

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
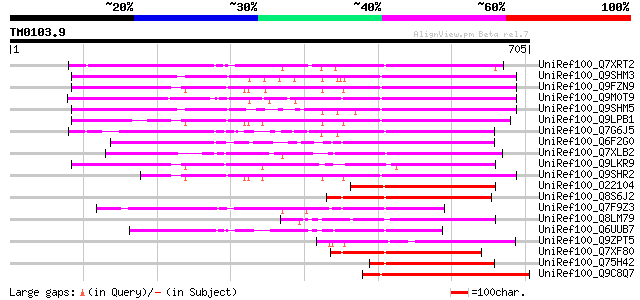
Query= TM0103.9
(705 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XRT2 OSJNBa0042F21.11 protein [Oryza sativa] 291 3e-77
UniRef100_Q9SHM3 F7F22.17 [Arabidopsis thaliana] 286 1e-75
UniRef100_Q9FZN9 Retroelement pol polyprotein-like [Arabidopsis ... 280 1e-73
UniRef100_Q9M0T9 Putative athila transposon protein [Arabidopsis... 271 6e-71
UniRef100_Q9SHM5 F7F22.15 [Arabidopsis thaliana] 265 3e-69
UniRef100_Q9LPB1 T32E20.9 [Arabidopsis thaliana] 263 2e-68
UniRef100_Q7G6J5 Putative polyprotein [Oryza sativa] 244 6e-63
UniRef100_Q6F2G0 Hypothetical protein OSJNBa0083K01.26 [Oryza sa... 216 2e-54
UniRef100_Q7XLB2 OSJNBa0011K22.17 protein [Oryza sativa] 209 3e-52
UniRef100_Q9LKR9 Hypothetical protein T15F17.a [Arabidopsis thal... 204 9e-51
UniRef100_Q9SHR2 F28L22.6 protein [Arabidopsis thaliana] 201 8e-50
UniRef100_O22104 Vicia faba mRNA expressed from retrotransposon-... 178 4e-43
UniRef100_Q8S6J2 Hypothetical protein OSJNBa0019N10.34 [Oryza sa... 174 6e-42
UniRef100_Q7F9Z3 OSJNBa0079C19.24 protein [Oryza sativa] 174 8e-42
UniRef100_Q8LM79 Hypothetical protein OSJNAa0082N11.11 [Oryza sa... 170 2e-40
UniRef100_Q6UUB7 Hypothetical protein [Oryza sativa] 166 2e-39
UniRef100_Q9ZPT5 Hypothetical protein At2g14040 [Arabidopsis tha... 165 5e-39
UniRef100_Q7XF80 Hypothetical protein [Oryza sativa] 165 5e-39
UniRef100_Q75H42 Hypothetical protein OSJNBa0006O14.11 [Oryza sa... 162 2e-38
UniRef100_Q9C8Q7 Athila ORF 1, putative; 43045-40843 [Arabidopsi... 162 4e-38
>UniRef100_Q7XRT2 OSJNBa0042F21.11 protein [Oryza sativa]
Length = 920
Score = 291 bits (746), Expect = 3e-77
Identities = 198/622 (31%), Positives = 319/622 (50%), Gaps = 48/622 (7%)
Query: 81 ELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQ 140
+ VRL+LFPFSL A++W + +I TW+ + F K FP L+ I F+Q
Sbjct: 298 DAVRLRLFPFSLLRRAKQWFYANC-AAINTWDKCSTSFLSKFFPIGKTNALRGRISRFQQ 356
Query: 141 EDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQ 200
E++ EA ER ++ + CP H + + ++ FY+GL +R +L+AAA G F + + Q
Sbjct: 357 TRDESIPEAWERLQEYVAACPHHGMDDWLILQNFYNGLTLMSRDHLDAAAGGAFFSKTVQ 416
Query: 201 AGWNLINKMAESAVNSTNDRQNR-RGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKIS 259
LI KM + S Q R RG+ ++ + + A L + + +++ Q +
Sbjct: 417 GAVELIEKMVSNTGWSKERLQTRQRGMHTVKETELLAAKLDLLMKCLDDHEKRPQGTVKA 476
Query: 260 NVDSIHCGTCGGP-HASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNN 318
+ C CGG H+ +C EE +G +NN Y R W +
Sbjct: 477 LDSHVTCEVCGGTGHSGNDC-LETHEEAMYMG--KNNEY--------RPQGGEGWNQPRP 525
Query: 319 FNQGGNNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMENFINKAD---TSFKNHE 375
+ GGNN +SNQ Q + ++++EN K D ++F+N
Sbjct: 526 YYPGGNNNGNFSNQPSLMDLVFAQAKTTDALRKKLAANDKILENINVKLDGFASAFQNQL 585
Query: 376 AAIKSLETQVGQMAKQMSERPPGMFPS--DTVINPKENCSAITLRNIE---------PLG 424
+ K +ETQ+ Q+A + G P D+ I EN AIT R + P G
Sbjct: 586 SFNKMIETQLAQLASLVPANESGRIPGQPDSSI---ENVKAITTRGGKSTRDPPYPNPAG 642
Query: 425 EN--KKKQKEKEEEKRKVE----LENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLR 478
N K+ + + ++++ + ++ PFP K ++KQF+ F+ + +K+
Sbjct: 643 TNGMSKETPSTDSDDKEIQPDKTVPQEYCDTRLLPFPQQSRKPSVDKQFACFVEVIQKIH 702
Query: 479 VELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSF 538
+ +P + ++ +P YA+++K+IL+ KR L E+++LTE CS ++ KLP K+KDPG
Sbjct: 703 INVPLLDAMQ-VPTYARYLKDILNNKRPLPT-TEVVKLTEHCSNLILHKLPEKKKDPGCP 760
Query: 539 TLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVE 598
T+ + GA + +ALCDLG+SV++MP +F++LN L PT M LQLAD S+ P G+ E
Sbjct: 761 TITCSIGAQQFDQALCDLGASVSVMPKDVFDKLNFTVLAPTPMRLQLADSSVHYPVGIAE 820
Query: 599 DVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRV--ADEKI 656
DV ++ +F PVDFV++DMD + PLILGRPFL+T+ A I+VG G+I +EK
Sbjct: 821 DVPAKIRDFFIPVDFVVLDMDTGKETPLILGRPFLSTAGANIDVGTGSIRFHTNGKEEKF 880
Query: 657 TF-------TIFDLKPKPVEKN 671
F T+ +K +P +N
Sbjct: 881 EFQPRMEQCTMVRIKYRPNPQN 902
>UniRef100_Q9SHM3 F7F22.17 [Arabidopsis thaliana]
Length = 1799
Score = 286 bits (732), Expect = 1e-75
Identities = 211/715 (29%), Positives = 328/715 (45%), Gaps = 123/715 (17%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A W + P SITTW+D K F K F +L+N+I F Q+
Sbjct: 70 KLRLFPFSLGDKAHIWEKNLPHDSITTWDDCKKVFLSKFFSNARTARLRNEISGFSQKTG 129
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E+ EA ERFK +CP H T + Y G+ R L+ A++G F+ + GW
Sbjct: 130 ESFCEAWERFKGYTNQCPHHGFTKASLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGW 189
Query: 204 NLINKMAESAVNSTNDRQNR-RGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKISNVD 262
L+ +A+S N D RG + S+ + +++ + + +S
Sbjct: 190 ELVENLAQSDGNYNEDCDRTVRGTAD---------SDDKHRKEIKALNDKLDRILLSQHK 240
