

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
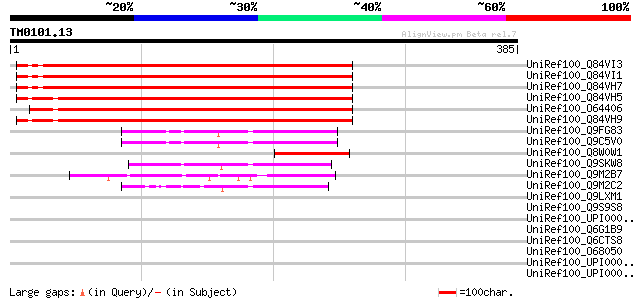
Query= TM0101.13
(385 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84VI3 Envelope-like protein [Glycine max] 272 1e-71
UniRef100_Q84VI1 Envelope-like protein [Glycine max] 270 4e-71
UniRef100_Q84VH7 Envelope-like protein [Glycine max] 270 7e-71
UniRef100_Q84VH5 Envelope-like protein [Glycine max] 269 1e-70
UniRef100_O64406 Envelope-like [Glycine max] 266 7e-70
UniRef100_Q84VH9 Envelope-like protein [Glycine max] 263 8e-69
UniRef100_Q9FG83 Gb|AAF18631.1 [Arabidopsis thaliana] 75 3e-12
UniRef100_Q9C5V0 Putative envelope protein [Arabidopsis thaliana] 75 3e-12
UniRef100_Q8W0W1 Putative envelope protein [Vicia faba] 73 2e-11
UniRef100_Q9SKW8 F5J5.2 [Arabidopsis thaliana] 69 2e-10
UniRef100_Q9M2B7 Hypothetical protein F23N14_70 [Arabidopsis tha... 53 2e-05
UniRef100_Q9M2C2 Hypothetical protein F23N14_20 [Arabidopsis tha... 52 3e-05
UniRef100_Q9LXM1 Hypothetical protein T10D17_60 [Arabidopsis tha... 40 0.14
UniRef100_Q9S9S8 F28J9.1 [Arabidopsis thaliana] 38 0.55
UniRef100_UPI000049995C UPI000049995C UniRef100 entry 37 1.2
UniRef100_Q6G1B9 Lysyl-tRNA synthetase [Bartonella quintana] 36 2.1
UniRef100_Q6CTS8 Kluyveromyces lactis strain NRRL Y-1140 chromos... 36 2.1
UniRef100_O68050 Potential regulatory protein [Rhodobacter capsu... 35 2.7
UniRef100_UPI000007FE0E UPI000007FE0E UniRef100 entry 35 4.6
UniRef100_UPI000031A45E UPI000031A45E UniRef100 entry 35 4.6
>UniRef100_Q84VI3 Envelope-like protein [Glycine max]
Length = 680
Score = 272 bits (695), Expect = 1e-71
Identities = 133/255 (52%), Positives = 185/255 (72%), Gaps = 5/255 (1%)
Query: 6 KKQKVSRIDLETESDVEGDVRDITASEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWK 65
K++++S D ++ DVE D++ S+K SGK++P +VP+ PLDN+S H+ N KWK
Sbjct: 290 KRKEISSSD--SDKDVE---LDVSTSKKAKTSGKKVPGNVPDAPLDNISFHSIGNVEKWK 344
Query: 66 YVYQRRVAKDREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNN 125
YVYQRR+A +RE+ D L CKEI+ LI AGL KTV +G CY+ LV+EFIVN+ D++N
Sbjct: 345 YVYQRRLAVERELGRDALDCKEIMDLIKAAGLLKTVSKLGDCYEGLVREFIVNIPSDISN 404
Query: 126 STSPEFRKVFVRGKCVNFSPAIINQTLERSVVEFVEEELSLDTVAEELTAGRVKKWPAKK 185
S E++KVFVRGKCV FSPA+IN+ L R ++ ++S +A+E+TA RV+ WP K
Sbjct: 405 RKSDEYQKVFVRGKCVKFSPAVINKYLGRPTDGVIDIDVSEHQIAKEITAKRVQHWPKKG 464
Query: 186 LLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLK 245
LS G LSVKYAIL+RIG NWVP+NHTS V+ L K +Y +GT+ ++FGN++F QT+K
Sbjct: 465 KLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIFDQTVK 524
Query: 246 HAETCAVKLPISFFT 260
H+E+ A+KLPI+F T
Sbjct: 525 HSESFAIKLPIAFPT 539
