

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
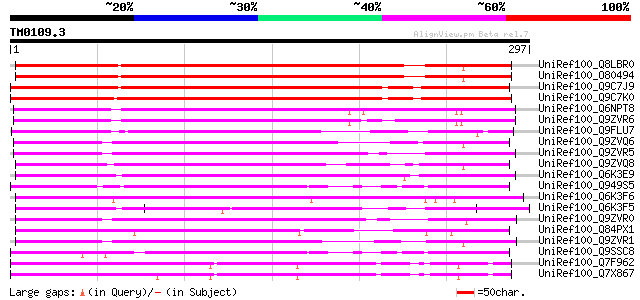
Query= TM0109.3
(297 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LBR0 Phloem-specific lectin PP2-like protein [Arabid... 280 3e-74
UniRef100_O80494 T12M4.17 protein [Arabidopsis thaliana] 279 6e-74
UniRef100_Q9C7J9 Hypothetical protein F14G9.15 [Arabidopsis thal... 262 7e-69
UniRef100_Q9C7K0 Hypothetical protein F14G9.14 [Arabidopsis thal... 258 1e-67
UniRef100_Q6NPT8 At2g02230 [Arabidopsis thaliana] 211 2e-53
UniRef100_Q9ZVR6 Putative phloem-specific lectin [Arabidopsis th... 210 4e-53
UniRef100_Q9FLU7 Phloem-specific lectin-like protein [Arabidopsi... 209 5e-53
UniRef100_Q9ZVQ6 Putative phloem-specific lectin [Arabidopsis th... 199 1e-49
UniRef100_Q9ZVR5 Lectin-like protein [Arabidopsis thaliana] 194 2e-48
UniRef100_Q9ZVQ8 Putative phloem-specific lectin [Arabidopsis th... 189 1e-46
UniRef100_Q6K3E9 F-box family protein-like [Oryza sativa] 183 4e-45
UniRef100_Q949S5 Hypothetical protein At1g80110 [Arabidopsis tha... 182 7e-45
UniRef100_Q6K3F6 Phloem-specific lectin-like [Oryza sativa] 179 1e-43
UniRef100_Q6K3F5 F-box family protein-like [Oryza sativa] 176 5e-43
UniRef100_Q9ZVR0 Putative phloem-specific lectin [Arabidopsis th... 164 2e-39
UniRef100_Q84PX1 Putative F-box protein [Oryza sativa] 155 2e-36
UniRef100_Q9ZVR1 Putative phloem-specific lectin [Arabidopsis th... 148 1e-34
UniRef100_Q9SSC8 F18B13.19 protein [Arabidopsis thaliana] 144 4e-33
UniRef100_Q7F962 OSJNBa0094P09.1 protein [Oryza sativa] 141 2e-32
UniRef100_Q7X867 OSJNBa0079F16.17 protein [Oryza sativa] 133 6e-30
>UniRef100_Q8LBR0 Phloem-specific lectin PP2-like protein [Arabidopsis thaliana]
Length = 288
Score = 280 bits (716), Expect = 3e-74
Identities = 150/290 (51%), Positives = 188/290 (64%), Gaps = 18/290 (6%)
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE CV+ ILS T+P D + VSS R A DSD VW FLP+DY ++SR+ +P +
Sbjct: 2 LPEACVATILSFTTPADTISSAAVSSVFRVAGDSDFVWEKFLPTDYCHVISRSTDPHRI- 60
Query: 64 FSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPE 123
FSS K+L+ LC +L+D G K FK++KLSGK SYILS+RDLSITWS +WSW +
Sbjct: 61 FSSKKELYRCLCESILIDNGRKIFKIEKLSGKISYILSSRDLSITWSDQRHYWSWSPRSD 120
Query: 124 SRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVGG 183
SRF E +L +WLEI GKI+TG L+PNT+YGAYLIMKV+ R YGLD PAE S+ VG
Sbjct: 121 SRFSEXVQLIMTDWLEIIGKIQTGALSPNTNYGAYLIMKVTSRAYGLDLVPAETSIKVGN 180
Query: 184 RVLRRGKAYLGQKDEKKVEMETLFYGNRRD-IFRNRVIQLEQLEEEGKDGGVIPVPGKRD 242
+ YL D KK +ME +FYG R + + V++ + E P RD
Sbjct: 181 GEKKIKSTYLSCLDNKKQQMERVFYGQREQRMATHEVVRSHRRE-----------PEVRD 229
Query: 243 DGWMEIELGEFFSGGG----DVEIKMGLREV-GYQLKGGLVVEGIEVRPK 287
DGWMEIELGEF +G G D E+ M L EV GYQLKGG+ ++GIEVRPK
Sbjct: 230 DGWMEIELGEFETGSGEGDDDKEVVMSLTEVKGYQLKGGIAIDGIEVRPK 279
>UniRef100_O80494 T12M4.17 protein [Arabidopsis thaliana]
Length = 288
Score = 279 bits (714), Expect = 6e-74
Identities = 150/290 (51%), Positives = 188/290 (64%), Gaps = 18/290 (6%)
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE CV+ ILS T+P D + VSS R A DSD VW FLP+DY ++SR+ +P +
Sbjct: 2 LPEACVATILSFTTPADTISSAAVSSVFRVAGDSDFVWEKFLPTDYCHVISRSTDPHRI- 60
Query: 64 FSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNPE 123
FSS K+L+ LC +L+D G K FK++KLSGK SYILS+RDLSITWS +WSW +
Sbjct: 61 FSSKKELYRCLCESILIDNGRKIFKIEKLSGKISYILSSRDLSITWSDQRHYWSWSPRSD 120