Query: 263 SIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNNFNQG 322
+H + ++ EEV + N+Q YN N+PN S+R N N
Sbjct: 241 HVHFLVDDEQYEVQDGEGNQLEEVSYINNNQGG--YKGYNYFKTNNPNLSYRSTNVANPQ 298
Query: 323 GN----NQRQYSNQRFQSQNSG--PQQN---------------QDQGSGNGKKSLEELME 361
Q+Q N+ F N G P+Q Q QG ++++++
Sbjct: 299 DQVYPPQQQQSQNKPFVPYNQGFVPKQQFQGNYQQQPPGFAPPQHQGPAAPDADMKQMLQ 358
Query: 362 NFI------------------NKADTSFKNHEAAIKSLETQV----GQMAKQMSERPPGM 399
+ NK D S+ + +++L+T+V G + +
Sbjct: 359 QLLHGQASCSMEMAKKISELHNKLDCSYNDLNVKMETLDTKVRYLEGHSTSSSATKQTSQ 418
Query: 400 FPSDTVINPKENCSAITLRNIEPL-----------------GENKKKQKEKEEEKRKVEL 442
P V NPKE AITLR+ + L GE+ +K++ ++ + L
Sbjct: 419 LPGKAVQNPKEYAHAITLRSGKALPTREEPKTVTEDSEDQDGEDLSLEKDQADKPLDLSL 478
Query: 443 E---------------NKFT-------------------------KVLFPP-------FP 455
E + FT KV PP FP
Sbjct: 479 EQPLDLSLQQSLDPPLDSFTRPTTRPVIPAASPTAPKPVAVKNKEKVFVPPPYKSQLPFP 538
Query: 456 TNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIE 515
K +K + F K++ + +P + L +P KF+K+++ + R+ E ++
Sbjct: 539 GRHKKALADKYRAMFAKNIKEVELRIPLVDALALIPDSHKFLKDLIVE--RIQEVQGMVV 596
Query: 516 LTEECSAILQRKLPPKR-KDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVG 574
L+ ECSAI+Q+K+ PK+ DPGSFTLP + G R LCDLG+SV+LMPLS+ +RL
Sbjct: 597 LSHECSAIIQKKIIPKKLSDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGFT 656
Query: 575 ELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLA 634
+ K + L LADRS+ P G++E++ +R+G E P DFV+++MDE+ K PLILGRPFLA
Sbjct: 657 QYKSCNISLILADRSVRIPHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFLA 716
Query: 635 TSQAKINVGNGTISLRVA-DEKITFTIFDLKPKPVEKNDVFLVEMMDEWSDEKLK 688
T+ A I+V G I L + D ++TF + D KP + +F +E MD+ +DE L+
Sbjct: 717 TAGAMIDVKKGKIDLNLGKDFRMTFDVKDAMKKPTIEGQLFWIEEMDQLADELLE 771
>UniRef100_Q9FZN9 Retroelement pol polyprotein-like [Arabidopsis thaliana]
Length = 1864
Score = 280 bits (715), Expect = 1e-73
Identities = 212/702 (30%), Positives = 319/702 (45%), Gaps = 113/702 (16%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A +W S QGSIT+W D K F K F + +L+NDI F Q +
Sbjct: 119 KLRLFPFSLGDKAHQWEKSLLQGSITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNN 178
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E EA ERFK +CP H + + Y G+ R L+ A++G F + GW
Sbjct: 179 ETFCEAWERFKGYQTQCPHHGFSKASLLSTLYRGVLPKIRMLLDTASNGNFLNKDVEDGW 238
Query: 204 NLINKMAESAVNSTNDRQNRRGVLEIEAYDRMVA----SNKQLSQQMTTIQRQFQAAKIS 259
L+ +A+S N D YDR V S+++ ++M + + +
Sbjct: 239 ELVENLAQSDGNYNED------------YDRSVRTSSDSDEKHRREMKAMNDKLDKLLLV 286
Query: 260 NVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQN-- 317
IH ++ T EEV + N Y+ +N +NHPN S+R N
Sbjct: 287 QQKHIHFLGDDETFQVQDGETMQSEEVSYVQNQGG--YNKGFNNFKQNHPNLSYRSTNVA 344
Query: 318 -------------------NFNQG----------GNNQRQYSNQRFQSQNSGPQQNQD-- 346
+NQG GN Q Q F Q P
Sbjct: 345 NPQDQVYPSQQQNQPRPFVPYNQGQGYVPKQQYQGNYQPQLPPPGFTQQQQQPASTTPDS 404
Query: 347 ----------QGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQVGQMAKQMSERP 396
QG G L + M NK D S+ + +++L +++ + Q
Sbjct: 405 DLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCSYNDINIKVEALTSKIRYIEGQTGSTA 464
Query: 397 PGMF--PS-DTVINPKENCSAITLRNIEPL-----------------GENKKKQKEKEEE 436
F PS ++ N +E AITLR+ + L GE+ + E+
Sbjct: 465 APKFTGPSRKSMSNSEEYAHAITLRSGKELPTKESPNQNTEDSVDQDGEDFCQNGNSAEK 524
Query: 437 KRKVELENKFTKVLFP----------------------------PFPTNIAKRRLEKQFS 468
+ + ++ T++L P PFP K ++K +
Sbjct: 525 AIEEPILDQPTRLLAPAASPLVEKPAATKTKDNVFVPPPYKPPLPFPGRFKKVMIQKYKA 584
Query: 469 KFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKL 528
K L V +P + L +P K++K+++++ R+ E ++ L+ ECSAI+Q+K+
Sbjct: 585 LLEKQLKNLEVTMPLVDCLALIPDSNKYVKDMITE--RIKEVQGMVVLSHECSAIIQQKI 642
Query: 529 PPKRK-DPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLAD 587
PK+ DPGSFTLP G + LCDLG+SV+LMPLS+ ++L + KP + L LAD
Sbjct: 643 IPKKLGDPGSFTLPCALGPLAFNKCLCDLGASVSLMPLSVAKKLGFNKYKPCNISLILAD 702
Query: 588 RSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTI 647
RS+ P G++ED+ V +G E P DFV+++MDE+ K PLILGRPFL T A I+V G I
Sbjct: 703 RSVRIPHGLLEDLPVMIGMVEVPTDFVVLEMDEEPKDPLILGRPFLVTVGAIIDVKKGKI 762
Query: 648 SLRVA-DEKITFTIFDLKPKPVEKNDVFLVEMMDEWSDEKLK 688
L + D K+TF I + KP + ++F +E MD +DE L+
Sbjct: 763 DLNLGRDLKMTFDITNTMKKPTIERNIFWIEEMDMLADEMLE 804
>UniRef100_Q9M0T9 Putative athila transposon protein [Arabidopsis thaliana]
Length = 866
Score = 271 bits (692), Expect = 6e-71
Identities = 204/628 (32%), Positives = 310/628 (48%), Gaps = 35/628 (5%)
Query: 79 NAELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVF 138
+A+ +L+LFPFSL D A W + P SI TW+D K F K F +L+N+I F
Sbjct: 82 SADGFKLRLFPFSLGDKAHIWEKNLPHDSIITWDDCKKAFLSKFFSNARTARLRNEISGF 141
Query: 139 KQEDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALS 198
