

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0112.15
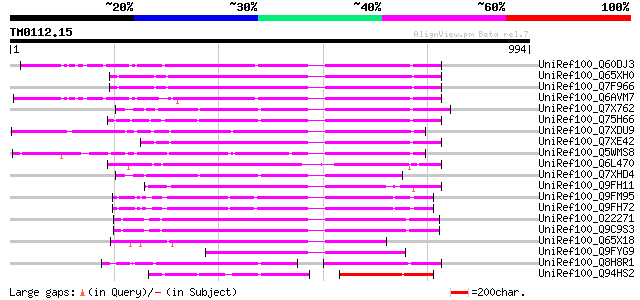
(994 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa] 365 4e-99
UniRef100_Q65XH0 Hypothetical protein OJ1126_B11.9 [Oryza sativa] 353 1e-95
UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa] 346 2e-93
UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa] 343 1e-92
UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa] 334 7e-90
UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sa... 330 1e-88
UniRef100_Q7XDU9 Putative mutator protein [Oryza sativa] 329 2e-88
UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa] 320 2e-85
UniRef100_Q5WMS8 Hypothetical protein OJ1333_C12.3 [Oryza sativa] 294 1e-77
UniRef100_Q6L470 Putative mutator transposable element [Solanum ... 288 4e-76
UniRef100_Q7XHD4 Putative mutator-like transposase [Oryza sativa] 283 2e-74
UniRef100_Q9FH11 Mutator-like transposase-like [Arabidopsis thal... 266 2e-69
UniRef100_Q9FM95 Similarity to mutator-like transposase [Arabido... 257 1e-66
UniRef100_Q9FH72 Arabidopsis thaliana genomic DNA, chromosome 5,... 256 2e-66
UniRef100_O22271 Putative MuDR-A-like transposon protein [Arabid... 249 3e-64
UniRef100_Q9C9S3 Putative Mutator-like transposase; 12516-14947 ... 248 8e-64
UniRef100_Q65X18 Putative polyprotein [Oryza sativa] 228 7e-58
UniRef100_Q9FYG9 F1N21.6 [Arabidopsis thaliana] 202 5e-50
UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa] 169 3e-40
UniRef100_Q94HS2 Putative transposon protein [Oryza sativa] 169 5e-40
>UniRef100_Q60DJ3 MuDR family transposase protein [Oryza sativa]
Length = 1030
Score = 365 bits (937), Expect = 4e-99
Identities = 237/812 (29%), Positives = 404/812 (49%), Gaps = 70/812 (8%)
Query: 22 SYVGGSIETLKGWDTDEISSLEVGKTVK---FFWNGSFKSLWYRP-PDIDNTLKPLNCDE 77
SY+ G + + D S L + + V+ + + K W P DI + L+ + D
Sbjct: 55 SYLNGKVSWFDNVEIDTWSPLWLDQFVEDLGYLRTPTLKIYWLLPGKDISDGLRVVVSDT 114
Query: 78 DVRNLLQDVKGHTGVCVYVEHDSEPEKARESDGDVEIVERSVNDAPNSAPVNDAPNSPEV 137
D + V+ + VY++HD +K D D +I++ P +PN
Sbjct: 115 DTNVMASMVEKFRTLVVYIDHD---DKVAGIDWD-DIIDNPGTPLPKVI----SPNKVNC 166
Query: 138 VHEVPNSAPVNDALNSSSALVHEVPNSAPVNDALNSSSALVHEVPNSAPVNEPLNESEDE 197
V ++ + ++ + E + S+ H N + +E +
Sbjct: 167 VEKI-----IGESSKQQEGNLKEQSSRQQQGSRQQQSNLKEHN--NREQQEDDCDEEDSG 219
Query: 198 SYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNG 257
+ SDE E D +DD D + D+ + E++I+ A + ++G G+
Sbjct: 220 TESDDSDE-EFYDSDYELDDGDDDLFVDYVD----ENVIDEGVA-KGKKILKGKKARGSR 273
Query: 258 ARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFK--AVQDGEEVKFQVGHTFS 315
+ E ED D +L++ D+E R K FK A +D F+VG F
Sbjct: 274 LKGNLAIVPRGDESEDTDEEELNAADSDDEGVRLK----FKSFAEEDLNNPNFKVGLVFP 329
Query: 316 SKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAGCPFYLRLSKTDSRSYWLLVSLEA 375
S + +QA+ +Y+++ + ++ ++D++R C GCP+ L S+ +++ + +
Sbjct: 330 SVEKLRQAITEYSVRNRVEIKMPRNDRRRIRAHCAEGCPWNLYASEDSRAKAFVVKTCDE 389
Query: 376 DHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVK 435
HTC + ++ +K+LA K++ R +++ + + +L+W + L + K RA+
Sbjct: 390 RHTCQKEWILKRCTSKWLAGKYIEAFRADEKMSLTSFAKTVQLQWNLTLSRSKLARARRI 449
Query: 436 AVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFA 495
A++ I G+ ++QY L Y EL RSNPGS+ +K G +F + Y+ A K F
Sbjct: 450 AMKTIHGDEVEQYKLLWDYGKELRRSNPGSSFFLK----LDGSLFSQCYMSMDACKRGFL 505
Query: 496 TTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVND 555
CRPLI LDGC +K YGGQLL AVG D N+ ++PIAFAVVE E+ A+W WF++ L ND
Sbjct: 506 NGCRPLICLDGCHIKTKYGGQLLTAVGMDPNDCIYPIAFAVVEVESLATWKWFLETLKND 565
Query: 556 LDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGD 615
L + W ++D+QK GL+P + ++ +
Sbjct: 566 LGIENTYPWTIMTDKQK-------------------------------GLIPAVQQVFPE 594
Query: 616 VEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNL 675
EHR C++HLY NF+ ++ G +K LW AR+S++ EW M+VM+ LN++A++ + L
Sbjct: 595 SEHRFCVRHLYSNFQLQFKGEVLKNQLWACARSSSVQEWNKNMDVMRNLNKSAYEWLEKL 654
Query: 676 PPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNR-IVKQKQ 734
PP W R+ +S +CD+ +NN CE FN+ ILE R+ PI+T+LE++K L R KQK+
Sbjct: 655 PPNTWVRAFFSEFPKCDILLNNNCEVFNKYILEARELPILTMLEKIKGQLMTRHFNKQKE 714
Query: 735 LMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLESRSCAC 794
L +++ +CP I++K+ + ++ +A G +F+V +Y V++ + C C
Sbjct: 715 LADQFQGLICPKIRKKVLKNADAANTCYALPAG---QGIFQVHEREYQYIVDINAMYCDC 771
Query: 795 RRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
RRWDL+GIPC HA++ + R E + + Y
Sbjct: 772 RRWDLTGIPCNHAISCLRHERINAESILPNCY 803
>UniRef100_Q65XH0 Hypothetical protein OJ1126_B11.9 [Oryza sativa]
Length = 792
Score = 353 bits (906), Expect = 1e-95
Identities = 212/637 (33%), Positives = 327/637 (51%), Gaps = 43/637 (6%)
Query: 192 NESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGS 251
+++ED S G E + E+ S DD + +EN + A + I + V S
Sbjct: 138 SDAEDSSETDGIAE---DDEERSEDDEEAQENREHANKVKKNPCATITVTSDNVVLVSDS 194
Query: 252 VDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVG 311
+ + +++ T + F D ++S D D E R R P++ + KF++G
Sbjct: 195 PNKQDDKGLDDD-TDTPFLDSSEEAS-YDDAEDGEAIRRKSRFPKYDS--KAHTPKFEIG 250
Query: 312 HTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICV-AGCPFYLRLSKTDSRSYWLL 370
TF+ + FK+AV Y L + + F KD+ K+ C CP+++ S + ++ +
Sbjct: 251 MTFAGRAKFKEAVIKYGLVTNRHIRFPKDEAKKIRARCSWKDCPWFIFASNGTNCDWFQV 310
Query: 371 VSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAY 430
+ H C + NR ++ +A K+ II+ P+ + +L + RL + K
Sbjct: 311 KTFNDVHNCPKRRDNRLVTSRRIADKYEHIIKSNPSWKLQSLKKTVRLDMFADVSISKVK 370
Query: 431 RAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAAT 490
RAK + I +Y+ + +Y AE+LRSNPGSTV I PVF+R+YVCF A
Sbjct: 371 RAKGIVMRRIYDACRGEYSKVFEYQAEILRSNPGSTVAICLDHEYNWPVFQRMYVCFDAC 430
Query: 491 KTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVD 550
K F CR +IGLDGCF KG G+LL A+G+D NNQM+PIA+AVVE ETK +WSWF+
Sbjct: 431 KKGFLAGCRKVIGLDGCFFKGACNGELLCALGRDPNNQMYPIAWAVVEKETKDTWSWFIG 490
Query: 551 LLVNDLDM-VERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTL 609
LL DL++ W ISDQQK GLV +
Sbjct: 491 LLQKDLNIDPHGAGWVIISDQQK-------------------------------GLVSAV 519
Query: 610 AELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAF 669
E + +EHR+C +H+Y N+RKKY ++ W+ A+AS P + + +L A