Query: 124 SRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVGG 183
SRF E +L +WLEI GKI+TG L+PNT+YGAYLIMKV+ R YGLD PAE S+ VG
Sbjct: 121 SRFSEGVQLIMTDWLEIIGKIQTGALSPNTNYGAYLIMKVTSRAYGLDLVPAETSIKVGN 180
Query: 184 RVLRRGKAYLGQKDEKKVEMETLFYGNRRD-IFRNRVIQLEQLEEEGKDGGVIPVPGKRD 242
+ YL D KK +ME +FYG R + + V++ + E P RD
Sbjct: 181 GEKKIKSTYLSCLDNKKQQMERVFYGQREQRMATHEVVRSHRRE-----------PEVRD 229
Query: 243 DGWMEIELGEFFSGGG----DVEIKMGLREV-GYQLKGGLVVEGIEVRPK 287
DGWMEIELGEF +G G D E+ M L EV GYQLKGG+ ++GIEVRPK
Sbjct: 230 DGWMEIELGEFETGSGEGDDDKEVVMSLTEVKGYQLKGGIAIDGIEVRPK 279
>UniRef100_Q9C7J9 Hypothetical protein F14G9.15 [Arabidopsis thaliana]
Length = 284
Score = 262 bits (670), Expect = 7e-69
Identities = 148/288 (51%), Positives = 189/288 (65%), Gaps = 10/288 (3%)
Query: 1 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPS 60
M+ LPE CV+ IL+ TSP DA S VSS R A DSD VW FLPS Y+ ++S++ +
Sbjct: 1 MMMLPEACVANILAFTSPADAFSSSEVSSVFRLAGDSDFVWEKFLPSHYKSLISQSTDHH 60
Query: 61 ALQFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS 120
+ FSS K+++ LC LL+D K FK++K SGK SYILSARD+SIT+S + SW +
Sbjct: 61 RI-FSSKKEIYRCLCDSLLIDNARKLFKINKFSGKISYILSARDISITYSDHASYCSWSN 119
Query: 121 NPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVA 180
+SRF E AEL T + LEI+GKI+T VL+PNT YGAYLIMKV++ YGLD PAE SV
Sbjct: 120 VSDSRFSESAELITTDRLEIKGKIQTTVLSPNTKYGAYLIMKVTNGAYGLDLVPAETSVK 179
Query: 181 VGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGK 240
+ YL DEKK +M+ LFYGNR + + +E + +GK P
Sbjct: 180 SKNGQNNKNTTYLCCLDEKKQQMKRLFYGNREE---RMAMTVEAVGGDGKR----REPKA 232
Query: 241 RDDGWMEIELGEFFS-GGGDVEIKMGLREV-GYQLKGGLVVEGIEVRP 286
RDDGW+EIELGEF + G D E+ M L EV GYQLKGG+V++GIEVRP
Sbjct: 233 RDDGWLEIELGEFVTREGEDDEVNMSLTEVKGYQLKGGIVIDGIEVRP 280
>UniRef100_Q9C7K0 Hypothetical protein F14G9.14 [Arabidopsis thaliana]
Length = 282
Score = 258 bits (660), Expect = 1e-67
Identities = 145/289 (50%), Positives = 187/289 (64%), Gaps = 10/289 (3%)
Query: 1 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPS 60
M+ LPE C++ IL+ TSP DA S VSS R A DSD VW FLPSDY+ ++S++ +
Sbjct: 1 MMMLPEACIANILAFTSPADAFSSSEVSSVFRLAGDSDFVWEKFLPSDYKSLISQSTDHH 60
Query: 61 ALQFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS 120
SS K+++ LC LL+D K FK++K SGK SY+LSARD+SIT S +WSW +
Sbjct: 61 -WNISSKKEIYRCLCDSLLIDNARKLFKINKFSGKISYVLSARDISITHSDHASYWSWSN 119
Query: 121 NPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVA 180
+SRF E AEL + LEIEGKI+T VL+ NT YGAYLI+KV+ YGLD PAE S+
Sbjct: 120 VSDSRFSESAELIITDRLEIEGKIQTRVLSANTRYGAYLIVKVTKGAYGLDLVPAETSIK 179
Query: 181 VGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGK 240
+ + YL DEKK +M+ LFYGNR + + +E + +GK P
Sbjct: 180 SKNGQISKSATYLCCLDEKKQQMKRLFYGNREE---RMAMTVEAVGGDGKR----REPKC 232
Query: 241 RDDGWMEIELGEFFS-GGGDVEIKMGLREV-GYQLKGGLVVEGIEVRPK 287
RDDGWMEIELGEF + G D E+ M L EV GYQLKGG++++GIEVRPK
Sbjct: 233 RDDGWMEIELGEFETREGEDDEVNMTLTEVKGYQLKGGILIDGIEVRPK 281
>UniRef100_Q6NPT8 At2g02230 [Arabidopsis thaliana]
Length = 336
Score = 211 bits (536), Expect = 2e-53
Identities = 118/307 (38%), Positives = 182/307 (58%), Gaps = 25/307 (8%)
Query: 3 SLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSAL 62
+LPEDC+S+++SHTSP DAC + VS +++SAA SD+VW FLPS+Y +V ++ N
Sbjct: 34 ALPEDCISKVISHTSPRDACVVASVSKSVKSAAQSDLVWEMFLPSEYSSLVLQSAN---- 89
Query: 63 QFSSYKQLFHALC-SPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSN 121