Q+ E+ EA ERFK +CP H T + Y G+ R L+ A++G F+
Sbjct: 142 SQKTGESFCEAWERFKGYTNQCPHHGFTKASMLSTLYRGVLPRIRMLLDTASNGNFQNKD 201
Query: 199 AQAGWNLINKMAESAVNSTNDRQNRRGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKI 258
+ GW L++ +A+S N N+ +R ++ D+ K L+ ++ I Q
Sbjct: 202 VEEGWELVDNLAQSDGN-YNEDCDRTVRDTADSDDKHRKEIKALNDKLDRILLNQQKHVH 260
Query: 259 SNVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNN 318
VD G G EEV + N+Q+ YN N+PN S+R N
Sbjct: 261 FLVDDEQFQVQDGE------GNQL-EEVSYINNNQSG--YKGYNNFKTNNPNLSYRSTNI 311
Query: 319 FNQGGN----NQRQYSNQRFQSQNSG--PQQNQDQGSGN-----GKKSLEELMENFINKA 367
N Q+Q N+ F N G P+Q Q QG+ G K+L E
Sbjct: 312 ANPQDQVYPPQQQQVQNKPFVPYNQGFVPKQ-QFQGNYQPPPPPGGKALPTREEPKTVTE 370
Query: 368 DTSFKNHEAAIKSLETQVG-----QMAKQMSERPPGMFPSDTVINPKENCSAITLRNIEP 422
D+ ++ E SLE Q Q E+P + + P +N + T R I P
Sbjct: 371 DSEDQDGEDL--SLEKDQADKPHEQPLDQSLEQPLDLSLEQPLDLPLDNVTRPTTRPIFP 428
Query: 423 LGENKKKQKEKEEEKRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELP 482
+ + K KV + + L PFP K +K + F K++ + +P
Sbjct: 429 AASATAPKPITVKNKEKVFVPPPYKPEL--PFPGRHKKALADKYRAMFAKNIKEVELRIP 486
Query: 483 FSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKR-KDPGSFTLP 541
+ L +P KF+K+++ + R+ E ++ L+ CSAI+Q+K+ PK+ DPGSFTLP
Sbjct: 487 LVDALALIPDSHKFLKDLIVE--RIQEVQGMVVLSHGCSAIIQKKIIPKKLSDPGSFTLP 544
Query: 542 VNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVL 601
+ G R LCDLG+SV+LM LS+ +RL + K + + L LADRS+ P G++++
Sbjct: 545 CSLGPLAFNRCLCDLGASVSLMALSVAKRLGFTQYKSSNISLILADRSVRIPHGLLKNFG 604
Query: 602 VRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVA-DEKITFTI 660
+ +G E P DFV+++MDE+ K PLILGRPFLAT+ A I+V G I L + D ++TF +
Sbjct: 605 ITIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKIDLSLGKDFRMTFDV 664
Query: 661 FDLKPKPVEKNDVFLVEMMDEWSDEKLK 688
D KP + +F ++ MD+ +DE L+
Sbjct: 665 KDAMKKPTIEGQLFWIKEMDQLADELLE 692
>UniRef100_Q9SHM5 F7F22.15 [Arabidopsis thaliana]
Length = 1862
Score = 265 bits (678), Expect = 3e-69
Identities = 202/637 (31%), Positives = 303/637 (46%), Gaps = 94/637 (14%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A W + P SITTW+D K F K F +L+N+I F Q+
Sbjct: 205 KLRLFPFSLGDKAHIWEKNLPHDSITTWDDCKKAFLSKFFSNARTARLRNEISGFSQKTG 264
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E+ EA ERFK +C H T + Y G+ R L+ A++G F+ + GW
Sbjct: 265 ESFCEAWERFKGYTNQCSHHGFTKASLLSTLYRGVLPRIRMLLDTASNGNFQNKDVEEGW 324
Query: 204 NLINKMAESAVN-STNDRQNRRGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKISNVD 262
L+ +A+S N + N + RG + S+ + +++ + + +S
Sbjct: 325 ELVENLAQSNGNYNENCDRTVRGTAD---------SDDKHRKEIKALNDKLDRILLSQHK 375
Query: 263 SIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNNFNQG 322
+H + ++ EEV + N+Q YN N+PN S+R N N
Sbjct: 376 HVHFLVDDEQYEVQDGEGNQLEEVSYINNNQGG--YKGYNNFKTNNPNLSYRSTNVANP- 432
Query: 323 GNNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLE 382
Q Q PQQ Q Q NK +
Sbjct: 433 ------------QDQVYPPQQQQSQ-----------------NKPFVPYN---------- 453
Query: 383 TQVGQMAKQMSERPPGMFPSDTVINPKENCSAITLRNIEPL-----------------GE 425
Q KQ S+ P V NPKE AITL + + L GE
Sbjct: 454 ----QATKQTSQ-----LPGKAVQNPKEYAHAITLHSGKALPTREEPKTVTEDSEDQDGE 504
Query: 426 NKKKQKEKEEEKRKVELEN----KFTKVLFPPFP--TNIAKRRLEKQFS----KFISMFK 475
+ +K++ ++ + LE + L PP T R + S K +++
Sbjct: 505 DLSLKKDQADKPLDLSLEQPLDLSLQQSLDPPLDSFTRPTTRPVIPAASPTAPKPVAVKN 564
Query: 476 KLRVEL--PFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKR- 532
K +VEL P + L +P KF+K+++ + R+ E ++ L+ ECSAI+Q+K+ PK+
Sbjct: 565 KEKVELRIPLVDALALIPDSHKFLKDLIVE--RIQEVQGMVVLSHECSAIIQKKIIPKKL 622
Query: 533 KDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVA 592
DPGSFTLP + G R LCDLG+SV+LMPLS+ +RL + K + L LADRS+
Sbjct: 623 SDPGSFTLPCSLGPLAFNRCLCDLGASVSLMPLSVAKRLGFTQYKSCNISLILADRSVRI 682
Query: 593 PWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVA 652
P G++E++ +R+G E P DFV+++MDE+ K PLILGRPFLAT+ A I+V G I L +
Sbjct: 683 PHGLLENLPIRIGAVEIPTDFVVLEMDEEPKDPLILGRPFLATAGAMIDVKKGKIDLNLG 742
Query: 653 -DEKITFTIFDLKPKPVEKNDVFLVEMMDEWSDEKLK 688
D ++TF + D KP + +F +E MD+ +DE L+
Sbjct: 743 KDFRMTFDVKDAMKKPTIEGQLFWIEEMDQLADELLE 779
>UniRef100_Q9LPB1 T32E20.9 [Arabidopsis thaliana]
Length = 1586
Score = 263 bits (671), Expect = 2e-68
Identities = 205/694 (29%), Positives = 312/694 (44%), Gaps = 132/694 (19%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A +W S PQGSIT+W D K F K F + +L+NDI F Q +
Sbjct: 119 KLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNN 178
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E +EA ERFK +CP H + L + A++G F + GW
Sbjct: 179 ETFYEAWERFKGYQTQCPHHEMLL-------------------DTASNGNFLNKDVEDGW 219
Query: 204 NLINKMAESAVNSTNDRQNRRGVLEIEAYDRMVA----SNKQLSQQMTTIQRQFQAAKIS 259
++ +A+S N D YDR + S+++ ++M + + +
