gaps = 25/313 (7%)
Query: 365 TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQT 424
T + P D P + +K + + ++ +PTP + ++ SS + P PS T
Sbjct: 281 TKEKPTPPHHDTTPCTKKKTTTSKTCTKK--TTTPVPTP-SSSTTESSSAPVPTPSSSTT 337
Query: 425 NSNEA----TTNLWHSSPLEQKKHETIRDE---FSSPTVRNAQSVLKESNSNTATTRLPP 477
S+ A +T S+P+ T +S T ++ + + S + +++ +P
Sbjct: 338 ESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPT 397
Query: 478 PLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTR--PVSVDSVQMFSGPLLPTPIP 535
P SS+ S+ ++ + S P+TS+ + PV+ + + S P+ T
Sbjct: 398 PSSSTTESSSAPVTSSTTE-----SSSAPVTSSTTESSSAPVTSSTTESSSAPV--TSST 450
Query: 536 QPPSSSPKVSPSASPTIVSS-PKISELHELPRPPTNFPSNSRLLGLVG--YSGPLVPRGQ 592
SS+P +PS+S T SS P S E P PS+S S
Sbjct: 451 TESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSA 510
Query: 593 KVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIAS 652
V P++ S+++P P P + S S P + + SSSA + SS +E ++
Sbjct: 511 PVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTS---STTESSSAPVPTPSSSTTESSST 567
Query: 653 PPLTPIALSNSRP 665
P + S+S P
Sbjct: 568 PVTSSTTESSSAP 580
Score = 47.4 bits (111), Expect = 0.001
Identities = 89/415 (21%), Positives = 159/415 (37%), Gaps = 53/415 (12%)
Query: 266 HVKSVTEQQHIDYHFNGLEEEDGDEGDEGYEDDDGYDEND---DGELSFDYG-QIEQEQD 321
HVK +++ID + + G G +G GY+EN D F ++ QD
Sbjct: 81 HVKG---KENIDLKYLWSLKIIGVTGPKGTVQLYGYNENTYLIDNPTDFTATFEVYATQD 137
Query: 322 VSTSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPDSS 381
V++ + M Q++ +GS A+ A Q SF + TG + DN+ S
Sbjct: 138 VNSCQVWMPNFQIQFEYLQGSAAQYASS----WQWGTTSFDLSTGCNNY----DNQGHSQ 189
Query: 382 EKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQ 441
++ + + S+ S + S+ T ++ + + +S +
Sbjct: 190 TDFPGFYWNIDCDNNCGGTKSSTTTSSTSESSTTTSSTSESSTTTSSTSESSTTTSSTSE 249
Query: 442 KKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRH 501
+ ++PT + + T+ T+ P +HD KK
Sbjct: 250 SSTSSSTTAPATPTTTSCTKEKPTPPTTTSCTKEKPT------PPHHDTTPCTKKKTTTS 303
Query: 502 AFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISEL 561
T+ P+PT S P+P P SS+ + S+S + SS S
Sbjct: 304 KTCTKKTTTPVPTPSSSTTESSS-------APVPTPSSSTTE---SSSAPVTSSTTESSS 353
Query: 562 HELPRPPTNFPSNSRLLGLVGYSGPLVPRG-QKVSAP-NNLVISSAASPLPIPPQAMARS 619
+P P ++ +S S P+ + SAP + S+++P+P P + S
Sbjct: 354 APVPTPSSSTTESS--------SAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTES 405
Query: 620 FSIP-------SSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
S P SS A VT+ S++ S+ ++S + + +S P+T S + SS
Sbjct: 406 SSAPVTSSTTESSSAPVTS--STTESSSAPVTSSTTESSSAPVTS---STTESSS 455
>UniRef100_Q8WML4 MUC1 protein precursor [Bos taurus]
Length = 580
Score = 60.1 bits (144), Expect = 2e-07
Identities = 86/335 (25%), Positives = 125/335 (36%), Gaps = 62/335 (18%)
Query: 387 MRPSLSRKFSSYVLPTPVDAKSSI----SSGSNNPK---------------PSKMQTNSN 427
M P + F S +L PV +++ +S S NP+ P+K +T+ +
Sbjct: 1 MTPDIQAPFLSLLLLFPVLTVANVPTLTTSDSINPRRTTPVSTTQSSPTSSPTK-ETSWS 59
Query: 428 EATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSN 487
TT L SSP D S+PT A S + A+T P S
Sbjct: 60 TTTTLLTASSPAPSPAASPGHDGASTPTSSPAPSPAASPGHDGASTPTSSPAPSPAASPG 119
Query: 488 HDYVSAYSKK-------IKRHAFSGPLTSNPMPTRPVSV--DSVQMFSGPLLPTPIPQP- 537
