

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
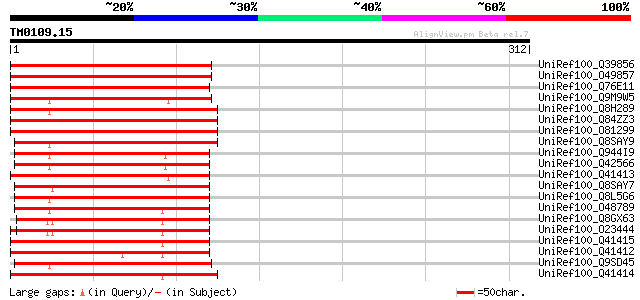
Query= TM0109.15
(312 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q39856 Epoxide hydrolase [Glycine max] 225 1e-57
UniRef100_O49857 Epoxide hydrolase [Glycine max] 225 1e-57
UniRef100_Q76E11 Soluble epoxide hydrolase [Citrus jambhiri] 193 6e-48
UniRef100_Q9M9W5 Putative epoxide hydrolase [Arabidopsis thaliana] 186 5e-46
UniRef100_Q8H289 Epoxide hydrolase [Ananas comosus] 182 1e-44
UniRef100_Q84ZZ3 Soluble epoxide hydrolase [Euphorbia lagascae] 181 2e-44
UniRef100_O81299 T14P8.15 protein [Arabidopsis thaliana] 180 4e-44
UniRef100_Q8SAY9 Putative hydrolase [Oryza sativa] 176 9e-43
UniRef100_Q944I9 At2g26740/F18A8.11 [Arabidopsis thaliana] 172 1e-41
UniRef100_Q42566 Putative epoxide hydrolase ATsEH [Arabidopsis t... 172 1e-41
UniRef100_Q41413 Epoxide hydrolase [Solanum tuberosum] 170 4e-41
UniRef100_Q8SAY7 Putative hydrolase [Oryza sativa] 169 7e-41
UniRef100_Q8L5G6 Soluble epoxide hydrolase [Brassica napus] 168 2e-40
UniRef100_O48789 Putative epoxide hydrolase [Arabidopsis thaliana] 167 3e-40
UniRef100_Q8GX63 Putative epoxide hydrolase [Arabidopsis thaliana] 166 1e-39
UniRef100_O23444 Putative epoxide hydrolase [Arabidopsis thaliana] 166 1e-39
UniRef100_Q41415 Epoxide hydrolase [Solanum tuberosum] 165 1e-39
UniRef100_Q41412 Epoxide hydrolase [Solanum tuberosum] 164 3e-39
UniRef100_Q9SD45 Epoxide hydrolase-like protein [Arabidopsis tha... 164 4e-39
UniRef100_Q41414 Epoxide hydrolase [Solanum tuberosum] 163 5e-39
>UniRef100_Q39856 Epoxide hydrolase [Glycine max]
Length = 341
Score = 225 bits (573), Expect = 1e-57
Identities = 101/121 (83%), Positives = 114/121 (93%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME I+HRTVEVNGIKMH+AEKG+GPVVLFLHGFPELWYSWRHQIL+LSS GY AVAPDLR
Sbjct: 26 MEQIKHRTVEVNGIKMHVAEKGEGPVVLFLHGFPELWYSWRHQILSLSSLGYRAVAPDLR 85
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDT++P SI++Y CFHIVGD+VALID LG +QVFLVAHDWG+IIGWYLCMFRP++VKA
Sbjct: 86 GYGDTEAPPSISSYNCFHIVGDLVALIDSLGVQQVFLVAHDWGAIIGWYLCMFRPDKVKA 145
Query: 121 Y 121
Y
Sbjct: 146 Y 146
>UniRef100_O49857 Epoxide hydrolase [Glycine max]
Length = 341
Score = 225 bits (573), Expect = 1e-57
Identities = 101/121 (83%), Positives = 114/121 (93%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME I+HRTVEVNGIKMH+AEKG+GPVVLFLHGFPELWYSWRHQIL+LSS GY AVAPDLR
Sbjct: 26 MEQIKHRTVEVNGIKMHVAEKGEGPVVLFLHGFPELWYSWRHQILSLSSLGYRAVAPDLR 85
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDT++P SI++Y CFHIVGD+VALID LG +QVFLVAHDWG+IIGWYLCMFRP++VKA