S K++F +L + +L++ G KSF ++K SGKK Y+LSA +L+I W P +W W +
Sbjct: 90 -HLSKKEIFLSLADNSVLVENGKKSFWVEKASGKKCYMLSAMELTIIWGDSPAYWKWITV 148
Query: 122 PESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSH-RGYGLDSAPAEVSVA 180
PES+F +VAELR V W E+ GKI G+L+ T Y Y++ K ++ R YG D P E V
Sbjct: 149 PESKFEKVAELRNVCWFEVRGKISCGMLSKGTHYSVYVVFKTANGRSYGFDLVPVEAGVG 208
Query: 181 VGGRVLRRGKAYL--GQKDEKKV---------EMETLFYGNRRDIFRNRVIQLEQLEEEG 229
G+V + Y G D + M + + RR + + + E++E E
Sbjct: 209 FVGKVATKKSVYFESGNADSRSATSHYSGISYAMVSRAFRMRRPWMQVQREEEEEVEGER 268
Query: 230 KDGGVIPVPGKRDDGWMEIELGEFF--SGG----GDVEIKMGLREV-GYQLKGGLVVEGI 282
+ G + P +R DGW E+ELG+F+ +GG G EI++ + E K GL+++GI
Sbjct: 269 ERGMNVVGPKERVDGWSEVELGKFYINNGGCGDDGSDEIEISIMETQNGNWKSGLIIQGI 328
Query: 283 EVRPKQT 289
E+RP+++
Sbjct: 329 EIRPERS 335
>UniRef100_Q9ZVR6 Putative phloem-specific lectin [Arabidopsis thaliana]
Length = 317
Score = 210 bits (534), Expect = 4e-53
Identities = 117/298 (39%), Positives = 178/298 (59%), Gaps = 26/298 (8%)
Query: 3 SLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSAL 62
+LPEDC+S+++SHTSP DAC + VS +++SAA SD+VW FLPS+Y +V ++ N
Sbjct: 34 ALPEDCISKVISHTSPRDACVVASVSKSVKSAAQSDLVWEMFLPSEYSSLVLQSAN---- 89
Query: 63 QFSSYKQLFHALC-SPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSN 121
S K++F +L + +L++ G KSF ++K SGKK Y+LSA +L+I W P +W W +
Sbjct: 90 -HLSKKEIFLSLADNSVLVENGKKSFWVEKASGKKCYMLSAMELTIIWGDSPAYWKWITV 148
Query: 122 PESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSH-RGYGLDSAPAEVSVA 180
PES+F +VAELR V W E+ GKI G+L+ T Y Y++ K ++ R YG D P E V
Sbjct: 149 PESKFEKVAELRNVCWFEVRGKISCGMLSKGTHYSVYVVFKTANGRSYGFDLVPVEAGVG 208
Query: 181 VGGRVLRRGKAYL--GQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVP 238
G+V + Y G D + T Y + + E++E E + G + P
Sbjct: 209 FVGKVATKKSVYFESGNADSRSA---TSHYSGISE-------EEEEVEGERERGMNVVGP 258
Query: 239 GKRDDGWMEIELGEFF--SGG----GDVEIKMGLREV-GYQLKGGLVVEGIEVRPKQT 289
+R DGW E+ELG+F+ +GG G EI++ + E K GL+++GIE+RP+++
Sbjct: 259 KERVDGWSEVELGKFYINNGGCGDDGSDEIEISIMETQNGNWKSGLIIQGIEIRPERS 316
>UniRef100_Q9FLU7 Phloem-specific lectin-like protein [Arabidopsis thaliana]
Length = 251
Score = 209 bits (533), Expect = 5e-53
Identities = 124/292 (42%), Positives = 168/292 (57%), Gaps = 51/292 (17%)
Query: 2 ISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSA 61
+ LPE+C++ ++S TSP DACR S VS LRSAADS+ W FLPSDY + +++
Sbjct: 4 LDLPEECIATMISFTSPFDACRISAVSKLLRSAADSNTTWERFLPSDYRMYIDNSLS--- 60
Query: 62 LQFSSYKQLFHALC-SPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS 120
+FS+ KQLF C SPLL++ G SF ++K SGKK ++LSAR L I W P FW W S
Sbjct: 61 -RFSN-KQLFLRFCESPLLIEDGRTSFWMEKRSGKKCWMLSARKLDIVWVDSPEFWIWVS 118
Query: 121 NPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRG-YGLDSAPAEVSV 179
P+SRF EVA L V W EI GKI T +L+ T+Y AYL+ K G +G +S P EVS
Sbjct: 119 IPDSRFEEVAGLLMVCWFEIRGKISTSLLSKATNYSAYLVFKEQEMGSFGFESLPLEVS- 177
Query: 180 AVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPG 239
F R +++ NR + L+ +E
Sbjct: 178 ---------------------------FRSTRTEVYNNRRVFLKSGTQE----------- 199
Query: 240 KRDDGWMEIELGEFFSGGGDVEIKMGL---REVGYQLKGGLVVEGIEVRPKQ 288
R+DGW+EIELGE++ G D EI+M + RE G+ KGG++V+GIE+RPK+
Sbjct: 200 SREDGWLEIELGEYYVGFDDEEIEMSVLETREGGW--KGGIIVQGIEIRPKE 249
>UniRef100_Q9ZVQ6 Putative phloem-specific lectin [Arabidopsis thaliana]
Length = 272