HD S + H + TS+P P+ S + +G P+P P
Sbjct: 120 HDGASTPTSSPAPSPAASPGHDGASTPTSSPAPSPAASPGHNGTSSPTGSPAPSPAASPG 179
Query: 538 ------PSSSPKVSPSASP----------TIVSSPKISELHELPRPPTNFPSNSRLLGLV 581
P+SSP SP+ASP + SP S H+ PT+ P+ S
Sbjct: 180 HDGASTPTSSPAPSPAASPGHNGTSSPTGSPAPSPAASPGHDGASTPTSSPAPSPAAS-P 238
Query: 582 GYSGPLVPRGQKVSAPN---------NLVISSAASPLPIPPQAMARSFSIPSSGARVTAL 632
G++G P G +P +L S A SP P Q A S + + + T
Sbjct: 239 GHNGTSSPTGSPAPSPTASPGHDSAPSLTSSPAPSPTASPGQHGASSPTSSDTSSMTTRS 298
Query: 633 HSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
SSS + S+H + S +P +S PSS
Sbjct: 299 MSSSMVTSAHKGTSSRATMTP------VSKGTPSS 327
>UniRef100_UPI0000337A4E UPI0000337A4E UniRef100 entry
Length = 902
Score = 57.0 bits (136), Expect = 2e-06
Identities = 46/157 (29%), Positives = 70/157 (44%), Gaps = 6/157 (3%)
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKV--SPSASPTIVSSPKISELHE 563
P++S P + + S P LP + PP+SSP SP+ +P + P ++
Sbjct: 362 PVSSGPHTPEDRNPPASSTPSAPTLPKELASPPASSPPPAESPTTAPKTRTPPALTPADG 421
Query: 564 L---PRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSF 620
P PPT+ + + + P RG+ V++P + SS +P P PP A
Sbjct: 422 PAPDPLPPTHVFQEGQAK-VSDTNQPKEGRGENVTSPTDTKQSSRTTPPPPPPPAYRTMV 480
Query: 621 SIPSSGARVTALHSSSALESSHISSISEDIASPPLTP 657
S P GA T+ SSS + + SS+S PP P
Sbjct: 481 SSPIPGAGGTSSGSSSPVRPATPSSVSPTPPPPPPRP 517
Score = 38.9 bits (89), Expect = 0.50
Identities = 83/358 (23%), Positives = 140/358 (38%), Gaps = 47/358 (13%)
Query: 282 GLEEEDGDEGDEGYED-DDGYDENDDGELSFDYGQIEQEQDVSTSRNSMELDQVELTLPR 340
G+ +++ D G D D G + +G + YG+I+ ++ NS E+D ++
Sbjct: 13 GIRKKEKDNDSTGSPDRDGGSQKKSNGAPNGFYGEIDWDR-----YNSPEVDDEGYSIRP 67
Query: 341 GSTAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPDSSEKLR-QMRPSLSRKFSSYV 399
+E + L KL NL ++ L + D +K + +++P L+ + +
Sbjct: 68 DDDSEG--DILTKLT-NLSHTKL-------DLLMYEEEDHRKKFKIKIKPLLADRVVA-- 115
Query: 400 LPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNA 459
PT + K+SI + S +P P N T + ++K R P V
Sbjct: 116 APTVDELKASIGNISLSPSPVVSPPNLTALTPPACLNVLPDEKIARPRRTVAPVPPVAPT 175
Query: 460 QSV--LKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTS-NPMPTRP 516
L+ +S +T PPL + V + S A++ PL+ + TRP
Sbjct: 176 APAPALQPPSSEDSTALFGPPLETAFGEQKTEAVLSESD-----AWAAPLSEPDSSMTRP 230
Query: 517 VSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSS---PKISELHELPRPPTNFPS 573
TP P PP + P SPS++ +S P I++L + P P+
Sbjct: 231 FPAG-----------TPPPLPPKNVPTTSPSSTDDAETSSRKPSIADLDNIFGPEQAPPA 279
Query: 574 -NSRLLGLVGYSGPLVPRGQKVSAPNNLVIS-----SAASPLPIPPQAMARSFSIPSS 625
+ V +S R QK L S S A PLP P + + +P+S
Sbjct: 280 GEDQGDAWVCFSEEAADRLQKQEPAPPLPTSPPPPESPAQPLPTSPLSPEQPAPLPTS 337
>UniRef100_Q7Q1W8 ENSANGP00000020885 [Anopheles gambiae str. PEST]
Length = 1521
Score = 56.6 bits (135), Expect = 2e-06
Identities = 71/345 (20%), Positives = 134/345 (38%), Gaps = 9/345 (2%)
Query: 324 TSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPDSSEK 383
+S +S E T P +T + E+ + RT S +A + SS +
Sbjct: 524 SSSSSTESTTASSTTP--ATTSSTTESTTVSSTTSETTSSRTESTTAITMTSDSTSSSTE 581
Query: 384 LRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKK 443
+ S ++ + PT + +S+SS + PS + S T + S
Sbjct: 582 STTASSTTSTSMTTSISPTIPSSPTSLSSPTTRTSPSAPTSPSTPETISSTTESTTASST 641