>UniRef100_Q84VI1 Envelope-like protein [Glycine max]
Length = 682
Score = 270 bits (691), Expect = 4e-71
Identities = 132/255 (51%), Positives = 184/255 (71%), Gaps = 5/255 (1%)
Query: 6 KKQKVSRIDLETESDVEGDVRDITASEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWK 65
K++++S D ++ DVE D++ S+K SGK++P +VP+ PLDN+S H+ N KWK
Sbjct: 292 KRKEISSSD--SDKDVE---LDVSTSKKAKTSGKKVPGNVPDAPLDNISFHSIGNVEKWK 346
Query: 66 YVYQRRVAKDREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNN 125
YVYQRR+A +RE+ D L CKEI+ LI GL KTV +G CY+ LV+EFIVN+ D++N
Sbjct: 347 YVYQRRLAVERELGRDALDCKEIMDLIKAGGLLKTVSKLGDCYEGLVREFIVNIPSDISN 406
Query: 126 STSPEFRKVFVRGKCVNFSPAIINQTLERSVVEFVEEELSLDTVAEELTAGRVKKWPAKK 185
S E++KVFVRGKCV FSPA+IN+ L R ++ ++S +A+E+TA RV+ WP K
Sbjct: 407 RKSDEYQKVFVRGKCVKFSPAVINKYLGRPTDGVIDIDVSEHQIAKEITAKRVQHWPKKG 466
Query: 186 LLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLK 245
LS G LSVKYAIL+RIG NWVP+NHTS V+ L K +Y +GT+ ++FGN++F QT+K
Sbjct: 467 KLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIFDQTVK 526
Query: 246 HAETCAVKLPISFFT 260
H+E+ A+KLPI+F T
Sbjct: 527 HSESFAIKLPIAFPT 541
>UniRef100_Q84VH7 Envelope-like protein [Glycine max]
Length = 680
Score = 270 bits (689), Expect = 7e-71
Identities = 131/255 (51%), Positives = 185/255 (72%), Gaps = 5/255 (1%)
Query: 6 KKQKVSRIDLETESDVEGDVRDITASEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWK 65
K++++S D ++ DVE D++ S+K SGK++P +VP+ PLDN+S H+ N KWK
Sbjct: 290 KRKEISSSD--SDKDVE---LDVSTSKKAKTSGKKVPGNVPDAPLDNISFHSIGNVEKWK 344
Query: 66 YVYQRRVAKDREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNN 125
YVYQRR+A +RE+ D L CKEI+ LI AGL KTV +G CY+ LV+EFIVN+ D++N
Sbjct: 345 YVYQRRLAVERELGRDALDCKEIMDLIKAAGLLKTVSKLGDCYEGLVREFIVNIPSDISN 404
Query: 126 STSPEFRKVFVRGKCVNFSPAIINQTLERSVVEFVEEELSLDTVAEELTAGRVKKWPAKK 185
S ++++VFVRGKCV FSPA+IN+ L R ++ ++S +A+E+TA RV+ WP K
Sbjct: 405 RKSDDYQRVFVRGKCVRFSPAVINKYLGRPTDGVIDIDVSEHQIAKEITAKRVQHWPKKG 464
Query: 186 LLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLK 245
LS G LSVKYAIL+RIG NWVP+NHTS V+ L K +Y +GT+ ++FGN++F QT+K
Sbjct: 465 KLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIFDQTVK 524
Query: 246 HAETCAVKLPISFFT 260
H+E+ A+KLPI+F T
Sbjct: 525 HSESFAIKLPIAFPT 539
>UniRef100_Q84VH5 Envelope-like protein [Glycine max]
Length = 685
Score = 269 bits (687), Expect = 1e-70
Identities = 133/255 (52%), Positives = 184/255 (72%), Gaps = 5/255 (1%)
Query: 6 KKQKVSRIDLETESDVEGDVRDITASEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWK 65
K++++S D ++ DVE DV DI ++ SGK++P +VP+ PLDN+S H+ +N +WK
Sbjct: 298 KRKEISSSD--SDDDVELDVPDIKRAKT---SGKKVPGNVPDAPLDNISFHSIDNVERWK 352