Sbjct: 520 EEFLPQIEHRMCTRHIYANWRKKYRDQAFQKPFWKCAKASCRPFFNFCRAKLAQLTPAGA 579
Query: 670 QDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRI 729
+DMM+ P W+R+ + + CD NNMCE+FN I++ R PII++ E ++ + RI
Sbjct: 580 KDMMSTEPMHWSRAWFRIGSNCDSVDNNMCESFNNWIIDIRAHPIISMFEGIRTKVYVRI 639
Query: 730 VKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLES 789
+ + W +CP I +KL S A W G + FEV+ +++Y+V+LE
Sbjct: 640 QQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIWNGKDG---FEVTDKDKRYTVDLEK 696
Query: 790 RSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
R+C+CR W L+GIPCAHA+ +++ + PEDY+ Y
Sbjct: 697 RTCSCRYWQLAGIPCAHAITALFVSSKQPEDYIADCY 733
>UniRef100_Q7F966 OSJNBa0091C07.2 protein [Oryza sativa]
Length = 879
Score = 346 bits (887), Expect = 2e-93
Identities = 209/637 (32%), Positives = 324/637 (50%), Gaps = 43/637 (6%)
Query: 192 NESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGS 251
+++ED S G E + E+ S DD + +EN + A + I + V S
Sbjct: 138 SDAEDSSETDGIAE---DDEERSEDDEEAQENREHANKVKKNPCATITVTSDNVVLVSDS 194
Query: 252 VDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVG 311
+ + +++ T + F D ++S D D E R R P++ + KF++G
Sbjct: 195 PNKQDDKGLDDD-TDTPFLDSSEEAS-YDDAEDGEAIRRKSRFPKYDS--KAHTPKFEIG 250
Query: 312 HTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICV-AGCPFYLRLSKTDSRSYWLL 370
TF+ + FK+AV Y L + + F KD+ K+ C CP+++ S + ++ +
Sbjct: 251 MTFAGRAKFKEAVIKYGLVTNRHIRFPKDEAKKIRARCSWKDCPWFIFASNGTNCDWFQV 310
Query: 371 VSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAY 430
+ H C + NR ++ +A K+ II+ P+ + +L + RL + K
Sbjct: 311 KTFNDVHNCPKRRDNRLVTSRRIADKYEHIIKSNPSWKLQSLKKTVRLDMFADVSISKVK 370
Query: 431 RAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAAT 490
RAK + I +Y+ + +Y AE+LRSNPGSTV I PVF+R+YVCF A
Sbjct: 371 RAKGIVMRRIYDACRGEYSKVFEYQAEILRSNPGSTVAICLDHEYNWPVFQRMYVCFDAC 430
Query: 491 KTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVD 550
K F CR +IGLDGCF KG G+LL A+G+D NNQM+PIA+AVVE ETK +WSWF+
Sbjct: 431 KKGFLAGCRKVIGLDGCFFKGACNGELLCALGRDPNNQMYPIAWAVVEKETKDTWSWFIG 490
Query: 551 LLVNDLDM-VERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTL 609
LL DL++ W ISDQQK GLV +
Sbjct: 491 LLQKDLNIDPHGAGWVIISDQQK-------------------------------GLVSAV 519
Query: 610 AELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAF 669
E + +EHR+C +H+Y N+RKKY ++ W+ A+AS P + + +L A
Sbjct: 520 EEFLPQIEHRMCTRHIYANWRKKYRDQAFQKPFWKCAKASCRPFFNFCRAKLAQLTPAGA 579
Query: 670 QDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRI 729
+ + P W+R+ + + CD NNMCE+FN I++ R PII++ E ++ + RI
Sbjct: 580 KXXXSTEPMHWSRAWFRIGSNCDSVDNNMCESFNNWIIDIRAHPIISMFEGIRTKVYVRI 639
Query: 730 VKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLES 789
+ + W +CP I +KL S A W G + FEV+ +++Y+V+LE
Sbjct: 640 QQNRSKAKGWLGRICPNILKKLNKYIDLSGNCEAIWNGKDG---FEVTDKDKRYTVDLEK 696
Query: 790 RSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
R+C+CR W L+GIPCAHA+ +++ + PEDY+ Y
Sbjct: 697 RTCSCRYWQLAGIPCAHAITALFVSSKQPEDYIADCY 733
>UniRef100_Q6AVM7 Putative polyprotein [Oryza sativa]
Length = 1006
Score = 343 bits (881), Expect = 1e-92
Identities = 232/833 (27%), Positives = 404/833 (47%), Gaps = 90/833 (10%)
Query: 7 MNIYFGGKITYEPTL-SYVGGSIETLKGWDTDEISSLEVGKTVK---FFWNGSFKSLWYR 62
+ ++ GG L SY+ G + + D S L + + V+ + + K W
Sbjct: 24 VKVHHGGFFVGHGNLRSYLNGKVSWFDNVEIDTWSPLWLDQFVEDLGYLRTPTLKIYWLL 83
Query: 63 P-PDIDNTLKPLNCDEDVRNLLQDVKGHTGVCVYVEHDSEPEKARESDGDVEIVERSVND 121
P DI + L+ + D + V+ + VY++HD +K D D +I++
Sbjct: 84 PGKDISDGLRVVVSDTYTNVMASMVEKFRTLVVYIDHD---DKVAGIDWD-DIIDNPGTP 139
Query: 122 APNSAPVNDAPNSPEVVHEVPNSAPVNDALNSSSALVHEVPNSAPVNDALNSSSALVHEV 181
P +PN V ++ + ++ + E + S+ H
Sbjct: 140 LPKVI----SPNKVNCVEKI-----IGESSKQQEGNLKEQSSRQQQGSRQQQSNLKEHN- 189
Query: 182 PNSAPVNEPLNESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATA 241
N + +E + + SDE E D +DD D + D+ + E++I+ A
Sbjct: 190 -NREQQEDDCDEEDSGTKSDDSDE-EFYDSDYELDDGDDDLFVDYVD----ENVIDEGVA 243
Query: 242 DRDPPFVQGSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQ 301
+ ++G G+ + E ED D +L++ D+E
Sbjct: 244 -KGKKILKGKKARGSRLKGNLAIVPRGDESEDTDEEELNAADSDDE-------------- 288
Query: 302 DGEEVKFQVGHTFSSKDL-------FKQAVKDYALQQKKDLTFKKDDKKRCVVICVAGCP 354
G +KF+ +F+ +DL +QA+ +Y+++ + ++ ++D++R C GCP
Sbjct: 289 -GVRLKFK---SFAEEDLNNPNFKKLRQAITEYSVRNRVEIKMPRNDRRRIRAHCEEGCP 344
Query: 355 FYLRLSKTDSRSYWLLVSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIE 414
+ L S+ +++ + + HTC + ++ +K+LA K++ R +++ + +
Sbjct: 345 WNLYTSEDSRAKAFVVKTYDERHTCQKEWILKRCTSKWLAGKYIEAFRADEKMSLTSFAK 404
Query: 415 EARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKG 474
+L+W + + K RA+ A++ I G+ ++QY L Y EL RSNPG++ +K
Sbjct: 405 TVQLQWNLTPSRSKLARARRIAMKTIHGDEVEQYKLLWDYGKELRRSNPGTSFFLK---- 460
Query: 475 AKGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAF 534
G +F + Y+ A K F CRPLI LDGC +K YGGQLL AVG D N+ ++PIAF
Sbjct: 461 LDGSLFSQCYMSMDACKRGFLNGCRPLICLDGCHIKTKYGGQLLTAVGMDPNDCIYPIAF 520
Query: 535 AVVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YL 594
AVVE E+ A+W WF++ L NDL + W ++D+QK
Sbjct: 521 AVVEVESLATWKWFLETLKNDLGIENTYPWTIMTDKQK---------------------- 558
Query: 595 SYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEW 654
GL+P + ++ + EHR C++HLY NF+ ++ G K LW AR+S++ EW
Sbjct: 559 ---------GLIPAVQQVFPESEHRFCVRHLYSNFQLQFKGEVPKNQLWACARSSSVQEW 609
Query: 655 YAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPI 714
M+VM+ LN++A++ + LPP W R+ +S +CD+ +NN CE FN+ ILE R+ PI
Sbjct: 610 NKNMDVMRNLNKSAYEWLEKLPPNTWVRAFFSEFPKCDILLNNNCEVFNKYILEARELPI 669
Query: 715 ITLLERLKFYLNNR-IVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSL 773
+T+LE++K L R KQK+L +++ +CP I++K+ + ++ +A G +
Sbjct: 670 LTMLEKIKGQLMTRHFNKQKELADQFQGLICPKIRKKVLKNADAANTCYALPAG---QGI 726
Query: 774 FEVSRDNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
F+V +Y V++ + C CRRWDL+GIPC HA++ + R E + + Y
Sbjct: 727 FQVHEREYQYIVDINAMHCDCRRWDLTGIPCNHAISCLRHERINAESILPNCY 779
>UniRef100_Q7X762 OSJNBb0118P14.3 protein [Oryza sativa]
Length = 939
Score = 334 bits (857), Expect = 7e-90
Identities = 211/642 (32%), Positives = 320/642 (48%), Gaps = 61/642 (9%)
Query: 204 DEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNGARIEEP 263
