

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
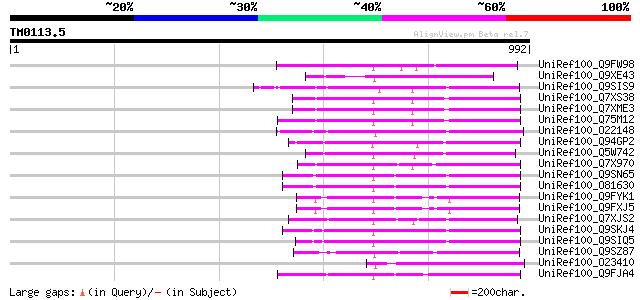
Query= TM0113.5
(992 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcrip... 224 1e-56
UniRef100_Q9XE43 Putative non-LTR retrolelement reverse transcri... 217 2e-54
UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcrip... 214 1e-53
UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa] 213 3e-53
UniRef100_Q7XME3 OSJNBa0061G20.16 protein [Oryza sativa] 213 3e-53
UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa] 210 1e-52
UniRef100_O22148 Putative non-LTR retroelement reverse transcrip... 203 2e-50
UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa] 202 3e-50
UniRef100_Q5W742 Hypothetical protein OJ1005_D04.4 [Oryza sativa] 189 4e-46
UniRef100_Q7X970 BZIP-like protein [Oryza sativa] 187 2e-45
UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis tha... 183 2e-44
UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana] 183 2e-44
UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana] 182 3e-44
UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana] 182 3e-44
UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcrip... 182 6e-44
UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcrip... 181 1e-43
UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcrip... 177 1e-42
UniRef100_Q9SZ87 RNA-directed DNA polymerase-like protein [Arabi... 174 9e-42
UniRef100_O23410 STRONG homology to reverse transcriptase [Arabi... 171 8e-41
UniRef100_Q9FJA4 Non-LTR retroelement reverse transcriptase-like... 171 8e-41
>UniRef100_Q9FW98 Putative non-LTR retroelement reverse transcriptase [Oryza sativa]
Length = 1382
Score = 224 bits (571), Expect = 1e-56
Identities = 141/479 (29%), Positives = 233/479 (48%), Gaps = 23/479 (4%)
Query: 510 LFSWNIICAGAQGVPLL--LKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVE 567
L +W C G + L+ +V R S + L E +++ +M +LGF F V
Sbjct: 6 LVTWGRNCRGLGSAATVGELRWLVKSLRPSLVFLSETKMRDKQARNLMWSLGFSGSFAVS 65
Query: 568 AEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLW 627
EG SGG+ + W+ + V + + F+ + + + +P + ++FVYG P +R+F W
Sbjct: 66 CEGLSGGLALFWTTAY-TVSLRGFNSHFIDVLVSTEE-LPPWRISFVYGEPKRELRHFFW 123
Query: 628 QELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGP 687
L+ + Q PW+ GDFN L + +G + M FR CLD C L DLGF GP
Sbjct: 124 NLLRRLHDQWRGPWLCCGDFNEVLCLDEHLGMRERSEPHMQHFRSCLDDCGLIDLGFVGP 183
Query: 688 PFTWKGR-----GVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDS 742
FTW + K R+D A+ N + F + V ++ DH I I L +
Sbjct: 184 KFTWSNKQDANSNSKVRLDRAVANGEFSRYFEDCLVENVITTSSDHYAISIDLSRRNHGQ 243
Query: 743 SQRP----FRFLAAWLTHEDFPRLVKSSWDSHHDSWI------PASEAFCEEAASWNRDV 792
+ P FRF AAWL ED+ +V++SW + + W++
Sbjct: 244 RRIPIQQGFRFEAAWLRAEDYREVVENSWRISSAGCVGLRGVWSVLQQVAVSLKDWSKAS 303
Query: 793 FGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNW 852
FG + R ++ R+L+ + + + + + +K + ++ + +EE++ RQ+SRV+W
Sbjct: 304 FGSVRRKILKMERKLKSLR---QSPVNDVVIQEEKLIEQQLCELFEKEEIMARQRSRVDW 360
Query: 853 LNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGE 912
L GDRNT FFH R+ N+I+ L D G Q+ ++ MA ++NL++S
Sbjct: 361 LREGDRNTAFFHARASARRRTNRIKELVRDDGSRCISQEGIKRMAEVFYENLFSSEPCDS 420
Query: 913 PYHVRNAFPSLPRETLDEIMG-PLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVI 970
V +A P+ + ++ +G EI+ A+F MG+ KAPGPD LF+Q+ W ++
Sbjct: 421 MEEVLDAIPNKVGDFINGELGKQYTNEEIKTALFQMGSTKAPGPDGFPALFYQTHWGIL 479
>UniRef100_Q9XE43 Putative non-LTR retrolelement reverse transcriptase [Arabidopsis
thaliana]
Length = 855
Score = 217 bits (552), Expect = 2e-54
Identities = 132/367 (35%), Positives = 187/367 (49%), Gaps = 47/367 (12%)
Query: 566 VEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNF 625
V+A G SGGIW+LW GDV I+ S QF+H ++ N + + VY +P+ S R+
Sbjct: 508 VDARGQSGGIWLLWKSEVGDVSIVESAEQFIHAKVG--NGLAAIHLLAVYAAPSVSRRSG 565
Query: 626 LWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFK 685
LW L I V +P + + D+GFK
Sbjct: 566 LWSLLSRIVQSVDEPII------------------------------------VGDMGFK 589
Query: 686 GPPFTWKGRG------VKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPE- 738
G FTWK RG V +R+D L + + EA+V HLP DH PI IQL PE
Sbjct: 590 GNKFTWK-RGRVESTFVAKRLDRVLCRPQTRLKWQEASVTHLPFFASDHAPIYIQLEPEV 648
Query: 739 HMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDSWIPASEAFCEEAASWNRDVFGEIGR 798
+ +RPFRF AAWLTH F L+++SW++ ++ + A A + WNR+VFG++ R
Sbjct: 649 RSNPLRRPFRFEAAWLTHSGFKDLLQASWNTEGETPV-ALAALKSKLKKWNREVFGDVNR 707
Query: 799 TKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDR 858
K LM ++ + L ++ + L ++ L KE+ VL +EE+LW QKSR W+ GDR
