

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= TM0100c.1
(773 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
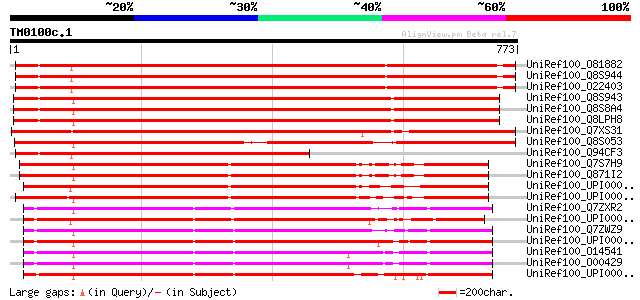
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O81882 Arabidopsis dynamin-like protein ADL2 [Arabidop... 1046 0.0
UniRef100_Q8S944 Dynamin like protein 2a [Arabidopsis thaliana] 1045 0.0
UniRef100_O22403 ADL2 protein [Arabidopsis thaliana] 1040 0.0
UniRef100_Q8S943 Dynamin like protein 2b [Arabidopsis thaliana] 1025 0.0
UniRef100_Q8S8A4 Dynamin-like protein [Arabidopsis thaliana] 1025 0.0
UniRef100_Q8LPH8 Dynamin-like protein [Arabidopsis thaliana] 1023 0.0
UniRef100_Q7XS31 OSJNBa0072D21.2 protein [Oryza sativa] 933 0.0
UniRef100_Q8S053 Putative dynamin-like protein ADL2 [Oryza sativa] 883 0.0
UniRef100_Q94CF3 Putative dynamin protein ADL2 [Arabidopsis thal... 689 0.0
UniRef100_Q7S7H9 Hypothetical protein [Neurospora crassa] 560 e-158
UniRef100_Q871I2 Probable VpsA protein [Neurospora crassa] 558 e-157
UniRef100_UPI000023F04A UPI000023F04A UniRef100 entry 556 e-157
UniRef100_UPI000021A7EC UPI000021A7EC UniRef100 entry 552 e-155
UniRef100_Q7ZXR2 MGC53884 protein [Xenopus laevis] 541 e-152
UniRef100_UPI0000362513 UPI0000362513 UniRef100 entry 536 e-150
UniRef100_Q7ZWZ9 Dnm1l-prov protein [Xenopus laevis] 535 e-150
UniRef100_UPI00003A9933 UPI00003A9933 UniRef100 entry 532 e-149
UniRef100_O14541 Dnm1p/Vps1p-like protein [Homo sapiens] 532 e-149
UniRef100_O00429 Dynamin-like protein [Homo sapiens] 531 e-149
UniRef100_UPI00003BFD07 UPI00003BFD07 UniRef100 entry 531 e-149
>UniRef100_O81882 Arabidopsis dynamin-like protein ADL2 [Arabidopsis thaliana]
Length = 808
Score = 1046 bits (2705), Expect = 0.0
Identities = 541/769 (70%), Positives = 639/769 (82%), Gaps = 18/769 (2%)
Query: 10 TAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGR 69
++++T A PLGSSVI +VN+LQDIF+++G+QSTI LPQV VVGSQSSGKSSVLE+LVGR
Sbjct: 24 SSSTTNAAPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVVVVGSQSSGKSSVLEALVGR 82
Query: 70 DFLPRGTDICTRRPLVLQLVHTKP-----SHPEFGEFLHLPGRKFHDFTQIRHEIQAETD 124
DFLPRG DICTRRPLVLQL+ TK S E+GEF HLP +F+DF++IR EI+AET+
Sbjct: 83 DFLPRGNDICTRRPLVLQLLQTKSRANGGSDDEWGEFRHLPETRFYDFSEIRREIEAETN 142
Query: 125 REAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKI 184
R GENKGV++ QIRLKI SPNVL+ITLVDLPGITKVPVGDQPSDIEARIRTMI+SYIK
Sbjct: 143 RLVGENKGVADTQIRLKISSPNVLNITLVDLPGITKVPVGDQPSDIEARIRTMILSYIKQ 202
Query: 185 PTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIP 244
TCLILAVTPAN+DLANSDALQ+A + DPDG+RTIGVITKLDIMD+GTDAR LLLG V+P
Sbjct: 203 DTCLILAVTPANTDLANSDALQIASIVDPDGHRTIGVITKLDIMDKGTDARKLLLGNVVP 262
Query: 245 LRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILA 304
LRLGYVGVVNR QEDI +NR++K+AL+AEE+FFRS PVY GLAD GVPQLAKKLN+IL
Sbjct: 263 LRLGYVGVVNRCQEDILLNRTVKEALLAEEKFFRSHPVYHGLADRLGVPQLAKKLNQILV 322
Query: 305 QHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGK 364
QHIK +LP LK+RIS +LVA AKEH SYGE+TES+AGQGAL+LN LSKYC+A+SS+LEGK
Sbjct: 323 QHIKVLLPDLKSRISNALVATAKEHQSYGELTESRAGQGALLLNFLSKYCEAYSSLLEGK 382
Query: 365 NKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEV 424
++EMST ELSGGARIHYIFQSI+V+SLE VDPCEDLTDDDIRTA+QNATGP+SALF+P+V
Sbjct: 383 SEEMSTSELSGGARIHYIFQSIFVKSLEEVDPCEDLTDDDIRTAIQNATGPRSALFVPDV 442
Query: 425 PFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRD 484
PFE+ VRRQISRLLDPSLQCARFI++EL+KISHRCM ELQRFP LRK MDEVIG+FLR+
Sbjct: 443 PFEVLVRRQISRLLDPSLQCARFIFEELIKISHRCMMNELQRFPVLRKRMDEVIGDFLRE 502
Query: 485 GLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LDALESD 542
GLEPSE MI II+MEMDYINTSHPNFIGG+KA+EAA+ Q K SR+ PV+ D +E D
Sbjct: 503 GLEPSEAMIGDIIDMEMDYINTSHPNFIGGTKAVEAAMHQVKSSRIPHPVARPKDTVEPD 562
Query: 543 KGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNR 602
+ +S KSR + RQANG+V D GV A D EK P+ NA + W I SIF G D R
Sbjct: 563 RTSSSTSQVKSRSFLGRQANGIVTDQGVVSA-DAEKAQPAANASDTRWGIPSIFRGGDTR 621
Query: 603 VYVKENTSTKPHAEPVHNVQSS-STIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYY 661
K++ KP +E V ++ + S I+L+EPP VLRP+E++SE E VEI +TKLLLRSYY
Sbjct: 622 AVTKDSLLNKPFSEAVEDMSHNLSMIYLKEPPAVLRPTETHSEQEAVEIQITKLLLRSYY 681
Query: 662 DIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRE 721
DI RKNIED VPKAIMHFLVN+TKRELHNVFI+KLYRE+LFEEMLQEP E+A KRKR +E
Sbjct: 682 DIVRKNIEDSVPKAIMHFLVNHTKRELHNVFIKKLYRENLFEEMLQEPDEIAVKRKRTQE 741
Query: 722 LLRAYQQAFRDLDELPLEAETVERGYGSPEATGLPRIRGLPSSSMYSSS 770
L QQA+R LDELPLEA++V + G+ + + L +SS YS+S
Sbjct: 742 TLHVLQQAYRTLDELPLEADSV--------SAGMSKHQELLTSSKYSTS 782
>UniRef100_Q8S944 Dynamin like protein 2a [Arabidopsis thaliana]
Length = 808
Score = 1045 bits (2703), Expect = 0.0
Identities = 541/769 (70%), Positives = 638/769 (82%), Gaps = 18/769 (2%)
Query: 10 TAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGR 69
++++T A PLGSSVI +VN+LQDIF+++G+QSTI LPQV VVGSQSSGKSSVLE+LVGR
Sbjct: 24 SSSTTNAAPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVVVVGSQSSGKSSVLEALVGR 82
Query: 70 DFLPRGTDICTRRPLVLQLVHTKP-----SHPEFGEFLHLPGRKFHDFTQIRHEIQAETD 124
DFLPRG DICTRRPLVLQL+ TK S E+GEF HLP +F+DF++IR EI+AET+
Sbjct: 83 DFLPRGNDICTRRPLVLQLLQTKSRANGGSDDEWGEFRHLPETRFYDFSEIRREIEAETN 142
Query: 125 REAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKI 184