Sbjct: 220 EVVENLAQSDGNYNED------------YDRSIRTSSDSDEKHRREMKAMNDKLDKLLLM 267
Query: 260 NVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQN-- 317
IH ++ T EEV + N Y+ +N +NHPN S+R N
Sbjct: 268 QQKHIHFLGDDETLQVQDGETLQLEEVSYVQNQGG--YNKGFNNFKQNHPNLSYRSTNVA 325
Query: 318 -------------------NFNQG----------GNNQRQYSNQRFQSQNSGPQ------ 342
+NQG GN Q+Q F Q P
Sbjct: 326 NRQDQVYPSQQQNQPKPFVPYNQGQGYVPKQQYQGNYQQQLPPPGFTQQQQQPALTTPDS 385
Query: 343 ------QNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQVGQMAKQMSERP 396
Q QG G L + M NK D S+ + +++L +++ + Q
Sbjct: 386 DLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCSYNDINIKVEALTSKIRYIEGQTGSTA 445
Query: 397 PGMF--PSD-TVINPKENCSAITLRNIEPL-----------------GENKKKQKEKEEE 436
F PS ++ N KE AITLR+ + L GE+ + E+
Sbjct: 446 APKFTGPSGKSMSNSKEYAHAITLRSGKELPTKESPNQNTEDSLDQDGEDFCQNGNSAEK 505
Query: 437 KRKVELENKFTKVLFP----------------------------PFPTNIAKRRLEKQFS 468
+ + ++ T+ L P PFP K ++K +
Sbjct: 506 AIEEPILHQPTRPLAPAASPLVEKPAAAKTKENVFIPPPYKPPLPFPGRFKKVMIQKYKA 565
Query: 469 KFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKL 528
K L V +P + L +P K++K+++++ R+ E ++ L+ ECSAI+Q+K+
Sbjct: 566 LLEKQLKNLEVTMPLVDCLALIPDSNKYVKDMITE--RIKEVQGMVVLSHECSAIIQQKI 623
Query: 529 PPKRK-DPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLAD 587
PK+ DPGSFTLP G + LCDLG+SV+LMPL + ++L + KP + L LAD
Sbjct: 624 IPKKLGDPGSFTLPCALGPLAFSKCLCDLGASVSLMPLPVAKKLGFNKYKPCNISLILAD 683
Query: 588 RSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTI 647
RS+ G++ED+ V +G E P DFV+++MDE+ K PLILGRPFLA ++A I+V G I
Sbjct: 684 RSVRISHGLLEDLPVMIGVVEVPTDFVVLEMDEEPKDPLILGRPFLARARAIIDVKKGKI 743
Query: 648 SLRVA-DEKITFTIFDLKPKPVEKNDVFLVEMMD 680
L + D K+TF I + KP + ++F +E MD
Sbjct: 744 DLNLGRDLKMTFDITNTMKKPTIEGNIFWIEEMD 777
>UniRef100_Q7G6J5 Putative polyprotein [Oryza sativa]
Length = 689
Score = 244 bits (623), Expect = 6e-63
Identities = 178/596 (29%), Positives = 293/596 (48%), Gaps = 85/596 (14%)
Query: 81 ELVRLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQ 140
++V+L+LFPFSL A++W + + ++ TW+ + F K FP
Sbjct: 75 DVVKLRLFPFSLLGRAKQWFYAN-RTAVNTWDKCSTAFLSKFFPM--------------- 118
Query: 141 EDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQ 200
EA ER ++ + CP + + ++ FY+GL +R +L+AAA G F + + Q
Sbjct: 119 -------EAWERLQEYVAACPHLGMDDWLILQNFYNGLTPMSRDHLDAAARGAFFSKTVQ 171
Query: 201 AGWNLINKMAESAVNSTNDRQNR-RGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKIS 259
LI KM + S Q R RG+ ++ + + A L +++ ++ Q +
Sbjct: 172 GAVELIEKMVSNMGWSEERLQTRQRGMHTVKETELLAAKLDLLMKRLDDHEKGPQRTVKA 231
Query: 260 NVDSIHCGTCGGP-HASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNN 318
+ C CG H+ +C EE +GN NN Y GW N P
Sbjct: 232 LDSHVMCEVCGNTGHSGNDC-LETCEEAMYMGN--NNGYCPQGGQGW-NQPR-------P 280
Query: 319 FNQGGNNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAI 378
+ QGGNN + NQ SL++L+ K +A
Sbjct: 281 YYQGGNNNSNFPNQ---------------------PSLKDLVF-------AQAKTTDALS 312
Query: 379 KSLETQVGQMAKQMSERPPGMFPSDTVINPK-ENCSAITLRNIE---------PLGENK- 427
K L ++A + G P +P EN AIT+R + P+G N+
Sbjct: 313 KKLAAN-DKLASLVPANETGRIPGQP--DPSIENVKAITMRRGKSTRDPPYPNPVGTNEI 369
Query: 428 -KKQKEKEEEKRKVELEN----KFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELP 482
K + ++V+ EN ++ FP + K +++QF++F+ + +K+ + +P
Sbjct: 370 SKGAPSNDSADKEVQPENTVPQEYCDTRLLSFPQRMRKPSVDEQFARFVEVIQKIHINVP 429
Query: 483 FSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPV 542
+ ++ +P YA+++K+IL+ KR L E+++LTE+CS ++ KLP K+KDPG T+
Sbjct: 430 LLDAMQ-VPTYARYLKDILNNKRPLPT-TEVVKLTEQCSNLILHKLPEKKKDPGCPTITC 487
Query: 543 NFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLV 602
+ GA + + C+LG+SV+++ +F++LN L PT M LQLAD S+ P G+ EDV V
Sbjct: 488 SIGAQQFDQVSCNLGASVSVVLKDVFDKLNFTVLAPTPMRLQLADSSVRYPAGIAEDVPV 547
Query: 603 RVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADEKITF 658
++ +F PVDFV++DMD + PLILGRPFL+T+ A I+VG G+I + +K F
Sbjct: 548 KIRDFFIPVDFVVLDMDTGKETPLILGRPFLSTAGANIDVGTGSIRFHINGKKEKF 603
>UniRef100_Q6F2G0 Hypothetical protein OSJNBa0083K01.26 [Oryza sativa]
Length = 585
Score = 216 bits (550), Expect = 2e-54
Identities = 156/523 (29%), Positives = 260/523 (48%), Gaps = 55/523 (10%)
Query: 138 FKQEDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEAL 197
F+Q E++ EA E ++ + CP H + + ++ FY+ L +R +L+AAA G F +
Sbjct: 14 FQQTRDESIPEAWELLQEYVAACPHHGMDDWLILQNFYNRLTPMSRDHLDAAAGGAFFSK 73
Query: 198 SAQAGWNLINKMAESAVNSTNDRQNR-RGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAA 256
+ Q LI KM + S Q R RG+ ++ + + A L +++ +++ Q
Sbjct: 74 TVQGTVELIEKMVSNMGWSKERLQTRQRGMHTVKETELLAAKLDLLMKRLDNHEKRPQGT 133
Query: 257 KISNVDSIHCGTCGG-PHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWRE 315
+ + C CGG H+ +C EE +GN+ N G+R W +