DE E +ED + DSDYE I+ D V+G +++ + EE
Sbjct: 172 DEEESRTEDPNFVDSDYE--------------IDHGDDDLFDNCVEGKIELAHNK--EEG 215
Query: 264 ATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQA 323
+ + EDE D+ S++EEE F+A D E +F+V F+ ++A
Sbjct: 216 SDYVSSEDERLMMPDI---SEEEEEKIKFSFKSFRANVDMETPEFKVSIMFADVVELRKA 272
Query: 324 VKDYALQQKKDLTFKKDDKKRCVVICVAGCPFYLRLSKTDSRSYWLLVSLEAD-HTCCRS 382
+ Y ++ + + ++ K R C GCP+ L S D+R L+V + HTC +
Sbjct: 273 IDQYTIKNQVAIKKTRNTKTRIEAKCAEGCPWMLSASM-DNRVKCLVVREYIEKHTCSKQ 331
Query: 383 ASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQG 442
+ KYLA++++ R +T+ + ++ + + + K RA+ A+ I G
Sbjct: 332 WEIKAVTAKYLAKRYIEEFRDNDKMTLMSFAKKIQKELHLTPSRHKLGRARRMAMRAIYG 391
Query: 443 ETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFATTCRPLI 502
+ + QY L Y EL SNPGS+ + G F +Y+ F A K F CRP+I
Sbjct: 392 DEISQYNQLWDYGQELRTSNPGSSFYLNLHFGC----FHTLYMSFDACKRGFMFGCRPII 447
Query: 503 GLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDMVERK 562
LDGC +K +GG +L AVG D N+ +FPIA AVVE E+ SWSWF+D L DL +
Sbjct: 448 CLDGCHIKTKFGGHILTAVGMDPNDCIFPIAIAVVEVESLKSWSWFLDTLKKDLGIENTS 507
Query: 563 RWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCI 622
W ++D+QK GLVP + D E R C+
Sbjct: 508 AWTVMTDRQK-------------------------------GLVPAVRREFSDAEQRFCV 536
Query: 623 KHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTR 682
+HLY NF+ + G +K LW AR+ST+PEW A ME MK L+ A++ + +PP W R
Sbjct: 537 RHLYQNFQVLHKGETLKNQLWAIARSSTVPEWNANMEKMKALSSEAYKYLEEIPPNQWCR 596
Query: 683 SAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRI-VKQKQLMLRWRT 741
+ +S +CD+ +NN E FN+ IL+ R+ PI+++LER++ + NR+ KQK+L W
Sbjct: 597 AFFSDFPKCDILLNNNSEVFNKYILDAREMPILSMLERIRNQIMNRLYTKQKELERNWPC 656
Query: 742 ALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLESRSCACRRWDLSG 801
+CP I++K+E + ++ + G M F+VS +Y V L + C CRRW L+G
Sbjct: 657 GICPKIKRKVEKNTEMANTCYVFPAG---MGAFQVSDIGSQYIVELNVKRCDCRRWQLTG 713
Query: 802 IPCAHAVASMWFGRRVPEDYVHSSYRHLLIHTPTSSYLIMDL 843
IPC HA++ + R PED V Y + SY IM L
Sbjct: 714 IPCNHAISCLRHERIKPEDVVSFCY-STRCYEQAYSYNIMPL 754
>UniRef100_Q75H66 Hypothetical protein OSJNBb0007E22.22 [Oryza sativa]
Length = 981
Score = 330 bits (846), Expect = 1e-88
Identities = 205/646 (31%), Positives = 328/646 (50%), Gaps = 56/646 (8%)
Query: 187 VNEPLNESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPP 246
V+E +E ED+ D E +D DSDYE D ++ E ++ D+ P
Sbjct: 172 VDEVADEIEDDVEY---DSDGSEVDDSDFVDSDYEFQDDDDDLF--EDNVDGDVVDQGPA 226
Query: 247 FVQGSVDVGNGARIEEPATFSDFEDEDGDSSD---LDSPSDDEEEGRSKRLPRFKAV--Q 301
+ + G +++ + E D +SSD L+ P + +E RFK+ +
Sbjct: 227 PKKFNQKKVAGGKLKGKRVIRE-EGSDEESSDEECLELPENPDEINL-----RFKSFNPE 280
Query: 302 DGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAGCPFYLRLSK 361
D F+VG F S +L ++ + +Y+L+ + D+ ++D+ R C GCP+ + S
Sbjct: 281 DMNNPVFKVGMVFPSVELLRKTITEYSLKNRVDIKMPRNDRTRIKAHCAEGCPWNMYAS- 339
Query: 362 TDSRSYWLLVSLEA-DHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRW 420
DSR + +V H C + ++ +LA K++ R +T+ + + + W
Sbjct: 340 LDSRVHGFIVKTYVPQHKCRKEWVLQRCTANWLALKYIESFRADSRMTLPSFAKTVQKEW 399
Query: 421 GILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVF 480
+ + K RA+ A++ I G+ + QY L Y EL RSNPGS+ +K G F
Sbjct: 400 NLTPSRSKLARARRLALKEIYGDEVAQYNMLWDYGNELRRSNPGSSFYLKLDDGK----F 455
Query: 481 ERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAE 540
+Y A K F + CRP+I LDGC +K +GGQLL AVG D N+ +FPIA AVVE E
Sbjct: 456 SCLYFSLDACKRGFLSGCRPIICLDGCHIKTKFGGQLLTAVGIDPNDCIFPIAMAVVEVE 515
Query: 541 TKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSC 600
+ ++WSWF+ L +D+ +V W ++D+QK
Sbjct: 516 SFSTWSWFLQTLKDDVGIVNTYPWTIMTDKQK---------------------------- 547
Query: 601 VMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEV 660
GL+P + +L D EHR C++HLY NF++ + G +K LW AR+S++ EW E
Sbjct: 548 ---GLIPAVQQLFPDSEHRFCVRHLYQNFQQSFKGEILKNQLWACARSSSVQEWNTKFEE 604
Query: 661 MKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLER 720
MK LNE A+ + + P W R+ +S +CD+ +NN CE FN+ ILE R+ PI+T+LE+
Sbjct: 605 MKALNEDAYNWLEQMAPNTWVRAFFSDFPKCDILLNNSCEVFNKYILEAREMPILTMLEK 664
Query: 721 LKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDN 780
+K L R +++ +W+ +CP I++KL ++ + G +F+V
Sbjct: 665 IKGQLMTRFFNKQKEAQKWQGPICPKIRKKLLKIAEQANICYVLPAG---KGVFQVEERG 721
Query: 781 EKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
KY V++ ++ C CRRWDL+GIPC HA+A + ED++ Y
Sbjct: 722 TKYIVDVVTKHCDCRRWDLTGIPCCHAIACIREDHLSEEDFLPHCY 767
>UniRef100_Q7XDU9 Putative mutator protein [Oryza sativa]
Length = 929
Score = 329 bits (844), Expect = 2e-88
Identities = 244/804 (30%), Positives = 377/804 (46%), Gaps = 71/804 (8%)
Query: 4 RIDMNIYFGGK-ITYEPTLSYVGGSIETLKGWDTDEISSLEVGKTVKFF---WNGSFKSL 59
++ + +F G+ +T Y GG E L D D+IS E+ ++ G+
Sbjct: 6 KLVVRFHFNGEFVTSGREKKYCGGR-EALSYIDRDKISLPEIFGHLRDHCKVMEGTMLHW 64
Query: 60 WYRPPDIDNTLKPLNCDEDVRNLLQDVKGHTGVC-VYVEHDSEPEKARESDGDVEIVERS 118
+ D+++ L+ L D+ V L+ D GV VY E + + SD D S
Sbjct: 65 LFPGKDLNSGLRAL-LDDTVCKLMADCIDDLGVAEVYAEEPTIVDLCDSSDDD------S 117
Query: 119 VNDAPNSAPVNDAPNSPEVVHEVPNSAPVNDALNSSSALVHEVPNSAPVNDALNSSSALV 178
+A S ++ E E +A + +EV ++ + + +V
Sbjct: 118 SYEADESEDDSEEMEVAEGGMEGTENAVALEG-GEIRTKTNEVRKGKAISSEIPENRLVV 176
Query: 179 HEVPNSAPVNEPLNESEDES-YIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLIN 237
+ N ++ +ESE S YIPG D + E++ + Y+ D + A L N
Sbjct: 177 YNCDNDPLASQAPSESESNSEYIPGDDAGSDDDEEIIEIEKHYK---DMKRKVKAGQLDN 233
Query: 238 IATADRDPPFV--QGSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLP 295
+ D ++ QG ++ N A E ++ + D S + +D E + + P
Sbjct: 234 L-----DDVYLGGQGWQNMNNEAGGEGDGIAAESPE---DESMEEIGTDGEVSTKENKFP 285
Query: 296 RFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICV-AGCP 354
R+K + G F++G FSSK FK+AV YAL ++K + F KDD KR C A CP
Sbjct: 286 RYKK-KVGTPC-FELGMKFSSKKQFKKAVTKYALAERKVINFIKDDLKRVRAKCDWASCP 343
Query: 355 FYLRLSKTDSRSYWLLVSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITV---NA 411
+ LSK W +V+LE+ H C N+ +A K+ II P + A
Sbjct: 344 WVCLLSKNTRSDSWQIVTLESLHACPPRRDNKMVTATRIAEKYGKIILANPGWNLAHMKA 403
Query: 412 LIEEARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKS 471
++E + K RAK ++ T QY L Y ELLRSNPGSTV++
Sbjct: 404 TVQEEMFGEASVP---KLKRAKAMVMKKAMDATKGQYQKLYNYQLELLRSNPGSTVVVNR 460