Query: 66 YVYQRRVAKDREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNN 125
+VYQRR+A +RE+ D L CKEI+ LI AGL KTV +G CY+ LV+EFIVN+ D+ N
Sbjct: 353 FVYQRRLALERELGRDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITN 412
Query: 126 STSPEFRKVFVRGKCVNFSPAIINQTLERSVVEFVEEELSLDTVAEELTAGRVKKWPAKK 185
S E++KVFVRGKCV FSPA+IN+ L R V+ +S +A+E+TA +V+ WP K
Sbjct: 413 RKSDEYQKVFVRGKCVRFSPAVINKYLGRPTDGVVDITVSEHQIAKEITAKQVQHWPKKG 472
Query: 186 LLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLK 245
LS G LSVKYAIL+RIG NWVP+NHTS V+ L K +Y +GT+ ++FGN++F QT+K
Sbjct: 473 KLSAGKLSVKYAILHRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIFDQTVK 532
Query: 246 HAETCAVKLPISFFT 260
H+E+ AVKLPI+F T
Sbjct: 533 HSESFAVKLPIAFPT 547
>UniRef100_O64406 Envelope-like [Glycine max]
Length = 648
Score = 266 bits (680), Expect = 7e-70
Identities = 130/245 (53%), Positives = 177/245 (72%), Gaps = 3/245 (1%)
Query: 16 ETESDVEGDVRDITASEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWKYVYQRRVAKD 75
+++ DVE DV DI ++K SGK++P +VP+ PLDN+S H+ N +WK+VYQRR+A +
Sbjct: 276 DSDDDVELDVPDIKRAKK---SGKKVPGNVPDAPLDNISFHSIGNVERWKFVYQRRLALE 332
Query: 76 REIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVF 135
RE+ D L CKEI+ LI AGL KTV +G CY+ LV+EFIVN+ D+ N S E++KVF
Sbjct: 333 RELGRDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITNRKSDEYQKVF 392
Query: 136 VRGKCVNFSPAIINQTLERSVVEFVEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVK 195
VRGKCV FSPA+IN+ L R V+ +S +A+E+TA +V+ WP K LS G LSVK
Sbjct: 393 VRGKCVRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITAKQVQHWPKKGKLSAGKLSVK 452
Query: 196 YAILNRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHAETCAVKLP 255
YAIL+RIG NWVP+NHTS V+ L K +Y +GT+ ++FG ++F QT+KH+E+ AVKLP
Sbjct: 453 YAILHRIGAANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGKYIFDQTVKHSESFAVKLP 512
Query: 256 ISFFT 260
I+F T
Sbjct: 513 IAFPT 517
>UniRef100_Q84VH9 Envelope-like protein [Glycine max]
Length = 656
Score = 263 bits (671), Expect = 8e-69
Identities = 131/255 (51%), Positives = 182/255 (71%), Gaps = 5/255 (1%)
Query: 6 KKQKVSRIDLETESDVEGDVRDITASEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWK 65
K++++S D ++ DVE DV DI ++K SGK++P +VP+ PLDN+S H+ N +WK
Sbjct: 276 KRKEISSSD--SDDDVELDVPDIKRAKK---SGKKVPGNVPDAPLDNISFHSIGNVERWK 330
Query: 66 YVYQRRVAKDREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNN 125
+VYQRR+A +RE+ D L CKEI+ LI AGL KTV +G CY+ LV+EFIVN+ D+ N
Sbjct: 331 FVYQRRLALERELGRDALDCKEIMDLIKAAGLLKTVTKLGDCYESLVREFIVNIPSDITN 390
Query: 126 STSPEFRKVFVRGKCVNFSPAIINQTLERSVVEFVEEELSLDTVAEELTAGRVKKWPAKK 185