R GENKGV++ QIRLKI SPNVL+ITLVDLPGITKVPVGDQPSDIEARIRTMI+SYIK
Sbjct: 143 RLVGENKGVADTQIRLKISSPNVLNITLVDLPGITKVPVGDQPSDIEARIRTMILSYIKQ 202
Query: 185 PTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIP 244
TCLILAVTPAN+DLANSDALQ+A + DPDG+RTIGVITKLDIMD+GTDAR LLLG V+P
Sbjct: 203 DTCLILAVTPANTDLANSDALQIASIVDPDGHRTIGVITKLDIMDKGTDARKLLLGNVVP 262
Query: 245 LRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILA 304
LRLGYVGVVNR QEDI +NR++K+AL+AEE+FFRS PVY GLAD GVPQLAKKLN+IL
Sbjct: 263 LRLGYVGVVNRCQEDILLNRTVKEALLAEEKFFRSHPVYHGLADRLGVPQLAKKLNQILV 322
Query: 305 QHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGK 364
QHIK +LP LK+RIS +LVA AKEH SYGE+TES+AGQGAL+LN LSKYC+A+SS+LEGK
Sbjct: 323 QHIKVLLPDLKSRISNALVATAKEHQSYGELTESRAGQGALLLNFLSKYCEAYSSLLEGK 382
Query: 365 NKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEV 424
++EMST ELSGGARIHYIFQSI+V+SLE VDPCEDLTDDDIRTA+QNATGP+SALF+P+V
Sbjct: 383 SEEMSTSELSGGARIHYIFQSIFVKSLEEVDPCEDLTDDDIRTAIQNATGPRSALFVPDV 442
Query: 425 PFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGNFLRD 484
PFE+ VRRQISRLLDPSLQCARFI++EL+KISHRCM ELQRFP LRK MDEVIG FLR+
Sbjct: 443 PFEVLVRRQISRLLDPSLQCARFIFEELIKISHRCMMNELQRFPVLRKRMDEVIGXFLRE 502
Query: 485 GLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LDALESD 542
GLEPSE MI II+MEMDYINTSHPNFIGG+KA+EAA+ Q K SR+ PV+ D +E D
Sbjct: 503 GLEPSEAMIGDIIDMEMDYINTSHPNFIGGTKAVEAAMHQVKSSRIPHPVARPKDTVEPD 562
Query: 543 KGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNR 602
+ +S KSR + RQANG+V D GV A D EK P+ NA + W I SIF G D R
Sbjct: 563 RTSSSTSQVKSRSFLGRQANGIVTDQGVVSA-DAEKAQPAANASDTRWGIPSIFRGGDTR 621
Query: 603 VYVKENTSTKPHAEPVHNVQSS-STIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYY 661
K++ KP +E V ++ + S I+L+EPP VLRP+E++SE E VEI +TKLLLRSYY
Sbjct: 622 AVTKDSLLNKPFSEAVEDMSHNLSMIYLKEPPAVLRPTETHSEQEAVEIQITKLLLRSYY 681
Query: 662 DIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRE 721
DI RKNIED VPKAIMHFLVN+TKRELHNVFI+KLYRE+LFEEMLQEP E+A KRKR +E
Sbjct: 682 DIVRKNIEDSVPKAIMHFLVNHTKRELHNVFIKKLYRENLFEEMLQEPDEIAVKRKRTQE 741
Query: 722 LLRAYQQAFRDLDELPLEAETVERGYGSPEATGLPRIRGLPSSSMYSSS 770
L QQA+R LDELPLEA++V + G+ + + L +SS YS+S
Sbjct: 742 TLHVLQQAYRTLDELPLEADSV--------SAGMSKHQELLTSSKYSTS 782
>UniRef100_O22403 ADL2 protein [Arabidopsis thaliana]
Length = 809
Score = 1040 bits (2690), Expect = 0.0
Identities = 541/770 (70%), Positives = 639/770 (82%), Gaps = 19/770 (2%)
Query: 10 TAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGR 69
++++T A PLGSSVI +VN+LQDIF+++G+QSTI LPQV VVGSQSSGKSSVLE+LVGR
Sbjct: 24 SSSTTNAAPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVVVVGSQSSGKSSVLEALVGR 82
Query: 70 DFLPRGTDICTRRPLVLQLVHTKP-----SHPEFGEFLHLPGRKFHDFTQIRHEIQAETD 124
DFLPRG DICTRRPLVLQL+ TK S E+GEF HLP +F+DF++IR EI+AET+
Sbjct: 83 DFLPRGNDICTRRPLVLQLLQTKSRANGGSDDEWGEFRHLPETRFYDFSEIRREIEAETN 142
Query: 125 REAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKI 184
R GENKGV++ QIRLKI SPNVL+ITLVDLPGITKVPVGDQPSDIEARIRTMI+SYIK
Sbjct: 143 RLVGENKGVADTQIRLKISSPNVLNITLVDLPGITKVPVGDQPSDIEARIRTMILSYIKQ 202
Query: 185 PTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIP 244
TCLILAVTPAN+DLANSDALQ+A + DPDG+RTIGVITKLDIMD+GTDAR LLLG V+P
Sbjct: 203 DTCLILAVTPANTDLANSDALQIASIVDPDGHRTIGVITKLDIMDKGTDARKLLLGNVVP 262
Query: 245 LRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILA 304
LRLGYVGVVNR QEDI +NR++K+AL+AEE+FFRS PVY GLAD GVPQLAKKLN+IL
Sbjct: 263 LRLGYVGVVNRCQEDILLNRTVKEALLAEEKFFRSHPVYHGLADRLGVPQLAKKLNQILV 322
Query: 305 QHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGK 364
QHIK +LP LK+RIS +LVA AKEH SYGE+TES+AGQGAL+LN LSKYC+A+SS+LEGK
Sbjct: 323 QHIKVLLPDLKSRISNALVATAKEHQSYGELTESRAGQGALLLNFLSKYCEAYSSLLEGK 382
Query: 365 NKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEV 424
++EMST ELSGGARIHYIFQSI+V+SLE VDPCEDLTDDDIRTA+QNATGP+SALF+P+V
Sbjct: 383 SEEMSTSELSGGARIHYIFQSIFVKSLEEVDPCEDLTDDDIRTAIQNATGPRSALFVPDV 442
Query: 425 PFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIG-NFLR 483
PFE+ VRRQISRLLDPSLQCARFI++EL+KISHRCM ELQRFP LRK MDE+ G +FLR
Sbjct: 443 PFEVLVRRQISRLLDPSLQCARFIFEELIKISHRCMMNELQRFPVLRKRMDELSGRDFLR 502
Query: 484 DGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LDALES 541
+GLEPSETMI II+MEMDYINTSHPNFIGG+KA+EAA+ Q K SR+ PV+ D +E
Sbjct: 503 EGLEPSETMIGDIIDMEMDYINTSHPNFIGGTKAVEAAMHQVKSSRIPHPVARPKDTVEP 562
Query: 542 DKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDN 601
D+ +S KSR + RQANGVV D GV A D EK P+ NA + W I SIF G D
Sbjct: 563 DRTSSSTSQVKSRSFLGRQANGVVTDQGVVSA-DAEKAQPAANANDTRWGIPSIFRGGDT 621
Query: 602 RVYVKENTSTKPHAEPVHNVQSS-STIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSY 660
R K++ KP +E V ++ + S I+L+EPP VLRP+E++SE E VEI +TKLLLRSY
Sbjct: 622 RAVTKDSLLNKPFSEAVEDMSHNLSMIYLKEPPAVLRPTETHSEQEAVEIQITKLLLRSY 681
Query: 661 YDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCR 720
YDI RKNIED VPKAIMHFLVN+TKRELHNVFI+KLYRE+LFEEMLQEP E+A KRKR +
Sbjct: 682 YDIVRKNIEDSVPKAIMHFLVNHTKRELHNVFIKKLYRENLFEEMLQEPDEIAVKRKRTQ 741
Query: 721 ELLRAYQQAFRDLDELPLEAETVERGYGSPEATGLPRIRGLPSSSMYSSS 770
E L QQA+R LDELPLEA++V + G+ + + L +SS YS+S
Sbjct: 742 ETLHVLQQAYRTLDELPLEADSV--------SAGMSKHQELLTSSKYSTS 783