Sbjct: 86 GYGDTEAPPSISSYNCFHIVGDLVALIDSLGVQQVFLVAHDWGAIIGWYLCMFRPDKVKA 145
Query: 121 Y 121
Y
Sbjct: 146 Y 146
>UniRef100_Q76E11 Soluble epoxide hydrolase [Citrus jambhiri]
Length = 316
Score = 193 bits (490), Expect = 6e-48
Identities = 83/120 (69%), Positives = 99/120 (82%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEH TV NGI MH+A G GP VLF+HGFPELWYSWR+Q+L LSSRGY A+APDLR
Sbjct: 1 MEKIEHTTVATNGINMHVASIGTGPAVLFIHGFPELWYSWRNQLLYLSSRGYRAIAPDLR 60
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGDTD+P S+T+YT H+VGD++ L+D LG QVFLV HDWG++I WY C+FRP+RVKA
Sbjct: 61 GYGDTDAPPSVTSYTALHLVGDLIGLLDKLGIHQVFLVGHDWGALIAWYFCLFRPDRVKA 120
>UniRef100_Q9M9W5 Putative epoxide hydrolase [Arabidopsis thaliana]
Length = 331
Score = 186 bits (473), Expect = 5e-46
Identities = 85/125 (68%), Positives = 102/125 (81%), Gaps = 4/125 (3%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPD 58
MEGI+HR V VNGI MHIAEKG +GPVVL LHGFP+LWY+WRHQI LSS GY AVAPD
Sbjct: 1 MEGIDHRMVSVNGITMHIAEKGPKEGPVVLLLHGFPDLWYTWRHQISGLSSLGYRAVAPD 60
Query: 59 LRGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQ--VFLVAHDWGSIIGWYLCMFRPE 116
LRGYGD+DSP S + YTC ++VGD+VAL+D + Q VFLV HDWG+IIGW+LC+FRPE
Sbjct: 61 LRGYGDSDSPESFSEYTCLNVVGDLVALLDSVAGNQEKVFLVGHDWGAIIGWFLCLFRPE 120
Query: 117 RVKAY 121
++ +
Sbjct: 121 KINGF 125
>UniRef100_Q8H289 Epoxide hydrolase [Ananas comosus]
Length = 318
Score = 182 bits (461), Expect = 1e-44
Identities = 83/128 (64%), Positives = 100/128 (77%), Gaps = 3/128 (2%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKG---QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAP 57
MEGI HRTVE+NGI +H+AEKG VL LHGFPELWYSWRHQI+ L++RGY A+AP
Sbjct: 1 MEGIVHRTVEINGIAVHVAEKGGDDAAAAVLLLHGFPELWYSWRHQIVGLAARGYRAIAP 60
Query: 58 DLRGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPER 117
DLRGYGDT +P S+ +YT FH+VGD+VAL+D L QVF+V HDWG+ I W LCM RP+R
Sbjct: 61 DLRGYGDTSAPPSVNSYTLFHLVGDVVALLDALELPQVFVVGHDWGAAIAWTLCMIRPDR 120
Query: 118 VKAYAPSS 125
VKA +S
Sbjct: 121 VKALVNTS 128
>UniRef100_Q84ZZ3 Soluble epoxide hydrolase [Euphorbia lagascae]
Length = 321
Score = 181 bits (459), Expect = 2e-44
Identities = 83/126 (65%), Positives = 96/126 (75%), Gaps = 1/126 (0%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKG-QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDL 59
ME IEH V NGI MHIA G QGPV+LFLHGFP+LWYSWRHQ+L LSS GY +APDL
Sbjct: 5 MEKIEHSMVSTNGINMHIASIGTQGPVILFLHGFPDLWYSWRHQLLYLSSVGYRCIAPDL 64
Query: 60 RGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVK 119
RGYGDTD+P +I YT FHI+GD+V L+D LG QVFLV HDWG+II WY C+ P R+K
Sbjct: 65 RGYGDTDAPPAINQYTVFHILGDLVGLLDSLGIDQVFLVGHDWGAIISWYFCLLMPFRIK 124