Query: 444 HETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAF 503
T SPT+ ++ + S + T + P SS + ++ + +
Sbjct: 642 ISTSMTTSISPTIPSSPTSPSSSTTRTTPSAPTSPSTPETTSSTTESTASSTTST---SM 698
Query: 504 SGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPS-SSPKVSPSASPTIVSSPKISELH 562
+ P TS +P+ P S S + P PT P + SS S +AS TI +S S
Sbjct: 699 TTP-TSPTIPSSPTSPSSPTTRTSPSAPTSPTTPETISSTTESTTASSTISTSMTTSISP 757
Query: 563 ELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSI 622
+P PT+ S++ + P P+ + + ++ + ++ S +
Sbjct: 758 TIPSSPTSPSSSTTRTSPSAPTSPSTPQTTSSTTESTTAVTMTSDSTSSSTESTTASSTT 817
Query: 623 PSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
P++ + T + S+ ISS +E ++ P + + S + P+S
Sbjct: 818 PATTSSTTLTTTVSSTTPETISSTTE--STTPSSTTSTSMTTPTS 860
Score = 50.4 bits (119), Expect = 2e-04
Identities = 65/280 (23%), Positives = 109/280 (38%), Gaps = 18/280 (6%)
Query: 396 SSYVLPTPVDAKSSISSGS-NNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSP 454
SS + T + +S S + +P T+S+ TT ++P ET S
Sbjct: 165 SSSIESTTASSMTSTSMTTPTSPMTPTSPTSSSSPTTRTSPTAPTSPSTPETTSSTTEST 224
Query: 455 TVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPT 514
TV S E+ S+ + + SS+ + +A S + F+ TS +PT
Sbjct: 225 TV---SSTTPETTSSITESTTAVTMTSDSSSSSTESTTASSTT---YYFNDTPTSPTIPT 278
Query: 515 RPVSVDSVQMFSGPLLPT-PIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPS 573
P S S + + P +PT P SSS S +AS T +S S +P PT+ S
Sbjct: 279 SPTSSSSPTIRTSPSVPTSPSTPQTSSSTTESTTASSTTSTSMTTSISPTIPSSPTSLSS 338
Query: 574 NSRLLGLVGYSGPLVPRGQKVSAPNNLVI------SSAASPLPIPPQAMARSFSIPSSGA 627
+ + P P+ + + + SS+++ P + S + P+S
Sbjct: 339 PTTRTSPSAPTSPSTPQTTISTTESTTAVTMNSDSSSSSTESTTPSSTTSTSMTTPTS-- 396
Query: 628 RVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
+ +S SS S + P TP S++ S+
Sbjct: 397 --PTIPTSPTSPSSPTIRTSPSATTSPSTPKTSSSTTEST 434
Score = 47.4 bits (111), Expect = 0.001
Identities = 58/283 (20%), Positives = 110/283 (38%), Gaps = 11/283 (3%)
Query: 389 PSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIR 448
P++ +S PT + S+ + + P+ S T S A++ S+ + TI
Sbjct: 274 PTIPTSPTSSSSPT-IRTSPSVPTSPSTPQTSSSTTESTTASSTT--STSMTTSISPTIP 330
Query: 449 DE---FSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSG 505
SSPT R + S ++ +T T + ++ N D S+ ++ + +
Sbjct: 331 SSPTSLSSPTTRTSPSA--PTSPSTPQTTISTTESTTAVTMNSDSSSSSTESTTPSSTTS 388
Query: 506 PLTSNPMPTRPVSVDSVQMFSGPLLPT-PIPQPPSSSPKVSPSASPTIVSSPKISELHEL 564
TS PT P S S P + T P S+PK S S + + +S S +
Sbjct: 389 --TSMTTPTSPTIPTSPTSPSSPTIRTSPSATTSPSTPKTSSSTTESTTASSTTSISPTI 446
Query: 565 PRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPS 624
P PT+ S + + P P + + ++ + ++ S + P
Sbjct: 447 PSSPTSLSSPTTRTSPSAPTSPSTPETTISTTESTTAVTMTSDSTSSSTESTTVSSTTPE 506
Query: 625 SGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
+ + +T ++ + S SS +E + TP S++ S+
Sbjct: 507 TTSSITESTTAVTMTSDSSSSSTESTTASSTTPATTSSTTEST 549
Score = 43.1 bits (100), Expect = 0.026
Identities = 61/270 (22%), Positives = 110/270 (40%), Gaps = 24/270 (8%)
Query: 406 AKSSISSG---SNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTV--RNAQ 460
A S+IS+ S +P T+ + +TT S+P +T S T +
Sbjct: 743 ASSTISTSMTTSISPTIPSSPTSPSSSTTRTSPSAPTSPSTPQTTSSTTESTTAVTMTSD 802