Score = 199 bits (505), Expect = 1e-49
Identities = 116/289 (40%), Positives = 159/289 (54%), Gaps = 41/289 (14%)
Query: 3 SLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSAL 62
S PEDC+S I+S T+P DAC + VS T S SD++W FLP+DYE ++ P +
Sbjct: 16 SFPEDCISYIISFTNPRDACVAATVSKTFESTVKSDIIWEKFLPADYESLI-----PPSR 70
Query: 63 QFSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSN 121
FSS K+L+ +LC+ P+L D KS L+K SGK+ +LSA +LSI W +P +W W
Sbjct: 71 VFSSKKELYFSLCNDPVLFDDDKKSVWLEKASGKRCLMLSAMNLSIIWGDNPQYWQWIPI 130
Query: 122 PESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAV 181
PESRF +VA+LR V W EI G+ T VL+P T Y AY++ K + YG + E +V V
Sbjct: 131 PESRFEKVAKLRDVCWFEIRGRTNTRVLSPRTRYSAYIVFKGVDKCYGFQNVAIEAAVGV 190
Query: 182 GGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKR 241
G+ R R+I + G+ V P +R
Sbjct: 191 VGQEPSR-----------------------------RLICFSEAIRRGRRNVV--KPKQR 219
Query: 242 DDGWMEIELGEFFSGGG---DVEIKMGLREV-GYQLKGGLVVEGIEVRP 286
+DGWMEIELGEFF+ GG + EI+M E K GL+++GIE+RP
Sbjct: 220 EDGWMEIELGEFFNDGGIMDNDEIEMSALETKQLNRKCGLIIQGIEIRP 268
>UniRef100_Q9ZVR5 Lectin-like protein [Arabidopsis thaliana]
Length = 305
Score = 194 bits (493), Expect = 2e-48
Identities = 113/290 (38%), Positives = 160/290 (54%), Gaps = 35/290 (12%)
Query: 3 SLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSAL 62
+LPEDC+S I+S TSP DAC + VS T SA +SD VW FLPSDY +V P +
Sbjct: 44 NLPEDCISNIISFTSPRDACVAASVSKTFESAVNSDSVWDKFLPSDYSSLV-----PPSR 98
Query: 63 QFSSYKQLFHALC-SPLLLDGGNKSFKLDKLSGKKSYILSAR-DLSITWSSDPMFWSWRS 120
FSS K+L+ A+C +P+L++ G KSF L+K +GKK ++LS + + ITW S P +W W S
Sbjct: 99 VFSSKKELYFAICDNPVLVEDGGKSFWLEKENGKKCFMLSPKKSMWITWVSTPQYWRWIS 158
Query: 121 NPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVA 180
PE+RF EV EL V W E+ G + T L+P T Y AY++ K + L P E +V
Sbjct: 159 IPEARFEEVPELLNVCWFEVRGGMNTKELSPGTRYSAYIVFKTKNGCPNLGDVPVEATVG 218
Query: 181 VGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGK 240
+ G+ + Y +++ + ET RD+ R P K
Sbjct: 219 LVGQESSQRHIYFVGPSDQRRDRET------RDVTR---------------------PTK 251
Query: 241 RDDGWMEIELGEFFSGGGDVEIKMGLREVGYQL-KGGLVVEGIEVRPKQT 289
R DGWME ELG+FF+ G + + E+ K GL+++GIE RP ++
Sbjct: 252 RKDGWMEAELGQFFNESGCDVVDTSILEIKTPYWKRGLIIQGIEFRPTKS 301
>UniRef100_Q9ZVQ8 Putative phloem-specific lectin [Arabidopsis thaliana]
Length = 305
Score = 189 bits (479), Expect = 1e-46
Identities = 112/288 (38%), Positives = 163/288 (55%), Gaps = 30/288 (10%)
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE+CVS I+S TSP DAC + VS T SA SD+VW F+P +YE ++S++ A +
Sbjct: 39 LPEECVSIIVSFTSPQDACVLASVSKTFASAVKSDIVWEKFIPPEYESLISQS---RAFK 95
Query: 64 FSSYKQLFHALCSP-LLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNP 122
F S K+L+ ALC +L+D G KS ++K + K+ ++SA +L+I W + P W W +P
Sbjct: 96 FLSKKELYFALCDKSVLIDDGKKSLWIEKANAKRCIMISAMNLAIAWGNSPQSWRWIPDP 155
Query: 123 ESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVG 182
++RF VAEL V EI G+I + V++P T Y AY++ K + YG ++ EV V V
Sbjct: 156 QARFETVAELLEVCLFEIRGRINSRVISPKTRYSAYIVYKKLNICYGFENVAVEVVVGV- 214
Query: 183 GRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKRD 242
+GQ E+ F + FR R + GK+ + P +R
Sbjct: 215 ----------VGQDLEESCRRYICFDETMDEQFRRR--------DRGKN---LVKPERRK 253
Query: 243 DGWMEIELGEFFSGGG---DVEIKM-GLREVGYQLKGGLVVEGIEVRP 286
DGWMEI++GEFF+ GG D EI+M L K GL+++GIE+RP
Sbjct: 254 DGWMEIKIGEFFNEGGLLNDDEIEMVALEAKQRHWKRGLIIQGIEIRP 301