Sbjct: 134 VKALDSHVTCEVCGGTDHSGNDC-PETREEAMYMGNNNN---------GYRPQGVQGWNQ 183
Query: 316 QNNFNQGGNNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMENFINKADTSFKNHE 375
++ QGGNN Q Q P + G S+E
Sbjct: 184 PRSYYQGGNNN---ETQLAQLAYLVPANETGRIPGQPYSSIEN----------------- 223
Query: 376 AAIKSLETQVGQMAKQMSERPPGMFPSDTVINPKENCSAITLRNIEPLGENKKKQKEKEE 435
+K++ T+ G+ + PP P+ T KE S N KE +
Sbjct: 224 --VKAITTRGGKSTRD----PPYPNPAGTNGMSKETPS------------NDSADKEIQP 265
Query: 436 EKRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAK 495
EK V E T++L PFP K +++ F++F+ + +++ + + + ++ +P YA+
Sbjct: 266 EK-TVPQEYCDTRLL--PFPQWSRKTSVDELFARFVEVIQRIHINVLLLDAMQ-VPIYAR 321
Query: 496 FMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCD 555
++K+IL+ KR L E+++LTE CS ++ KLP K+K T+ + GA + +ALCD
Sbjct: 322 YLKDILNNKRPL-PTTEVVKLTEHCSNVILHKLPEKKKYSECPTITCSIGAQQFDQALCD 380
Query: 556 LGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVI 615
LG+SV++MP +F+RLN L PT M LQ+AD S+ G+ EDV +++ +F PVDFV+
Sbjct: 381 LGASVSVMPKDVFDRLNFTVLAPTPMRLQMADSSVRYQAGIAEDVPIKIWDFFIPVDFVV 440
Query: 616 IDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADEKITF 658
+DMD + PLILGRPFL+T+ A I+V +I + +++ F
Sbjct: 441 LDMDTRKETPLILGRPFLSTAGANIDVETSSICFHINEKEEKF 483
>UniRef100_Q7XLB2 OSJNBa0011K22.17 protein [Oryza sativa]
Length = 551
Score = 209 bits (531), Expect = 3e-52
Identities = 150/545 (27%), Positives = 263/545 (47%), Gaps = 68/545 (12%)
Query: 131 LKNDILVFKQEDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAA 190
L+ I F+Q E++ EA ER ++ + CP H + + ++ FY+GL + +L+AAA
Sbjct: 7 LRGRISSFQQTRDESIPEAWERLQEYMAACPHHGMDDWLILQNFYNGLTPMSHDHLDAAA 66
Query: 191 SGEFEALSAQAGWNLINKMAESAVNSTNDRQNR-RGVLEIEAYDRMVASNKQLSQQMTTI 249
G F + + Q +LI KM + + +D + +G +++
Sbjct: 67 RGAFFSKTVQGAVDLIEKMLDLLMKRLDDHDKKPQGTVKV-------------------- 106
Query: 250 QRQFQAAKISNVDS-IHCGTCGGP-HASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRN 307
+DS + C CG H+ +C E +GN NN Y GW
Sbjct: 107 -----------LDSHVTCEVCGNTGHSGNDC--LDTREAMYMGN--NNGYCPQRGQGWNQ 151
Query: 308 HPNFSWREQNNFNQGGNNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKSLEELMENFINKA 367
+ R+ + + +Q + + N K ++EN K
Sbjct: 152 PCPYVDRKHEAWEICLTPVQPSLKDLVFAQAKTIDALSKKLATNDK-----ILENINVKL 206
Query: 368 D---TSFKNHEAAIKSLETQVGQMAKQMSERPPGMFPSDTVINPKENCSAITLRNIEPLG 424
D ++F+N K LETQ+ Q A + G + ++
Sbjct: 207 DGFASAFQNQLRFNKMLETQLAQFASLVPANETGTNGIAKEVPSSDSA------------ 254
Query: 425 ENKKKQKEKEEEKRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELPFS 484
+KE + K+ + ++ F K +++QF++F + +K+ + +P
Sbjct: 255 -------DKEVQLEKI-VPQEYCDTWLLSFHQRSRKPSVDEQFARFAEVIQKIHINVPLL 306
Query: 485 EVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNF 544
+ ++ +P YA+++K++L+ KR L E+++LTE+CS+++ KLP K+KDPG T+ +
Sbjct: 307 DAMQ-VPTYARYLKDMLNNKRLLPTM-EVVKLTEQCSSVILHKLPEKKKDPGCPTITCSI 364
Query: 545 GASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRV 604
GA + +A CDLG+S+++MP + ++LN L PT+M LQLAD S+ P G+ EDV V++
Sbjct: 365 GAQQFDQAFCDLGASISVMPKDVPDKLNFTVLTPTLMCLQLADLSVHYPTGIAEDVPVKI 424
Query: 605 GEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADEKITFTIFDLK 664
+F PVDFV++DMD + PLILGRPFL+T+ A I+VG G+I + ++ + + +
Sbjct: 425 RDFFIPVDFVVLDMDMGKETPLILGRPFLSTAGANIDVGMGSIRFDINGKEKSLSFIRGR 484
Query: 665 PKPVE 669
+P E
Sbjct: 485 AEPTE 489
>UniRef100_Q9LKR9 Hypothetical protein T15F17.a [Arabidopsis thaliana]
Length = 703
Score = 204 bits (518), Expect = 9e-51
Identities = 164/592 (27%), Positives = 265/592 (44%), Gaps = 55/592 (9%)
Query: 84 RLQLFPFSLRDTAEEWLNSQPQGSITTWEDLAKKFTKKLFPRTLLRKLKNDILVFKQEDT 143
+L+LFPFSL D A +W S PQGSIT+W D K F K F + +L+NDI F Q +
Sbjct: 87 KLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTARLRNDISGFTQTNN 146
Query: 144 ENLHEALERFKKLLRKCPQHNLTLGVQVERFYDGLADSARSNLEAAASGEFEALSAQAGW 203
E EA E FK +CP H + + Y + R L+ A++G F + GW
Sbjct: 147 ETFCEAWECFKGYQTQCPHHGFSKASLLSTLYRRVLPKIRMLLDTASNGNFLNKDVEDGW 206
Query: 204 NLINKMAESAVNSTNDRQNRRGVLEIEAYDRMVA----SNKQLSQQMTTIQRQFQAAKIS 259
L+ +A+S N D YDR + S+++ ++M + + +
Sbjct: 207 ELVENLAQSDGNYNED------------YDRSIRTSSDSDEKHRREMKAMNDKLDKLLLV 254
Query: 260 NVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNN- 318
IH ++ T EEV + N Y+ +N +NHP +
Sbjct: 255 QQKHIHFLGDDETFQVQDGETLQSEEVSYVQN--QGGYNKGFNNFKQNHPERCKPAGPSL 312
Query: 319 -FNQGGNNQRQYSNQRFQSQNSGP---QQNQDQGSGNGKKSLEELMENFINKADTSFKNH 374
F + Q S Q Q P QQ Q S L+ +++ + +
Sbjct: 313 PFTAAESTQDLCSIQPRSRQLPPPGFTQQQQQPASTTPDSDLKNMLQQILQGQAAGAMDL 372
Query: 375 EAAIKSLETQVGQMAKQMSERPPGMFPSDTVINPKENCSAITLRNIEPLGENKKKQKEKE 434
+ + + G+ + Q+ G+ S + + + IT R + K+ K
Sbjct: 373 AKKMAEIHNKNGEASTQIEISVVGLDHSAGSRSNLDEKATITERMV-------KRFKPAP 425
Query: 435 EEKRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYA 494