Sbjct: 708 RKESLMNEIKVVQELLEINQTDNLLSKEEELIKEFDVVLEQEEVLWFQKSREKWVELGDR 767
Query: 859 NTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRN 918
NT +FHT T+V R+RN+IE L D G ++ Q +L MAV + LY+ E
Sbjct: 768 NTKYFHTMTVVRRRRNRIEMLKADDGSWVSQQQELEKMAVDYYSRLYSMEDVDEVVERLP 827
Query: 919 AFPSLPR 925
P PR
Sbjct: 828 TNPPNPR 834
>UniRef100_Q9SIS9 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1715
Score = 214 bits (544), Expect = 1e-53
Identities = 152/520 (29%), Positives = 246/520 (47%), Gaps = 28/520 (5%)
Query: 467 SNAPGDMGGRTFRERDRSLSPRKHRSS*RAFPSISTLFMMLYNLFSWNIICAGAQGVPLL 526
+NA GD+ G + L RKH R +T M + WN C G G PL
Sbjct: 326 ANATGDILGPV-ELLSQELRKRKHNFESRGGAGTTTSPM---RVGFWN--CQGL-GQPLT 378
Query: 527 LKDIVMRHRV---SCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHW 583
++ + RV L L E + Q N + +GFED ++ G SGG+ V W KH
Sbjct: 379 VRRLEEVQRVYFLDMLFLIETKQQDNYTRDLGVKMGFEDMCIISPRGLSGGLVVYWKKHL 438
Query: 584 GDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVT 643
++++S + V L + KN NF ++ +YG P S R+ LW++LQ +++ + PW+
Sbjct: 439 S-IQVISHDVRLVDLYVEYKNF--NFYLSCIYGHPIPSERHHLWEKLQRVSAHRSGPWMM 495
Query: 644 VGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKGRGVKERIDWA 703
GDFN L ++K GG ++ S+ F + ++ C++ DL KG P++W G+ E I+
Sbjct: 496 CGDFNEILNLNEKKGGRRRSIGSLQNFTNMINCCNMKDLKSKGNPYSWVGKRQNETIESC 555
Query: 704 LG----NARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDF 759
L N+ W +SFP LP DH P++I + E + + + FR+ EDF
Sbjct: 556 LDRVFINSDWQASFPAFETEFLPIAGSDHAPVIIDIA-EEVCTKRGQFRYDRRHFQFEDF 614
Query: 760 PRLVKSSWD----SHHDSWIPASEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLR 815
V+ W+ H + +E A W R L R++
Sbjct: 615 VDSVQRGWNRGRSDSHGGYYEKLHCCRQELAKWKRRTKTNTAEKIETLKYRVDAAERDHT 674
Query: 816 MSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNK 875
+ H L +L++ L + Y+ EEL W KSR W+ GDRNT FF+ +T + + RN+
Sbjct: 675 LPHQTIL-RLRQDLNQAYRD----EELYWHLKSRNRWMLLGDRNTMFFYASTKLRKSRNR 729
Query: 876 IEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAF-PSLPRETLDEIMGP 934
I+A+ + G D + +A F +L+T+ + + + P + + E++
Sbjct: 730 IKAITDAQGIENFRDDTIGKVAENYFADLFTTTQTSDWEEIISGIAPKVTEQMNHELLQS 789
Query: 935 LEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSV 974
+ E+R AVF++GA +APG D F+ W +IG V
Sbjct: 790 VTDQEVRDAVFAIGADRAPGFDGFTAAFYHHLWDLIGNDV 829
>UniRef100_Q7XS38 OSJNBa0081G05.2 protein [Oryza sativa]
Length = 1711
Score = 213 bits (541), Expect = 3e-53
Identities = 130/449 (28%), Positives = 210/449 (45%), Gaps = 23/449 (5%)
Query: 541 LYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRI 600
L E R + ++ + L F F V +EG SGG+ + W + V++ +++++ +
Sbjct: 581 LCETRQSVEKMSRLRRKLAFRGFVGVSSEGMSGGLALYWDESVS-VDVKDINKRYIDAYV 639
Query: 601 HPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGG 660
++ P + +TFVYG P R+ +W L+ I PW+ +GDFN L +
Sbjct: 640 RLSSDEPQWHITFVYGEPRVENRHRMWSLLRTIRQSSALPWMVIGDFNETLWQFEHFSKN 699
Query: 661 PPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG-----RGVKERIDWALGNARWISSFPE 715
P M FRD L C L DLGFKG P T+ R VK R+D A+ + +W FPE
Sbjct: 700 PRCETQMQNFRDALYDCDLQDLGFKGVPHTYDNRRDGWRNVKVRLDRAVADDKWRDLFPE 759
Query: 716 ATVLHLPKLKYDHKPILIQ-LGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSW-----DS 769
A V HL DH PIL++ + + Q+ + W + ++++ +W +
Sbjct: 760 AQVSHLVSPCSDHSPILLEFIVKDTTRPRQKCLHYEIVWEREPESVQVIEEAWINAGVKT 819
Query: 770 HHDSWIPASEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGI--NNHLRMSHSPYLEKLQK 827
A SW + +GR + ++LE + +N R S + + +
Sbjct: 820 DLGDINIALGRVMSALRSWGKTKVKNVGRELEKARKKLEDLIASNAARSSIRQATDHMNE 879
Query: 828 RLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELI 887
L+ REE+LW Q+SRVNWL GDRNT FFH+ + K+NKI L ++ G +
Sbjct: 880 MLY--------REEMLWLQRSRVNWLKEGDRNTRFFHSRAVWRAKKNKISKLRDENGAIH 931
Query: 888 TDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAFPSLPRETLDE-IMGPLEAAEIRVAVFS 946
+ L +MA FQ +Y + P V F + ++E + + EI A+F
Sbjct: 932 SMTSVLETMATEYFQEVYKADPSLNPESVTRLFQEKVTDAMNEKLCQEFKEEEIAQAIFQ 991
Query: 947 MGAMKAPGPDDLNPLFFQSQWQVIGPSVV 975
+G +K+PGPD F+Q W + ++
Sbjct: 992 IGPLKSPGPDGFPARFYQRNWGTLKSDII 1020
>UniRef100_Q7XME3 OSJNBa0061G20.16 protein [Oryza sativa]
Length = 1285
Score = 213 bits (541), Expect = 3e-53
Identities = 130/449 (28%), Positives = 210/449 (45%), Gaps = 23/449 (5%)
Query: 541 LYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRI 600
L E R + ++ + L F F V +EG SGG+ + W + V++ +++++ +
Sbjct: 581 LCETRQSVEKMSRLRRKLAFRGFVGVSSEGMSGGLALYWDESVS-VDVKDINKRYIDAYV 639
Query: 601 HPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGG 660
++ P + +TFVYG P R+ +W L+ I PW+ +GDFN L +
Sbjct: 640 RLSSDEPQWHITFVYGEPRVENRHRMWSLLRTIRQSSALPWMVIGDFNETLWQFEHFSKN 699