>UniRef100_Q6K3E9 F-box family protein-like [Oryza sativa]
Length = 297
Score = 183 bits (465), Expect = 4e-45
Identities = 114/291 (39%), Positives = 164/291 (56%), Gaps = 13/291 (4%)
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE+ +S +S SP DAC + VS R+AADSD VW SFLP + D+ ++P+
Sbjct: 14 LPEELLSAAISRASPRDACHAAAVSPAFRAAADSDAVWASFLPRNLPDLADGELSPAP-- 71
Query: 64 FSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNP 122
+S K+LF L P LL S LD+ +G K Y+LSAR L I W P +W W
Sbjct: 72 -ASKKELFLRLSDGPYLLSDRLMSMWLDRETGAKCYMLSARSLVIIWGDTPHYWRWIPLT 130
Query: 123 ESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVG 182
+SRF E AEL V WLEI G+I + +L+PN++Y AY++ K++ YGLD+ E SV++G
Sbjct: 131 DSRFAEGAELIDVCWLEIRGRIHSKMLSPNSTYAAYMVFKIADEFYGLDAPFQEASVSLG 190
Query: 183 GRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQ--LEEEGKDGGVIPVPGK 240
GR + E + E + + + R R + ++ + +EG + VP K
Sbjct: 191 GRGSTKIVCVQSYDSEDEEVPENYWPMSIGPLLRRRARRRDRRLVLDEG-----VTVPQK 245
Query: 241 RDDGWMEIELGEFFS-GGGDVEIKMGLREV-GYQLKGGLVVEGIEVRPKQT 289
R D WME+E+GEF + G D E+ L E G K GL+V+GIE+R K++
Sbjct: 246 RTDEWMELEMGEFINEEGEDGEVCFSLMETKGGNWKRGLIVQGIEIRLKKS 296
>UniRef100_Q949S5 Hypothetical protein At1g80110 [Arabidopsis thaliana]
Length = 257
Score = 182 bits (463), Expect = 7e-45
Identities = 118/289 (40%), Positives = 165/289 (56%), Gaps = 37/289 (12%)
Query: 1 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPS 60
M +LPEDC+++ILS T+P D CR S VSS RSAA SD VW FLP+D+ + P+
Sbjct: 1 MNNLPEDCIAKILSLTTPLDVCRLSAVSSIFRSAAGSDDVWNHFLPADFP---AGFAAPA 57
Query: 61 ALQFSSYKQLFHALC-SPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWR 119
L + KQLF +L +PLL++G SF L++ SG K Y+++AR L+I W + +W W
Sbjct: 58 GLP--TRKQLFFSLVDNPLLINGTLLSFSLERKSGNKCYMMAARALNIVWGHEQRYWHWI 115
Query: 120 SNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSV 179
S P +RF EVAEL V WLEI GKI +L+ +T Y AY + K +H YG P E S+
Sbjct: 116 SLPNTRFGEVAELIMVWWLEITGKINITLLSDDTLYAAYFVFKWNHSPYGF-RQPVETSL 174
Query: 180 AVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPG 239
+ D + + ++ + +I L +++ G + G PV
Sbjct: 175 VLA--------------DTESTD----------NVVQPSMISL--MQDSGGEEGQSPV-- 206
Query: 240 KRDDGWMEIELGEFFSGGGDV-EIKMGLREV-GYQLKGGLVVEGIEVRP 286
R DGW E+ELG+FF GD+ EI+M L+E G K GL+V GIE+RP
Sbjct: 207 LRRDGWYEVELGQFFKRRGDLGEIEMSLKETKGPYEKKGLIVYGIEIRP 255
>UniRef100_Q6K3F6 Phloem-specific lectin-like [Oryza sativa]
Length = 332
Score = 179 bits (453), Expect = 1e-43
Identities = 122/316 (38%), Positives = 164/316 (51%), Gaps = 25/316 (7%)
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVN--PSA 61
LPE+CV+ +S T+P DAC S VS R+AADSD VW SFLP D+ I++RA + +A
Sbjct: 14 LPEECVAYAISMTTPGDACHSSAVSPAFRAAADSDAVWDSFLPPDHAAILARADDGIAAA 73
Query: 62 LQFSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS 120
+ +S K LF LC P+LLD SF LD+ SG K +LSAR LSI W DP W W
Sbjct: 74 GECASKKDLFARLCGRPVLLDDATMSFGLDRRSGAKCVMLSARALSIAWGDDPSRWRWTP 133
Query: 121 N-PESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLD-------- 171
P SRF EVAEL V WLEI GK++ +L+P T+Y AYL+ + GL+
Sbjct: 134 GLPGSRFPEVAELLDVCWLEITGKLQLSLLSPATTYAAYLVYSFADYTTGLECNIGMPTP 193
Query: 172 SAPAEVSVAVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKD 231
A V GG R A ++ ++ G I + + + + G+
Sbjct: 194 MATVTVVSGAGGTTSRPPAAPATTTTTEQHKICLQHMGEEETIMHRQELVIRLRKAFGRT 253