R + P K +E+ S ++ +P EVL +P
Sbjct: 426 LPSRAL--------------PWKFRKAWIERYNSLAEKQLDEMEAVMPLIEVLNLIPDPH 471
Query: 495 KFMKEILSKKRRLSEENEIIELTEECSAI-----LQRKLPPKRKDPGSFTLPVNFGASKQ 549
K +++ + ++ ++ +++E ++C AI ++R + K +DPG+FT P + G
Sbjct: 472 KDVRKSILERIKIHQDSE-----DKCDAIPSRTTVKRSVQEKLEDPGTFTQPCSIGQLVF 526
Query: 550 VRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEF 609
LCDLG+SV+LMPLS+ +L + KP + L LADRS P+G+++D+ V + E
Sbjct: 527 SNCLCDLGASVSLMPLSVARKLEFTQYKPCDLTLILADRSSRKPFGLLQDLPVMINGVEV 586
Query: 610 PVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADE-KITFTI 660
P DFV++DM+ + K LILGRPFLA+ A I+V +G ISL + K+ F I
Sbjct: 587 PTDFVVLDMEAEPKDLLILGRPFLASVGAMIDVKDGRISLNLGKHIKLQFDI 638
>UniRef100_Q9SHR2 F28L22.6 protein [Arabidopsis thaliana]
Length = 719
Score = 201 bits (510), Expect = 8e-50
Identities = 171/608 (28%), Positives = 270/608 (44%), Gaps = 113/608 (18%)
Query: 178 LADSARSNLEAAASGEFEALSAQAGWNLINKMAESAVNSTNDRQNRRGVLEIEAYDRMVA 237
LA S L+ A++G F + GW ++ +A+S N D YDR +
Sbjct: 91 LALSTEMLLDTASNGNFLNKDVEDGWEVVENLAQSDGNYNED------------YDRSIR 138
Query: 238 ----SNKQLSQQMTTIQRQFQAAKISNVDSIHCGTCGGPHASEECGTYFDEEVKVLGNSQ 293
S+++ ++M + + + IH ++ T EEV + N
Sbjct: 139 TSSDSDEKHRREMKAMNDKLDKLLLMQQKHIHFLGDDETLQVQDGETLQLEEVSYVQNQG 198
Query: 294 NNPYSNTYNLGWRNHPNFSWREQN---------------------NFNQG---------- 322
Y+ +N +NHPN S+R N +NQG
Sbjct: 199 G--YNKGFNNFKQNHPNLSYRSTNVANRQDQVYPSQQQNQPKPFVPYNQGQGYVPKQQYQ 256
Query: 323 GNNQRQYSNQRFQSQNSGPQ------------QNQDQGSGNGKKSLEELMENFINKADTS 370
GN Q+Q F Q P Q QG G L + M NK D S
Sbjct: 257 GNYQQQLPPPGFTQQQQQPALTTPDSDLKNMLQQILQGQATGAMDLSKRMAEIHNKVDCS 316
Query: 371 FKNHEAAIKSLETQVGQMAKQMSERPPGMF--PSD-TVINPKENCSAITLRNIEPL---- 423
+ + +++L +++ + Q F PS ++ N KE AITLR+ + L
Sbjct: 317 YNDINIKVEALTSKIRYIEGQTGSTAAPKFTGPSGKSMSNSKEYAHAITLRSGKELPTKE 376
Query: 424 -------------GENKKKQKEKEEEKRKVELENKFTKVLFP------------------ 452
GE+ + E+ + + ++ T+ L P
Sbjct: 377 SPNQNTEDSLDQDGEDFCQNGNSAEKAIEEPILHQPTRPLAPAASPLVEKPAAAKTKENV 436
Query: 453 ----------PFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILS 502
PFP K ++K + K L V +P + L +P K++K++++
Sbjct: 437 FIPPPYKPPLPFPGRFKKVMIQKYKALLEKQLKNLEVTMPLVDCLALIPDSNKYVKDMIT 496
Query: 503 KKRRLSEENEIIELTEECSAILQRKLPPKRK-DPGSFTLPVNFGASKQVRALCDLGSSVN 561
+ R+ E ++ L+ ECSAI+Q+K+ PK+ DPGSFTLP G + LCDLG+SV+
Sbjct: 497 E--RIKEVQGMVVLSHECSAIIQQKIIPKKLGDPGSFTLPCALGPLAFSKCLCDLGASVS 554
Query: 562 LMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDED 621
LMPL + ++L + KP + L LADRS+ G++ED+ V +G E P DFV+++MDE+
Sbjct: 555 LMPLPVAKKLGFNKYKPCNISLILADRSVRISHGLLEDLPVMIGVVEVPTDFVVLEMDEE 614
Query: 622 SKIPLILGRPFLATSQAKINVGNGTISLRVA-DEKITFTIFDLKPKPVEKNDVFLVEMMD 680
K PLILGRPFLA ++A I+V G I L + D K+TF I + KP + ++F +E MD
Sbjct: 615 PKDPLILGRPFLARARAIIDVKKGKIDLNLGRDLKMTFDITNTMKKPTIEGNIFWIEEMD 674
Query: 681 EWSDEKLK 688
+D+ L+
Sbjct: 675 MLADKMLE 682
>UniRef100_O22104 Vicia faba mRNA expressed from retrotransposon-like gene [Vicia
faba]
Length = 238
Score = 178 bits (452), Expect = 4e-43
Identities = 91/198 (45%), Positives = 134/198 (66%), Gaps = 3/198 (1%)
Query: 464 EKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAI 523
EK F F+ MFKKL + + E LEKMP YAKFMK+I+SKKR + + + I TE CS+I
Sbjct: 11 EKNFEIFLEMFKKLELNILVLEALEKMPTYAKFMKDIISKKRTI--DTDPIIHTETCSSI 68
Query: 524 LQ-RKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMM 582
LQ K+P K+KD G T P G + +LG+SV+L+P ++++L + ++ T M
Sbjct: 69 LQGMKIPVKKKDRGFVTTPCTIGDRSFKKYFINLGASVSLLPFYIYKKLGISNVQNTRMT 128
Query: 583 LQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINV 642
L+ AD S+ P+G+VEDVLV++ +F FPVDFV+++M ED +I LILGRPFL T + I++
Sbjct: 129 LRFADHSVKIPYGIVEDVLVKIDKFVFPVDFVVLEMPEDEEITLILGRPFLETRRCLIDI 188
Query: 643 GNGTISLRVADEKITFTI 660
GT++L+V DE++ +
Sbjct: 189 EEGTVTLKVYDEELKIDV 206
>UniRef100_Q8S6J2 Hypothetical protein OSJNBa0019N10.34 [Oryza sativa]
Length = 237
Score = 174 bits (442), Expect = 6e-42
Identities = 92/224 (41%), Positives = 147/224 (65%), Gaps = 4/224 (1%)
Query: 431 KEKEEEKRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKM 490
K K ++ V E +T++L PFP K +++QF+ F+ + +K+ +++P + ++ +
Sbjct: 14 KAKGLPEKTVPQEYCYTRLL--PFPQRSRKPSVDEQFAHFVEVIQKIHIDVPLLDAMQ-V 70
Query: 491 PQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQV 550
P YA ++K+IL+ KR L E+++LT +CS ++ KLP K+KDPG T+ + GA +
Sbjct: 71 PTYACYLKDILNNKRPLPT-TEVVKLTLQCSNVILHKLPEKKKDPGCPTITCSIGAQQFD 129
Query: 551 RALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFP 610
+ALCDLG+SV++MP +F++LN L PT M LQLAD S+ P G+ EDV V++ +F
Sbjct: 130 QALCDLGASVSVMPKYVFDKLNFTVLAPTPMRLQLADSSVRYPAGIAEDVPVKIRDFFIL 189
Query: 611 VDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADE 654