S E++ VFVRGK + FSPA+IN+ L R V+ +S +A+E+TA +V+ WP K
Sbjct: 391 RKSDEYQTVFVRGKGIRFSPAVINKYLGRPTEGVVDIAVSEHQIAKEITAKQVQHWPKKG 450
Query: 186 LLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLK 245
LS G LSVKYAIL+RIG NWVP+NHTS V+ L K +Y +GT+ ++FGN++F QT+K
Sbjct: 451 KLSAGKLSVKYAILHRIGTANWVPTNHTSTVATGLGKFLYAVGTKSKFNFGNYIFDQTVK 510
Query: 246 HAETCAVKLPISFFT 260
H+E+ AVKLPI+F T
Sbjct: 511 HSESFAVKLPIAFPT 525
>UniRef100_Q9FG83 Gb|AAF18631.1 [Arabidopsis thaliana]
Length = 515
Score = 75.1 bits (183), Expect = 3e-12
Identities = 54/170 (31%), Positives = 82/170 (47%), Gaps = 12/170 (7%)
Query: 86 KEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSP 145
++++ +I L TV I +++ EF N N T KVFVRGK FSP
Sbjct: 203 RDMMKVIEDNHLLDTVAEIDPFVKEVIWEFYANF-HIFNEETKKS--KVFVRGKMYEFSP 259
Query: 146 AIINQTLERSVV------EFVEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAIL 199
+IN+T + EF +S +A+ L+ G+V W K L T V+ A L
Sbjct: 260 QMINKTFRSPAIKWHAKAEFERVTMSKHALAKYLSDGKVVTW---KDLRTPVMPTHLAGL 316
Query: 200 NRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHAET 249
I +NW+PS + S V+ AK++Y + I ++FG V+ Q K A +
Sbjct: 317 YLICCINWIPSKNNSNVTVDRAKMLYLLNERIPFNFGKLVYDQVEKAARS 366
>UniRef100_Q9C5V0 Putative envelope protein [Arabidopsis thaliana]
Length = 461
Score = 75.1 bits (183), Expect = 3e-12
Identities = 54/170 (31%), Positives = 82/170 (47%), Gaps = 12/170 (7%)
Query: 86 KEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSP 145
++++ +I L TV I +++ EF N N T KVFVRGK FSP
Sbjct: 203 RDMMKVIEDNHLLDTVAEIDPFVKEVIWEFYANF-HIFNEETKKS--KVFVRGKMYEFSP 259
Query: 146 AIINQTLERSVV------EFVEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAIL 199
+IN+T + EF +S +A+ L+ G+V W K L T V+ A L
Sbjct: 260 QMINKTFRSPAIKWHAKAEFERVTMSKHALAKYLSDGKVVTW---KDLRTPVMPTHLAGL 316
Query: 200 NRIGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHAET 249
I +NW+PS + S V+ AK++Y + I ++FG V+ Q K A +
Sbjct: 317 YLICCINWIPSKNNSNVTVDRAKMLYLLNERIPFNFGKLVYDQVEKAARS 366
>UniRef100_Q8W0W1 Putative envelope protein [Vicia faba]
Length = 59
Score = 72.8 bits (177), Expect = 2e-11
Identities = 31/57 (54%), Positives = 41/57 (71%)
Query: 202 IGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHAETCAVKLPISF 258
IG NWVP+NH S +S L +LIY +GT +D+G ++F QTLKHA + AVK PI+F
Sbjct: 1 IGAANWVPTNHKSTISTGLGRLIYAVGTRTEFDYGRYIFDQTLKHAGSYAVKGPIAF 57
>UniRef100_Q9SKW8 F5J5.2 [Arabidopsis thaliana]
Length = 463
Score = 69.3 bits (168), Expect = 2e-10
Identities = 48/161 (29%), Positives = 76/161 (46%), Gaps = 12/161 (7%)
Query: 91 LIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQ 150
L+ GL TV I ++V EF NL + VFVR FS AIINQ