>UniRef100_Q8S943 Dynamin like protein 2b [Arabidopsis thaliana]
Length = 780
Score = 1025 bits (2650), Expect = 0.0
Identities = 534/752 (71%), Positives = 629/752 (83%), Gaps = 14/752 (1%)
Query: 6 DEAVTAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLES 65
D+ ++++A TPLGSSVI +VN+LQDIF+++G+QSTI LPQVAVVGSQSSGKSSVLE+
Sbjct: 4 DDLPPSSASAVTPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVAVVGSQSSGKSSVLEA 62
Query: 66 LVGRDFLPRGTDICTRRPLVLQLVHTKPSHP-----EFGEFLHL-PGRKFHDFTQIRHEI 119
LVGRDFLPRG DICTRRPL LQLV TKPS E+GEFLH P R+ +DF++IR EI
Sbjct: 63 LVGRDFLPRGNDICTRRPLRLQLVQTKPSSDGGPDEEWGEFLHHDPVRRIYDFSEIRREI 122
Query: 120 QAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIM 179
+AET+R +GENKGVS+ I LKIFSPNVLDI+LVDLPGITKVPVGDQPSDIEARIRTMI+
Sbjct: 123 EAETNRVSGENKGVSDIPIGLKIFSPNVLDISLVDLPGITKVPVGDQPSDIEARIRTMIL 182
Query: 180 SYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLL 239
+YIK P+CLILAV+PAN+DLANSDALQ+A ADPDG+RTIGVITKLDIMDRGTDARN LL
Sbjct: 183 TYIKEPSCLILAVSPANTDLANSDALQIAGNADPDGHRTIGVITKLDIMDRGTDARNHLL 242
Query: 240 GKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKL 299
GK IPLRLGYVGVVNRSQEDI MNRSIKDALVAEE+FFRSRPVYSGL D GVPQLAKKL
Sbjct: 243 GKTIPLRLGYVGVVNRSQEDILMNRSIKDALVAEEKFFRSRPVYSGLTDRLGVPQLAKKL 302
Query: 300 NKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSS 359
N++L QHIKA+LP LK+RI+ +L A AKE+ SYG+ITES+ GQGAL+L+ ++KYC+A+SS
Sbjct: 303 NQVLVQHIKALLPSLKSRINNALFATAKEYESYGDITESRGGQGALLLSFITKYCEAYSS 362
Query: 360 MLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSAL 419
LEGK+KEMST ELSGGARI YIFQS++V+SLE VDPCEDLT DDIRTA+QNATGP+SAL
Sbjct: 363 TLEGKSKEMSTSELSGGARILYIFQSVFVKSLEEVDPCEDLTADDIRTAIQNATGPRSAL 422
Query: 420 FIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIG 479
F+P+VPFE+ VRRQISRLLDPSLQCARFI+DEL+KISH+CM ELQRFP L+K MDEVIG
Sbjct: 423 FVPDVPFEVLVRRQISRLLDPSLQCARFIFDELVKISHQCMMKELQRFPVLQKRMDEVIG 482
Query: 480 NFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LD 537
NFLR+GLEPS+ MI +IEMEMDYINTSHPNFIGG+KA+E A+Q K SR+ PV+ D
Sbjct: 483 NFLREGLEPSQAMIRDLIEMEMDYINTSHPNFIGGTKAVEQAMQTVKSSRIPHPVARPRD 542
Query: 538 ALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWR-ISSIF 596
+E ++ +S K+R + RQANG++ D V A+D E+ AP AG +SW SSIF
Sbjct: 543 TVEPERTASSGSQIKTRSFLGRQANGIITDQAVPTAADAERPAP---AGSTSWSGFSSIF 599
Query: 597 GGRDNRVYVKENTSTKPHAEPVHNV-QSSSTIHLREPPPVLRPSESNSEMEGVEITVTKL 655
G D + K N KP +E V Q+ STI+L+EPP +L+ SE++SE E VEI +TKL
Sbjct: 600 RGSDGQAAAKNNLLNKPFSETTQEVYQNLSTIYLKEPPTILKSSETHSEQESVEIEITKL 659
Query: 656 LLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAK 715
LL+SYYDI RKN+EDLVPKAIMHFLVN TKRELHNVFIEKLYRE+L EE+L+EP E+A K
Sbjct: 660 LLKSYYDIVRKNVEDLVPKAIMHFLVNYTKRELHNVFIEKLYRENLIEELLKEPDELAIK 719
Query: 716 RKRCRELLRAYQQAFRDLDELPLEAETVERGY 747
RKR +E LR QQA R LDELPLEAE+VERGY
Sbjct: 720 RKRTQETLRILQQANRTLDELPLEAESVERGY 751
>UniRef100_Q8S8A4 Dynamin-like protein [Arabidopsis thaliana]
Length = 780
Score = 1025 bits (2650), Expect = 0.0
Identities = 534/752 (71%), Positives = 629/752 (83%), Gaps = 14/752 (1%)
Query: 6 DEAVTAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLES 65
D+ ++++A TPLGSSVI +VN+LQDIF+++G+QSTI LPQVAVVGSQSSGKSSVLE+
Sbjct: 4 DDLPPSSASAVTPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVAVVGSQSSGKSSVLEA 62
Query: 66 LVGRDFLPRGTDICTRRPLVLQLVHTKPSHP-----EFGEFLHL-PGRKFHDFTQIRHEI 119
LVGRDFLPRG DICTRRPL LQLV TKPS E+GEFLH P R+ +DF++IR EI
Sbjct: 63 LVGRDFLPRGNDICTRRPLRLQLVQTKPSSDGGSDEEWGEFLHHDPVRRIYDFSEIRREI 122
Query: 120 QAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIM 179
+AET+R +GENKGVS+ I LKIFSPNVLDI+LVDLPGITKVPVGDQPSDIEARIRTMI+
Sbjct: 123 EAETNRVSGENKGVSDIPIGLKIFSPNVLDISLVDLPGITKVPVGDQPSDIEARIRTMIL 182
Query: 180 SYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLL 239
+YIK P+CLILAV+PAN+DLANSDALQ+A ADPDG+RTIGVITKLDIMDRGTDARN LL
Sbjct: 183 TYIKEPSCLILAVSPANTDLANSDALQIAGNADPDGHRTIGVITKLDIMDRGTDARNHLL 242
Query: 240 GKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKL 299
GK IPLRLGYVGVVNRSQEDI MNRSIKDALVAEE+FFRSRPVYSGL D GVPQLAKKL
Sbjct: 243 GKTIPLRLGYVGVVNRSQEDILMNRSIKDALVAEEKFFRSRPVYSGLTDRLGVPQLAKKL 302
Query: 300 NKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSS 359
N++L QHIKA+LP LK+RI+ +L A AKE+ SYG+ITES+ GQGAL+L+ ++KYC+A+SS
Sbjct: 303 NQVLVQHIKALLPSLKSRINNALFATAKEYESYGDITESRGGQGALLLSFITKYCEAYSS 362
Query: 360 MLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSAL 419
LEGK+KEMST ELSGGARI YIFQS++V+SLE VDPCEDLT DDIRTA+QNATGP+SAL
Sbjct: 363 TLEGKSKEMSTSELSGGARILYIFQSVFVKSLEEVDPCEDLTADDIRTAIQNATGPRSAL 422
Query: 420 FIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIG 479
F+P+VPFE+ VRRQISRLLDPSLQCARFI+DEL+KISH+CM ELQRFP L+K MDEVIG
Sbjct: 423 FVPDVPFEVLVRRQISRLLDPSLQCARFIFDELVKISHQCMMKELQRFPVLQKRMDEVIG 482
Query: 480 NFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LD 537
NFLR+GLEPS+ MI +IEMEMDYINTSHPNFIGG+KA+E A+Q K SR+ PV+ D
Sbjct: 483 NFLREGLEPSQAMIRDLIEMEMDYINTSHPNFIGGTKAVEQAMQTVKSSRIPHPVARPRD 542
Query: 538 ALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWR-ISSIF 596
+E ++ +S K+R + RQANG++ D V A+D E+ AP AG +SW SSIF
Sbjct: 543 TVEPERTASSGSQIKTRSFLGRQANGIITDQAVPTAADAERPAP---AGSTSWSGFSSIF 599
Query: 597 GGRDNRVYVKENTSTKPHAEPVHNV-QSSSTIHLREPPPVLRPSESNSEMEGVEITVTKL 655