Query: 472 TKGAKGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFP 531
G PVF+RIY+C A K F CR ++GLDGCF KG G+LL A+G+D NNQM+P
Sbjct: 461 EIGTDPPVFKRIYICLDACKRGFVDGCRKVVGLDGCFFKGATNGELLCAIGRDANNQMYP 520
Query: 532 IAFAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I 591
IA+AVV E W WF+DLL +DL +++ W FISDQQK
Sbjct: 521 IAWAVVHKENNEEWDWFLDLLCSDLKVLDGTGWVFISDQQK------------------- 561
Query: 592 *YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTL 651
G++ + + EHR C +H+Y N+++ + E ++ WR A+A
Sbjct: 562 ------------GIINAVEKWAPQAEHRNCARHIYANWKRHFSDKEFQKKFWRCAKAPCR 609
Query: 652 PEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRD 711
+ A + ++ A Q ++N P+ W+R+ + + CD NNMCE+FN+ ILE R
Sbjct: 610 MLFNLARAKLAQVTPAGAQAILNTHPEHWSRAWFKLGSNCDSVDNNMCESFNKWILEARF 669
Query: 712 KPIITLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYM 771
PIIT+LE ++ + RI +Q+++ RW T +CP I +KL S A G +
Sbjct: 670 FPIITMLETIRRKVMVRISEQRKVSARWNTVVCPGILKKLNVYITESAYCHAICNGAD-- 727
Query: 772 SLFEVSRDNEKYSVNLESRSCACR 795
FEV +++V L+ + C+CR
Sbjct: 728 -SFEVKHHTNRFTVQLDKKECSCR 750
>UniRef100_Q7XE42 Putative mutator-like transposase [Oryza sativa]
Length = 1005
Score = 320 bits (819), Expect = 2e-85
Identities = 186/579 (32%), Positives = 298/579 (51%), Gaps = 40/579 (6%)
Query: 250 GSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQ 309
G + +GN ++ E +++ D DGD+S D SD R R P F + D + F
Sbjct: 232 GDIPLGNN-QVLEGGDGAEYFDSDGDAS-YDEDSDGVFTRRKCRFPIFDSFADTPQ--FA 287
Query: 310 VGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICV-AGCPFYLRLSKTDSRSYW 368
V F KD K A++ YAL++K ++ + K+++KR +C GCP+ L S ++
Sbjct: 288 VDMCFRGKDQLKDAIERYALKKKINIRYVKNEQKRIRAVCRWKGCPWLLYASHNSRSDWF 347
Query: 369 LLVSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWK 428
+V+ +H CC N++ T+ + ++ I+ P+ + E + G+ +
Sbjct: 348 QIVTYNPNHACCPELKNKRLSTRRICDRYESTIKANPSWKAREMKETVQEDMGVDVSITM 407
Query: 429 AYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFA 488
RAK ++ I +Y+ L YA +L RSNPG++V I + VF+R YVCF
Sbjct: 408 IKRAKTHVMKKIMDTQTGEYSKLFDYALKLQRSNPGNSVHIALDPEEEDHVFQRFYVCFD 467
Query: 489 ATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWF 548
A + F CR +IGLDGCFLKG G+LL+A+G+D NNQ++PIA+AVVE E K SW+WF
Sbjct: 468 ACRRGFLEGCRRIIGLDGCFLKGPLKGELLSAIGRDANNQLYPIAWAVVEYENKDSWNWF 527
Query: 549 VDLLVNDLDM-VERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVP 607
+ L D+++ V W FI+DQQK GL+
Sbjct: 528 LGHLQKDINIPVGAAGWVFITDQQK-------------------------------GLLS 556
Query: 608 TLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEA 667
++ L EHR+C +H+Y N+RKK+ E ++ W+ A+A + +
Sbjct: 557 IVSTLFPFAEHRMCARHIYANWRKKHRLQEYQKRFWKIAKAPNEQLFNHYKRKLAAKTPR 616
Query: 668 AFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNN 727
+QD+ P W+R+ + + C+ NNM E+FN I+E R KPIIT+LE ++ +
Sbjct: 617 GWQDLEKTNPIHWSRAWFRLGSNCESVDNNMSESFNSWIIESRFKPIITMLEDIRIQVTR 676
Query: 728 RIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNL 787
RI + + RW +CP I +K + + W GD + FEV +++V+L
Sbjct: 677 RIQENRSNSERWTMTVCPNIIRKFNKIRHRTQFCHVLWNGD---AGFEVRDKKWRFTVDL 733
Query: 788 ESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
S++C+CR W +SGIPC HA A+++ + P + +H +
Sbjct: 734 TSKTCSCRYWQVSGIPCQHACAALFKMAQEPNNCIHECF 772
>UniRef100_Q5WMS8 Hypothetical protein OJ1333_C12.3 [Oryza sativa]
Length = 892
Score = 294 bits (752), Expect = 1e-77
Identities = 236/805 (29%), Positives = 366/805 (45%), Gaps = 89/805 (11%)
Query: 5 IDMNIYFGGKI--TYEPTLSYVGGSIETLKGWDTDEISSLEVGKTVKFFWN---GSFKSL 59
+ + +FGG+ + T YVGGS E + + D+IS E+ +K G
Sbjct: 7 LSVRFHFGGEFINNWRETF-YVGGS-EGMSYIERDKISLPELIGYLKDHCEVLPGMLLHW 64
Query: 60 WYRPPDIDNTLKPLNCDEDVRNLLQDVKGHTGVC-VYVE-----HDSEPEKARESDGDVE 113
+ ++ + L+ L D+ V + + GV +YVE +E + +SD + E
Sbjct: 65 LFPGKELADGLRVL-VDDKVCDYMSKCVVDGGVAEIYVEAVVGGESNESGSSDDSDFEDE 123
Query: 114 IVERSVNDAPNSAPVNDAPNSPEVVHEVPNSAPVNDALNSSSALVHEVPNSAPVNDALNS 173
+ + S+ D +ND ++PE + S L S P
Sbjct: 124 LEDMSLAD-DEWDDLNDDASTPE---------------KTKSDLDRLRAWSTPSPKDKGK 167
Query: 174 SSALVHEVPNSAPVNEPLNESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAE 233
+ L E SA ++ D Y+ G GE ++V Y+E + + L
Sbjct: 168 AVVLCEERVKSADLS-----GSDNDYVGGDSCSSGEDQEVEEIRKAYKE---FKKKLKDG 219
Query: 234 SLINIATADRDPPFVQGSVDVGNGA--RIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRS 291
+ N+ D ++ S + +GA +I+ S +E+ D + SD + +
Sbjct: 220 EVGNLD----DVIYMGSSRQINDGALVQIQGEEDNSPYENSSADGDSYEEDSDGQLVRKK 275
Query: 292 KRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICV- 350
+ P F A Q V+ +G F K FK+AV ALQ K+ + F KD+ R C
Sbjct: 276 SKYPIFNANQP--HVRLALGMKFDGKKEFKEAVVQLALQNKRFIRFPKDEGYRTRAKCDW 333
Query: 351 AGCPFYLRLSKTDSRSYWLLVSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVN 410
A CP+ LS+ + W + SL +H C N K +A+K+ I P ++
Sbjct: 334 ATCPWSFLLSRNSRTNSWQIASLVDEHNCPPRKDNNLVTYKRIAQKYEKTIIDNPTWSIQ 393
Query: 411 ALIEEARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIK 470
++ + + K RAK ++ I D+Y+ + Y ELLRSNPGS V++K
Sbjct: 394 SMQSTVSEQ---MFANCK--RAKAYVLKKIYESRRDEYSRIFDYQLELLRSNPGSIVVVK 448
Query: 471 STKGAKGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMF 530
PVF+RIYVC AA K F CR ++GLDGCF KG G+LL AVG+D NN M+
Sbjct: 449 LDTDQPSPVFKRIYVCLAACKNGFLLGCRKVVGLDGCFFKGSNNGELLCAVGRDANNSMY 508
Query: 531 PIAFAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY* 590
PIA+AV ET SW WF DLL DL E W FISDQQK
Sbjct: 509 PIAWAV---ETNDSWDWFCDLLCKDLGFGEGDGWVFISDQQK------------------ 547
Query: 591 I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARAST 650
G+V + EHR C +H+Y N++KK+ E ++ WR A+A
Sbjct: 548 -------------GIVNAVQHWAPSAEHRNCARHIYANWKKKFSKKEWQKKFWRCAKAPN 594
Query: 651 LPEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYR 710
+ + A + + + +MN P W+R+ + + D NN+CE FN+ I++ R
Sbjct: 595 VMLFNLAKARLAQETVEGARAIMNTDPSHWSRAWFRFGSNYDSVDNNICETFNKWIVQAR 654
Query: 711 DKPIITLLERLKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEY 770
PII++LE ++ + RI ++ LM +W ++CP I +KL +S A G
Sbjct: 655 FLPIISMLEAIRRKVMVRIHEKITLMDKWLGSICPNIHKKLNAYIIDSGNCHAICNG--- 711
Query: 771 MSLFEVSRDNEKYSVNLESRSCACR 795
M FEV N +++V+LE ++C+CR
Sbjct: 712 MDKFEVKHQNHRFTVDLERKTCSCR 736
>UniRef100_Q6L470 Putative mutator transposable element [Solanum demissum]