Query: 461 SVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVD 520
S + S TA++ P L++ VS+ + + ++S T P S
Sbjct: 803 STSSSTESTTASSTTPATTSSTTLTTT---VSSTTPET--------ISSTTESTTPSSTT 851
Query: 521 SVQMFS--GPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLL 578
S M + P +PT P S + + SPSA PT S+P+ + ++ S +
Sbjct: 852 STSMTTPTSPTIPTSPTSPSSPTIRTSPSA-PTSPSTPETTSSTTESTTVSSTTSETTSS 910
Query: 579 GLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSAL 638
+ + S+ + ++A+S P + S ++PS+ T+ + S
Sbjct: 911 TTESTTAVTITSDSSSSSTES---TTASSTTPATTSSTTESTTVPSTTPETTSSTTESTT 967
Query: 639 ESSHISSISEDIASPPLTPIA-LSNSRPSS 667
SS +S S + P+TP + S+S P++
Sbjct: 968 ASS-TTSTSMTTPTSPMTPTSPASSSSPTT 996
Score = 42.7 bits (99), Expect = 0.034
Identities = 67/307 (21%), Positives = 109/307 (34%), Gaps = 23/307 (7%)
Query: 365 TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQT 424
T + +AP + + SS L + + SS + T V + + S+ S + T
Sbjct: 30 TTTTNAPSSSSSSSASSSTLTTLSSTTPATTSSTTVSTTVSSTAPESTSSITESTTPSST 89
Query: 425 NSNEATTNLWHSSPLEQKKHE--TIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDG 482
S TT + P TIR S+PT + + +T + P
Sbjct: 90 TSTSMTTPTSPTIPTSPTSPSPPTIRTSPSAPTSPSTPETTSSTTESTTVSSTTPETTSS 149
Query: 483 LLSSNHDYV----SAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPT--PIPQ 536
+ S S+ S A S TS PT P++ S S P T P
Sbjct: 150 ITESTTAVTMTSDSSSSSIESTTASSMTSTSMTTPTSPMTPTSPTSSSSPTTRTSPTAPT 209
Query: 537 PPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSA 596
PS+ S + T VSS P ++ ++ + + S S+
Sbjct: 210 SPSTPETTSSTTESTTVSS-------TTPETTSSITESTTAVTMTSDSSSSSTESTTASS 262
Query: 597 PNNLVISSAASP-LPIPPQA-----MARSFSIPSSGARVTALHSSSALESSHISSISEDI 650
+ SP +P P + + S S+P+S + T SSS ES+ SS +
Sbjct: 263 TTYYFNDTPTSPTIPTSPTSSSSPTIRTSPSVPTSPS--TPQTSSSTTESTTASSTTSTS 320
Query: 651 ASPPLTP 657
+ ++P
Sbjct: 321 MTTSISP 327
Score = 41.2 bits (95), Expect = 0.10
Identities = 62/299 (20%), Positives = 111/299 (36%), Gaps = 16/299 (5%)
Query: 365 TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQT 424
T S +A + SS + + + SS T +S SS S+ + T
Sbjct: 478 TESTTAVTMTSDSTSSSTESTTVSSTTPETTSSITESTTAVTMTSDSSSSSTESTTASST 537
Query: 425 NSNEATTNLWHSSPLEQKKHETI--RDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDG 482
T++ S+ + ET R E ++ + S + S TA++ +
Sbjct: 538 TP-ATTSSTTESTTVSSTTSETTSSRTESTTAITMTSDSTSSSTESTTASSTTSTSMTTS 596
Query: 483 L---LSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPS 539
+ + S+ +S+ + + A + P T + + S + S + + P PS
Sbjct: 597 ISPTIPSSPTSLSSPTTRTSPSAPTSPSTPETISSTTESTTASSTISTSMTTSISPTIPS 656
Query: 540 SSPKVSPSASPTIVSSPKISELHELPRPPT-NFPSNSRLLGLVGYSGPLVPRGQKVSAPN 598
S S S + T S+P E T + S++ + + P +P +
Sbjct: 657 SPTSPSSSTTRTTPSAPTSPSTPETTSSTTESTASSTTSTSMTTPTSPTIPSSPTSPSSP 716
Query: 599 NLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSALESSHISSISEDIASPPLTP 657
S +A P P+ +I S+ TA SS + +S +SIS I S P +P
Sbjct: 717 TTRTSPSAPTSPTTPE------TISSTTESTTA---SSTISTSMTTSISPTIPSSPTSP 766
Score = 40.8 bits (94), Expect = 0.13
Identities = 45/213 (21%), Positives = 83/213 (38%), Gaps = 28/213 (13%)
Query: 455 TVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPT 514