G D + K N KP +E V Q+ STI+L+EPP +L+ SE++SE E VEI +TKL
Sbjct: 600 RGSDGQAAAKNNLLNKPFSETTQEVYQNLSTIYLKEPPTILKSSETHSEQESVEIEITKL 659
Query: 656 LLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAK 715
LL+SYYDI RKN+EDLVPKAIMHFLVN TKRELHNVFIEKLYRE+L EE+L+EP E+A K
Sbjct: 660 LLKSYYDIVRKNVEDLVPKAIMHFLVNYTKRELHNVFIEKLYRENLIEELLKEPDELAIK 719
Query: 716 RKRCRELLRAYQQAFRDLDELPLEAETVERGY 747
RKR +E LR QQA R LDELPLEAE+VERGY
Sbjct: 720 RKRTQETLRILQQANRTLDELPLEAESVERGY 751
>UniRef100_Q8LPH8 Dynamin-like protein [Arabidopsis thaliana]
Length = 780
Score = 1023 bits (2645), Expect = 0.0
Identities = 533/752 (70%), Positives = 629/752 (82%), Gaps = 14/752 (1%)
Query: 6 DEAVTAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLES 65
D+ ++++A TPLGSSVI +VN+LQDIF+++G+QSTI LPQVAVVGSQSSGKSSVLE+
Sbjct: 4 DDLPPSSASAVTPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVAVVGSQSSGKSSVLEA 62
Query: 66 LVGRDFLPRGTDICTRRPLVLQLVHTKPSHP-----EFGEFLHL-PGRKFHDFTQIRHEI 119
LVGRDFLPRG DICTRRPL LQLV TKPS E+GEFLH P R+ +DF++IR EI
Sbjct: 63 LVGRDFLPRGNDICTRRPLRLQLVQTKPSSDGGSDEEWGEFLHHDPVRRIYDFSEIRREI 122
Query: 120 QAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIM 179
+AET+R +GENKGVS+ I LKIFSPNVLDI+LVDLPGITKVPVGDQPSDIEARIRTMI+
Sbjct: 123 EAETNRVSGENKGVSDIPIGLKIFSPNVLDISLVDLPGITKVPVGDQPSDIEARIRTMIL 182
Query: 180 SYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLL 239
+YIK P+CLILAV+PAN+DLA+SDALQ+A ADPDG+RTIGVITKLDIMDRGTDARN LL
Sbjct: 183 TYIKEPSCLILAVSPANTDLASSDALQIAGNADPDGHRTIGVITKLDIMDRGTDARNHLL 242
Query: 240 GKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKL 299
GK IPLRLGYVGVVNRSQEDI MNRSIKDALVAEE+FFRSRPVYSGL D GVPQLAKKL
Sbjct: 243 GKTIPLRLGYVGVVNRSQEDILMNRSIKDALVAEEKFFRSRPVYSGLTDRLGVPQLAKKL 302
Query: 300 NKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSS 359
N++L QHIKA+LP LK+RI+ +L A AKE+ SYG+ITES+ GQGAL+L+ ++KYC+A+SS
Sbjct: 303 NQVLVQHIKALLPSLKSRINNALFATAKEYESYGDITESRGGQGALLLSFITKYCEAYSS 362
Query: 360 MLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSAL 419
LEGK+KEMST ELSGGARI YIFQS++V+SLE VDPCEDLT DDIRTA+QNATGP+SAL
Sbjct: 363 TLEGKSKEMSTSELSGGARILYIFQSVFVKSLEEVDPCEDLTADDIRTAIQNATGPRSAL 422
Query: 420 FIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIG 479
F+P+VPFE+ VRRQISRLLDPSLQCARFI+DEL+KISH+CM ELQRFP L+K MDEVIG
Sbjct: 423 FVPDVPFEVLVRRQISRLLDPSLQCARFIFDELVKISHQCMMKELQRFPVLQKRMDEVIG 482
Query: 480 NFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVS--LD 537
NFLR+GLEPS+ MI +IEMEMDYINTSHPNFIGG+KA+E A+Q K SR+ PV+ D
Sbjct: 483 NFLREGLEPSQAMIRDLIEMEMDYINTSHPNFIGGTKAVEQAMQTVKSSRIPHPVARPRD 542
Query: 538 ALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWR-ISSIF 596
+E ++ +S K+R + RQANG++ D V A+D E+ AP AG +SW SSIF
Sbjct: 543 TVEPERTASSGSQIKTRSFLGRQANGIITDQAVPTAADAERPAP---AGSTSWSGFSSIF 599
Query: 597 GGRDNRVYVKENTSTKPHAEPVHNV-QSSSTIHLREPPPVLRPSESNSEMEGVEITVTKL 655
G D + K N KP +E V Q+ STI+L+EPP +L+ SE++SE E VEI +TKL
Sbjct: 600 RGSDGQAAAKNNLLNKPFSETTQEVYQNLSTIYLKEPPTILKSSETHSEQESVEIEITKL 659
Query: 656 LLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAK 715
LL+SYYDI RKN+EDLVPKAIMHFLVN TKRELHNVFIEKLYRE+L EE+L+EP E+A K
Sbjct: 660 LLKSYYDIVRKNVEDLVPKAIMHFLVNYTKRELHNVFIEKLYRENLIEELLKEPDELAIK 719
Query: 716 RKRCRELLRAYQQAFRDLDELPLEAETVERGY 747
RKR +E LR QQA R LDELPLEAE+VERGY
Sbjct: 720 RKRTQETLRILQQANRTLDELPLEAESVERGY 751
>UniRef100_Q7XS31 OSJNBa0072D21.2 protein [Oryza sativa]
Length = 800
Score = 933 bits (2412), Expect = 0.0
Identities = 494/780 (63%), Positives = 598/780 (76%), Gaps = 27/780 (3%)
Query: 3 VTDDEAVTAASTAATPLGSSVISLVNRLQDIFSRV-GNQSTIIDLPQVAVVGSQSSGKSS 61
+ + A AA+ A +G +VI LVNRLQDI +R+ G ++LPQVA +G QSSGKSS
Sbjct: 1 MAEPAAAAAAAEAPATVGQAVIPLVNRLQDIVARLDGGGGGGLELPQVAAIGGQSSGKSS 60
Query: 62 VLESLVGRDFLPRGTDICTRRPLVLQLV-HTKPSHPEFGEFLHLPGRKFHDFTQIRHEIQ 120
VLE+LVGRDFLPRG DICTRRPLVLQLV H+ P E+GEFLH P R+FHDF QI+ EIQ
Sbjct: 61 VLEALVGRDFLPRGPDICTRRPLVLQLVRHSAPE--EWGEFLHAPARRFHDFDQIKREIQ 118
Query: 121 AETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMS 180
ETD+EAG NKGVSEKQIRLKIFSPNVLDITLVDLPGIT+VPVGDQPSDIE+RIR+MIM
Sbjct: 119 LETDKEAGGNKGVSEKQIRLKIFSPNVLDITLVDLPGITRVPVGDQPSDIESRIRSMIMQ 178
Query: 181 YIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLG 240
YIK P+C+ILAVTPAN+DLANSDALQ+A++ADPDG+RTIGVITKLDIMDRGTDARN LLG
Sbjct: 179 YIKHPSCIILAVTPANADLANSDALQLAKLADPDGSRTIGVITKLDIMDRGTDARNFLLG 238
Query: 241 KVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLN 300
VIPL+LGYVGVVNRSQEDI RS+KDAL EE+FF + P Y GL CGVPQLAKKLN
Sbjct: 239 NVIPLKLGYVGVVNRSQEDINFKRSVKDALAFEEKFFSTLPAYHGLTHCCGVPQLAKKLN 298
Query: 301 KILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSM 360
IL +HI +LPGLK+RI++ LVA+AKEHA+YG+ ES AGQG +LNIL KYC+AFSSM
Sbjct: 299 TILLKHITYMLPGLKSRINSQLVAVAKEHAAYGDTAESTAGQGVKLLNILRKYCEAFSSM 358
Query: 361 LEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALF 420
+EGKNK +ST ELSGGARIHYIFQSI+V+SLE VDPC+ +TD+DIRTA+QN+ GPK +F
Sbjct: 359 VEGKNK-VSTDELSGGARIHYIFQSIFVKSLEEVDPCKSITDEDIRTAIQNSDGPKGPMF 417
Query: 421 IPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVIGN 480
+PE+PFEI VRRQISRLLDPSLQCA FIYDEL+KIS C+ +ELQ++P L+K M E + N
Sbjct: 418 LPELPFEILVRRQISRLLDPSLQCANFIYDELVKISRGCLTSELQKYPILKKRMGEAVSN 477