Query: 120 AYAPSS 125
A +S
Sbjct: 125 ALVNAS 130
>UniRef100_O81299 T14P8.15 protein [Arabidopsis thaliana]
Length = 324
Score = 180 bits (457), Expect = 4e-44
Identities = 76/125 (60%), Positives = 98/125 (77%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEH T+ NGI MH+A G GPV+LF+HGFP+LWYSWRHQ+++ ++ GY A+APDLR
Sbjct: 1 MEKIEHTTISTNGINMHVASIGSGPVILFVHGFPDLWYSWRHQLVSFAALGYRAIAPDLR 60
Query: 61 GYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
GYGD+D+P S +YT HIVGD+V L+D LG +VFLV HDWG+I+ W+LCM RP+RV A
Sbjct: 61 GYGDSDAPPSRESYTILHIVGDLVGLLDSLGVDRVFLVGHDWGAIVAWWLCMIRPDRVNA 120
Query: 121 YAPSS 125
+S
Sbjct: 121 LVNTS 125
>UniRef100_Q8SAY9 Putative hydrolase [Oryza sativa]
Length = 338
Score = 176 bits (445), Expect = 9e-43
Identities = 78/126 (61%), Positives = 97/126 (76%), Gaps = 4/126 (3%)
Query: 4 IEHRTVEVNGIKMHIAEKG----QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDL 59
I HRTVE NGI +H+AE G G VLFLHGFPELWYSWRHQ+ L+ RG+ +APDL
Sbjct: 11 IRHRTVEANGISIHVAEAGGEGGDGAAVLFLHGFPELWYSWRHQMEHLAGRGFRCLAPDL 70
Query: 60 RGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVK 119
RGYGDTD+P I +Y+ FH+VGD+VAL+D LG +VF+V HDWG+II WY+C+FRP+RV
Sbjct: 71 RGYGDTDAPPEIESYSAFHVVGDLVALLDALGLAKVFVVGHDWGAIIAWYMCLFRPDRVT 130
Query: 120 AYAPSS 125
A +S
Sbjct: 131 ALVNTS 136
>UniRef100_Q944I9 At2g26740/F18A8.11 [Arabidopsis thaliana]
Length = 211
Score = 172 bits (436), Expect = 1e-41
Identities = 78/122 (63%), Positives = 100/122 (81%), Gaps = 5/122 (4%)
Query: 4 IEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
+EHR V NGI +H+A +G GP+VL LHGFPELWYSWRHQI L++RGY AVAPDLRG
Sbjct: 1 MEHRKVRGNGIDIHVAIQGPSDGPIVLLLHGFPELWYSWRHQIPGLAARGYRAVAPDLRG 60
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHLGA---KQVFLVAHDWGSIIGWYLCMFRPERV 118
YGD+D+P I++YTCF+IVGD++A+I L A ++VF+V HDWG++I WYLC+FRP+RV
Sbjct: 61 YGDSDAPAEISSYTCFNIVGDLIAVISALTASEDEKVFVVGHDWGALIAWYLCLFRPDRV 120
Query: 119 KA 120
KA
Sbjct: 121 KA 122
>UniRef100_Q42566 Putative epoxide hydrolase ATsEH [Arabidopsis thaliana]
Length = 321
Score = 172 bits (436), Expect = 1e-41
Identities = 78/122 (63%), Positives = 100/122 (81%), Gaps = 5/122 (4%)
Query: 4 IEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
+EHR V NGI +H+A +G GP+VL LHGFPELWYSWRHQI L++RGY AVAPDLRG
Sbjct: 1 MEHRKVRGNGIDIHVAIQGPSDGPIVLLLHGFPELWYSWRHQIPGLAARGYRAVAPDLRG 60
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHLGA---KQVFLVAHDWGSIIGWYLCMFRPERV 118
YGD+D+P I++YTCF+IVGD++A+I L A ++VF+V HDWG++I WYLC+FRP+RV
Sbjct: 61 YGDSDAPAEISSYTCFNIVGDLIAVISALTASEDEKVFVVGHDWGALIAWYLCLFRPDRV 120
Query: 119 KA 120