Query: 661 PPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG-----RGVKERIDWALGNARWISSFPE 715
P M FRD L C L DLGFKG P T+ R VK R+D A+ + +W FPE
Sbjct: 700 PRCETQMQNFRDALYDCDLQDLGFKGVPHTYDNRRDGWRNVKVRLDRAVADDKWRDLFPE 759
Query: 716 ATVLHLPKLKYDHKPILIQ-LGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSW-----DS 769
A V HL DH PIL++ + + Q+ + W + ++++ +W +
Sbjct: 760 AQVSHLVSPCSDHSPILLEFIVKDTTRPRQKCLHYEIVWEREPESVQVIEEAWINAGVKT 819
Query: 770 HHDSWIPASEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGI--NNHLRMSHSPYLEKLQK 827
A SW + +GR + ++LE + +N R S + + +
Sbjct: 820 DLGDINIALGRVMSALRSWGKTKVKNVGRELEKARKKLEDLIASNAARSSIRQATDHMNE 879
Query: 828 RLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELI 887
L+ REE+LW Q+SRVNWL GDRNT FFH+ + K+NKI L ++ G +
Sbjct: 880 MLY--------REEMLWLQRSRVNWLKEGDRNTRFFHSRAVWRAKKNKISKLRDENGAIH 931
Query: 888 TDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAFPSLPRETLDE-IMGPLEAAEIRVAVFS 946
+ L +MA FQ +Y + P V F + ++E + + EI A+F
Sbjct: 932 SMTSVLETMATEYFQEVYKADPSLNPESVTRLFQEKVTDAMNEKLCQEFKEEEIAQAIFQ 991
Query: 947 MGAMKAPGPDDLNPLFFQSQWQVIGPSVV 975
+G +K+PGPD F+Q W + ++
Sbjct: 992 IGPLKSPGPDGFPARFYQRNWGTLKSDII 1020
>UniRef100_Q75M12 Hypothetical protein P0676G05.2 [Oryza sativa]
Length = 1936
Score = 210 bits (535), Expect = 1e-52
Identities = 132/478 (27%), Positives = 219/478 (45%), Gaps = 23/478 (4%)
Query: 512 SWNIICAGAQGVPLLLKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGF 571
+WN G L+ ++ + + L E R + ++ + L F F V +EG
Sbjct: 640 AWNCRGLGNTATVQDLRALIQKAGSQLVFLCETRQSVEKMSRLRRKLAFRGFVGVSSEGK 699
Query: 572 SGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQ 631
SGG+ + W + V++ +++++ + + P + +TFVYG P R+ +W L+
Sbjct: 700 SGGLALYWDESVS-VDVKDINKRYIDAYVRLSPDEPQWHITFVYGEPRVENRHRMWSLLR 758
Query: 632 NIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTW 691
I PW+ +GDFN L + P M FRD L C L DLGFKG P T+
Sbjct: 759 TIRQSSALPWMVIGDFNETLWQFEHFSKNPRCETQMQNFRDALYDCDLQDLGFKGVPHTY 818
Query: 692 KG-----RGVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQ-LGPEHMDSSQR 745
R VK R+D A+ + +W FPEA V HL DH PIL++ + + Q+
Sbjct: 819 DNRRDGWRNVKVRLDRAVADDKWRDLFPEAQVSHLVSPCSDHSPILLEFIVKDTTRPRQK 878
Query: 746 PFRFLAAWLTHEDFPRLVKSSW-----DSHHDSWIPASEAFCEEAASWNRDVFGEIGRTK 800
+ W + ++++ +W + A SW++ +G+
Sbjct: 879 CLHYEIVWEREPESVQVIEEAWINAGVKTDLGDINIALGRVMSALRSWSKTKVKNVGKEL 938
Query: 801 RRLMRRLEGI--NNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDR 858
+ ++LE + +N R S + + + L+ REE+LW Q+SRVNWL GDR
Sbjct: 939 EKARKKLEDLIASNAARSSIRQATDHMNEMLY--------REEMLWLQRSRVNWLKEGDR 990
Query: 859 NTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRN 918
NT FFH+ + K+NKI L ++ G + + L +MA FQ +Y + P V
Sbjct: 991 NTRFFHSRAVWRAKKNKISKLRDENGAIHSTTSVLETMATEYFQGVYKADPSLNPESVTR 1050
Query: 919 AFPSLPRETLDE-IMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVV 975
F + ++E + + EI A+F +G +K+P PD F+Q W + ++
Sbjct: 1051 LFQEKVTDAMNEKLCQEFKEEEIAQAIFQIGPLKSPRPDGFPARFYQRNWGTLKSDII 1108
>UniRef100_O22148 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1374
Score = 203 bits (517), Expect = 2e-50
Identities = 131/484 (27%), Positives = 228/484 (47%), Gaps = 22/484 (4%)
Query: 510 LFSWNIICAGAQGVPLL--LKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVE 567
+ SWN C G P + L++I + + L E + + N L ++ LGF D VE
Sbjct: 3 ILSWN--CQGVGNTPTVRHLREIRGLYFPEVIFLCETKKRRNYLENVVGHLGFFDLHTVE 60
Query: 568 AEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLW 627
G SGG+ ++W K +++L S ++ + + ++ F +T +YG P + R LW
Sbjct: 61 PIGKSGGLALMW-KDSVQIKVLQSDKRLIDALLIWQDK--EFYLTCIYGEPVQAERGELW 117
Query: 628 QELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGP 687
+ L + + PW+ GDFN + S+K+GG S +FR L++C L ++ G
Sbjct: 118 ERLTRLGLSRSGPWMLTGDFNELVDPSEKIGGPARKESSCLEFRQMLNSCGLWEVNHSGY 177
Query: 688 PFTWKGRGVKE----RIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSS 743
F+W G E R+D + N W+ FP+A +L K+ DH P++ L ++
Sbjct: 178 QFSWYGNRNDELVQCRLDRTVANQAWMELFPQAKATYLQKICSDHSPLINNLVGDNW-RK 236
Query: 744 QRPFRFLAAWLTHEDFPRLVKSSWDSHH---DSWIPASEAFCE-EAASWNRDVFGEIGRT 799
F++ W+ E F L+ + W ++ + A C E + W R
Sbjct: 237 WAGFKYDKRWVQREGFKDLLCNFWSQQSTKTNALMMEKIASCRREISKWKRVSKPSSAVR 296
Query: 800 KRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRN 859
+ L +L+ + L +L+K L +EY EE W++KSR+ W+ +GDRN
Sbjct: 297 IQELQFKLDAATKQIPFDRRE-LARLKKELSQEYNN----EEQFWQEKSRIMWMRNGDRN 351
Query: 860 THFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPY-HVRN 918
T +FH T R +N+I+ L ++ G T + L +A F+ L+ S G + N
Sbjct: 352 TKYFHAATKNRRAQNRIQKLIDEEGREWTSDEDLGRVAEAYFKKLFASEDVGYTVEELEN 411