T+ + + + +NS T+TT P S++ ++ S S +++ T
Sbjct: 13 TLSSTSTTMMSTNSITSTTTTNAPSSSSSSSASSSTLTTLSSTTPATTSSTTVSTTVSST 72
Query: 515 RPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSN 574
P S S+ S++P + S S T +SP I P PPT S
Sbjct: 73 APESTSSI--------------TESTTPSSTTSTSMTTPTSPTIPTSPTSPSPPTIRTSP 118
Query: 575 SRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHS 634
S P P S P ++ ++ + S + ++ +T+ S
Sbjct: 119 ---------SAPTSP-----STPETTSSTTESTTVSSTTPETTSSITESTTAVTMTSDSS 164
Query: 635 SSALESSHISSISEDIASPPLTPIALSNSRPSS 667
SS++ES+ SS++ + P +P+ ++ SS
Sbjct: 165 SSSIESTTASSMTSTSMTTPTSPMTPTSPTSSS 197
Score = 37.4 bits (85), Expect = 1.4
Identities = 58/278 (20%), Positives = 105/278 (36%), Gaps = 16/278 (5%)
Query: 406 AKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKE 465
A S+ S+ P T+ +++ +SP T + S+ S E
Sbjct: 968 ASSTTSTSMTTPTSPMTPTSPASSSSPTTRTSPTALTSPTTPKTTSSTTESTTVSSTTPE 1027
Query: 466 SNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFS--GPLTSNPMPTRPVSVDSVQ 523
+ S+T + + SS+ + +A S + S TS+ T PV+ +
Sbjct: 1028 TTSSTTESTTATTMTSDSTSSSTESTTASSSSTVSISISTSSSTTSSSSVTTPVTPRTTS 1087
Query: 524 MFSGPLL------PTPIPQPPSSSPKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRL 577
F+ P + PT +P + + S P SS S P T +
Sbjct: 1088 QFTLPTIRTSPSNPTTLPTSSTIESTTASSTIPATTSSTTESTTVSSTTPETTSSTTEST 1147
Query: 578 LGLVGYSGPLVPRGQKVSA-PNNLVI----SSAASPLPIPPQAMARSFSIPSS-GARVTA 631
+ S + + P VI ++ +S + + SF+IP+S T+
Sbjct: 1148 TAVTMTSDSTSSSTESTTVLPTTSVITTESTTTSSTTSDSLTSSSESFTIPTSLSTEATS 1207
Query: 632 LHSSSALE-SSHISSISEDIAS-PPLTPIALSNSRPSS 667
+SS+ E S+ +S+ SE +S + +++S S SS
Sbjct: 1208 SSASSSTEFSTSLSTTSETTSSTTSSSTVSISISTSSS 1245
>UniRef100_Q6FSJ1 Similarities with sp|P47179 Saccharomyces cerevisiae YJR151c
[Candida glabrata]
Length = 577
Score = 56.6 bits (135), Expect = 2e-06
Identities = 52/168 (30%), Positives = 75/168 (43%), Gaps = 6/168 (3%)
Query: 500 RHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTIVSSPKIS 559
R A +G +T+ P P+ S S P+P P P SSP SPS SP+ SP S
Sbjct: 109 RLADAGIVTAWPSPSPSPSPSPSPSPSPSPSPSPSPSPSPSSPSPSPSPSPSPSPSPSPS 168
Query: 560 ELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARS 619
P+ PS S P P S+ ++ + S++S P + + S
Sbjct: 169 PSPSPSPSPSPSPSPSPSPSPSPSPSPKSPSPSPSSSSSSSSMPSSSSSSSSMPSSSSSS 228
Query: 620 FSIPSSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSS 667
S+PSS + SSS++ SS SS S +S +TP ++ PSS
Sbjct: 229 SSMPSSSS------SSSSMPSSSSSSSSMPSSSSSMTPSQKASIIPSS 270
Score = 50.4 bits (119), Expect = 2e-04
Identities = 55/180 (30%), Positives = 82/180 (45%), Gaps = 38/180 (21%)
Query: 510 NPMPTRPVSVDSVQMFSGPLLPTPIPQP-PSSSPKVSPSASPTIVSSPKISELHELPRPP 568
+P P+ P S P P+P P P PS SP SPS SP+ SP S P+ P
Sbjct: 145 SPSPSSPSPSPSPSPSPSPS-PSPSPSPSPSPSPSPSPSPSPSPSPSPSPS-----PKSP 198
Query: 569 TNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIP---------PQAMARS 619
+ PS+S S +P S+ ++ + SS++S +P P + + S
Sbjct: 199 SPSPSSSS-------SSSSMPSS---SSSSSSMPSSSSSSSSMPSSSSSSSSMPSSSSSS 248
Query: 620 FSIPSSGARVTALHSSSALESS----------HISSISEDIASP--PLTPIALSNSRPSS 667
S+PSS + +T +S + SS SSIS ASP P + ++ S++ PSS
Sbjct: 249 SSMPSSSSSMTPSQKASIIPSSAAPSSSSSIVTTSSISSADASPVLPSSVVSSSSTEPSS 308