KA
Sbjct: 121 KA 122
>UniRef100_Q41413 Epoxide hydrolase [Solanum tuberosum]
Length = 321
Score = 170 bits (431), Expect = 4e-41
Identities = 77/124 (62%), Positives = 102/124 (82%), Gaps = 4/124 (3%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEH+ V VNG+ MHIAE GQGP +LF+HGFPELWYSWRHQ++ L+ RGY AVAPDLR
Sbjct: 1 MEKIEHKMVAVNGLNMHIAELGQGPTILFIHGFPELWYSWRHQMVYLAERGYRAVAPDLR 60
Query: 61 GYGD-TDSPIS-ITTYTCFHIVGDIVALIDHLGAKQ--VFLVAHDWGSIIGWYLCMFRPE 116
GYGD T +PI+ + ++ FH+VGD+VAL++ + + VF+VAHDWG++I W+LC+FRP+
Sbjct: 61 GYGDTTGAPINDPSKFSIFHLVGDVVALLEAIAPNEDKVFVVAHDWGALIAWHLCLFRPD 120
Query: 117 RVKA 120
+VKA
Sbjct: 121 KVKA 124
>UniRef100_Q8SAY7 Putative hydrolase [Oryza sativa]
Length = 333
Score = 169 bits (429), Expect = 7e-41
Identities = 76/121 (62%), Positives = 93/121 (76%), Gaps = 4/121 (3%)
Query: 4 IEHRTVEVNGIKMHIAEKGQG----PVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDL 59
+ HRTVE NGI MH+AE G G P VLF+HGFPELWYSWRHQ+ L++RGY VAPDL
Sbjct: 7 VRHRTVEANGISMHVAEAGPGSGTAPAVLFVHGFPELWYSWRHQMGHLAARGYRCVAPDL 66
Query: 60 RGYGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVK 119
RGYG T +P T+YT FH+VGD+VAL+D L QVF+V HDWG+I+ W LC+ RP+RV+
Sbjct: 67 RGYGGTTAPPEHTSYTIFHLVGDLVALLDALELPQVFVVGHDWGAIVSWNLCLLRPDRVR 126
Query: 120 A 120
A
Sbjct: 127 A 127
>UniRef100_Q8L5G6 Soluble epoxide hydrolase [Brassica napus]
Length = 318
Score = 168 bits (425), Expect = 2e-40
Identities = 72/119 (60%), Positives = 96/119 (80%), Gaps = 2/119 (1%)
Query: 4 IEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
+EHR + NG+ +H+A +G GPVVL +HGFP LWYSWRHQI L++ GY AVAPDLRG
Sbjct: 1 MEHRKLRGNGVDIHVAIQGPSDGPVVLLIHGFPTLWYSWRHQIPGLAALGYRAVAPDLRG 60
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKA 120
YGD+D+P I++YTCFH+VGD++A+I L +VF+V HDWG++I WYLC+FRP++VKA
Sbjct: 61 YGDSDAPSEISSYTCFHLVGDMIAVISALTEDKVFVVGHDWGALIAWYLCLFRPDKVKA 119
>UniRef100_O48789 Putative epoxide hydrolase [Arabidopsis thaliana]
Length = 320
Score = 167 bits (423), Expect = 3e-40
Identities = 76/121 (62%), Positives = 98/121 (80%), Gaps = 4/121 (3%)
Query: 4 IEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
+EHR V NGI +H+A +G G +VL LHGFPELWYSWRHQI L++RGY AVAPDLRG
Sbjct: 1 MEHRNVRGNGIDIHVAIQGPSDGTIVLLLHGFPELWYSWRHQISGLAARGYRAVAPDLRG 60
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMFRPERVK 119
YGD+D+P I+++TCF+IVGD+VA+I L K+VF+V HDWG++I WYLC+FRP++VK
Sbjct: 61 YGDSDAPAEISSFTCFNIVGDLVAVISTLIKEDKKVFVVGHDWGALIAWYLCLFRPDKVK 120
Query: 120 A 120
A
Sbjct: 121 A 121
>UniRef100_Q8GX63 Putative epoxide hydrolase [Arabidopsis thaliana]
Length = 178
Score = 166 bits (419), Expect = 1e-39