Query: 919 AFPSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVVKLA 978
P + + + ++ P+ E++ A FS+ K PGPD +N +Q W+ +G + ++
Sbjct: 412 LTPLVSDQMNNNLLAPITKEEVQRATFSINPHKCPGPDGMNGFLYQQFWETMGDQITEMV 471
Query: 979 MTVY 982
+
Sbjct: 472 QAFF 475
>UniRef100_Q94GP2 Putative reverse transcriptase [Oryza sativa]
Length = 1833
Score = 202 bits (515), Expect = 3e-50
Identities = 123/453 (27%), Positives = 213/453 (46%), Gaps = 20/453 (4%)
Query: 534 HRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHR 593
HR + L E R + + +G F V G GG+ + W + +V + S +
Sbjct: 10 HRPKIVFLAETR---QDQYMVQNRIGLRRCFTVNGVGKGGGLALFWDESM-NVVLKSYNS 65
Query: 594 QFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQA 653
+ + + I + + + TFVYG P A R+ +W L+ I S PW+ +GDFN +
Sbjct: 66 RHIDVLI-TELDCCTWRATFVYGEPRAQDRHLMWSLLRRIRSNSGDPWLMIGDFNEAMWQ 124
Query: 654 SDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTW-----KGRGVKERIDWALGNAR 708
++ + M FR+ L C L D+GF+G P+T+ +GR VK R+D + +
Sbjct: 125 TEHKSHRKRSESQMRDFREVLSECDLHDIGFQGAPWTFCNMQREGRNVKVRLDRGVASPA 184
Query: 709 WISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWD 768
W S FP+A + HL DH P+L++ + + R+ W P +++ +W
Sbjct: 185 WSSRFPQAVITHLTTPSSDHAPLLLEREETTLARPMKIMRYEEVWERESSLPEVIQEAWT 244
Query: 769 SHHDSWIPAS-----EAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLE 823
D+ + + SW++D G + + + L +L + N + +
Sbjct: 245 MGADASTLGDINDKMKVTMTKLVSWSKDKIGNVRKKIKDLREKLGELRNIGLLDTDNEVH 304
Query: 824 KLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDV 883
++K L + +L REE+ W+Q+SR+ WL GD NT +FH K+NKI+ L +
Sbjct: 305 SVKKEL----EEMLHREEIWWKQRSRITWLKEGDLNTRYFHLKASWRAKKNKIKKLKKND 360
Query: 884 GELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAFPSLPRETLD-EIMGPLEAAEIRV 942
G ++ +++ + + FQ LYT P ++ N F E ++ +++ P EI
Sbjct: 361 GSTTMNKKEMKEINRSFFQQLYTKDDNLNPVNLLNMFHEKISEQMNADLIKPFTNEEISD 420
Query: 943 AVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVV 975
A+F +G +KAPGPD F Q W ++ V+
Sbjct: 421 ALFQIGPLKAPGPDGFPARFLQRNWGLLKGEVI 453
>UniRef100_Q5W742 Hypothetical protein OJ1005_D04.4 [Oryza sativa]
Length = 796
Score = 189 bits (480), Expect = 4e-46
Identities = 118/414 (28%), Positives = 201/414 (48%), Gaps = 18/414 (4%)
Query: 566 VEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNF 625
+ G GGI + W + VE+ S + + + + I +++ + TFVYG P + R
Sbjct: 387 MNGRGKGGGIALFWDESI-KVELKSYNVRHIDVLI-TESDGERWRGTFVYGEPRSQDRLK 444
Query: 626 LWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFK 685
+W L+ I S PW+ +GDFN + S+ + M FR+ L C L DLGF
Sbjct: 445 MWTLLRRIKSNAALPWLMIGDFNEAMWQSEHWSRVRRSERQMKDFREALADCDLHDLGFH 504
Query: 686 GPPFTWKGR-----GVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQ-LGPEH 739
G P+T+ V+ R+D A+ + W+ F +A++ HL DH P+L++ L E
Sbjct: 505 GVPWTYDNNQRGANNVRVRLDRAVASPAWLGRFRQASITHLVTPSSDHVPLLLERLEEEL 564
Query: 740 MDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHH-----DSWIPASEAFCEEAASWNRDVFG 794
+ +S + R+ A W E F +V+ +W + +E W++ G
Sbjct: 565 VHNSVKISRYEAIWEREESFDSVVQQAWGEGEVVRNLGDFKTKMAYTMDELVKWSKSKIG 624
Query: 795 EIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLN 854
I + ++L + + P ++K++++L Q +L REE+ WRQ+S++ WL
Sbjct: 625 NIKKGIESRRKKLGELRMAGMLDSEPEVKKIKEQL----QEMLRREEIWWRQRSQITWLK 680
Query: 855 HGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPY 914
GD+NT +FH ++NK++ L G L++++ ++ +A FQNLY +P
Sbjct: 681 EGDKNTKYFHLKASWRARKNKVKKLRRPNGSLVSNEKEMGEVARDFFQNLYMKDENLDPT 740
Query: 915 HVRNAFPS-LPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQW 967
+ N F + + E + +M P EI +F +G +KAPG D FFQ W
Sbjct: 741 ELFNLFQTKITAEMNESLMKPFSEEEIGDVLFQIGHLKAPGSDGFPAHFFQRNW 794
>UniRef100_Q7X970 BZIP-like protein [Oryza sativa]
Length = 2367
Score = 187 bits (474), Expect = 2e-45
Identities = 123/439 (28%), Positives = 196/439 (44%), Gaps = 23/439 (5%)
Query: 551 LPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFL 610
+ ++ LG F V +EG SGG+ + W + V++ +++++ + P +
Sbjct: 773 MSRLRGRLGLRGFTGVSSEGMSGGLALYWDESVS-VDVKDINKRYIDAYVQLSPEEPQWH 831
Query: 611 VTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKF 670
VTFVYG P R+ +W L+ I + PW +GDFN + + P M F
Sbjct: 832 VTFVYGEPRVENRHRMWSLLRTIHQSSSLPWAVIGDFNETMWQFEHFSRTPRGEPQMQDF 891
Query: 671 RDCLDACSLSDLGFKGPPFTWKG-----RGVKERIDWALGNARWISSFPEATVLHLPKLK 725
RD L C L DLGFKG P T+ R VK R+D + + +W + A V+HL
Sbjct: 892 RDVLQDCELHDLGFKGVPHTYDNKREGWRNVKVRLDRVVADDKWRDIYSTAQVVHLVSPC 951
Query: 726 YDHKPILIQL---GPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSW-----DSHHDSWIPA 777
DH PIL+ L P + Q+ + W + ++++ +W + A
Sbjct: 952 SDHCPILLNLVVKDPHQL--RQKCLHYEIVWEREPEATQVIEEAWVVAGEKADLGDINKA 1009