Query: 232 GGVIP-----VPGKRD-----DGWMEIELGEF---FSGGGDVEIKMGLREVGYQLKGGLV 278
P P RD GW E+ELGEF +GG D +++ +E + K GL+
Sbjct: 254 VRFDPDMDIRCPRPRDGGGGGGGWREVELGEFAVPAAGGEDGVVEVSFKEETGRWKTGLI 313
Query: 279 VEGIEVRPKQTQSTQK 294
V+GIE+RPK T K
Sbjct: 314 VQGIELRPKCTSKLIK 329
>UniRef100_Q6K3F5 F-box family protein-like [Oryza sativa]
Length = 479
Score = 176 bits (447), Expect = 5e-43
Identities = 113/296 (38%), Positives = 158/296 (53%), Gaps = 30/296 (10%)
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE+ + LS TSP DAC + V R+AADSD VW FLP D + ++P
Sbjct: 15 LPEELLVAALSLTSPRDACSAAAVCRDFRAAADSDAVWSRFLPRDLPRLADGELSPPP-- 72
Query: 64 FSSYKQLFHAL-CSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNP 122
S K LF L +PLLL S L++ G K Y+LSAR L ITW P +W W
Sbjct: 73 -PSTKGLFLRLSAAPLLLPHELTSMWLEREKGGKCYMLSARALQITWGDTPRYWRWIPLT 131
Query: 123 ESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVG 182
+SR E AEL +V WLEI GKI + +L+ NT+Y AYL+ +++ R YGLD E SV++G
Sbjct: 132 DSRL-EGAELLSVCWLEIHGKILSKMLSRNTNYAAYLVYRIADRSYGLDFPFQEASVSIG 190
Query: 183 GRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKRD 242
G R + ++ +++ + ++ E +E P KR
Sbjct: 191 GSTTTRQVGSVERRLKRRCS--------------HALVLAEDIEH----------PQKRS 226
Query: 243 DGWMEIELGEFFS-GGGDVEIKMGLREVGYQLKGGLVVEGIEVRPKQTQSTQKTKC 297
DGWME++LGE ++ G D E+ + RE K GLVV+GIE+RPK+T ++ C
Sbjct: 227 DGWMELKLGELYNEEGDDGEVCISFRETEGHWKRGLVVQGIEIRPKKTLTSNCLAC 282
Score = 58.2 bits (139), Expect = 3e-07
Identities = 59/193 (30%), Positives = 82/193 (41%), Gaps = 34/193 (17%)
Query: 78 LLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRS---NPESRFREVAELRT 134
+ L G S LD +G K Y+LSAR L + S+D +WR SRF EV EL
Sbjct: 302 IFLTDGLTSMWLDMETGFKCYMLSARALQLANSTD----TWRLISLTGASRFSEVIELPA 357
Query: 135 VNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVGGRVLRRGKAYLG 194
L I GKI +L NT+Y AY++ V +GL + + SV+VGG + +
Sbjct: 358 CYELVICGKIPCKMLPGNTNYAAYIVFVVVDDSFGL-ATILDASVSVGGSLCTTRQVCFD 416
Query: 195 QKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKRDDGWMEIELGEFF 254
+ +E E +DG VI +P +RDDG
Sbjct: 417 STSSLSADEHF----------------VEDNIEVPQDGSVI-LPQERDDG---------L 450
Query: 255 SGGGDVEIKMGLR 267
G G ++GLR
Sbjct: 451 DGAGGGASRLGLR 463
>UniRef100_Q9ZVR0 Putative phloem-specific lectin [Arabidopsis thaliana]
Length = 307
Score = 164 bits (416), Expect = 2e-39
Identities = 98/292 (33%), Positives = 152/292 (51%), Gaps = 37/292 (12%)
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPEDC+S I+S TSP D C + VS + A D +W FLPS+YE ++ P
Sbjct: 48 LPEDCISNIISFTSPRDVCVSASVSKSFAHAVQCDSIWEKFLPSEYESLI-----PPWRV 102
Query: 64 FSSYKQLFHALC-SPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSNP 122
FSS K L+ LC P+L++ G KSF L+ SGKK +L+A++L IT ++P +W W
Sbjct: 103 FSSKKDLYFTLCYDPVLVEDGKKSFWLETASGKKCVLLAAKELWITGGNNPEYWQWIELC 162
Query: 123 ESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAVG 182
ES F +V EL ++ G + T +L+ T Y Y++ K+ +GL P +V V
Sbjct: 163 ESSFEKVPELLNNRSFQMGGSMSTQILSLGTHYSVYIVYKIKDERHGLRDLPIQVGVGFK 222
Query: 183 GRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKRD 242
G+ + + + +K E ++ + +++ KR
Sbjct: 223 GQEMPKQFICFDESTDKTKEWP------KKKLMKSK---------------------KRG 255
Query: 243 DGWMEIELGEFFSGGGDV---EIKMGLREV-GYQLKGGLVVEGIEVRPKQTQ 290
DGWME E+G+FF+ GG + E+++ + +V LK G+++EGIE RPK Q
Sbjct: 256 DGWMEAEIGDFFNDGGLMGFDEVEVSIVDVTSPNLKCGVMIEGIEFRPKDCQ 307
>UniRef100_Q84PX1 Putative F-box protein [Oryza sativa]