Identities = 76/127 (59%), Positives = 97/127 (75%), Gaps = 11/127 (8%)
Query: 5 EHRTVEVNGIKMHIAEK-----GQG----PVVLFLHGFPELWYSWRHQILTLSSRGYHAV 55
+H V+VNGI MH+AEK G G PV+LFLHGFPELWY+WRHQ++ LSS GY +
Sbjct: 6 DHSFVKVNGITMHVAEKSPSVAGNGAIRPPVILFLHGFPELWYTWRHQMVALSSLGYRTI 65
Query: 56 APDLRGYGDTDSPISITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMF 113
APDLRGYGDTD+P S+ YT H+VGD++ LID + ++VF+V HDWG+II W+LC+F
Sbjct: 66 APDLRGYGDTDAPESVDAYTSLHVVGDLIGLIDAVVGDREKVFVVGHDWGAIIAWHLCLF 125
Query: 114 RPERVKA 120
RP+RVKA
Sbjct: 126 RPDRVKA 132
>UniRef100_O23444 Putative epoxide hydrolase [Arabidopsis thaliana]
Length = 536
Score = 166 bits (419), Expect = 1e-39
Identities = 76/127 (59%), Positives = 97/127 (75%), Gaps = 11/127 (8%)
Query: 5 EHRTVEVNGIKMHIAEK-----GQG----PVVLFLHGFPELWYSWRHQILTLSSRGYHAV 55
+H V+VNGI MH+AEK G G PV+LFLHGFPELWY+WRHQ++ LSS GY +
Sbjct: 371 DHSFVKVNGITMHVAEKSPSVAGNGAIRPPVILFLHGFPELWYTWRHQMVALSSLGYRTI 430
Query: 56 APDLRGYGDTDSPISITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMF 113
APDLRGYGDTD+P S+ YT H+VGD++ LID + ++VF+V HDWG+II W+LC+F
Sbjct: 431 APDLRGYGDTDAPESVDAYTSLHVVGDLIGLIDAVVGDREKVFVVGHDWGAIIAWHLCLF 490
Query: 114 RPERVKA 120
RP+RVKA
Sbjct: 491 RPDRVKA 497
Score = 155 bits (393), Expect = 1e-36
Identities = 70/127 (55%), Positives = 94/127 (73%), Gaps = 7/127 (5%)
Query: 1 MEGIEHRTVEVNGIKMHIAEK-----GQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAV 55
++G+EH+T++VNGI MH+AEK G+ P++LFLHGFPELWY+WRHQ++ LSS GY +
Sbjct: 51 LDGVEHKTLKVNGINMHVAEKPGSGSGEDPIILFLHGFPELWYTWRHQMVALSSLGYRTI 110
Query: 56 APDLRGYGDTDSPISITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMF 113
APDLRGYGDT++P + YT +VALI + G K V +V HDWG++I W LC +
Sbjct: 111 APDLRGYGDTEAPEKVEDYTLLKRGRSVVALIVAVTGGDKAVSVVGHDWGAMIAWQLCQY 170
Query: 114 RPERVKA 120
RPE+VKA
Sbjct: 171 RPEKVKA 177
>UniRef100_Q41415 Epoxide hydrolase [Solanum tuberosum]
Length = 321
Score = 165 bits (418), Expect = 1e-39
Identities = 73/124 (58%), Positives = 102/124 (81%), Gaps = 4/124 (3%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEH+ V VNG+ MH+AE G+GP +LF+HGFPELWYSWRHQ++ L+ RGY AVAPDLR
Sbjct: 1 MEKIEHKMVAVNGLNMHLAELGEGPTILFIHGFPELWYSWRHQMVYLAERGYRAVAPDLR 60
Query: 61 GYGD-TDSPIS-ITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMFRPE 116
GYGD T +P++ + ++ H+VGD+VAL++ + ++VF+VAHDWG++I W+LC+FRP+
Sbjct: 61 GYGDTTGAPLNDPSKFSILHLVGDVVALLEAIAPNEEKVFVVAHDWGALIAWHLCLFRPD 120
Query: 117 RVKA 120
+VKA
Sbjct: 121 KVKA 124
>UniRef100_Q41412 Epoxide hydrolase [Solanum tuberosum]
Length = 321
Score = 164 bits (415), Expect = 3e-39