Query: 778 SEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVL 837
SW+R +GR + ++L + S + + +L
Sbjct: 1010 LAKVMTALRSWSRAKVKNVGRELEKARKKL------AELIESNADRTVIRNATDHMNELL 1063
Query: 838 IREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMA 897
REE+LW Q+SRVNWL DRNT FFH+ + K+NKI L + + + +L SMA
Sbjct: 1064 YREEMLWLQRSRVNWLKDEDRNTKFFHSRAVWRAKKNKISKLRDANETVHSSTMKLESMA 1123
Query: 898 VTSFQNLYTSPGPGEPYHVRNAFPSLPRETLDE-IMGPLEAAEIRVAVFSMGAMKAPGPD 956
FQ++YT+ P V + ++E + EI A+F +G +K+PGPD
Sbjct: 1124 TEYFQDVYTADPNLNPETVTRLIQEKVTDIMNEKLCEDFTEDEISQAIFQIGPLKSPGPD 1183
Query: 957 DLNPLFFQSQWQVIGPSVV 975
F+Q W I ++
Sbjct: 1184 GFPARFYQRNWGTIKADII 1202
>UniRef100_Q9SN65 Hypothetical protein F25I24.40 [Arabidopsis thaliana]
Length = 1294
Score = 183 bits (465), Expect = 2e-44
Identities = 130/467 (27%), Positives = 209/467 (43%), Gaps = 20/467 (4%)
Query: 522 GVPLL---LKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVL 578
GVPL L ++ + L L E + + + LGF + +G SGG+ +L
Sbjct: 373 GVPLTQSQLSNLCKVFKFDVLFLIETLNKCEVISNLASVLGFPNVITQPPQGHSGGLALL 432
Query: 579 WSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVT 638
W V + + ++ H+ +H N NF ++ VYG P S R+ LW +N++
Sbjct: 433 WKD---SVRLSNLYQDDRHIDVHISINNINFYLSRVYGHPCQSERHSLWTHFENLSKTRN 489
Query: 639 KPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG----R 694
PW+ +GDFN L ++K+GG + + FR+ + C L D+ G F+W G
Sbjct: 490 DPWILIGDFNEILSNNEKIGGPQRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGERHSH 549
Query: 695 GVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWL 754
VK +D A N+ FP A + L DHKP+ + L + RPFRF L
Sbjct: 550 TVKCCLDRAFINSEGAFLFPFAELEFLEFTGSDHKPLFLSL-EKTETRKMRPFRFDKRLL 608
Query: 755 THEDFPRLVKSSWD---SHHDSWIPASEAFCEEAASWNRDVFGEIGRTK-RRLMRRLEGI 810
F VK+ W+ + +P C +A + + R + +L L+
Sbjct: 609 EVPHFKTYVKAGWNKAINGQRKHLPDQVRTCRQAMAKLKHKSNLNSRIRINQLQAALDKA 668
Query: 811 NNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVH 870
+ + + + +Q+ L Y+ EE W+QKSR W+ GDRNT FFH T
Sbjct: 669 MSSVNRTERRTISHIQRELTVAYRD----EERYWQQKSRNQWMKEGDRNTEFFHACTKTR 724
Query: 871 RKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPG-PGEPYHVRNAFPSLPRETLD 929
N++ + ++ G + ++ A F +Y S G P P + + D
Sbjct: 725 FSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTKVYESNGRPVSIIDFAGFKPIVTEQIND 784
Query: 930 EIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVVK 976
++ L EI A+ +G KAPGPD L F++S W+++GP V+K
Sbjct: 785 DLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIK 831
>UniRef100_O81630 F8M12.22 protein [Arabidopsis thaliana]
Length = 1662
Score = 183 bits (465), Expect = 2e-44
Identities = 130/467 (27%), Positives = 209/467 (43%), Gaps = 20/467 (4%)
Query: 522 GVPLL---LKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVL 578
GVPL L ++ + L L E + + + LGF + +G SGG+ +L
Sbjct: 393 GVPLTQSQLSNLCKVFKFDVLFLIETLNKCEVISNLASVLGFPNVITQPPQGHSGGLALL 452
Query: 579 WSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVT 638
W V + + ++ H+ +H N NF ++ VYG P S R+ LW +N++
Sbjct: 453 WKD---SVRLSNLYQDDRHIDVHISINNINFYLSRVYGHPCQSERHSLWTHFENLSKTRN 509
Query: 639 KPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG----R 694
PW+ +GDFN L ++K+GG + + FR+ + C L D+ G F+W G
Sbjct: 510 DPWILIGDFNEILSNNEKIGGPQRDEWTFRGFRNMVSTCDLKDIRSIGDRFSWVGERHSH 569
Query: 695 GVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWL 754
VK +D A N+ FP A + L DHKP+ + L + RPFRF L
Sbjct: 570 TVKCCLDRAFINSEGAFLFPFAELEFLEFTGSDHKPLFLSL-EKTETRKMRPFRFDKRLL 628
Query: 755 THEDFPRLVKSSWD---SHHDSWIPASEAFCEEAASWNRDVFGEIGRTK-RRLMRRLEGI 810
F VK+ W+ + +P C +A + + R + +L L+
Sbjct: 629 EVPHFKTYVKAGWNKAINGQRKHLPDQVRTCRQAMAKLKHKSNLNSRIRINQLQAALDKA 688
Query: 811 NNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVH 870
+ + + + +Q+ L Y+ EE W+QKSR W+ GDRNT FFH T
Sbjct: 689 MSSVNRTERRTISHIQRELTVAYRD----EERYWQQKSRNQWMKEGDRNTEFFHACTKTR 744
Query: 871 RKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPG-PGEPYHVRNAFPSLPRETLD 929
N++ + ++ G + ++ A F +Y S G P P + + D
Sbjct: 745 FSVNRLVTIKDEEGMIYRGDKEIGVHAQEFFTKVYESNGRPVSIIDFAGFKPIVTEQIND 804
Query: 930 EIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVVK 976
++ L EI A+ +G KAPGPD L F++S W+++GP V+K
Sbjct: 805 DLTKDLSDLEIYNAICHIGDDKAPGPDGLTARFYKSCWEIVGPDVIK 851
>UniRef100_Q9FYK1 F21J9.30 [Arabidopsis thaliana]
Length = 1270
Score = 182 bits (463), Expect = 3e-44
Identities = 132/451 (29%), Positives = 198/451 (43%), Gaps = 50/451 (11%)
Query: 549 NNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKH------WGDVEILSSHRQFVHLRIHP 602
++L I L ++ + VE G GG+ +LW + D ++ + QF
Sbjct: 5 DDLVDIQSWLEYDQVYTVEPVGKCGGLALLWKSSVQVDLKFVDKNLMDAQVQF------- 57