Query: 481 FLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSL---- 536
FLRDGL P+ETMITHIIEMEMDYINTSHPNF+GG+K +E A Q+ P + V++
Sbjct: 478 FLRDGLRPAETMITHIIEMEMDYINTSHPNFVGGNKVVELARQEILPPKAPTSVTIPKDG 537
Query: 537 DALESDKGLASERNGKSRGIIARQA-NGVVADHGVRGASDVEKVAPSGNAGGSSWRISSI 595
A+ + L S+R+ KSR I AR A G +D GV+ +D E S G + R S+
Sbjct: 538 TAISPEIQLTSDRSQKSRAIFARDATRGATSDQGVQPDADTENTGTS--VAGRNQRGHSL 595
Query: 596 FGGRDNRVYVKENTSTKPHAEPVHNVQS-SSTIHLREPPPVLRPSESNSEMEGVEITVTK 654
G +S+ VH++ + S I LREPP L+PSE+ + E+ + K
Sbjct: 596 VAG----------SSSSKSVARVHSLDNLISIIQLREPPITLKPSENQPAQDATEVAIVK 645
Query: 655 LLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAA 714
LL++SYYDI RK+IED VPKAIMHFLVN+TKRELHNV I KLYRE+L +EML+E EV
Sbjct: 646 LLIKSYYDIVRKSIEDAVPKAIMHFLVNHTKRELHNVLIRKLYRENLLDEMLRETDEVII 705
Query: 715 KRKRCRELLRAYQQAFRDLDELPLEAETVERGYGSPE-ATGLPRIRGLPS---SSMYSSS 770
+R+R +E L+ +QA R L+E LEAE VE+GY E ATGLP+I GL + S +Y+SS
Sbjct: 706 RRQRIQETLQVLEQAHRTLEEFSLEAEKVEKGYSPAEYATGLPKIHGLSNGDPSIIYASS 765
>UniRef100_Q8S053 Putative dynamin-like protein ADL2 [Oryza sativa]
Length = 786
Score = 883 bits (2282), Expect = 0.0
Identities = 482/777 (62%), Positives = 579/777 (74%), Gaps = 76/777 (9%)
Query: 8 AVTAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLV 67
+ A+ AA +GSSVI +VN+LQDIFS++G+ STI DLPQVAVVGSQSSGKSSVLE+LV
Sbjct: 13 STATAAAAAAAVGSSVIPIVNKLQDIFSQLGSSSTI-DLPQVAVVGSQSSGKSSVLEALV 71
Query: 68 GRDFLPRGTDICTRRPLVLQLVHTKPSHP------EFGEFLHLPGRKFHDFTQIRHEIQA 121
GRDFLPRG+DICTRRPLVLQLVH +P P E+GEFLHLPGR+F+DF +IR EIQA
Sbjct: 72 GRDFLPRGSDICTRRPLVLQLVH-QPRRPADAEADEWGEFLHLPGRRFYDFREIRREIQA 130
Query: 122 ETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSY 181
ETDREAG NKGVS+KQIRLKI+SPNVL+ITLVDLPGITKVPVGDQP+DIEARIRTMI+SY
Sbjct: 131 ETDREAGGNKGVSDKQIRLKIYSPNVLNITLVDLPGITKVPVGDQPTDIEARIRTMILSY 190
Query: 182 IKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGK 241
IK TC+ILAV+PAN+DL+NSDALQ+A+ ADPDG+RTIGVITKLDIMDRGTDARN LLG
Sbjct: 191 IKHKTCIILAVSPANADLSNSDALQIARNADPDGSRTIGVITKLDIMDRGTDARNFLLGN 250
Query: 242 VIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPV---YSGLADSCGVPQLAKK 298
VIPLRLGYVGVVNRSQ+ + R I + + + +R++ V Y+GLA CG+PQLAKK
Sbjct: 251 VIPLRLGYVGVVNRSQQFSRRCR-ISNQISVSRKLWRAKKVSSAYNGLAQYCGIPQLAKK 309
Query: 299 LNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFS 358
LN+IL QHIK VLPGLK+RIS+ L AKE + YG+ ESKAGQGA +LNIL+KYC+
Sbjct: 310 LNQILVQHIKTVLPGLKSRISSQLTTTAKELSFYGDPVESKAGQGAKLLNILAKYCEVI- 368
Query: 359 SMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSA 418
E+ T + VDPCED+TD+DIR A+QNATGP+SA
Sbjct: 369 --------ELFT---------------------QDVDPCEDVTDEDIRMAIQNATGPRSA 399
Query: 419 LFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTELQRFPFLRKHMDEVI 478
LF+PEVPFE+ VRRQISRLLDPSLQCA FIYDEL+K+SHRC+ ELQ+FP LR+ MDEVI
Sbjct: 400 LFVPEVPFEVLVRRQISRLLDPSLQCAGFIYDELVKMSHRCLAVELQQFPLLRRSMDEVI 459
Query: 479 GNFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDA 538
G FLRDGL+P++ MI HIIEME DYINTSHPNFIGGSKA+E A QQ + SR++ +
Sbjct: 460 GRFLRDGLKPAQDMIAHIIEMEADYINTSHPNFIGGSKAVEQAQQQVRSSRLAAVARREG 519
Query: 539 LESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGG 598
+++DK AS++ K R SG+ S W SIF
Sbjct: 520 VDADKSQASDKTQKPR---------------------------SGST--SFWGSISIFSS 550
Query: 599 -RDNRVY--VKENTSTKPH-AEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTK 654
D+R + K+N+S K + A H S STI LREPP VL+PSES SE E +EI +TK
Sbjct: 551 TSDDRTHSSAKDNSSNKSYTASTSHLEHSLSTIQLREPPVVLKPSESQSEQEALEIAITK 610
Query: 655 LLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAA 714
LLL+SYY+I RKN+ED VPKAIMHFLVN+TKRELHN I KLYR+DLF +ML+EP E+
Sbjct: 611 LLLKSYYNIVRKNVEDFVPKAIMHFLVNHTKRELHNYLITKLYRDDLFADMLREPDEITI 670
Query: 715 KRKRCRELLRAYQQAFRDLDELPLEAETVERGYG-SPEATGLPRIRGLPSSSMYSSS 770
KR++ R+ L+ QQA++ LDE+PLEA+TVERGY +ATGLPR GL SS SS
Sbjct: 671 KRRQIRDTLKVLQQAYKTLDEIPLEADTVERGYSLDADATGLPRAHGLSSSFQDGSS 727
>UniRef100_Q94CF3 Putative dynamin protein ADL2 [Arabidopsis thaliana]
Length = 480
Score = 689 bits (1779), Expect = 0.0
Identities = 348/452 (76%), Positives = 406/452 (88%), Gaps = 6/452 (1%)
Query: 10 TAASTAATPLGSSVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGR 69
++++T A PLGSSVI +VN+LQDIF+++G+QSTI LPQV VVGSQSSGKSSVLE+LVGR
Sbjct: 24 SSSTTNAAPLGSSVIPIVNKLQDIFAQLGSQSTIA-LPQVVVVGSQSSGKSSVLEALVGR 82
Query: 70 DFLPRGTDICTRRPLVLQLVHTKP-----SHPEFGEFLHLPGRKFHDFTQIRHEIQAETD 124
DFLPRG DICTRRPLVLQL+ TK S E+GEF HLP +F+DF++IR EI+AET+
Sbjct: 83 DFLPRGNDICTRRPLVLQLLQTKSRANGGSDDEWGEFRHLPETRFYDFSEIRREIEAETN 142
Query: 125 REAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKI 184
R GENKGV++ QIRLKI SPNVL+ITLVDLPGITKVPVGDQPSDIEARIRTMI+SYIK
Sbjct: 143 RLVGENKGVADTQIRLKISSPNVLNITLVDLPGITKVPVGDQPSDIEARIRTMILSYIKQ 202
Query: 185 PTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIP 244
TCLILAVTPAN+DLANSDALQ+A + DPDG+RTIGVITKLDIMD+GTDAR LLLG V+P
Sbjct: 203 DTCLILAVTPANTDLANSDALQIASIVDPDGHRTIGVITKLDIMDKGTDARKLLLGNVVP 262
Query: 245 LRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILA 304
LRLGYVGVVNR QEDI +NR++K+AL+AEE+FFRS PVY GLAD GVPQLAKKLN+IL
Sbjct: 263 LRLGYVGVVNRCQEDILLNRTVKEALLAEEKFFRSHPVYHGLADRLGVPQLAKKLNQILV 322
Query: 305 QHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGK 364