Identities = 75/124 (60%), Positives = 98/124 (78%), Gaps = 4/124 (3%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEH+ V VNG+ MHIAE GQGP +LFLHGFPELWYSWRHQ++ L+ GY AVAPDLR
Sbjct: 1 MEKIEHKMVAVNGLNMHIAELGQGPTILFLHGFPELWYSWRHQMVYLAECGYRAVAPDLR 60
Query: 61 GYGDTD--SPISITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMFRPE 116
GYGDT S + ++ H+VGD+VAL++ + ++VF+VAHDWG++I W+LC+FRP+
Sbjct: 61 GYGDTTGASLNDPSKFSILHLVGDVVALLEAIAPNEEKVFVVAHDWGALIAWHLCLFRPD 120
Query: 117 RVKA 120
+VKA
Sbjct: 121 KVKA 124
>UniRef100_Q9SD45 Epoxide hydrolase-like protein [Arabidopsis thaliana]
Length = 323
Score = 164 bits (414), Expect = 4e-39
Identities = 71/120 (59%), Positives = 91/120 (75%), Gaps = 2/120 (1%)
Query: 4 IEHRTVEVNGIKMHIAEKG--QGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLRG 61
+ + ++ NGI +++AEKG +GP+VL LHGFPE WYSWRHQI LSS GYH VAPDLRG
Sbjct: 5 VREKKIKTNGIWLNVAEKGDEEGPLVLLLHGFPETWYSWRHQIDFLSSHGYHVVAPDLRG 64
Query: 62 YGDTDSPISITTYTCFHIVGDIVALIDHLGAKQVFLVAHDWGSIIGWYLCMFRPERVKAY 121
YGD+DS S +YT H+V D++ L+DH G Q F+ HDWG+IIGW LC+FRP+RVK +
Sbjct: 65 YGDSDSLPSHESYTVSHLVADVIGLLDHYGTTQAFVAGHDWGAIIGWCLCLFRPDRVKGF 124
>UniRef100_Q41414 Epoxide hydrolase [Solanum tuberosum]
Length = 321
Score = 163 bits (413), Expect = 5e-39
Identities = 76/129 (58%), Positives = 102/129 (78%), Gaps = 4/129 (3%)
Query: 1 MEGIEHRTVEVNGIKMHIAEKGQGPVVLFLHGFPELWYSWRHQILTLSSRGYHAVAPDLR 60
ME IEH+ V VNG+ MHIAE GQGP +LFLHGFPELWYSWRHQ++ L+ RGY AVAP LR
Sbjct: 1 MEKIEHKMVAVNGLNMHIAELGQGPTILFLHGFPELWYSWRHQMVYLAERGYRAVAPVLR 60
Query: 61 GYGD-TDSPIS-ITTYTCFHIVGDIVALIDHL--GAKQVFLVAHDWGSIIGWYLCMFRPE 116
GYGD T +P++ + ++ +VGD+VAL++ + ++VF+VAHDWG++I W+LC+FRP+
Sbjct: 61 GYGDTTGAPLNDPSKFSILQLVGDVVALLEAIAPNEEKVFVVAHDWGALIAWHLCLFRPD 120
Query: 117 RVKAYAPSS 125
+VKA SS
Sbjct: 121 KVKALVNSS 129
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.340 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 470,942,300
Number of Sequences: 2790947
Number of extensions: 17259980
Number of successful extensions: 50660
Number of sequences better than 10.0: 2004
Number of HSP's better than 10.0 without gapping: 815
Number of HSP's successfully gapped in prelim test: 1189
Number of HSP's that attempted gapping in prelim test: 48560
Number of HSP's gapped (non-prelim): 2073
length of query: 312
length of database: 848,049,833
effective HSP length: 127
effective length of query: 185
effective length of database: 493,599,564
effective search space: 91315919340
effective search space used: 91315919340
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 74 (33.1 bits)
Lotus: description of TM0109.15