Length = 299
Score = 155 bits (391), Expect = 2e-36
Identities = 103/299 (34%), Positives = 145/299 (48%), Gaps = 40/299 (13%)
Query: 4 LPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYEDIVSRAVNPSALQ 63
LPE C++++++ TSP DACR + VS + R+AA+SD VW FLP DY I P+
Sbjct: 23 LPEACLADVIALTSPRDACRLAAVSPSFRAAAESDAVWDRFLPPDYRAIAPLPPPPATAA 82
Query: 64 FSSYKQL----FHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSW 118
S K++ + LC P+ +D G+ L+K SG K + L AR LS+ W W W
Sbjct: 83 ASGGKRMKKGVYLGLCDKPVPVDDGSMMVWLEKESGAKCFALPARKLSLPWEDGEFSWRW 142
Query: 119 RSNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVS-----HRGYGLDSA 173
+P SRF EVA+L L+I G++ LTP T Y AYL+ + HR GL
Sbjct: 143 TPHPLSRFEEVAQLVDCTCLDIYGRLPAAALTPATPYAAYLVFGTAAAAEGHR--GLSFP 200
Query: 174 PAEVSVAVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGG 233
E +V+ GRV+ R L D + + G G
Sbjct: 201 DQETTVSAAGRVVARHAVCLRPDD----------------------AEARKFRGVGLAGA 238
Query: 234 VIPV--PGKRDDGWMEIELG----EFFSGGGDVEIKMGLREVGYQLKGGLVVEGIEVRP 286
+PV P +R DGW E+ELG + +G G ++ +G+ K GLVVE +E RP
Sbjct: 239 GVPVRRPARRGDGWSEMELGRVAADEVAGAGGEDVVASFEVLGWYPKRGLVVECMEFRP 297
>UniRef100_Q9ZVR1 Putative phloem-specific lectin [Arabidopsis thaliana]
Length = 284
Score = 148 bits (374), Expect = 1e-34
Identities = 98/296 (33%), Positives = 143/296 (48%), Gaps = 57/296 (19%)
Query: 4 LPEDCVSEILSHTSPP-DACRFSLVSSTLRSAADSDMVWRSFLPS-DYEDIVSRAVNPSA 61
LP+DC++ I S TS P DA +LVS + +SD VW FLP DY ++ P +
Sbjct: 37 LPDDCLAIISSFTSTPRDAFLAALVSKSFGLQFNSDSVWEKFLPPPDYVSLL-----PKS 91
Query: 62 LQFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDPMFWSWRSN 121
FSS K+L+ ALC P G SF+LDK SGKK +LSA+ L I+ +P +W W S
Sbjct: 92 RVFSSKKELYFALCDPFPNHNGKMSFRLDKASGKKCVMLSAKKLLISRVVNPKYWKWISI 151
Query: 122 PESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLDSAPAEVSVAV 181
PESRF EV EL ++ +I G + T +++P T Y AY++ + G ++P + V
Sbjct: 152 PESRFDEVPELLNIDSFDIRGVLNTRIISPGTHYSAYIVYTKTSHFNGFQTSPIQAGV-- 209
Query: 182 GGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKDGGVIPVPGKR 241
G +R I+ + KR
Sbjct: 210 ---------------------------GFQRHGMSKTFIRFDS--------------KKR 228
Query: 242 DDGWMEIELGEFFSGGG-------DVEIKMGLREVGYQLKGGLVVEGIEVRPKQTQ 290
DGWME ++G+F++ GG +V + R +K GL++EGIE RPK ++
Sbjct: 229 QDGWMEAKIGDFYNEGGLIGFNLIEVSVVDVARYPHMNMKSGLIIEGIEFRPKDSR 284
>UniRef100_Q9SSC8 F18B13.19 protein [Arabidopsis thaliana]
Length = 264
Score = 144 bits (362), Expect = 4e-33
Identities = 104/297 (35%), Positives = 151/297 (50%), Gaps = 46/297 (15%)
Query: 1 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMV----WRSFLPSDYEDIV--- 53
M +LPEDC+++ILS T+P D CR S VSS M +R P ++
Sbjct: 1 MNNLPEDCIAKILSLTTPLDVCRLSAVSSISDPPPAPTMSGIISYRLISPPALRHLLVFP 60
Query: 54 --SRAVNPSALQFSSYKQLFHALCSPLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSS 111
S + +PS++ LF L + +SF L++ SG K Y+++AR L+I W
Sbjct: 61 PGSNSSSPSSITLFLSTALF------CLTELYKQSFSLERKSGNKCYMMAARALNIVWGH 114
Query: 112 DPMFWSWRSNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKVSHRGYGLD 171
+ +W W S P +RF EVAEL V WLEI GKI +L+ +T Y AY + K +H YG
Sbjct: 115 EQRYWHWISLPNTRFGEVAELIMVWWLEITGKINITLLSDDTLYAAYFVFKWNHSPYGF- 173
Query: 172 SAPAEVSVAVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEEGKD 231
P E S+ + D + + ++ + +I L +++ G +
Sbjct: 174 RQPVETSLVLA--------------DTESTD----------NVVQPSMISL--MQDSGGE 207