QHIK +LP LK+RIS +LVA AKEH SYGE+TES+AGQGAL+LN LSKYC+A+SS+LEGK
Sbjct: 323 QHIKVLLPDLKSRISNALVATAKEHQSYGELTESRAGQGALLLNFLSKYCEAYSSLLEGK 382
Query: 365 NKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEV 424
++EMST ELSGGARIHYIFQSI+V+SLE VDPCEDLTDDDIRTA+QNATGP+SALF+P+V
Sbjct: 383 SEEMSTSELSGGARIHYIFQSIFVKSLEEVDPCEDLTDDDIRTAIQNATGPRSALFVPDV 442
Query: 425 PFEIFVRRQISRLLDPSLQCARFIYDELMKIS 456
PFE+ VRRQISRLLDPSLQCARFI++EL+K+S
Sbjct: 443 PFEVLVRRQISRLLDPSLQCARFIFEELIKVS 474
>UniRef100_Q7S7H9 Hypothetical protein [Neurospora crassa]
Length = 706
Score = 560 bits (1444), Expect = e-158
Identities = 317/736 (43%), Positives = 457/736 (62%), Gaps = 70/736 (9%)
Query: 16 ATPLGSS---VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFL 72
+TP G S +I LVN+LQD+F+ VG + I DLPQ+ VVGSQSSGKSSVLE++VGRDFL
Sbjct: 11 STPGGISDPGLIKLVNKLQDVFTTVGVNNPI-DLPQIVVVGSQSSGKSSVLENIVGRDFL 69
Query: 73 PRGTDICTRRPLVLQLVHTKPSH-----------------PEFGEFLHLPGRKFHDFTQI 115
PRG+ I TRRPLVLQL++ + E+GEFLH+PG+KF+DF +I
Sbjct: 70 PRGSGIVTRRPLVLQLINRTATQNGFGNELDDNTDKAANTDEWGEFLHIPGQKFYDFNKI 129
Query: 116 RHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIR 175
R EI ET+ + G N G+S I L+I+SPNVL++TLVDLPG+T+VPVGDQP DIE +IR
Sbjct: 130 RDEISRETEAKVGRNAGISPAPINLRIYSPNVLNLTLVDLPGLTRVPVGDQPRDIEKQIR 189
Query: 176 TMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDAR 235
MI+ YI+ +ILAVT AN DLANSD L++A+ DP+G RTIGV+TK+D+MD GTD
Sbjct: 190 DMILKYIQKSNAIILAVTAANVDLANSDGLKLAREVDPEGQRTIGVLTKVDLMDEGTDVV 249
Query: 236 NLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQL 295
++L G++IPLRLGYV VVNR Q DI + I +L AE+ FF + Y + CG P L
Sbjct: 250 DILAGRIIPLRLGYVPVVNRGQRDIDNKKPINASLEAEKNFFENHKAYRNKSSYCGTPYL 309
Query: 296 AKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGA-LVLNILSKYC 354
A+KLN IL HIK LP +KARIS+SL +E S G S G A +VLNI++++
Sbjct: 310 ARKLNLILMMHIKQTLPDIKARISSSLQKYTQELESLG---PSILGNSANIVLNIITEFT 366
Query: 355 DAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATG 414
+ + ++L+GK+ E+S+ ELSGGARI ++F +Y ++AVDP + + D DIR L N++G
Sbjct: 367 NEWRTVLDGKSTELSSQELSGGARISFVFHELYSNGVKAVDPFDQVKDSDIRVILYNSSG 426
Query: 415 PKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTEL-QRFPFLRKH 473
P ALF+ FE+ V++QI RL +PSL+CA +YDEL++I + +L +R+P L++
Sbjct: 427 PSPALFVGTAAFELIVKQQIKRLEEPSLKCASLVYDELVRILTTVLSKQLYRRYPGLKEK 486
Query: 474 MDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPP 533
+ +V+ +F + +EP+ ++ ++ ME YINT HP+F+ G +A+ ++ SR P
Sbjct: 487 IHQVVISFFKKAMEPTNKLVKDLVAMEACYINTGHPDFLNGHRAMAIVNERHNGSR---P 543
Query: 534 VSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRIS 593
V +D + GK I + A A S++++ GGS+
Sbjct: 544 VQVD----------PKTGKP---IPQSATPARAASPTLAGSNLDE-------GGSN---- 579
Query: 594 SIFGGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVT 653
GG + +N EP PPP L+ S + SE E +E+ V
Sbjct: 580 ---GGFFGSFFAAKNKKKAAAMEP--------------PPPSLKASGTLSERENIEVEVI 622
Query: 654 KLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVA 713
KLL++SYY+I ++ + D+VPKAIM LV TK E+ +E +YR D +++L+E
Sbjct: 623 KLLIQSYYNIVKRTMIDMVPKAIMLNLVQLTKDEMQKELLENMYRADELDDLLKESDYTV 682
Query: 714 AKRKRCRELLRAYQQA 729
+RK C++++ + +A
Sbjct: 683 RRRKECQQMVESLSKA 698
>UniRef100_Q871I2 Probable VpsA protein [Neurospora crassa]
Length = 706
Score = 558 bits (1439), Expect = e-157
Identities = 316/736 (42%), Positives = 457/736 (61%), Gaps = 70/736 (9%)
Query: 16 ATPLGSS---VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFL 72
+TP G S +I LVN+LQ++F+ VG + I DLPQ+ VVGSQSSGKSSVLE++VGRDFL
Sbjct: 11 STPGGISDPGLIKLVNKLQNVFTTVGVNNPI-DLPQIVVVGSQSSGKSSVLENIVGRDFL 69
Query: 73 PRGTDICTRRPLVLQLVHTKPSH-----------------PEFGEFLHLPGRKFHDFTQI 115
PRG+ I TRRPLVLQL++ + E+GEFLH+PG+KF+DF +I
Sbjct: 70 PRGSGIVTRRPLVLQLINRTATQNGFGNELDDNTDKAANTDEWGEFLHIPGQKFYDFNKI 129
Query: 116 RHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIR 175
R EI ET+ + G N G+S I L+I+SPNVL++TLVDLPG+T+VPVGDQP DIE +IR
Sbjct: 130 RDEISRETEAKVGRNAGISPAPINLRIYSPNVLNLTLVDLPGLTRVPVGDQPRDIEKQIR 189
Query: 176 TMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDAR 235
MI+ YI+ +ILAVT AN DLANSD L++A+ DP+G RTIGV+TK+D+MD GTD
Sbjct: 190 DMILKYIQKSNAIILAVTAANVDLANSDGLKLAREVDPEGQRTIGVLTKVDLMDEGTDVV 249
Query: 236 NLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQL 295
++L G++IPLRLGYV VVNR Q DI + I +L AE+ FF + Y + CG P L
Sbjct: 250 DILAGRIIPLRLGYVPVVNRGQRDIDNKKPINASLEAEKNFFENHKAYRNKSSYCGTPYL 309
Query: 296 AKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGA-LVLNILSKYC 354
A+KLN IL HIK LP +KARIS+SL +E S G S G A +VLNI++++
Sbjct: 310 ARKLNLILMMHIKQTLPDIKARISSSLQKYTQELESLG---PSILGNSANIVLNIITEFT 366
Query: 355 DAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATG 414
+ + ++L+GK+ E+S+ ELSGGARI ++F +Y ++AVDP + + D DIR L N++G
Sbjct: 367 NEWRTVLDGKSTELSSQELSGGARISFVFHELYSNGVKAVDPFDQVKDSDIRVILYNSSG 426
Query: 415 PKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTEL-QRFPFLRKH 473
P ALF+ FE+ V++QI RL +PSL+CA +YDEL++I + +L +R+P L++
Sbjct: 427 PSPALFVGTAAFELIVKQQIKRLEEPSLKCASLVYDELVRILTTVLSKQLYRRYPGLKEK 486
Query: 474 MDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPP 533
+ +V+ +F + +EP+ ++ ++ ME YINT HP+F+ G +A+ ++ SR P