Query: 603 KNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPP 662
NF V+ VYG P+ S R+ W+ + I W GDFN L +K GG
Sbjct: 58 --GAVNFCVSCVYGDPDRSKRSQAWERISRIGVGRRDKWCMFGDFNDILHNGEKNGGPRR 115
Query: 663 NLLSMAKFRDCLDACSLSDLGFKGPPFTWKGR----GVKERIDWALGNARWISSFPEATV 718
+ L F + + C L ++ G FTW GR ++ R+D A GN W FP +
Sbjct: 116 SDLDCKAFNEMIKGCDLVEMPAHGNGFTWAGRRGDHWIQCRLDRAFGNKEWFCFFPVSNQ 175
Query: 719 LHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWD-SHHDSWIPA 777
L DH+P+LI+L DS + FRF +L ED + +W H + I
Sbjct: 176 TFLDFRGSDHRPVLIKL-MSSQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGKHGTNISV 234
Query: 778 SE---AFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQ 834
++ A + +SW K+ + L+ IN LEK Q +W +Q
Sbjct: 235 ADRLRACRKSLSSWK----------KQNNLNSLDKIN-----QLEAALEKEQSLVWPIFQ 279
Query: 835 TVLI----------REELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVG 884
V + EE W+QKSR WL G+RN+ +FH +R+R +IE L + G
Sbjct: 280 RVSVLKKDLAKAYREEEAYWKQKSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNG 339
Query: 885 ELITDQDQLRSMAVTSFQNLYTSPGP-GEPYHVRNAFPSLPRETLDEIMGPLEAAEIRVA 943
+ T + +A F NL+ S P G P + + ++G + A EI+ A
Sbjct: 340 NMQTSEAAKGEVAAAYFGNLFKSSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEA 399
Query: 944 VFSMGAMKAPGPDDLNPLFFQSQWQVIGPSV 974
VFS+ APGPD ++ LFFQ W +G V
Sbjct: 400 VFSIKPASAPGPDGMSALFFQHYWSTVGNQV 430
>UniRef100_Q9FXJ5 F5A9.24 [Arabidopsis thaliana]
Length = 1254
Score = 182 bits (463), Expect = 3e-44
Identities = 132/451 (29%), Positives = 198/451 (43%), Gaps = 50/451 (11%)
Query: 549 NNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKH------WGDVEILSSHRQFVHLRIHP 602
++L I L ++ + VE G GG+ +LW + D ++ + QF
Sbjct: 8 DDLVDIQSWLEYDQVYTVEPVGKCGGLALLWKSSVQVDLKFVDKNLMDAQVQF------- 60
Query: 603 KNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPP 662
NF V+ VYG P+ S R+ W+ + I W GDFN L +K GG
Sbjct: 61 --GAVNFCVSCVYGDPDRSKRSQAWERISRIGVGRRDKWCMFGDFNDILHNGEKNGGPRR 118
Query: 663 NLLSMAKFRDCLDACSLSDLGFKGPPFTWKGR----GVKERIDWALGNARWISSFPEATV 718
+ L F + + C L ++ G FTW GR ++ R+D A GN W FP +
Sbjct: 119 SDLDCKAFNEMIKGCDLVEMPAHGNGFTWAGRRGDHWIQCRLDRAFGNKEWFCFFPVSNQ 178
Query: 719 LHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWD-SHHDSWIPA 777
L DH+P+LI+L DS + FRF +L ED + +W H + I
Sbjct: 179 TFLDFRGSDHRPVLIKL-MSSQDSYRGQFRFDKRFLFKEDVKEAIIRTWSRGKHGTNISV 237
Query: 778 SE---AFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQ 834
++ A + +SW K+ + L+ IN LEK Q +W +Q
Sbjct: 238 ADRLRACRKSLSSWK----------KQNNLNSLDKIN-----QLEAALEKEQSLVWPIFQ 282
Query: 835 TVLI----------REELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVG 884
V + EE W+QKSR WL G+RN+ +FH +R+R +IE L + G
Sbjct: 283 RVSVLKKDLAKAYREEEAYWKQKSRQKWLRSGNRNSKYFHAAVKQNRQRKRIEKLKDVNG 342
Query: 885 ELITDQDQLRSMAVTSFQNLYTSPGP-GEPYHVRNAFPSLPRETLDEIMGPLEAAEIRVA 943
+ T + +A F NL+ S P G P + + ++G + A EI+ A
Sbjct: 343 NMQTSEAAKGEVAAAYFGNLFKSSNPSGFTDWFSGLVPRVSEVMNESLVGEVSAQEIKEA 402
Query: 944 VFSMGAMKAPGPDDLNPLFFQSQWQVIGPSV 974
VFS+ APGPD ++ LFFQ W +G V
Sbjct: 403 VFSIKPASAPGPDGMSALFFQHYWSTVGNQV 433
>UniRef100_Q7XJS2 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1344
Score = 182 bits (461), Expect = 6e-44
Identities = 130/448 (29%), Positives = 207/448 (46%), Gaps = 22/448 (4%)
Query: 534 HRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHR 593
H L L E + + + K+ LG++ VE EG SGG+ + W H ++E L + +
Sbjct: 5 HFPDILFLMETKNSQDFVYKVFCWLGYDFIHTVEPEGRSGGLAIFWKSHL-EIEFLYADK 63
Query: 594 QFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQA 653
+ L++ +N V + ++ VYG P +R LW+ L +I + + W +GDFN
Sbjct: 64 NLMDLQVSSRNKV--WFISCVYGLPVTHMRPKLWEHLNSIGLKRAEAWCLIGDFNDIRSN 121
Query: 654 SDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG----RGVKERIDWALGNARW 709
+K+GG + S F L CS+ +LG G FTW G + V+ ++D GN W
Sbjct: 122 DEKLGGPRRSPSSFQCFEHMLLNCSMHELGSTGNSFTWGGNRNDQWVQCKLDRCFGNPAW 181
Query: 710 ISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDS 769
S FP A L K DH+P+L++ ++ + + FR+ ++ SW+S
Sbjct: 182 FSIFPNAHQWFLEKFGSDHRPVLVKFTNDN-ELFRGQFRYDKRLDDDPYCIEVIHRSWNS 240
Query: 770 H-----HDSWIPASEAFCEEAAS-WNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLE 823
H S+ E C A S W +RL + L+ +++ P +E
Sbjct: 241 AMSQGTHSSFFSLIE--CRRAISVWKHSSDTNAQSRIKRLRKDLDA-EKSIQIPCWPRIE 297
Query: 824 KLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDV 883
++ +L Y EEL WRQKSR WL GD+NT FFH T R +N++ L ++
Sbjct: 298 YIKDQLSLAYGD----EELFWRQKSRQKWLAGGDKNTGFFHATVHSERLKNELSFLLDEN 353
Query: 884 GELITDQDQLRSMAVTSFQNLYTSPG-PGEPYHVRNAFPSLPRETLDEIMGPLEAAEIRV 942
+ T +A + F+NL+TS H+ + E ++ + E+