Sbjct: 194 LLFNGGLLPTVTEIDSYVHEVVMEFYANLPDGEEGDNLAY--SVFVRRNMYEFSLAIINQ 251
Query: 151 TLERSVVEF-------VEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIG 203
+ + ++ S+D VA L+ G+ W K+L++ +LS + A+LN+I
Sbjct: 252 MFQLPNPSYPLDGMSEIQVPESMDEVAIALSNGKANSW---KMLTSRLLSPELALLNKIC 308
Query: 204 VVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTL 244
NW P+ + SV+ L+Y + + ++FG +F + L
Sbjct: 309 CHNWSPTVNRSVLKPERMTLLYMVAKALPFNFGKLIFDENL 349
>UniRef100_Q9M2B7 Hypothetical protein F23N14_70 [Arabidopsis thaliana]
Length = 539
Score = 52.8 bits (125), Expect = 2e-05
Identities = 52/215 (24%), Positives = 100/215 (46%), Gaps = 25/215 (11%)
Query: 46 PNVPLDNVSIHAPENAFKWKYVYQRRVA--KDREIDSDVLACKEIIALIAKAGLRKTVMN 103
P VP + A ++K++ +R K +D +VL + + +G+ +TV+
Sbjct: 233 PLVPPHTKRFLSAAAAERYKHIAKRDFIFQKTLPLDPEVLTATKYF--LEHSGMAQTVVA 290
Query: 104 IGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQ--TLERSVVEFVE 161
+ + ++V+EF NL E + V+VRGK FSPA+IN +++ S ++ E
Sbjct: 291 VEQFVPEVVREFYANLPEMEYRECGLDL--VYVRGKMYEFSPALINHMFSIDDSALD-PE 347
Query: 162 EELSLDTVAEE-----LTAGRVKKW----PAKKLLSTGVLSVKYAILNRIGVVNWVPSNH 212
++L T + + +T G ++W PA L + +L+++ NW P+ +
Sbjct: 348 APVTLSTASRDDLALMMTGGTTRRWLRLQPADHLDT-------MKMLHKVCCGNWFPTTN 400
Query: 213 TSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHA 247
TS + +LI +++ G V + T+ A
Sbjct: 401 TSTLRVDRLRLIDMGTHGKSFNLGKLVVTHTMSLA 435
>UniRef100_Q9M2C2 Hypothetical protein F23N14_20 [Arabidopsis thaliana]
Length = 428
Score = 52.0 bits (123), Expect = 3e-05
Identities = 43/161 (26%), Positives = 82/161 (50%), Gaps = 17/161 (10%)
Query: 86 KEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSP 145
+ ++A+ + G K + N +C++ + F L D NN+ S + + +F G F+
Sbjct: 164 RRLLAVRDRLGRYKVIGN--RCFNDM--RF---LPLDGNNTASTQ-QLLFNAGLLPTFTE 215
Query: 146 AIINQTLERSVVEFV----EEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNR 201
I+ + V+EF + S+D VA L+ G+ W K+L++ +LS + A+LN+
Sbjct: 216 --IDSYVHEVVMEFYANLPDVPESMDEVAIALSNGKANSW---KMLTSRLLSPELALLNK 270
Query: 202 IGVVNWVPSNHTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQ 242
I NW P+ + SV+ L+Y + + ++FG +F +
Sbjct: 271 ICCHNWSPTVNHSVLKPERMTLLYMVAKALPFNFGKLIFDE 311
>UniRef100_Q9LXM1 Hypothetical protein T10D17_60 [Arabidopsis thaliana]
Length = 324
Score = 39.7 bits (91), Expect = 0.14
Identities = 42/156 (26%), Positives = 66/156 (41%), Gaps = 27/156 (17%)
Query: 92 IAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQT 151
I AGL +TV + ++LV EF NL +STS +++ Q
Sbjct: 147 IYDAGLIRTVTELKPFEEELVFEFWANLPTVKVDSTSV----------------SVMQQM 190
Query: 152 LERSVVEFVEEELSLDTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIGVVNWVPSN 211