Length = 707
Score = 288 bits (738), Expect = 4e-76
Identities = 201/660 (30%), Positives = 325/660 (48%), Gaps = 84/660 (12%)
Query: 188 NEPLNESEDESYIPG-SDEYEGESEDVSVDDSDYEENWD----WAEVLPA-ESL-INIAT 240
N+ NE + S + ++E+ E +D + +DSDY D + V P+ ES INI
Sbjct: 61 NQCGNEMKKNSLLANDTEEHSDEFDDENFNDSDYSLEEDDLLFFKNVDPSVESFGINITV 120
Query: 241 ADRDPPFVQGSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAV 300
+ Q V +++ SD D D D+ L+S D E E + P+
Sbjct: 121 KKKKNDGNQNYVSEEMHRKMQNEEGDSDCVDFD-DTKSLNSDCDSEFEDCN--FPKHNPK 177
Query: 301 QDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAGCPFYLRLS 360
+ ++G +K FK+AV + K + + KDD++R C + L
Sbjct: 178 IGAFNPELELGMVLDNKKEFKEAVVANQAKIGKSIRWTKDDRERARAKCRTNACKWRILG 237
Query: 361 KTDSR--SYWLLVSLEADHTCCR-SASNRQAKTKYLARKFMPIIRHTPNITVNALIEEAR 417
R S + + + ++HTC + +N+ + ++ARK++ ++ N + +
Sbjct: 238 SLMQRDISTFQIKTFVSEHTCFGWNYNNKTINSSWIARKYVDRVKSNKNWRTSEFRDTLS 297
Query: 418 LRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGA-- 475
+ +A RAK KA+ MI G+ DQ+ L Y E++R+NPG++V +K T
Sbjct: 298 RELKFHVSMHQARRAKEKAIAMIDGDINDQFGILWNYCNEIVRTNPGTSVFMKLTPNEIL 357
Query: 476 -KGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKG-MYGGQLLAAVGKDGNNQMFPIA 533
K F+R Y+CFAA K F CR +IG++GC+LK MYG QLL+AV DGNN +FPIA
Sbjct: 358 NKPMRFQRFYICFAACKAGFKADCRKIIGVNGCWLKNIMYGAQLLSAVTLDGNNNIFPIA 417
Query: 534 FAVVEAETKASWSWFVDLLVNDLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*Y 593
+A+VE E K W WF+ L+NDL++ E+
Sbjct: 418 YAIVEKENKEIWQWFLTYLMNDLEIEEQ-------------------------------- 445
Query: 594 LSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNFRKK-YPGAEMKQALWRAARASTLP 652
YL+ C++HL+ NF++ Y G +K ALW+AA A+T+
Sbjct: 446 --YLF----------------------CVRHLHNNFKRAGYSGMALKNALWKAASATTID 481
Query: 653 EWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDK 712
+ A M + EL++ A+ + P W+RS +S +CD+ +NN CE FN+ IL+ RDK
Sbjct: 482 RFDACMTDLFELDKDAYAWLSAKVPSEWSRSHFSPLPKCDILLNNQCEVFNKFILDARDK 541
Query: 713 PIITLLERLKFYLNNRIVKQKQLMLRWR-TALCPMIQQKLEDSKRNSDKWFAN----WMG 767
PI+ LLE ++ L RI ++ +W +CP I++KL + + + + W
Sbjct: 542 PIVKLLETIRHLLMTRINSIREKAEKWNLNDICPTIKKKLAKTMKKAANYIPQRSNMW-- 599
Query: 768 DEYMSLFEVSRDNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSYR 827
Y + V DN ++V+L +R+C+CR+W+LSG+PC HA++S+W +YV Y+
Sbjct: 600 -NYEVIGPVEGDN--WAVDLYNRTCSCRQWELSGVPCKHAISSIWLKNDEVLNYVDDCYK 656
>UniRef100_Q7XHD4 Putative mutator-like transposase [Oryza sativa]
Length = 812
Score = 283 bits (724), Expect = 2e-74
Identities = 175/551 (31%), Positives = 273/551 (48%), Gaps = 55/551 (9%)
Query: 204 DEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNGARIEEP 263
DE E +ED+ DSDYE I+ D V+G +++ + EE
Sbjct: 172 DEEESRTEDLDFVDSDYE--------------IDHGDDDLFDNCVEGQIELAHNK--EEG 215
Query: 264 ATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKDLFKQA 323
+ + EDE D+ ++EEE + F+A D E +F+V F+ ++A
Sbjct: 216 SDYGSSEDERLMMPDISEEEEEEEEIKFS-FKSFRADVDMETPEFKVSMMFADAVELRKA 274
Query: 324 VKDYALQQKKDLTFKKDDKKRCVVICVAGCPFYLRLSKTDSRSYWLLVSLEAD-HTCCRS 382
+ Y ++ + + ++ K R C GCP+ L S D+R L+V + HT +
Sbjct: 275 IDQYTIKNRVAIKKTRNTKTRIEAKCAEGCPWMLSASM-DNRVKCLVVREYIEKHTRSKQ 333
Query: 383 ASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQG 442
+ KYL ++++ R +T+ + ++ + + + K RA+ A+ I G
Sbjct: 334 WEIKAVTAKYLTKRYIEEFRDNDKMTLMSFAKKIQKELHLTPSRHKLGRARRMAMRAIYG 393
Query: 443 ETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFATTCRPLI 502
+ + QY L Y EL SNP S+ + G F +Y+ F A K F + CRP+I
Sbjct: 394 DEISQYNQLWDYGQELRTSNPRSSFYLNLLFGC----FHTLYMSFDACKRGFLSGCRPII 449
Query: 503 GLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDMVERK 562
LDGC +K +GG +L AVG D N+ +FPIA VVE E+ SWSWF+D L DL +
Sbjct: 450 CLDGCHIKTKFGGHILTAVGMDPNDCIFPIAIVVVEVESLKSWSWFLDTLKKDLGIENTS 509
Query: 563 RWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCI 622
W ++D+QK GLVP + E R C+
Sbjct: 510 AWTVMTDRQK-------------------------------GLVPAVRREFSHAEQRFCV 538
Query: 623 KHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTR 682
+HLY NF+ + G +K LW AR+ST+PEW A ME MK L+ A++ + +PP W R
Sbjct: 539 RHLYQNFQVLHKGETLKNQLWAIARSSTVPEWNANMEKMKALSSEAYKYLEEIPPNQWCR 598
Query: 683 SAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRI-VKQKQLMLRWRT 741
+ +S +CD+ +NN E FN+ IL+ R+ PI+++LER++ + NR+ KQK+L W
Sbjct: 599 AFFSDFPKCDILLNNNSEVFNKYILDAREMPILSMLERIRNQIMNRLYTKQKELERNWPC 658
Query: 742 ALCPMIQQKLE 752
LCP I++K+E
Sbjct: 659 GLCPKIKRKVE 669
>UniRef100_Q9FH11 Mutator-like transposase-like [Arabidopsis thaliana]
Length = 825
Score = 266 bits (681), Expect = 2e-69
Identities = 170/588 (28%), Positives = 301/588 (50%), Gaps = 75/588 (12%)
Query: 258 ARIEEPATFSDFEDEDGDSSDLDSP-SDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSS 316
AR+E+ + F DE+ +D +P + D EEG + + K D ++ F S
Sbjct: 155 ARVEQ--NLAGFVDEEEHDNDESTPLNSDCEEGERRYVRCKKGSGD-----IRIEQVFDS 207
Query: 317 KDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAG--CPFYLRLSKTDSRSYWLLVSLE 374
FK+AV DYAL++ ++ F + ++ V C G C F + + + +++ +
Sbjct: 208 IAEFKEAVIDYALKKGVNIKFTRWGSEKSEVRCSIGGNCKFRIYCAYDEKIGVYMVKTFI 267
Query: 375 ADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKV 434
+H C + + K++ + F+ IR P V A+ + R+ ++ + +A+
Sbjct: 268 DEHACTKDGFCKVLKSRIIVDMFLNDIRQDPTFKVKAMQKAIEERYSLIASSDQCRKARG 327
Query: 435 KAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAF 494
KA++MIQ E +Q+A + Y +LL +N GSTV +++ F++ YVCF A + +
Sbjct: 328 KALKMIQDEYDEQFARIHDYKEQLLETNLGSTVEVETITIDGIVKFDKFYVCFDALRKTW 387
Query: 495 ATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVN 554
CRP+IG+DGCFLK GQLLAAVG+D NNQ +P+A+AVVE E SW WF+ L +
Sbjct: 388 LAYCRPIIGIDGCFLKNNMKGQLLAAVGRDANNQFYPVAWAVVETENIDSWLWFIRKLKS 447
Query: 555 DLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVG 614
DL + + + ISD+QK GL+ T+ + +
Sbjct: 448 DLKLQDGDGFTLISDRQK-------------------------------GLLNTVDQELP 476
Query: 615 DVEHRLCIKHLYGNFRKKYPGAEMKQALWRA-ARASTLPEWYAAMEVMKELNEAAFQDMM 673
VEHR+C +H+YGN R+ YPG ++ + L+ A A++ E+ A++ +K+ ++ + +M
Sbjct: 477 KVEHRMCARHIYGNLRRVYPGKDLSKNLFCAVAKSFNDYEYNRAIDELKQFDQGVYDAVM 536
Query: 674 NLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQK 733
P+ +R+ + + C+ NN E++N AI + R+ P++ +LE ++ +