VDFV++DMD ++ +ILGRPFL T+ A I+VG G+I + E
Sbjct: 190 VDFVVLDMDTGKEMSIILGRPFLGTAGANIHVGTGSIRFQYQRE 233
>UniRef100_Q7F9Z3 OSJNBa0079C19.24 protein [Oryza sativa]
Length = 461
Score = 174 bits (441), Expect = 8e-42
Identities = 135/483 (27%), Positives = 229/483 (46%), Gaps = 35/483 (7%)
Query: 118 FTKKLFPRTLLRKLKNDILVFKQEDTENLHEALERFKKLLRKCPQHNLTLGVQVERFYDG 177
F K FP L+ I F+Q E++ EA ER P H + + ++ FY+G
Sbjct: 3 FLSKFFPMGKTNALRGRISNFQQTRDESIPEAWER--------PHHGMDNSLILQNFYNG 54
Query: 178 LADSARSNLEAAASGEFEALSAQAGWNLINKMAESAVNSTNDRQNRR-GVLEIEAYDRMV 236
L + +L+AAA G F + + + +LI KM + S Q R+ G+ ++ + +
Sbjct: 55 LTTMSHDHLDAAAGGAFFSKTVRGAIDLIEKMVSNMGWSKERLQTRQQGIHTVKETELLA 114
Query: 237 ASNKQLSQQMTTIQRQFQAAKISNVDSIHCGTCGGP-HASEECGTYFDEEVKVLGNSQNN 295
A L +++ + Q + + C CGG H+ +C EE +G + N
Sbjct: 115 AKLDLLMKRLDDHDNRPQDTVKALDLHVTCEVCGGTGHSGNDCPETH-EEAMYMGKNNNE 173
Query: 296 PYSNTYNLGWRNHPNFSWREQNNFNQGGNNQRQYSNQRFQSQNSGPQQNQDQGSGNGKKS 355
+R W + + QG NN +SNQ Q +
Sbjct: 174 ---------YRPQGGQGWNQPRPYYQGENNNSNFSNQPSLKDLIFVQAKTTDALSKKLAA 224
Query: 356 LEELMENFINKAD---TSFKNHEAAIKSLETQVGQMAKQMSERPPGMFP----SDTVINP 408
++++EN + D +SF+N K +ETQ+ Q+A + G P T P
Sbjct: 225 NDKILENINVQLDGFTSSFQNQLIFNKMIETQLAQLASLVPANEIGRIPRRGGKSTRDRP 284
Query: 409 KENCSAIT-LRNIEPLGENKKKQKEKEEEKRKVELENKFTKVLFPPFPTNIAKRRLEKQF 467
N + + P ++ K+ + E + V E T++L FP K +++QF
Sbjct: 285 YPNPAGTNGVAREAPSSDSANKEVQPE---KAVPQEYYDTRLL--SFPQRSRKPSVDEQF 339
Query: 468 SKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRK 527
++F+ + +K+ + + + ++ +P YA+++K+IL+ KR L E+IELTE+CS ++ K
Sbjct: 340 ARFVEVIQKIHINVSLFDAMQ-VPTYARYLKDILNNKRPLPT-TEVIELTEQCSNVILHK 397
Query: 528 LPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLAD 587
L KRK PG T+ + GA + +AL DLG SV+++P+ +F++LN L PT M LQLAD
Sbjct: 398 LSEKRKHPGCPTITCSIGAQQFNQALRDLGVSVSVIPMDVFDKLNFTVLAPTPMRLQLAD 457
Query: 588 RSI 590
S+
Sbjct: 458 SSV 460
>UniRef100_Q8LM79 Hypothetical protein OSJNAa0082N11.11 [Oryza sativa]
Length = 322
Score = 170 bits (430), Expect = 2e-40
Identities = 107/298 (35%), Positives = 170/298 (56%), Gaps = 26/298 (8%)
Query: 369 TSFKNHEAAIKSLETQVGQMAKQM----SERPPGMFPSDTVINPKENCSAITLRNIEPLG 424
++F+N + K +ETQ+ Q+A + SER PG P ++ EN AIT R G
Sbjct: 35 SAFQNQLSFNKMIETQLAQLASLVPANESERIPGQ-PDSSI----ENIKAITTRG----G 85
Query: 425 ENKKKQKEKEEEKRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELPFS 484
N ++ + E++ + T + R+ QF++ + + +K+ + +P
Sbjct: 86 TNGMSKETPSTDLADKEIQPEKTVPQEYCDTRGVGNRQ---QFARSVEVIQKIHINVPLL 142
Query: 485 EVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNF 544
+ ++ +P YA+++K+IL+ KR L TE CS ++ KLP K+KDP T+ +
Sbjct: 143 DAMQ-VPTYARYLKDILNNKRPLPT-------TEHCSNVILHKLPEKKKDPRCPTITCSI 194
Query: 545 GASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRV 604
GA + +ALCDLG+SV++MP +F++LN L PT M LQLAD S+ P G+ EDV V++
Sbjct: 195 GAQQFDQALCDLGASVSVMPKDVFDKLNFTVLAPTPMRLQLADSSVHCPVGIEEDVPVKI 254
Query: 605 GEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRV--ADEKITFTI 660
+F PVDFV++DMD + LILG PFL+T+ A I+VG G+I + +EK F +
Sbjct: 255 WDFFIPVDFVVLDMDTGKETSLILGHPFLSTAGANIDVGMGSIRFHINGKEEKFEFQL 312
>UniRef100_Q6UUB7 Hypothetical protein [Oryza sativa]
Length = 450
Score = 166 bits (421), Expect = 2e-39
Identities = 126/430 (29%), Positives = 211/430 (48%), Gaps = 49/430 (11%)
Query: 164 NLTLGVQVE-RFYDGLADSARSNLEAAASGEFEALSAQAGWNLINKMAESAVNSTNDRQN 222
N+ +G+Q+ FY+GL +R +L+AAA G F + Q +L+ KM + S Q
Sbjct: 63 NVAIGLQINMNFYNGLTPMSRDHLDAAAGGAFFSKMVQGAVDLVEKMVSNMGWSEEQLQT 122
Query: 223 -RRGVLEIEAYDRMVASNKQLSQQMTTIQRQFQAAKISNVDSIHCGTCGGP-HASEECGT 280
+RG+ ++ + + A L +++ +++ Q + + C C H+ +C
Sbjct: 123 CQRGMHTVKETELLAAKLDLLMKRLDNHEKRPQGTVKALDSHVTCEVCSSTGHSGNDCPE 182
Query: 281 YFDEEVKVLGNSQNNPYSNTYNLGWRNHPNFSWREQNNFNQGGNNQRQYSNQRFQSQNSG 340
EE +GN NN Y R W + + QGGNN +SNQ
Sbjct: 183 T-REEAMYMGN--NNWY--------RPQGGQGWSQLRPYYQGGNNNGNFSNQ-------- 223
Query: 341 PQQNQDQGSGNGKKSLEELMENFINKADTSFKNHEAAIKSLETQ-VGQMAKQMSERPPGM 399
SL++L+ + D K K LE + Q+A + G
Sbjct: 224 -------------PSLKDLVFSQAKTTDALSKKLATNDKILENNNLAQLASLVPVNETGR 270
Query: 400 FPS--DTVINPKENCSAITLRNIEPLGENKKKQKEKEEEKRKVELENKFTKVLFPPFPTN 457
P D+ I EN AIT+R G + +KE++ K + ++ FP
Sbjct: 271 IPGQPDSSI---ENVKAITMR-----GASSSDSADKEDQPEKT-VPQEYCDTWLLLFPQR 321
Query: 458 IAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELT 517
+ K +++QF++F+ + +K+ + +P + ++ MP YA+++K IL+ KR L E+++LT
Sbjct: 322 MRKPLVDEQFARFVEVIQKIHINVPLLDAMQ-MPTYARYLKGILNNKRPLPT-TEVVKLT 379
Query: 518 EECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELK 577
E+ S ++ KLP K+KDPG T+ + GA + +ALCDLG+SV++MP ++F++LN L
Sbjct: 380 EQWSNVILHKLPEKKKDPGCPTITCSIGAQQFDQALCDLGASVSVMPKNVFDKLNFTMLA 439
Query: 578 PTMMMLQLAD 587