++E D VA+ LT G+VK K L S+ L ++ NW P++
Sbjct: 191 DMAGLLE--------DEVAQFLTDGKVK---LLKRLLLSAFSLPGRELFKLCCTNWSPTS 239
Query: 212 HTSVVSAMLAKLIYRIGTEIAYDFGNFVFSQTLKHA 247
+ S AKL+Y I ++ ++F F L+ A
Sbjct: 240 NNSYAQPDRAKLVYMIMHKMPFNFVRLAFDHILQLA 275
>UniRef100_Q9S9S8 F28J9.1 [Arabidopsis thaliana]
Length = 505
Score = 37.7 bits (86), Expect = 0.55
Identities = 26/92 (28%), Positives = 40/92 (43%), Gaps = 3/92 (3%)
Query: 167 DTVAEELTAGRVKKWPAKKLLSTGVLSVKYAILNRIGVVNWVPSNHTSVVSAMLAKLIYR 226
+ VA LT GRV+ + ++ S L +I NW P + S +L+YR
Sbjct: 173 EEVARYLTDGRVQNL---QNIAMNKFSENCKSLFKISCRNWSPQTNEGYASIDRGRLVYR 229
Query: 227 IGTEIAYDFGNFVFSQTLKHAETCAVKLPISF 258
I ++ DFG V+ L+ A K + F
Sbjct: 230 IAHKMPIDFGEMVYDHVLQLALMSVSKFFLMF 261
>UniRef100_UPI000049995C UPI000049995C UniRef100 entry
Length = 1319
Score = 36.6 bits (83), Expect = 1.2
Identities = 44/167 (26%), Positives = 73/167 (43%), Gaps = 16/167 (9%)
Query: 106 KCYDKL-VKEFIVNLSEDVNNST-SPEFRKVFVRGKCV---NFSPAIINQTLERSVVEFV 160
K DK V + NL +D + S + E KV ++ K + + + I Q L++S +
Sbjct: 848 KAVDKTNVISVLCNLIDDKDCSDYATELMKVLMKEKELFIESLNVEIFQQLLQKSFIN-- 905
Query: 161 EEELSLDTVAEELTAGRVKKWPAKKLLSTGV------LSVKYAILNRIGVVNWVPSNHTS 214
EL V +++ K KLLS + L+ I + +VNW+P T
Sbjct: 906 --ELVYSLVIDQINTNETIKPTLLKLLSNTIIEDILFLTETKNIPKVLQLVNWIPLLSTI 963
Query: 215 VVSAMLAKLIYRIGTEIAYDFGNFV-FSQTLKHAETCAVKLPISFFT 260
++ + I+ + D NF+ F Q LK E ++ L +SF T
Sbjct: 964 NLTQFIKSCIHYLPNLNTKDINNFISFLQKLKPQELESLTLNVSFNT 1010
>UniRef100_Q6G1B9 Lysyl-tRNA synthetase [Bartonella quintana]
Length = 553
Score = 35.8 bits (81), Expect = 2.1
Identities = 27/104 (25%), Positives = 48/104 (45%), Gaps = 4/104 (3%)
Query: 99 KTVMNIGKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQTLERSVVE 158
+T++ I CYDK++ + L E + S GK + +I + +E+ V
Sbjct: 168 ETLLKILACYDKIMAIVLPTLGEKRQATYSLFLPISPFSGKVLQV--PMIARNVEKGTVT 225
Query: 159 FVEEELSLDTVAEELTAGRVK-KWPAKKLLSTGVLSVKYAILNR 201
++E E +T+ E+T GRVK +W + L V Y + +
Sbjct: 226 YIEPETG-ETIETEITGGRVKCQWKVDWAMRWSALGVDYEMAGK 268
>UniRef100_Q6CTS8 Kluyveromyces lactis strain NRRL Y-1140 chromosome C of strain NRRL
Y- 1140 of Kluyveromyces lactis [Kluyveromyces lactis]
Length = 526
Score = 35.8 bits (81), Expect = 2.1
Identities = 44/188 (23%), Positives = 84/188 (44%), Gaps = 9/188 (4%)
Query: 8 QKVSRIDLETESDVEGD-VRDITASEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWKY 66
+ VS + +ES++E D D + KK K + + V ++N++ A +A K
Sbjct: 123 EDVSESEATSESEIEKDREEDEETAMKKEKLAKIVQTQTKQV-INNLAHSAQRDASKGYA 181
Query: 67 VYQRRVAKDREIDSDVLACKEIIALIAKAGLRKTVMNIGKCYDKLVKEFIVNLSEDVNNS 126