Sbjct: 537 MRNPRNCSRAFFGCKSSCEDVSNNFSESYNNAINKAREMPLVEMLETIR----------R 586
Query: 734 QLMLRWRTALCPMIQQKLEDSKRNSDKW---FANWMGDEYM------------SLFEVSR 778
Q M+R I +L + ++ K+ AN + +E + FEV
Sbjct: 587 QTMIR--------IDMRLRKAIKHQGKFTLKVANTIKEETIYRKYCQVVPCGNGQFEVLE 638
Query: 779 DNEKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
+N + V++ +++CACRRW+++GIPC H + + + PE+ V+S +
Sbjct: 639 NNHGFRVDMNAKTCACRRWNMTGIPCRHVLRIILNKKVNPEELVNSDW 686
>UniRef100_Q9FM95 Similarity to mutator-like transposase [Arabidopsis thaliana]
Length = 733
Score = 257 bits (656), Expect = 1e-66
Identities = 180/622 (28%), Positives = 298/622 (46%), Gaps = 74/622 (11%)
Query: 198 SYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNG 257
SY P + G++ DSD E L +++L + D P++ DV G
Sbjct: 19 SYFPSGEV--GQNRKDLCGDSDVAEMITCTHELESKNLYVVRA---DDPYLD---DVLIG 70
Query: 258 ARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSK 317
R EE E+ +S LD DD E DG + F VG F SK
Sbjct: 71 EREEE---------EELESEQLDFYRDDYVESDDSD-------DDGARIDFYVGQEFVSK 114
Query: 318 DLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAGCPFYLRLSKTDSRSYWLLV-SLEAD 376
+ K ++ YA+++K ++ FK+ ++ + +CV C + + SRS ++V S +
Sbjct: 115 EKCKDTIEKYAIREKVNIHFKRSERNKIEGVCVQDCCKWRIYASITSRSDKMVVQSYKGI 174
Query: 377 HTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKA 436
H+C +A F+ R N++ +++ L G+ + K K A+
Sbjct: 175 HSCYPIGVVDLYSAPKIAADFINEFRTNSNLSAGQIMQRLYLN-GLRVTKTKCQSARQIM 233
Query: 437 VEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFAT 496
+I E ++Q+ + Y EL ++NPGST+I+ G K +FE+ Y CF A KT + T
Sbjct: 234 KHIISDEYVEQFTRMYDYVEELRKTNPGSTIIL----GTKDRIFEKFYTCFEAQKTGWKT 289
Query: 497 TCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDL 556
CR +I L G FLKG GQLL AVG+D N+ M+ IA+A+V E K W WF++LL DL
Sbjct: 290 ACRRVIHLGGTFLKGRMKGQLLTAVGRDPNDAMYIIAWAIVPVENKVYWQWFMELLGEDL 349
Query: 557 DMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDV 616
+ A SDQQK GL+ + ++
Sbjct: 350 RLELGNGLALSSDQQK-------------------------------GLIYAIKNVLPYA 378
Query: 617 EHRLCIKHLYGNFRKKYPG-AEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNL 675
EHR+C +H++ N +K+Y + + W+ ARA ++ +E MK + A+ ++
Sbjct: 379 EHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKMKTIKFEAYDEVKRS 438
Query: 676 PPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFY-LNNRIVKQKQ 734
W+R+ +S T+ NN+ E++N + + R+KP++ LLE ++ + + + +VK K+
Sbjct: 439 VGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDIRRHIMASNLVKLKE 498
Query: 735 LMLRWRTALCPMIQQKLEDSKRNSDKW---FANWMGDEYMSLFEVSRDNEKYSVNLESR- 790
++ T L + + ++ S KW F+N G ++EV KY V++ +
Sbjct: 499 --MQNVTGLITPKAIAIMEKRKKSLKWCYPFSNGRG-----IYEVDHGKNKYVVHVRDKT 551
Query: 791 SCACRRWDLSGIPCAHAVASMW 812
SC CR +D+SGIPC H +++MW
Sbjct: 552 SCTCREYDVSGIPCCHIMSAMW 573
>UniRef100_Q9FH72 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MRD20
[Arabidopsis thaliana]
Length = 733
Score = 256 bits (655), Expect = 2e-66
Identities = 180/622 (28%), Positives = 296/622 (46%), Gaps = 74/622 (11%)
Query: 198 SYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNG 257
SY P + G++ DSD E L +++L + D P++ DV G
Sbjct: 19 SYFPSGEV--GQNRKDLCGDSDVAEMITCTHELESKNLYVVRA---DDPYLD---DVLIG 70
Query: 258 ARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSK 317
R EE E+ +S LD DD E DG + F VG F SK
Sbjct: 71 EREEE---------EEPESEQLDFYRDDYVESDDSD-------DDGAIIDFYVGQEFVSK 114
Query: 318 DLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAG-CPFYLRLSKTDSRSYWLLVSLEAD 376
+ K ++ YA+++K ++ FK+ ++ + +CV C + + S T ++ S +
Sbjct: 115 EKCKDTIEKYAIREKVNIHFKRSERNKIEGVCVQDYCKWRIYASITSRSDKMVVQSYKGI 174
Query: 377 HTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKA 436
H+C +A F+ R N++ +++ L G+ + K K A+
Sbjct: 175 HSCYPIGVVDLYSAPKIAADFINEFRTNSNLSAGQIMQRLYLN-GLRVTKTKCQSARQIM 233
Query: 437 VEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFAT 496
+I E +Q+ + Y EL ++NPGSTVI+ G K +FE+ Y CF A KT + T
Sbjct: 234 KHIISDEYAEQFTRMYDYVEELRKTNPGSTVIL----GTKDRIFEKFYTCFEAQKTGWKT 289
Query: 497 TCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDL 556
CR +I LDG FLKG GQLL AVG+D N+ M+ IA+A+V E K W WF++LL DL
Sbjct: 290 ACRRVIHLDGTFLKGRMKGQLLTAVGRDPNDAMYIIAWAIVPVENKVYWQWFMELLGEDL 349
Query: 557 DMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDV 616
+ A SDQQK GL+ + ++
Sbjct: 350 RLELGNGLALSSDQQK-------------------------------GLIYAIKNVLPYA 378
Query: 617 EHRLCIKHLYGNFRKKYPG-AEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNL 675
EHR+C +H++ N +K+Y + + W+ ARA ++ +E MK + A+ ++
Sbjct: 379 EHRMCARHIFANLQKRYKQMGPLHKVFWKCARAYNETVFWKQLEKMKTIKFEAYDEVKRS 438
Query: 676 PPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFY-LNNRIVKQKQ 734
W+R+ +S T+ NN+ E++N + + R+KP++ LLE ++ + + + +VK K+
Sbjct: 439 VGSNWSRAFFSDITKSAAVENNISESYNAVLKDAREKPVVALLEDIRRHIMASNLVKIKE 498
Query: 735 LMLRWRTALCPMIQQKLEDSKRNSDKW---FANWMGDEYMSLFEVSRDNEKYSVNLESR- 790
++ T L + + ++ S KW F+N G ++EV KY V++ +
Sbjct: 499 --MQNVTGLITPKAIAIMEKRKKSLKWCYPFSNGRG-----IYEVDHGKNKYVVHVRDKT 551
Query: 791 SCACRRWDLSGIPCAHAVASMW 812
SC CR +D+SGIPC H +++MW
Sbjct: 552 SCTCREYDVSGIPCCHIMSAMW 573
>UniRef100_O22271 Putative MuDR-A-like transposon protein [Arabidopsis thaliana]
Length = 761
Score = 249 bits (636), Expect = 3e-64
Identities = 173/628 (27%), Positives = 301/628 (47%), Gaps = 50/628 (7%)
Query: 199 YIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNGA 258
Y G EY G +E S + EE ++ ++ D + P + +V +G
Sbjct: 12 YSDGDVEYNGGNESES--EERIEEEIVRSKKNTKKNKKEKKKKDEEDPLERFDFEVDDGV 69
Query: 259 RIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKD 318
++FEDE D+ S DE+E L R + ++ KF +G TF +
Sbjct: 70 --------ANFEDEIPIGRDIIPESSDEDE--DPILVRDRRIRRAMGDKFVIGSTFFTGI 119
Query: 319 LFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAG--CPFYLRLSKTDSRSYWLLVSLEAD 376
FK+A+ + L+ ++ + +K++ C G C + + S R +++ +
Sbjct: 120 EFKEAILEVTLKHGHNVVQDRWEKEKISFKCGMGGKCKWRVYCSYDPDRQLFVVKTSCTW 179
Query: 377 HTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKA 436
H+C + + K+ +AR F+ +R + E + RW ++ + R ++ A
Sbjct: 180 HSCSPNGKCQILKSPVIARLFLDKLRLNEKFMPMDIQEYIKERWKMVSTIPQCQRGRLLA 239
Query: 437 VEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGP-VFERIYVCFAATKTAFA 495