>UniRef100_UPI000035F9F4 UPI000035F9F4 UniRef100 entry
Length = 437
Score = 55.1 bits (131), Expect = 7e-06
Identities = 68/319 (21%), Positives = 124/319 (38%), Gaps = 51/319 (15%)
Query: 365 TGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSI--SSGSNNPKPSKM 422
T S++ + P S L + P+ + +SS V + S + S+ P P K
Sbjct: 41 TKSKTLTNRSHTSPMPSPPLASISPTAGKYYSSGETDKNVLSASKVHPSTAQTMPTPHKP 100
Query: 423 QTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDG 482
+T T+ W P + K + E S+P+ AQ+++ ++TA +P D
Sbjct: 101 ETPGGPTTSESW---PSAEAKEADAKTEISAPSPNPAQAIITSDQTSTA---VPVLSQDA 154
Query: 483 LLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQ--PPSS 540
L + ++ ++A PT PV S ++P+ +P P +
Sbjct: 155 LQTDDNSSIAA------------------KPTIPVPATDQSHSSTSVVPSNVPALLPATP 196
Query: 541 SPKVSPSASPTIVSSPKISELHEL------------PRPPTNF---PSNSR-------LL 578
+P VS +SP + +P +S+ + P PPT PS ++ +L
Sbjct: 197 TPGVSKPSSPPVEQAPIVSQTVKTGPESIQTVTDATPAPPTTASVSPSETQTSITVPDIL 256
Query: 579 GLVGYSGPLVPR-GQKVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSSSA 637
V + +P+ + SAP + + + P A++ S+P + + T +A
Sbjct: 257 PSVTTTDASLPQPSARTSAPGSASLEQVSISAPAAVPQTAKASSLPPASTQATLTCGPAA 316
Query: 638 LESSHISSISEDIASPPLT 656
+S S S S T
Sbjct: 317 AGASQPPSTSRPTVSDSST 335
>UniRef100_UPI000030431E UPI000030431E UniRef100 entry
Length = 289
Score = 55.1 bits (131), Expect = 7e-06
Identities = 77/288 (26%), Positives = 122/288 (41%), Gaps = 47/288 (16%)
Query: 378 PDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSS 437
P S+++ Q R +L SS TP + SS+S+ S P PS S ++ ++ S
Sbjct: 37 PSQSDRMLQSRVALE---SSSTSRTP-SSSSSVSALSPTPSPSL----SLDSVESVAFQS 88
Query: 438 PLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKK 497
L SPT + SV+ E++ A+ P P S + D +++ K
Sbjct: 89 EL-------------SPTPSPSVSVVSEASLGNASPLSPTPSP----SVSADSLASSGK- 130
Query: 498 IKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPK---VSPSASPTIVS 554
+ PL+ P P+ VSV SV S +PIP P +S+ VS ++ ++
Sbjct: 131 ------ASPLSPTPSPS--VSVSSVASESNSSALSPIPSPSASADSLASVSKASVSSLTP 182
Query: 555 SPKISELHELP----RPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLP 610
SP S P T PS+S V P+ VS+ ++ S+ S +P
Sbjct: 183 SPSQSTSRSQPASSAESSTPSPSSS-----VSALSPVPSPSVSVSSESSRGAQSSESAVP 237
Query: 611 IPPQAMARSFSIPSSGARVTALHSS-SALESSHISSISEDIASPPLTP 657
P +++ +PS V++ SS A SS I S S ++ P P
Sbjct: 238 SPSSSVSALSPVPSPSVSVSSDESSGKASLSSPIPSPSSSVSRPSCRP 285
Score = 46.6 bits (109), Expect = 0.002
Identities = 57/225 (25%), Positives = 92/225 (40%), Gaps = 12/225 (5%)
Query: 453 SPTVRNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPM 512
S + R QS + +S+T+ T V L + +S S ++ AF L+ P
Sbjct: 38 SQSDRMLQSRVALESSSTSRTPSSSSSVSALSPTPSPSLSLDS--VESVAFQSELSPTPS 95
Query: 513 PTRPVSVDSVQMFSGPLLPTPIPQPPSSS-------PKVSPSASPTIVSSPKISELHELP 565
P+ V ++ + PL PTP P + S +SP+ SP++ S SE +
Sbjct: 96 PSVSVVSEASLGNASPLSPTPSPSVSADSLASSGKASPLSPTPSPSVSVSSVASESNSSA 155
Query: 566 RPPTNFPSNSR--LLGLVGYSGPLVPRGQKVSAPNNLVISSAASPLPIPPQAMARSFSIP 623
P PS S L + S + S + SSA S P P +++ +P
Sbjct: 156 LSPIPSPSASADSLASVSKASVSSLTPSPSQSTSRSQPASSAESSTPSPSSSVSALSPVP 215
Query: 624 SSGARVTALHSSSALESSHISSISEDIASPPLTPIALSNSRPSSD 668
S V++ SS +SS + S + L+P+ + SSD
Sbjct: 216 SPSVSVSS-ESSRGAQSSESAVPSPSSSVSALSPVPSPSVSVSSD 259