Sbjct: 354 DQEFTRNSDKGKIASSFFENLFTSTYILTHNNHLEGLQAKVTSEMNHNLIQEVTELEVYN 413
Query: 943 AVFSMGAMKAPGPDDLNPLFFQSQWQVI 970
AVFS+ APGPD LFFQ W ++
Sbjct: 414 AVFSINKESAPGPDGFTALFFQQHWDLV 441
>UniRef100_Q9SKJ4 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1750
Score = 181 bits (458), Expect = 1e-43
Identities = 127/466 (27%), Positives = 218/466 (46%), Gaps = 20/466 (4%)
Query: 522 GVPLLLKDIVMRHRV---SCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVL 578
G+PL + R+ L L E Q + + K+ LGF + G SGG+ ++
Sbjct: 393 GMPLTQSRLFRLFRMYNYDILFLVETLNQCDKVCKLAYDLGFPNVITQPPNGRSGGLALM 452
Query: 579 WSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVT 638
W K+ + ++S + + H N +F ++ VYG P S R+ LWQ L++I+
Sbjct: 453 W-KNNVSLSLISQDERLIDS--HVTFNNKSFYLSCVYGHPTQSERHQLWQTLEHISDNRN 509
Query: 639 KPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTWKG----R 694
W+ VGDFN L ++K+GG + FR+ + C + D+ KG F+W G
Sbjct: 510 AEWLLVGDFNEILSNAEKIGGPMREEWTFRNFRNMVSHCDIEDMRSKGDRFSWVGERHTH 569
Query: 695 GVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWL 754
VK +D N+ W ++FP A + L DHKP+L+ E + FRF +
Sbjct: 570 TVKCCLDRVFINSAWTATFPYAEIEFLDFTGSDHKPVLVHFN-ESFPRRSKLFRFDNRLI 628
Query: 755 THEDFPRLVKSSWDSHHDS-WIPASEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNH 813
F R+V++SW ++ +S P +E + R +++R+ + +N
Sbjct: 629 DIPTFKRIVQTSWRTNRNSRSTPITERISSCRQAMARLKHASNLNSEQRIKKLQSSLNRA 688
Query: 814 L---RMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFHTTTMVH 870
+ R + +LQ+ L K + EE+ W+QKSR W+ GD+NT +FH T
Sbjct: 689 MESTRRVDRQLIPQLQESLAKAFSD----EEIYWKQKSRNQWMKEGDQNTGYFHACTKTR 744
Query: 871 RKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYHVRNAFPSLPRETLD- 929
+N++ + +D G + T ++ + A F N++++ G F S T++
Sbjct: 745 YSQNRVNTIMDDQGRMFTGDKEIGNHAQDFFTNIFSTNGIKVSPIDFADFKSTVTNTVNL 804
Query: 930 EIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVV 975
++ EI A+ +G KAPGPD L F+++ W ++G V+
Sbjct: 805 DLTKEFSDTEIYDAICQIGDDKAPGPDGLTARFYKNCWDIVGYDVI 850
>UniRef100_Q9SIQ5 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1524
Score = 177 bits (450), Expect = 1e-42
Identities = 120/438 (27%), Positives = 207/438 (46%), Gaps = 17/438 (3%)
Query: 547 QTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNV 606
Q + + K+ LGF + G SGG+ ++W K+ + ++S + + H N
Sbjct: 195 QCDKVCKLAYDLGFPNVITQPPNGRSGGLALMW-KNNVSLSLISQDERLIDS--HVTFNN 251
Query: 607 PNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLS 666
+F ++ VYG P S R+ LWQ L++I+ W+ VGDFN L ++K+GG +
Sbjct: 252 KSFYLSCVYGHPTQSERHQLWQTLEHISDNRNAEWLLVGDFNEILSNAEKIGGPMREEWT 311
Query: 667 MAKFRDCLDACSLSDLGFKGPPFTWKG----RGVKERIDWALGNARWISSFPEATVLHLP 722
FR+ + C + D+ KG F+W G VK +D N+ W ++FP A L
Sbjct: 312 FRNFRNMVSHCDIEDMRSKGDRFSWVGERHTHTVKCCLDRVFINSAWTATFPYAETEFLD 371
Query: 723 KLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDS-WIPASEAF 781
DHKP+L+ E + FRF + F R+V++SW ++ +S P +E
Sbjct: 372 FTGSDHKPVLVHFN-ESFPRRSKLFRFDNRLIDIPTFKRIVQTSWRTNRNSRSTPITERI 430
Query: 782 CEEAASWNRDVFGEIGRTKRRLMRRLEGINNHL---RMSHSPYLEKLQKRLWKEYQTVLI 838
+ R +++R+ + +N + R + +LQ+ L K +
Sbjct: 431 SSCRQAMARLKHASNLNSEQRIKKLQSSLNRAMESTRRVDRQLIPQLQESLAKAFSD--- 487
Query: 839 REELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAV 898
EE+ W+QKSR W+ GD+NT +FH T +N++ + +D G + T ++ + A
Sbjct: 488 -EEIYWKQKSRNQWMKEGDQNTGYFHACTKTRYSQNRVNTIMDDQGRMFTGDKEIGNHAQ 546
Query: 899 TSFQNLYTSPGPGEPYHVRNAFPSLPRETLD-EIMGPLEAAEIRVAVFSMGAMKAPGPDD 957
F N++++ G F S T++ ++ EI A+ +G KAPGPD
Sbjct: 547 DFFTNIFSTNGIKVSPIDFADFKSTVTNTVNLDLTKEFSDTEIYDAICQIGDDKAPGPDG 606
Query: 958 LNPLFFQSQWQVIGPSVV 975
L F+++ W ++G V+
Sbjct: 607 LTARFYKNCWDIVGYDVI 624
>UniRef100_Q9SZ87 RNA-directed DNA polymerase-like protein [Arabidopsis thaliana]
Length = 1274
Score = 174 bits (442), Expect = 9e-42
Identities = 120/438 (27%), Positives = 196/438 (44%), Gaps = 37/438 (8%)
Query: 543 EPRVQTNNLPKIMKTLGFEDFFVVEAEGFSGGIWVLWSKHWGDVEILSSHRQFVHLRIHP 602
E + Q + K +G+ F + EG SGG+ + W ++ +VEIL +
Sbjct: 2 ETKNQDEFISKTFDWMGYAHRFTIPPEGLSGGLALYWKENV-EVEILEA----------- 49
Query: 603 KNNVPNFLVTFVYGSPNASIRNFLWQELQNIASQVTKPWVTVGDFNTYLQASDKMGGGPP 662
PNF+ R+ W ++ ++ +Q + W+ GDFN L S+K GG
Sbjct: 50 ---APNFIDN----------RSVFWDKISSLGAQRSSAWLLTGDFNDILDNSEKQGGPLR 96
Query: 663 NLLSMAKFRDCLDACSLSDLGFKGPPFTWKGRG----VKERIDWALGNARWISSFPEATV 718
FR + L D+ G +W+G +K R+D ALGN W FP +
Sbjct: 97 WEGFFLAFRSFVSQNGLWDINHTGNSLSWRGTRYSHFIKSRLDRALGNCSWSELFPMSKC 156
Query: 719 LHLPKLKYDHKPILIQLGPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWD-SHHDSWIPA 777
+L DH+P++ G + S +PFRF E+ LVK W+ + DS +
Sbjct: 157 EYLRFEGSDHRPLVTYFGAPPLKRS-KPFRFDRRLREKEEIRALVKEVWELARQDSVLYK 215