Sbjct: 487 IHQVVISFFKKAMEPTNKLVKDLVAMEACYINTGHPDFLNGHRAMAIVNERHNGSR---P 543
Query: 534 VSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRIS 593
V +D + GK I + A A S++++ GGS+
Sbjct: 544 VQVD----------PKTGKP---IPQSATPARAASPTLAGSNLDE-------GGSN---- 579
Query: 594 SIFGGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVT 653
GG + +N EP PPP L+ S + SE E +E+ V
Sbjct: 580 ---GGFFGSFFAAKNKKKAAAMEP--------------PPPSLKASGTLSERENIEVEVI 622
Query: 654 KLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVA 713
KLL++SYY+I ++ + D+VPKAIM LV TK E+ +E +YR D +++L+E
Sbjct: 623 KLLIQSYYNIVKRTMIDMVPKAIMLNLVQLTKDEMQKELLENMYRADELDDLLKESDYTV 682
Query: 714 AKRKRCRELLRAYQQA 729
+RK C++++ + +A
Sbjct: 683 RRRKECQQMVESLSKA 698
>UniRef100_UPI000023F04A UPI000023F04A UniRef100 entry
Length = 695
Score = 556 bits (1434), Expect = e-157
Identities = 308/728 (42%), Positives = 444/728 (60%), Gaps = 75/728 (10%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I+LVN+LQD+F+ VG + I DLPQ+AVVGSQSSGKSSVLE++VGRDFLPRG+ I TR
Sbjct: 15 ALITLVNKLQDVFATVGVNNPI-DLPQIAVVGSQSSGKSSVLENIVGRDFLPRGSGIVTR 73
Query: 82 RPLVLQLVHT------------------KPSHPEFGEFLHLPGRKFHDFTQIRHEIQAET 123
RPLVLQL++ + + E+GEFLH PG+KF+DF++IR EI ET
Sbjct: 74 RPLVLQLINRPAESNSASAEEIDTSNDKQANADEWGEFLHAPGQKFYDFSKIRDEISRET 133
Query: 124 DREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIK 183
+ + G+N G+S I L+I+SPNVL +TLVDLPG+TKVPVGDQP DIE +IR M++ +I
Sbjct: 134 EAKVGKNAGISPAPINLRIYSPNVLTLTLVDLPGLTKVPVGDQPRDIERQIREMVLKHIG 193
Query: 184 IPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVI 243
+ILAVT AN DLANSD L++A+ DP+G RTIGV+TK+D+MD GTD ++L +VI
Sbjct: 194 KSNAIILAVTAANQDLANSDGLKLAREVDPEGQRTIGVLTKVDLMDEGTDVIDILSNRVI 253
Query: 244 PLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKIL 303
PLRLGYV VVNR Q DI ++I AL AE+ FF + Y + CG P LA+KLN IL
Sbjct: 254 PLRLGYVPVVNRGQRDIDNRKAINQALEAEKNFFENHKAYRNKSSYCGTPYLARKLNLIL 313
Query: 304 AQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGA-LVLNILSKYCDAFSSMLE 362
HIK LP +KARIS+SL E S G S G A +VLNI++++ + + ++L+
Sbjct: 314 MMHIKQTLPDIKARISSSLQKYTAELESLG---PSMLGNSANIVLNIITEFTNEWRTVLD 370
Query: 363 GKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIP 422
G N E+S+ ELSGGARI ++F +Y ++A+DP + + D DIRT L N++G ALF+
Sbjct: 371 GNNTELSSTELSGGARISFVFHELYSNGVKAIDPFDVVKDVDIRTILYNSSGSSPALFVG 430
Query: 423 EVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTEL-QRFPFLRKHMDEVIGNF 481
FE+ V++QI RL DPSL+C +YDEL++I + + +L +R+P L++ M V+ F
Sbjct: 431 TTAFELIVKQQIKRLEDPSLKCVSLVYDELVRILSQLLAKQLYRRYPSLKEKMHGVVIAF 490
Query: 482 LRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALES 541
+ +EP+ ++ ++ ME Y+NT HP+F+ G +A+ ++ P PV +D
Sbjct: 491 FKKAMEPTNKLVRDLVSMESCYVNTGHPDFLNGHRAMAMVNERYNPK----PVQVD---- 542
Query: 542 DKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDN 601
+ GK R A+ V P G G+S S F ++
Sbjct: 543 ------PKTGKPLAGTPRAASPTV---------------PDGENAGNSGFFGSFFAAKNK 581
Query: 602 RVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYY 661
+ + PPP L+ S + SE E +E+ V KLL+ SYY
Sbjct: 582 K----------------------KAAAMEAPPPTLKASGTLSERENIEVEVIKLLISSYY 619
Query: 662 DIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRE 721
+I ++ + D+VPKA+M LV TK E+ +E +YR D +++L+E +RK C++
Sbjct: 620 NIVKRTMIDMVPKAVMLNLVQFTKDEMQRELLENMYRTDTLDDLLKESDFTIRRRKECQQ 679
Query: 722 LLRAYQQA 729
++ + +A
Sbjct: 680 MVESLGKA 687
>UniRef100_UPI000021A7EC UPI000021A7EC UniRef100 entry
Length = 698
Score = 552 bits (1423), Expect = e-155
Identities = 314/745 (42%), Positives = 457/745 (61%), Gaps = 79/745 (10%)
Query: 9 VTAASTAATPLGSS---VISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLES 65
+ A ++ +TP G S +I+LVN+LQD+F+ VG + I DLPQ+ VVGSQSSGKSSVLE+
Sbjct: 1 MAAQTSLSTPGGISDPALITLVNKLQDVFATVGVTNPI-DLPQIVVVGSQSSGKSSVLEN 59
Query: 66 LVGRDFLPRGTDICTRRPLVLQLVHTKPSHP-------------------EFGEFLHLPG 106
+VGRDFLPRG+ I TRRPLVLQL H +P+ E+GEFLH+ G
Sbjct: 60 IVGRDFLPRGSGIVTRRPLVLQL-HNRPASQSNGVNEEIAGGTDKHANADEWGEFLHITG 118
Query: 107 RKFHDFTQIRHEIQAETDREAGENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQ 166
+KF+DF +IR EI ET+ + G N G+S + I L+I+SPNVL +TLVDLPG+TKVPVGDQ
Sbjct: 119 QKFYDFNKIRDEITRETEAKVGRNAGISPQPINLRIYSPNVLTLTLVDLPGLTKVPVGDQ 178
Query: 167 PSDIEARIRTMIMSYIKIPTCLILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLD 226
P DIE +IR M+M YI P +ILAVTPANSDLANSD L+MA+ DP+G RTIGV+TK+D
Sbjct: 179 PRDIEKQIREMVMKYISKPNAIILAVTPANSDLANSDGLKMAREVDPEGQRTIGVLTKVD 238
Query: 227 IMDRGTDARNLLLGKVIPLRLGYVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGL 286
+MD GTD ++L G++IPLRLGYV VVNR Q DI + I+ AL E+ FF + Y
Sbjct: 239 LMDEGTDVVDILAGRIIPLRLGYVPVVNRGQRDIDNKKPIQAALENEKNFFDNHKAYRNK 298
Query: 287 ADSCGVPQLAKKLNKILAQHIKAVLPGLKARISTSLVALAKEHASYGEITESKAGQGA-L 345
+ CG P LA+KLN IL HIK LP +KARIS+SL + E S G S G +
Sbjct: 299 SSYCGTPYLARKLNLILMMHIKQTLPDIKARISSSLQKYSAELESLG---PSILGDSTNI 355
Query: 346 VLNILSKYCDAFSSMLEGKNKEMSTCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDI 405
VL +++ + + +L+G N E+S ELSGGARI ++F +Y ++AVDP + + D +I
Sbjct: 356 VLKVITDFSGEWKDVLDGNNTEISGQELSGGARISFVFHELYANGIKAVDPFDVVKDQEI 415
Query: 406 RTALQNATGPKSALFIPEVPFEIFVRRQISRLLDPSLQCARFIYDELMKISHRCMFTEL- 464
RT + N++GP ALF+ FE V++QI+RL +PSL+C +YDEL++I + + +L