Score = 44.7 bits (104), Expect = 0.009
Identities = 67/261 (25%), Positives = 107/261 (40%), Gaps = 47/261 (18%)
Query: 378 PDSSEKLRQMRPSLSRK----------FSSYVLPTP---VDAKSSISSGSNNP-KPSKMQ 423
P SS + + P+ S F S + PTP V S S G+ +P P+
Sbjct: 59 PSSSSSVSALSPTPSPSLSLDSVESVAFQSELSPTPSPSVSVVSEASLGNASPLSPTPSP 118
Query: 424 TNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDGL 483
+ S ++ + +SPL SP+V + SV ESNS+ A + +P P
Sbjct: 119 SVSADSLASSGKASPLSPTP---------SPSV-SVSSVASESNSS-ALSPIPSP----- 162
Query: 484 LSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRPVSVDSVQMFSGPLLPTPIPQPPSSSPK 543
S++ D +++ SK A LT +P + S + S P P SS
Sbjct: 163 -SASADSLASVSK-----ASVSSLTPSPSQSTSRSQPASSAESST------PSPSSSVSA 210
Query: 544 VSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQKVSAPNNLVIS 603
+SP SP++ S + S + PS+S V P+ VS+ + +
Sbjct: 211 LSPVPSPSVSVSSESSRGAQSSESAVPSPSSS-----VSALSPVPSPSVSVSSDESSGKA 265
Query: 604 SAASPLPIPPQAMARSFSIPS 624
S +SP+P P +++R PS
Sbjct: 266 SLSSPIPSPSSSVSRPSCRPS 286
Score = 41.2 bits (95), Expect = 0.10
Identities = 42/156 (26%), Positives = 68/156 (42%), Gaps = 8/156 (5%)
Query: 401 PTPVDAKSSISSGSNN----PKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTV 456
P+P + SS++S SN+ P PS + + A+ + S L ++ S P
Sbjct: 138 PSPSVSVSSVASESNSSALSPIPSPSASADSLASVSKASVSSLTPSPSQSTSR--SQPAS 195
Query: 457 RNAQSVLKESNSNTATTRLPPPLVDGLLSSNHDYVSAYSKKIKRHAFSGPLTSNPMPTRP 516
S S+S +A + +P P V +SS + S+ S +P+P+
Sbjct: 196 SAESSTPSPSSSVSALSPVPSPSVS--VSSESSRGAQSSESAVPSPSSSVSALSPVPSPS 253
Query: 517 VSVDSVQMFSGPLLPTPIPQPPSSSPKVSPSASPTI 552
VSV S + L +PIP P SS + S SP++
Sbjct: 254 VSVSSDESSGKASLSSPIPSPSSSVSRPSCRPSPSV 289
>UniRef100_UPI0000364562 UPI0000364562 UniRef100 entry
Length = 1027
Score = 54.3 bits (129), Expect = 1e-05
Identities = 93/367 (25%), Positives = 135/367 (36%), Gaps = 68/367 (18%)
Query: 308 ELSFDYGQIEQEQDVSTSRNSMELDQVELTLPRGSTAEAAKENLDKLQRNLFSFRVRTGS 367
EL Y +++ ++ +NS Q EL R + + +L+ L+ +
Sbjct: 214 ELRKLYQELQIRNKLNHEQNSKLQQQKELLNKRNMEVTLMDKRISELRERLYKKKAEARQ 273
Query: 368 -QSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPTPVDAKSSISSGSN-NPKPSKMQTN 425
++ PL N P S Q PS + ++ V A G N P+P K QT
Sbjct: 274 KENLPLNRANGPPSP----QPAPSTLGRVAAVGPYIQVPAPGQQEGGYNIPPEPLKPQTL 329
Query: 426 SNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSVLKESNSNTATTRLPPPLVDG--- 482
N S S VR K S+ + +PPPL D
Sbjct: 330 GVNNQANHGRSK---------------SDGVRKPSGPWKVSDLDIVVDPVPPPLPDSHPG 374
Query: 483 -LLSSNHDYVSAYSKKIKRHAFSGPLTSNPMP----TRPVSVDSVQMFS----GPLLPTP 533
+ S+ D VS YSK A S P S P P + +S++S P P P
Sbjct: 375 SVASTGSDSVS-YSKL----AVSSPAISKPQPPPYGSHLISMNSASSLERRKDAPPPPRP 429
Query: 534 IPQPPSSS-PKVSPSASPTIVSSPKISELHELPRPPTNFPSNSRLLGLVGYSGPLVPRGQ 592
+P PP+S+ P+V PS T SS +I + +P PT P + P P G
Sbjct: 430 LPNPPASAWPRVPPS---TGSSSQQIQQRISVPPSPTFQP-----------NAPFFPPG- 474
Query: 593 KVSAPNNLVISSAASPLPIPPQAMARSFSIPSSGARVTALHSS-SALESSHISSISEDIA 651
A+ L PP R F IP G+R + + SS I + A
Sbjct: 475 ------------ASERLDPPPAVAVRPF-IPDRGSRPQSPRKGPPTMNSSSIYHMYLQQA 521
Query: 652 SPPLTPI 658
+P P+
Sbjct: 522 APKNQPL 528
>UniRef100_UPI00003A9848 UPI00003A9848 UniRef100 entry
Length = 883