PT M LQLAD
Sbjct: 440 PTPMHLQLAD 449
>UniRef100_Q9ZPT5 Hypothetical protein At2g14040 [Arabidopsis thaliana]
Length = 1048
Score = 165 bits (417), Expect = 5e-39
Identities = 114/286 (39%), Positives = 156/286 (53%), Gaps = 28/286 (9%)
Query: 418 RNIEP-LGENKKKQKEK------EEE---KRKVELENKFTKVLFPP------FPTNIAKR 461
R + P L + K K+K K EE+ K LE K K + PP FP K
Sbjct: 48 RELNPILKKEKAKEKSKAASDDLEEDNTGKESDTLEEKAKKTVLPPYVPKLPFPGRQRKI 107
Query: 462 RLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECS 521
+ EK++S F + ++L+V+LPF ++++ + Y K +K+IL+ KR L E + +I + ECS
Sbjct: 108 QREKEYSLFDEIMRQLQVKLPFLDLVQNVSIYRKHLKDILTNKRTLEEGHVLI--SHECS 165
Query: 522 AILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMM 581
AILQ R G +T R LCDLG+ V+LMP S+ +RL PT M
Sbjct: 166 AILQNVALFSRMI-GEYTFD---------RCLCDLGAGVSLMPFSVAKRLGDTNFTPTKM 215
Query: 582 MLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKIN 641
L L DRSI P GV EDV VRVG F P DFVII++DE+ + LILGRPFL A I+
Sbjct: 216 SLVLGDRSISFPVGVAEDVQVRVGNFYIPTDFVIIELDEEPRHRLILGRPFLNIVAALID 275
Query: 642 VGNGTISLRVADEKITFTIFDLKPKPVEKNDVFLVEMMDEWSDEKL 687
V I+LR+ D F + + KP + F V++MDE +E L
Sbjct: 276 VRKSKINLRIGDIVQEFNMERIMSKPTTECQTFWVDIMDELVNELL 321
>UniRef100_Q7XF80 Hypothetical protein [Oryza sativa]
Length = 725
Score = 165 bits (417), Expect = 5e-39
Identities = 85/205 (41%), Positives = 138/205 (66%), Gaps = 4/205 (1%)
Query: 437 KRKVELENKFTKVLFPPFPTNIAKRRLEKQFSKFISMFKKLRVELPFSEVLEKMPQYAKF 496
++ V E +T++L PFP K +++QF+ F+ + +K+ +++P + ++ +P YA +
Sbjct: 51 EKTVPQEYCYTRLL--PFPQRSRKPSVDEQFAHFVEVIQKIHIDVPLLDAMQ-VPTYACY 107
Query: 497 MKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASKQVRALCDL 556
+K+IL+ KR L E+++LT +CS ++ KLP K+KDPG T+ + GA + +ALCDL
Sbjct: 108 LKDILNNKRPLPT-TEVVKLTLQCSNVILHKLPEKKKDPGCPTITCSIGAQQFDQALCDL 166
Query: 557 GSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFEFPVDFVII 616
G+SV++MP +F++LN L PT M LQLAD S+ P G+ EDV V++ +F VDFV++
Sbjct: 167 GASVSVMPKYVFDKLNFTVLAPTPMRLQLADSSVRYPAGIAEDVPVKIRDFFILVDFVVL 226
Query: 617 DMDEDSKIPLILGRPFLATSQAKIN 641
DMD ++ +ILGRPFL T+ A I+
Sbjct: 227 DMDTGKEMSIILGRPFLGTAGANIH 251
>UniRef100_Q75H42 Hypothetical protein OSJNBa0006O14.11 [Oryza sativa]
Length = 256
Score = 162 bits (411), Expect = 2e-38
Identities = 82/170 (48%), Positives = 119/170 (69%), Gaps = 1/170 (0%)
Query: 489 KMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKLPPKRKDPGSFTLPVNFGASK 548
++P YA+++K IL+ KR L E+++LTE CS ++ KL K+KDPG T+ + GA +
Sbjct: 2 QVPTYARYLKVILNNKRPLPT-TEVVKLTEHCSNLILHKLQEKKKDPGCPTITCSIGAQQ 60
Query: 549 QVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVEDVLVRVGEFE 608
V ALCDLG+SV++MP ++F++LN L PT M LQLAD S+ P G+VEDV V++ F
Sbjct: 61 FVHALCDLGASVSVMPKNVFDKLNFMVLAPTPMCLQLADSSVRYPAGIVEDVPVKIRNFF 120
Query: 609 FPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADEKITF 658
PVDFV++DMD + PLILGRPFL+T+ A +VG G+IS + +++ F
Sbjct: 121 IPVDFVVLDMDMGKETPLILGRPFLSTAGANSDVGTGSISFHINGKELKF 170
>UniRef100_Q9C8Q7 Athila ORF 1, putative; 43045-40843 [Arabidopsis thaliana]
Length = 530
Score = 162 bits (409), Expect = 4e-38
Identities = 91/228 (39%), Positives = 144/228 (62%), Gaps = 4/228 (1%)
Query: 480 ELPFSEVLEKMPQYAKFMKEILSKKRRLSEENEIIELTEECSAILQRKL-PPKRKDPGSF 538
++P + L +P K++K+++++ R+ E ++ L+ ECSAI+Q+K+ K +DPGSF
Sbjct: 120 DMPLVDCLALIPNEHKYVKDLITE--RIKEVQGMVVLSHECSAIIQQKIVQEKLEDPGSF 177
Query: 539 TLPVNFGASKQVRALCDLGSSVNLMPLSMFERLNVGELKPTMMMLQLADRSIVAPWGVVE 598
TLP + +LCDLG+SV++MPLSM +L + KP + L LADR+ P+G++E
Sbjct: 178 TLPCSIRQLTFSNSLCDLGASVSIMPLSMARKLGFVQYKPCDLTLILADRTSRRPFGLLE 237
Query: 599 DVLVRVGEFEFPVDFVIIDMDEDSKIPLILGRPFLATSQAKINVGNGTISLRVADE-KIT 657
DV V + E P+DFV+++MDE+SK PLILGRPFLA++ A I+V G I+L + ++ K+
Sbjct: 238 DVPVMINGVEVPIDFVVLEMDEESKDPLILGRPFLASAGAVIDVKQGKINLNLGEDFKMK 297
Query: 658 FTIFDLKPKPVEKNDVFLVEMMDEWSDEKLK*FFLKEKADASNKKKKE 705
F I + KP + FLVE M + ++E L+ K+ + K E
Sbjct: 298 FEIRNTMKKPTIEGQTFLVEEMGQLANELLEELIDKDHLQTALTKNSE 345
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,124,963,622
Number of Sequences: 2790947
Number of extensions: 48624296
Number of successful extensions: 255003
Number of sequences better than 10.0: 1652
Number of HSP's better than 10.0 without gapping: 525
Number of HSP's successfully gapped in prelim test: 1210
Number of HSP's that attempted gapping in prelim test: 231788
Number of HSP's gapped (non-prelim): 8492
length of query: 705
length of database: 848,049,833
effective HSP length: 135
effective length of query: 570
effective length of database: 471,271,988
effective search space: 268625033160
effective search space used: 268625033160
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 78 (34.7 bits)
Lotus: description of TM0103.9