+ Q+ D+ +DS + K AL A L + + + D+ + + + + +
Sbjct: 182 ILQQNKFFDKILDSRIKLQK---ALSASNQLPLSQQSWDELLDENNSKLLTSTFKVLEKV 238
Query: 127 TSP--EFRKVFVRGKCVNFS--PAIINQTLERSVVEFVEEELSLDTVAEELTAGRVKKWP 182
+ R F G +N S P+ + + +RS E V E ++LD+ + + + KW
Sbjct: 239 LTQCVTVRHKFQIGDHINQSENPSDFDNSKKRSFKELVHETVTLDSDLKSYRSAVLNKWS 298
Query: 183 AKKLLSTG 190
AK S+G
Sbjct: 299 AKVSASSG 306
>UniRef100_O68050 Potential regulatory protein [Rhodobacter capsulatus]
Length = 142
Score = 35.4 bits (80), Expect = 2.7
Identities = 24/84 (28%), Positives = 43/84 (50%), Gaps = 3/84 (3%)
Query: 9 KVSRIDLETESDVEGDVRDITASEKKMFSGKRIPSDVPNVPLDNVSIHAPENAFKWKYVY 68
++ R +ET+ DVEG+VR IT E + +++ + +P + ++HA A W V
Sbjct: 49 RIRRFTIETDVDVEGEVRAITLVELQGKLSRQVIRTLTGLP-EVSTVHATNGA--WDLVV 105
Query: 69 QRRVAKDREIDSDVLACKEIIALI 92
+ R + D + A +EI +I
Sbjct: 106 EIRTGSLVDFDRVLRAIREIPGVI 129
>UniRef100_UPI000007FE0E UPI000007FE0E UniRef100 entry
Length = 448
Score = 34.7 bits (78), Expect = 4.6
Identities = 22/76 (28%), Positives = 40/76 (51%), Gaps = 4/76 (5%)
Query: 105 GKCYDKLVKEFIVNLSEDVNNSTSPEFRKVFVRGKCVNFSPAIINQTLERSVVEFVEEEL 164
G+C + KE+I ++E ++ + + K+F+ K F+PA +NQ +++ E L
Sbjct: 355 GRC---VCKEYIRQITELLHGGINDK-EKLFLTVKITKFTPAGVNQKFSTGTAKYIGELL 410
Query: 165 SLDTVAEELTAGRVKK 180
T +LTAG + K
Sbjct: 411 EGRTDETKLTAGCIGK 426
>UniRef100_UPI000031A45E UPI000031A45E UniRef100 entry
Length = 273
Score = 34.7 bits (78), Expect = 4.6
Identities = 34/136 (25%), Positives = 64/136 (47%), Gaps = 14/136 (10%)
Query: 1 KKTTGK--KQKVSRIDLETESDVEGD---VRDITASEKKMFSGKRIPSDVPNVPLDNVSI 55
K+ +G+ K K++++ L+TE D V D+T K++ ++ PN ++I
Sbjct: 8 KENSGEMGKFKIAKV-LKTEKHPNADKLKVCDVTIGGKEIL---KVVCGAPNAREGLITI 63
Query: 56 HAPENAFKWKYVYQRRVAKDREIDSDVLACKEIIALIAKAG-----LRKTVMNIGKCYDK 110
+AP A K ++ +VAK R ++S + C E ++ L+ IGK Y K
Sbjct: 64 YAPPGAVIPKNNFELKVAKIRGVESKGMLCSESELNLSDESDGIIELKNKEREIGKSYFK 123
Query: 111 LVKEFIVNLSEDVNNS 126
+ +++S N +
Sbjct: 124 KNSQKAIDISITPNRA 139
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.339 0.147 0.497
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 582,124,949
Number of Sequences: 2790947
Number of extensions: 23072813
Number of successful extensions: 87333
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 87308
Number of HSP's gapped (non-prelim): 41
length of query: 385
length of database: 848,049,833
effective HSP length: 129
effective length of query: 256
effective length of database: 488,017,670
effective search space: 124932523520
effective search space used: 124932523520
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 76 (33.9 bits)
Lotus: description of TM0101.13