++M++ E +Q+A++R Y E+ NPGS I + + KG VF R YVCF +T +A
Sbjct: 240 LKMLKKEYEEQFAHIRGYVEEIHSQNPGSVAFIDTYRNEKGEDVFNRFYVCFNILRTQWA 299
Query: 496 TTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVND 555
+CRP+IGLDG FLK + G LL AVG D NNQ++PIA+AVV++E +W WFV + D
Sbjct: 300 GSCRPIIGLDGTFLKVVVKGVLLTAVGHDPNNQIYPIAWAVVQSENAENWLWFVQQIKKD 359
Query: 556 LDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGD 615
L++ + R+ +SD+ K GL+ + + + +
Sbjct: 360 LNLEDGSRFVILSDRSK-------------------------------GLLSAVKQELPN 388
Query: 616 VEHRLCIKHLYGNFRKKYPGAEM-KQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMN 674
EHR+C+KH+ N +K + +M K +W+ A + E+ + ++ +EA + D++N
Sbjct: 389 AEHRMCVKHIVENLKKNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDEALYNDVLN 448
Query: 675 LPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQ 734
P W+R Y + C+ NN E+FN I + R K +I +LE ++ RIVK+ +
Sbjct: 449 EEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGMTRIVKRNK 508
Query: 735 LMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLESRSCAC 794
LR + L K ++D+ + + +FEV D + V++ C+C
Sbjct: 509 KSLRHEGRFTKYALKMLALEKTDADR---SKVYRCTHGVFEVYIDENGHRVDIPKTQCSC 565
Query: 795 RRWDLSGIPCAHAVASMWFGRRVPEDYV 822
+W +SGIPC H+ +M E+Y+
Sbjct: 566 GKWQISGIPCEHSYGAMIEAGLDAENYI 593
>UniRef100_Q9C9S3 Putative Mutator-like transposase; 12516-14947 [Arabidopsis
thaliana]
Length = 761
Score = 248 bits (632), Expect = 8e-64
Identities = 174/628 (27%), Positives = 302/628 (47%), Gaps = 50/628 (7%)
Query: 199 YIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESLINIATADRDPPFVQGSVDVGNGA 258
Y G EY G +E S + EE + ++ D + P + +V +G
Sbjct: 12 YSDGDVEYNGGNESES--EERIEEEIVRRKKNTKKNKKEKKKKDEEDPLERFDFEVDDGV 69
Query: 259 RIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAVQDGEEVKFQVGHTFSSKD 318
++FEDE D+ S DE+E L R + ++ KF +G TF +
Sbjct: 70 --------ANFEDEIPIGRDIIPESSDEDE--DPILVRDRRIRRAMGDKFVIGSTFFTGI 119
Query: 319 LFKQAVKDYALQQKKDLTFKKDDKKRCVVIC-VAG-CPFYLRLSKTDSRSYWLLVSLEAD 376
FK+A+ + L+ ++ + +K++ C ++G C + + S R +++ +
Sbjct: 120 EFKEAILEVTLKHGHNVVQDRWEKEKISFKCGMSGKCKWRVYCSYDPDRQLFVVKTSCTW 179
Query: 377 HTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKA 436
H+C + + K+ +AR F+ +R + E + RW ++ + R ++ A
Sbjct: 180 HSCSPNGKCQILKSPVIARLFLDKLRLNEKFMPMDIQEYIKERWKMVSTIPQCQRGRLLA 239
Query: 437 VEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGP-VFERIYVCFAATKTAFA 495
++M++ E +Q+A++R Y E+ NPGS I + + KG VF R YVCF +T +A
Sbjct: 240 LKMLKKEYEEQFAHIRGYVEEIHSQNPGSVAFIDTYRNEKGEDVFNRFYVCFNILRTQWA 299
Query: 496 TTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVND 555
+CRP+IGLDG FLK + G LL AVG D NNQ++PIA+AVV++E +W WFV + D
Sbjct: 300 GSCRPIIGLDGTFLKVVVKGVLLTAVGHDPNNQIYPIAWAVVQSENAENWLWFVQQIKKD 359
Query: 556 LDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGD 615
L++ + R+ +SD+ K GL+ + + + +
Sbjct: 360 LNLEDGSRFVILSDRSK-------------------------------GLLSAVKQELPN 388
Query: 616 VEHRLCIKHLYGNFRKKYPGAEM-KQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMN 674
EHR+C+KH+ N +K + +M K +W+ A + E+ + ++ +EA + D++N
Sbjct: 389 AEHRMCVKHIVENLKKNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDEALYNDVLN 448
Query: 675 LPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQ 734
P W+R Y + C+ NN E+FN I + R K +I +LE ++ RIVK+ +
Sbjct: 449 EEPHTWSRCFYKLGSCCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGMTRIVKRNK 508
Query: 735 LMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLESRSCAC 794
LR + L K ++D+ + + +FEV D + V++ C+C
Sbjct: 509 KSLRHEGRFTKYALKMLALEKTDADR---SKVYRCTHGVFEVYIDGNGHRVDIPKTQCSC 565
Query: 795 RRWDLSGIPCAHAVASMWFGRRVPEDYV 822
+W +SGIPC HA +M E+Y+
Sbjct: 566 GKWQISGIPCEHAYGAMIEAGLDAENYI 593
>UniRef100_Q65X18 Putative polyprotein [Oryza sativa]
Length = 1040
Score = 228 bits (581), Expect = 7e-58
Identities = 165/561 (29%), Positives = 266/561 (47%), Gaps = 64/561 (11%)
Query: 194 SEDESYIPGSDEYEGESED---VSVDDSDYE-ENWDWAEVL-----PAESLINIATADRD 244
S+ E I + E G ++D +S D+D+E E D+ E L E + D D
Sbjct: 364 SQQEDNIDYNPELIGVADDQHSISSGDTDHEQEKVDYVEKLNEMKRQREDPMTHCEGDTD 423
Query: 245 PPFV----QGSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSKRLPRFKAV 300
+ VD+G + SD EDG S L+ +D+E + + R K +
Sbjct: 424 IDDLYTPEDADVDLGVADIDDMSCELSDPPSEDGGLSTLECETDEENKPPPGKRRRGKGI 483
Query: 301 QDGEEVKFQV-----------GHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVIC 349
Q + + F G F+ F++A+K Y + + +D + ++ R V C
Sbjct: 484 QV-QRIYFDESNMMDTSQLCKGMCFTDAGQFRKALKSYHIVKGRDYKYTRNKADRIKVKC 542
Query: 350 VAG---CPFYLRLSKTDSRSYWLLVSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPN 406
C F++R S+ S +++ + A HTC + + + +L+ ++ PN
Sbjct: 543 SQDKVKCDFFIRASQVGSEKTFMVREMIAPHTCPSHRNCTRVDSTWLSERYEDDFWSDPN 602
Query: 407 ITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGST 466
V A + G + K KAYRA+ KA E + G QY +R Y L+ +NPG+T
Sbjct: 603 WKVEAFMARCLRETGTYISKSKAYRARRKATEKVLGNKEKQYKRIRDYLQTLIDTNPGTT 662
Query: 467 VIIKSTKG---AKGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGK 523
++ + P F +++CF+A K F CRP I +DGCF+K G Q+LAA +
Sbjct: 663 AVVTTINRDVLGLAPRFSGLFICFSAQKEGFINGCRPFISIDGCFVKLTNGAQVLAASAR 722
Query: 524 DGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDM-VERKRWAFISDQQKVMLN*SCIH* 582
D NN MFPIAFAVV E +W+WF+++L + E W F+SD+QK
Sbjct: 723 DENNNMFPIAFAVVGKEDTDNWTWFLEMLKCAIGSGEEHGGWTFMSDRQK---------- 772
Query: 583 SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVGDVEHRLCIKHLYGNF-RKKYPGAEMKQA 641
GL+ + + D EHR C HL N K + G + K
Sbjct: 773 ---------------------GLMNAIPIVFPDSEHRYCKMHLLQNMGNKGWRGEKYKGF 811
Query: 642 LWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEA 701
+ A A+T+ ++ AME +K+LN A++ ++ + + ++R A+S + DL VNN+ E
Sbjct: 812 VDAAIYATTVWDYDKAMEDLKKLNLKAWEWLIAIGKEHFSRHAFSPKAKSDLVVNNLSEV 871
Query: 702 FNRAILEYRDKPIITLLERLK 722
FN+ IL+ RDKPI+T++E ++
Sbjct: 872 FNKYILDARDKPIVTMVEHIR 892
>UniRef100_Q9FYG9 F1N21.6 [Arabidopsis thaliana]
Length = 901
Score = 202 bits (513), Expect = 5e-50
Identities = 116/384 (30%), Positives = 192/384 (49%), Gaps = 32/384 (8%)
Query: 376 DHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVK 435
+H S R K +AR F R P + + +E +R+ I + KW +A+
Sbjct: 360 EHEHAISGKARMLKQGVIARLFRDEARRRPTLRWTDIKDEIMMRYTISVSKWICQKARRL 419