Query: 778 SEAFCEEAASWNRDVFGEIGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVL 837
+ W ++ + ++ + LE +S L + +E +
Sbjct: 216 ISRCRQSIIKWTKEQNSNSAKAIKKAQQALESA-----LSADIPDPSLIGSITQELEAAY 270
Query: 838 IREELLWRQKSRVNWLNHGDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMA 897
+EEL W+Q SRV WLN GDRN +FH TT R N + + + G+ +++Q+ S
Sbjct: 271 RQEELFWKQWSRVQWLNSGDRNKGYFHATTRTRRMLNNLSVIEDGSGQEFHEEEQIASTI 330
Query: 898 VTSFQNLYTSPGPGEPYHVRNAF-PSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPD 956
+ FQN++T+ + V+ A P + +E++ EI+ A+FS+ A KAPGPD
Sbjct: 331 SSYFQNIFTTSNNSDLQVVQEALSPIISSHCNEELIKISSLLEIKEALFSISADKAPGPD 390
Query: 957 DLNPLFFQSQWQVIGPSV 974
+ FF + W +I V
Sbjct: 391 GFSASFFHAYWDIIEADV 408
>UniRef100_O23410 STRONG homology to reverse transcriptase [Arabidopsis thaliana]
Length = 929
Score = 171 bits (434), Expect = 8e-41
Identities = 106/311 (34%), Positives = 162/311 (52%), Gaps = 26/311 (8%)
Query: 682 LGFKGPPFTWKGRGVKE------RIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQL 735
+GFKG FTW+ RG+ E R+D L A + EA +
Sbjct: 1 MGFKGNRFTWR-RGLVESTFVAKRLDRVLFCAHARLKWQEALLCPA-------------- 45
Query: 736 GPEHMDSSQRPFRFLAAWLTHEDFPRLVKSSWDSHHDSWIPASEAFCEEAASWNRDVFGE 795
+++D+ +RPFRF AAWL+HE F L+ +SWD+ + + A + WN++VFG
Sbjct: 46 --QNVDARRRPFRFEAAWLSHEGFKELLTASWDTGLSTPV-ALNRLRWQLKKWNKEVFGN 102
Query: 796 IGRTKRRLMRRLEGINNHLRMSHSPYLEKLQKRLWKEYQTVLIREELLWRQKSRVNWLNH 855
I K +++ L+ + + L + + L + L KE+ +L +EE LW QKSR L
Sbjct: 103 IHVRKEKVVSDLKAVQDLLEVVQTDDLLMKEDTLLKEFDVLLHQEETLWFQKSREKLLAL 162
Query: 856 GDRNTHFFHTTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTSPGPGEPYH 915
GDRNT FFHT+T++ R+RN+IE L + +T+++ L +A+ ++ LY+
Sbjct: 163 GDRNTTFFHTSTVIRRRRNRIEMLKDSEDRWVTEKEALEKLAMDYYRKLYSLEDVSVVRG 222
Query: 916 V--RNAFPSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPS 973
FP L RE + + P E+ VAV SMG KAPGPD P+F+Q W+ +G S
Sbjct: 223 TLPTEGFPRLTREEKNNLNRPFTRDEVVVAVRSMGRFKAPGPDGYQPVFYQQCWETVGES 282
Query: 974 VVKLAMTVYSN 984
V K M + +
Sbjct: 283 VSKFVMEFFES 293
>UniRef100_Q9FJA4 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 1197
Score = 171 bits (434), Expect = 8e-41
Identities = 127/476 (26%), Positives = 214/476 (44%), Gaps = 21/476 (4%)
Query: 512 SWNIICAGAQGVPLLLKDIVMRHRVSCLALYEPRVQTNNLPKIMKTLGFEDFFVVEAEGF 571
SWN G +++I+ + + L E Q + K + L + F+V G
Sbjct: 2 SWNCQGIGNPKTVRRVREILKKLSPDVIFLMETMNQDEVVLKKLDFLKRYNHFLVSPSGQ 61
Query: 572 SGGIWVLWSKHWGDVEILSSHRQFVHLRIHPKNNVPNFLVTFVYGSPNASIRNFLWQELQ 631
G L+ KH ++ I+SS F+ R+ K +FL T+VYG P S + W++L+
Sbjct: 62 GKGGLALFWKHDVEMAIVSSCDNFIDARVSFKRI--SFLATYVYGEPEQSKKGATWEKLK 119
Query: 632 NIASQVTKPWVTVGDFNTYLQASDKMGGGPPNLLSMAKFRDCLDACSLSDLGFKGPPFTW 691
+A +PW GDFN L S+K G P S +FR + C L DL G +W
Sbjct: 120 CLADNREEPWFLTGDFNEILDNSEKSEGPPRAEGSFVEFRSFMSECGLFDLKHLGNSLSW 179
Query: 692 KG----RGVKERIDWALGNARWISSFPEATVLHLPKLKYDHKPILIQLGPEHMDSSQRPF 747
+G V+ R+D A+ N + F + +H+PI+ L + + F
Sbjct: 180 RGVRHTHLVRARLDRAMSNGACLELFSRGRCDYRKFEGSNHRPIITFL-RQDKQRKKGLF 238
Query: 748 RFLAAWLTHEDFPRLVKSSWDSHHDSWIPASEAFCEEA-ASWNRDVFGEIGRTKRRLMRR 806
R+ +E+ L+ ++W+ + + C A SW ++ T+R +
Sbjct: 239 RYDRRLKENEEIKDLIAAAWNEDLSDSVEKKLSRCRRAIISWTKE-------TQRNSQKI 291
Query: 807 LEGINNHLRMSHSPYLEK--LQKRLWKEYQTVLIREELLWRQKSRVNWLNHGDRNTHFFH 864
+E + L + + L ++ E + EE W+Q+SR NWL+ GD+N+ +FH
Sbjct: 292 IEALKVKLEQAMVIPVNDVCLLVQINSELSAAYLAEEAFWKQRSRQNWLSLGDQNSGYFH 351
Query: 865 TTTMVHRKRNKIEALNNDVGELITDQDQLRSMAVTSFQNLYTS--PGPGEPYHV--RNAF 920
T N + D+GE + + Q + T +Q L+TS P E + +
Sbjct: 352 AVTKGRSCLNNLSVRETDLGEKVFEDSQFAEVISTCYQALFTSQLSNPEEISRIVQESVH 411
Query: 921 PSLPRETLDEIMGPLEAAEIRVAVFSMGAMKAPGPDDLNPLFFQSQWQVIGPSVVK 976
P + E D ++ A EI+ A+F++ KAPG D + FFQ+ W ++G S+V+
Sbjct: 412 PKVIEEENDFLISIPSAMEIKEALFNVHPDKAPGYDGFSASFFQTNWTIVGESIVQ 467
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.328 0.141 0.459
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,760,067,972
Number of Sequences: 2790947
Number of extensions: 77411765
Number of successful extensions: 215007
Number of sequences better than 10.0: 477
Number of HSP's better than 10.0 without gapping: 178
Number of HSP's successfully gapped in prelim test: 307
Number of HSP's that attempted gapping in prelim test: 213511
Number of HSP's gapped (non-prelim): 955
length of query: 992
length of database: 848,049,833
effective HSP length: 137
effective length of query: 855
effective length of database: 465,690,094
effective search space: 398165030370
effective search space used: 398165030370
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0113.5