Score = 53.9 bits (128), Expect = 1e-05
Identities = 79/326 (24%), Positives = 125/326 (38%), Gaps = 74/326 (22%)
Query: 283 LEEEDGDEGDEGYEDDDGYDENDDGELSFDYGQIEQEQDVSTSRNSMELDQVELTLPRGS 342
LEEEDG+E + +++ D + Y E+E D ++ E T P+ S
Sbjct: 400 LEEEDGEEVGKARSEENRSDATEPKS----YNPFEEEDD----------EEEESTAPQKS 445
Query: 343 TAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPT 402
T E + T ++ + P SS K+++ RP+ +S +
Sbjct: 446 TPEQEQSE--------------TAAKPLHPWYGITPTSSPKVKK-RPAPRAPNASPLAQH 490
Query: 403 PVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSV 462
PV S S+ P P+ + N ++ + L ++ S PTV + +
Sbjct: 491 PVSRLSHSEPSSSTPSPALSLESINSESS----AKVLGDTDEASVPKSSSEPTV-HTPTA 545
Query: 463 LKESNSNTATTRLPP---PLVDGLLSSNHDYVSAYSKKIKRHAFSGPL-TSNPMPTRPVS 518
K S+++T T + P LS+N S++S + +FSG + S P +R +S
Sbjct: 546 TKTSHTDTPLTSISSSKSPAAPASLSTN----SSFSSSSELASFSGEVQNSTPHASRSIS 601
Query: 519 VDSVQMFSGPLLPTP----IPQP-------------PSSSPKV---------------SP 546
S++ L P P P P PSSSPK SP
Sbjct: 602 AGSLKTSPSRLPPKPPAGTSPTPILLASEGGTGSPKPSSSPKPQLKSSCKENPFNRKPSP 661
Query: 547 SASPTIVSSPKISELHELPRPPTNFP 572
ASP+ PK S+ P P FP
Sbjct: 662 VASPSAKKPPKGSKPARPPAPGHGFP 687
>UniRef100_UPI00003A9847 UPI00003A9847 UniRef100 entry
Length = 889
Score = 53.9 bits (128), Expect = 1e-05
Identities = 79/326 (24%), Positives = 125/326 (38%), Gaps = 74/326 (22%)
Query: 283 LEEEDGDEGDEGYEDDDGYDENDDGELSFDYGQIEQEQDVSTSRNSMELDQVELTLPRGS 342
LEEEDG+E + +++ D + Y E+E D ++ E T P+ S
Sbjct: 406 LEEEDGEEVGKARSEENRSDATEPKS----YNPFEEEDD----------EEEESTAPQKS 451
Query: 343 TAEAAKENLDKLQRNLFSFRVRTGSQSAPLFADNKPDSSEKLRQMRPSLSRKFSSYVLPT 402
T E + T ++ + P SS K+++ RP+ +S +
Sbjct: 452 TPEQEQSE--------------TAAKPLHPWYGITPTSSPKVKK-RPAPRAPNASPLAQH 496
Query: 403 PVDAKSSISSGSNNPKPSKMQTNSNEATTNLWHSSPLEQKKHETIRDEFSSPTVRNAQSV 462
PV S S+ P P+ + N ++ + L ++ S PTV + +
Sbjct: 497 PVSRLSHSEPSSSTPSPALSLESINSESS----AKVLGDTDEASVPKSSSEPTV-HTPTA 551
Query: 463 LKESNSNTATTRLPP---PLVDGLLSSNHDYVSAYSKKIKRHAFSGPL-TSNPMPTRPVS 518
K S+++T T + P LS+N S++S + +FSG + S P +R +S
Sbjct: 552 TKTSHTDTPLTSISSSKSPAAPASLSTN----SSFSSSSELASFSGEVQNSTPHASRSIS 607
Query: 519 VDSVQMFSGPLLPTP----IPQP-------------PSSSPKV---------------SP 546
S++ L P P P P PSSSPK SP
Sbjct: 608 AGSLKTSPSRLPPKPPAGTSPTPILLASEGGTGSPKPSSSPKPQLKSSCKENPFNRKPSP 667
Query: 547 SASPTIVSSPKISELHELPRPPTNFP 572
ASP+ PK S+ P P FP
Sbjct: 668 VASPSAKKPPKGSKPARPPAPGHGFP 693
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.129 0.361
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,089,291,188
Number of Sequences: 2790947
Number of extensions: 48929641
Number of successful extensions: 294733
Number of sequences better than 10.0: 3080
Number of HSP's better than 10.0 without gapping: 220
Number of HSP's successfully gapped in prelim test: 3095
Number of HSP's that attempted gapping in prelim test: 271016
Number of HSP's gapped (non-prelim): 12248
length of query: 669
length of database: 848,049,833
effective HSP length: 134
effective length of query: 535
effective length of database: 474,062,935
effective search space: 253623670225
effective search space used: 253623670225
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0117b.11