Query: 436 AVEMIQGETLDQYANLRKYAAELLRSNPG-STVIIKSTKGAKGPVFERIYVCFAATKTAF 494
A +M+ +Q+A L Y AEL RSN T I+ + F++ Y+CF + +
Sbjct: 420 AFDMVIQTQREQFAKLWDYEAELQRSNKDIHTEIVTIPQDCGKQQFDKFYICFENMRRTW 479
Query: 495 ATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVN 554
CRP++GLDG FLK G++LAAVG+D +N+++PIA+A+V E SW+WFV+ L
Sbjct: 480 KECCRPIVGLDGAFLKWELKGEILAAVGRDADNRIYPIAWAIVRVEDNDSWAWFVEHLKT 539
Query: 555 DLDMVERKRWAFISDQQKVMLN*SCIH*SIIHLYY*I*YLSYLYSCVMQGLVPTLAELVG 614
DL + ISD++K GL+ +A+L+
Sbjct: 540 DLGLGLGSLLTVISDKKK-------------------------------GLINVVADLLP 568
Query: 615 DVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMN 674
EHR C +H+Y N+RK Y + W A +ST ++ M+ ++ + A D++
Sbjct: 569 QAEHRHCARHIYANWRKVYSDYSHESYFWAIAYSSTNGDYRWNMDALRLYDPQAHDDLLK 628
Query: 675 LPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNRIVKQKQ 734
P+ W R+ +S ++C+ NN+CE+FNR I + R+ P+I +LE ++ RI K
Sbjct: 629 TDPRTWCRAFFSTHSRCEDVSNNLCESFNRTIRDARNLPVINMLEEVRRTSMKRIAKLCG 688
Query: 735 LMLRWRTALCPMIQQKLEDSKRNS 758
++ T P I + LE +++++
Sbjct: 689 KTMKCETRFPPKIMEILEGNRKSA 712
>UniRef100_Q8H8R1 Putative mutator-like transposase [Oryza sativa]
Length = 746
Score = 169 bits (429), Expect = 3e-40
Identities = 82/226 (36%), Positives = 136/226 (59%), Gaps = 3/226 (1%)
Query: 601 VMQGLVPTLAELVGDVEHRLCIKHLYGNFRKKYPGAEMKQALWRAARASTLPEWYAAMEV 660
V++GL+P + + D EHR C++HLY NF Y G +K LW AR++T+PEW E
Sbjct: 391 VVEGLIPAIKDEFPDSEHRFCVRHLYQNFAVLYKGEALKNQLWAIARSTTVPEWNVNTEK 450
Query: 661 MKELNEAAFQDMMNLPPKMWTRSAYSRDTQCDLQVNNMCEAFNRAILEYRDKPIITLLER 720
MK +N+ A+ + +PP W R+ + ++CD+ +NN E R ILE R+ I+++LE+
Sbjct: 451 MKAVNKDAYGYLEEIPPNQWCRAFFRDFSKCDILLNNNLECHVRYILEARELTILSMLEK 510
Query: 721 LKFYLNNRIVKQKQLMLRWRTALCPMIQQKLEDSKRNSDKWFANWMGDEYMSLFEVSRDN 780
++ L NRI +++ +W +CP I+QK+E + S+ +A M +F+V+ +
Sbjct: 511 IRSKLMNRIYTKQEECKKWVFDICPKIKQKVEKNIEMSNTCYAL---PSRMGVFQVTDRD 567
Query: 781 EKYSVNLESRSCACRRWDLSGIPCAHAVASMWFGRRVPEDYVHSSY 826
+++ V+++++ C CRRW L GIPC HA++ + R PED V Y
Sbjct: 568 KQFVVDIKNKQCDCRRWQLIGIPCNHAISCLRHERIKPEDEVSFCY 613
Score = 141 bits (355), Expect = 1e-31
Identities = 109/376 (28%), Positives = 167/376 (43%), Gaps = 22/376 (5%)
Query: 176 ALVHEVPNSAPVNEPLNESEDESYIPGSDEYEGESEDVSVDDSDYEENWDWAEVLPAESL 235
A+ + P S N N DE + ED DSDYE + D ++ +
Sbjct: 51 AMTSQAPPSVAANPECN----------IDEEDNTMEDPDFVDSDYEVDRDDDDLY--DKY 98
Query: 236 INIATADRDPPFVQGSVDVGNGARIEEPATFSDFEDEDGDSSDLDSPSDDEEEGRSK-RL 294
I+ T + V+G V G + A+ F ++ L P E+EG K
Sbjct: 99 ID-GTVHDELALVKGKAVVKEGVGNIQDASDDIFYSDER----LFLPESSEDEGEIKLNF 153
Query: 295 PRFKAVQDGEEVKFQVGHTFSSKDLFKQAVKDYALQQKKDLTFKKDDKKRCVVICVAGCP 354
F+ D + +F G F + + ++AV Y++ + + +++KKR C CP
Sbjct: 154 KTFRPDVDMQSPEFYPGMVFGTIEELRKAVSQYSITNRVAVKPDRNNKKRYEAHCAENCP 213
Query: 355 FYLRLSKTDSRSYWLLVSLEADHTCCRSASNRQAKTKYLARKFMPIIRHTPNITVNALIE 414
+ L S + +++ HTC + + +YLA +F+ R +T+ + +
Sbjct: 214 WKLVASVDSRANCFMVKQYVGSHTCRKEWELKAVTARYLAGRFIEEFRGDDKMTLASFAK 273
Query: 415 EARLRWGILLGKWKAYRAKVKAVEMIQGETLDQYANLRKYAAELLRSNPGSTVIIKSTKG 474
+ + I + K RA+ A E I G+ + QY L YA EL RSNPGS G
Sbjct: 274 KVQKSLNITPSRHKLGRARQIAREAIYGDEIAQYDQLWDYAQELRRSNPGSKFFSNLHNG 333
Query: 475 AKGPVFERIYVCFAATKTAFATTCRPLIGLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAF 534
F +YV A+K F + CRPLI DGC +K +GG +L AVG D N+ ++PIA
Sbjct: 334 C----FHTLYVSMDASKRGFLSGCRPLICFDGCHIKTKFGGHILTAVGMDPNDCIYPIAI 389
Query: 535 AVVEAETKASWSWFVD 550
AVVE A F D
Sbjct: 390 AVVEGLIPAIKDEFPD 405
>UniRef100_Q94HS2 Putative transposon protein [Oryza sativa]
Length = 841
Score = 169 bits (427), Expect = 5e-40
Identities = 108/311 (34%), Positives = 162/311 (51%), Gaps = 25/311 (8%)
Query: 267 SDFEDEDGDSSDLDSP-SDDEEEGRSKRLPRFKA--VQDGEEVKFQVGHTFSSKDLFKQA 323
+D ++ D DL P SDD+ E R K FKA +D + + F+VG F S ++ ++A
Sbjct: 140 ADRKEVSTDDEDLQLPDSDDDGEVRLK----FKAFMAEDVKNLVFKVGMVFPSVEVLRKA 195
Query: 324 VKDYALQQKKDLTFKKDDKKRCVVICVAGCPFYLRLSKTDSRSYWLLVSLE-ADHTCCRS 382
+ +Y+L+ + D+ ++++KR CV GCP+ L S DSRS ++V +H C +
Sbjct: 196 ITEYSLKARVDIKMPRNEQKRLRAHCVEGCPWNLYAS-FDSRSKSMMVKTYLGEHKCQKE 254
Query: 383 ASNRQAKTKYLARKFMPIIRHTPNITVNALIEEARLRWGILLGKWKAYRAKVKAVEMIQG 442
++ K+L+ K++ R +T+ K K RA+ A + I G
Sbjct: 255 WVLKRCTAKWLSEKYIETFRANDKMTLGGF------------AKSKLARARRLAFKDIYG 302
Query: 443 ETLDQYANLRKYAAELLRSNPGSTVIIKSTKGAKGPVFERIYVCFAATKTAFATTCRPLI 502
+ + QY L Y AEL RSNPGS + F Y+ F A K + CRPLI
Sbjct: 303 DEIQQYNQLWNYGAELRRSNPGSCFFLNLVDSC----FSTCYMSFDACKRGSLSGCRPLI 358
Query: 503 GLDGCFLKGMYGGQLLAAVGKDGNNQMFPIAFAVVEAETKASWSWFVDLLVNDLDMVERK 562
LDGC +K +GGQ+L AVG D N+ ++PIA VVE E+ SW WF+ L DL +
Sbjct: 359 CLDGCHIKTKFGGQILTAVGIDPNDCIYPIAIVVVETESLRSWRWFLQTLKEDLGIDNTY 418
Query: 563 RWAFISDQQKV 573
W ++D+QKV
Sbjct: 419 PWTIMTDKQKV 429
Score = 146 bits (368), Expect = 3e-33
Identities = 70/180 (38%), Positives = 113/180 (61%), Gaps = 4/180 (2%)
Query: 633 YPGAEMKQALWRAARASTLPEWYAAMEVMKELNEAAFQDMMNLPPKMWTRSAYSRDTQCD 692
+ G +K LW AR+S+ EW A ME MK LN+ A++ + +PPK W ++ +S +CD
Sbjct: 431 FKGENLKNQLWACARSSSEVEWNANMEEMKSLNQDAYEWLQKMPPKTWVKAYFSEFPKCD 490
Query: 693 LQVNNMCEAFNRAILEYRDKPIITLLERLKFYLNNR-IVKQKQLMLRWRTALCPMIQQKL 751
+ +NN CE FN+ ILE R+ PI+++ E++K L +R KQK++ +W +CP I++K+
Sbjct: 491 ILLNNNCEVFNKYILEARELPILSMFEKIKSQLISRHYSKQKEVAEQWHGPICPKIRKKV 550
Query: 752 EDSKRNSDKWFANWMGDEYMSLFEVSRDNEKYSVNLESRSCACRRWDLSGIPCAHAVASM 811
+N+D ++ +F+V N KY V+L ++ C CRRWDL+GIPC HA++ +
Sbjct: 551 ---LKNADMANTCYVLPAGKGIFQVEDRNFKYIVDLSAKHCDCRRWDLTGIPCNHAISCL 607
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,630,368,125
Number of Sequences: 2790947
Number of extensions: 70235752
Number of successful extensions: 284508
Number of sequences better than 10.0: 988
Number of HSP's better than 10.0 without gapping: 297
Number of HSP's successfully gapped in prelim test: 743
Number of HSP's that attempted gapping in prelim test: 277331
Number of HSP's gapped (non-prelim): 3688
length of query: 994
length of database: 848,049,833
effective HSP length: 137
effective length of query: 857
effective length of database: 465,690,094
effective search space: 399096410558
effective search space used: 399096410558
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 80 (35.4 bits)
Lotus: description of TM0112.15