Query: 232 GGVIPVPGKRDDGWMEIELGEFFSGGGDV-EIKMGLREV-GYQLKGGLVVEGIEVRP 286
G PV R DGW E+ELG+FF GD+ EI+M L+E G K GL+V GIE+RP
Sbjct: 208 EGQSPV--LRRDGWYEVELGQFFKRRGDLGEIEMSLKETKGPYEKKGLIVYGIEIRP 262
>UniRef100_Q7F962 OSJNBa0094P09.1 protein [Oryza sativa]
Length = 308
Score = 141 bits (356), Expect = 2e-32
Identities = 109/306 (35%), Positives = 152/306 (49%), Gaps = 32/306 (10%)
Query: 1 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYE-DIVSRAVNP 59
M LP DC++ I S TSP DACR + ++ LR ADSD VW SFLP ++ D + P
Sbjct: 15 MCDLPMDCIACIASLTSPGDACRLAAAAAALRPVADSDDVWGSFLPPEWAGDGDALDGKP 74
Query: 60 SALQFSSYKQLFHALCS-PLLLDGGNKSFKLDKLSGKKSYILSARDLSITWSSDP---MF 115
+ S K++F LC P+LLDGG SF L+K SG K Y++ AR L WS P +
Sbjct: 75 GGREGESKKEMFLRLCDLPVLLDGGKLSFSLEKRSGAKKYMMRARALGFGWSGYPYGGLV 134
Query: 116 WSWRSNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLIMKV-------SHRGY 168
W +++P+SRF EVA L + WL+I G T L+ TSYGAYL+ V + GY
Sbjct: 135 WI-QNHPDSRFSEVAILSHLCWLDIYGIFNTKHLSNGTSYGAYLVYNVQFLHTEDQNGGY 193
Query: 169 GLDSAPAEVSVAVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRDIFRNRVIQLEQLEEE 228
A A S + + QK + + + G+ + Q ++L+
Sbjct: 194 KEQDATASGSSSTSSICSHECNHLMPQKHLRSLLFNMDYDGS--SFVKTNNNQKKELKYV 251
Query: 229 GKDGGVIPVPGKRDDGWMEIELGEFFS-------GGGDVEIKMGLREVGYQLKGGLVVEG 281
G I V R DGWME E+ S GD+ I+ +Q + ++VEG
Sbjct: 252 G-----ICV---RSDGWMEQEISTEISVVKQNNEENGDISIEFRGLTGSHQCQ--IIVEG 301
Query: 282 IEVRPK 287
IE+RPK
Sbjct: 302 IEIRPK 307
>UniRef100_Q7X867 OSJNBa0079F16.17 protein [Oryza sativa]
Length = 323
Score = 133 bits (334), Expect = 6e-30
Identities = 109/321 (33%), Positives = 154/321 (47%), Gaps = 47/321 (14%)
Query: 1 MISLPEDCVSEILSHTSPPDACRFSLVSSTLRSAADSDMVWRSFLPSDYE-DIVSRAVNP 59
M LP DC++ I S TSP DACR + ++ LR ADSD VW SFLP ++ D + P
Sbjct: 15 MCDLPMDCIACIASLTSPGDACRLAAAAAALRPVADSDDVWGSFLPPEWAGDGDALDGKP 74
Query: 60 SALQFSSYKQLFHALCS-PLLLDGG---------------NKSFKLDKLSGKKSYILSAR 103
+ S K++F LC P+LLDGG ++SF L+K SG K Y++ AR
Sbjct: 75 GGREGESKKEMFLRLCDLPVLLDGGKLYWAFSSLISLLSLHQSFSLEKRSGAKKYMMRAR 134
Query: 104 DLSITWSSDP---MFWSWRSNPESRFREVAELRTVNWLEIEGKIRTGVLTPNTSYGAYLI 160
L WS P + W +++P+SRF EVA L + WL+I G T L+ TSYGAYL+
Sbjct: 135 ALGFGWSGYPYGGLVWI-QNHPDSRFSEVAILSHLCWLDIYGIFNTKHLSNGTSYGAYLV 193
Query: 161 MKV-------SHRGYGLDSAPAEVSVAVGGRVLRRGKAYLGQKDEKKVEMETLFYGNRRD 213
V + GY A A S + + QK + + + G+
Sbjct: 194 YNVQFLHTEDQNGGYKEQDATASGSSSTSSICSHECNHLMPQKHLRSLLFNMDYDGS--S 251
Query: 214 IFRNRVIQLEQLEEEGKDGGVIPVPGKRDDGWMEIELGEFFS-------GGGDVEIKMGL 266
+ Q ++L+ G I V R DGWME E+ S GD+ I+
Sbjct: 252 FVKTNNNQKKELKYVG-----ICV---RSDGWMEQEISTEISVVKQNNEENGDISIEFRG 303
Query: 267 REVGYQLKGGLVVEGIEVRPK 287
+Q + ++VEGIE+RPK
Sbjct: 304 LTGSHQCQ--IIVEGIEIRPK 322
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.136 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 522,072,529
Number of Sequences: 2790947
Number of extensions: 22674223
Number of successful extensions: 47890
Number of sequences better than 10.0: 92
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 47653
Number of HSP's gapped (non-prelim): 125
length of query: 297
length of database: 848,049,833
effective HSP length: 126
effective length of query: 171
effective length of database: 496,390,511
effective search space: 84882777381
effective search space used: 84882777381
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0109.3