Sbjct: 416 RTFIYNSSGPSPALFVGTGAFEQIVKKQIARLEEPSLKCISLVYDELVRILSQLLAKQLY 475
Query: 465 QRFPFLRKHMDEVIGNFLRDGLEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQ 524
+R+P L++ +V+ F + +EPS ++ ++ ME+ YINT HP+F+ G +A+ ++
Sbjct: 476 RRYPQLKEKFHQVVVAFFKKAMEPSVKLVKDLVAMEVCYINTGHPDFLNGHRAMAIVNER 535
Query: 525 TKPSRVSPPVSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGN 584
P PV+ ++ K +++ AR A+ V D G
Sbjct: 536 MNPK----PVTQVDPKTGKAISTTP--------ARTASPGVPDSG--------------- 568
Query: 585 AGGSSWRISSIFGGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSE 644
G+S S F G++ K+ + +P PPP+L+ S + SE
Sbjct: 569 -DGNSGFFGSFFAGKNK----KKAAAMEP------------------PPPMLKASGNLSE 605
Query: 645 MEGVEITVTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEE 704
E +E+ V KLL+ SY++I ++ + D+VPKAIM LV TK +H + +Y+ + ++
Sbjct: 606 REAMEVEVIKLLIASYFNIVKRTMIDMVPKAIMLNLVQYTKENMHAELLASMYKNEQLDD 665
Query: 705 MLQEPAEVAAKRKRCRELLRAYQQA 729
+L+E +RK C++++ + +A
Sbjct: 666 LLKESDYTIRRRKECQQMVESLSRA 690
>UniRef100_Q7ZXR2 MGC53884 protein [Xenopus laevis]
Length = 698
Score = 541 bits (1393), Expect = e-152
Identities = 307/731 (41%), Positives = 444/731 (59%), Gaps = 57/731 (7%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I ++N+LQD+F+ VG S +I LPQ+ VVG+QSSGKSSVLESLVGRD LPRGT + TR
Sbjct: 3 ALIPVINKLQDVFNTVG--SDVIQLPQIVVVGTQSSGKSSVLESLVGRDLLPRGTGVVTR 60
Query: 82 RPLVLQLVHTKPSHP-------------EFGEFLHLPGRKFHDFTQIRHEIQAETDREAG 128
RPL+LQLVH + E+G+FLH + F DF +IR EI++ET+R +G
Sbjct: 61 RPLILQLVHVPSDYGRKTSADENGVETNEWGKFLHTKNKIFTDFDEIRQEIESETERISG 120
Query: 129 ENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCL 188
NKG+S I LK+FSPNV+++TLVDLPG+TKVPVGDQP DIE +IR +I+ YI P C+
Sbjct: 121 NNKGISSDPIHLKVFSPNVVNLTLVDLPGMTKVPVGDQPKDIEIQIRELILRYISNPNCI 180
Query: 189 ILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLG 248
ILAVT AN+D+A S+AL++A+ +DPDG RT+ VITKLD+MD GTDA ++LLG+VIP++LG
Sbjct: 181 ILAVTAANTDMATSEALKIARESDPDGRRTLAVITKLDLMDAGTDAMDVLLGRVIPVKLG 240
Query: 249 YVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIK 308
+GVVNRSQ DI + + D++ E F + + Y LA G LA+ LN++L HI+
Sbjct: 241 IIGVVNRSQLDINNKKIVADSIRDEYGFLQKK--YPSLATRNGTKYLARTLNRLLMHHIR 298
Query: 309 AVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEM 368
LP LK RI+ SYGE E Q + +L +++K+ + + +EG+ K +
Sbjct: 299 DCLPELKTRINVLAAQYQSLLNSYGEPVED---QSSTLLQLITKFATEYCNTIEGRAKYI 355
Query: 369 STCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEI 428
T EL GGARI YIF + R+LE+VDP LT DI TA++NATGP+ ALF+PE+ FE+
Sbjct: 356 ETSELCGGARISYIFYETFGRTLESVDPLGGLTTIDILTAIRNATGPRPALFVPEISFEL 415
Query: 429 FVRRQISRLLDPSLQCARFIYDELMKISHRC---MFTELQRFPFLRKHMDEVIGNFLRDG 485
V+RQ+ RL +PSL+C +++E+ +I C EL RFP L + EV+ + LR
Sbjct: 416 LVKRQVKRLEEPSLRCVELVHEEMQRIIQHCSNYSTQELLRFPKLHDAIVEVVTSLLRKR 475
Query: 486 LEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGL 545
L + M+ +++ +E+ YINT HP+F + +++ + +R++ V A DK
Sbjct: 476 LPVTNEMVHNLVAIELAYINTKHPDFADACGVMNNNIEEQRRNRIARDVPPPAASRDKAA 535
Query: 546 ASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNRVYV 605
S G +GV +AG +WR + R V
Sbjct: 536 PSGSTG----------DGV------------------PDAGTGNWR--GMLKARGEEAPV 565
Query: 606 KENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYYDIAR 665
+E P PV ++ + L PV R S E + V + L++SY+ I R
Sbjct: 566 EERNKAPP-TVPVSPLRGHAVNLLDVSVPVAR---KLSAREQRDCEVIERLIKSYFLIVR 621
Query: 666 KNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRELLRA 725
KNI+D VPKA+MHFLVN+ K L + + +LY+ L E++L E ++A +R ++L+A
Sbjct: 622 KNIQDSVPKAVMHFLVNHVKDTLQSELVGQLYKSLLLEDLLTESEDIAQRRNEAADMLKA 681
Query: 726 YQQAFRDLDEL 736
Q+A + + E+
Sbjct: 682 LQRASQIIAEI 692
>UniRef100_UPI0000362513 UPI0000362513 UniRef100 entry
Length = 700
Score = 536 bits (1380), Expect = e-150
Identities = 311/733 (42%), Positives = 450/733 (60%), Gaps = 65/733 (8%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I +VN+LQD+F+ VG + II LPQ+AVVG+QSSGKSSVLESLVGRD LPRGT I TR
Sbjct: 3 ALIPVVNKLQDVFNTVG--ADIIQLPQIAVVGTQSSGKSSVLESLVGRDLLPRGTGIVTR 60
Query: 82 RPLVLQLVH-----------TKPSHPEFGEFLHLPGRKFHDFTQIRHEIQAETDREAGEN 130
RPL+LQLVH T E+G+FLH + F DF +IR EI+ ET+R +G N
Sbjct: 61 RPLILQLVHVDAGDARKNDDTGRQGEEWGKFLHTKNKIFTDFDEIRQEIENETERLSGNN 120
Query: 131 KGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCLIL 190
KG+S++ I LKIFSP+V+++TLVDLPGITKVPVGDQP DIE +IR +I+ +I P C+IL
Sbjct: 121 KGISDEPIHLKIFSPHVVNLTLVDLPGITKVPVGDQPKDIEVQIRDLILKHISNPNCIIL 180
Query: 191 AVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLGYV 250
AVT AN+D+A S+AL++A+ DPDG RT+ V+TKLD+MD GTDA ++L+G+VIP++LG +
Sbjct: 181 AVTAANTDMATSEALKVAREVDPDGRRTLAVVTKLDLMDAGTDAMDVLMGRVIPVKLGLI 240
Query: 251 GVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIKAV 310
GVVNRSQ DI +S+ D++ E F + + Y LA+ G LAK LN++L HI+
Sbjct: 241 GVVNRSQLDINNRKSVADSIRDEYVFLQKK--YPSLANRNGTKYLAKTLNRLLMHHIRDC 298
Query: 311 LPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEMST 370
LP LK RI+ +SYGE E Q A +L +++K+ + + +EG K + T
Sbjct: 299 LPELKTRINVLAAQYQSLLSSYGEPVED---QSATLLQLITKFASEYCNTIEGTAKYIET 355
Query: 371 CELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEIFV 430
EL GGARI YIF + R+LE+VDP L+ DI TA++NATGP+ +LF+PEV FE+ V
Sbjct: 356 AELCGGARICYIFHETFGRTLESVDPLGGLSTIDILTAIRNATGPRPSLFVPEVSFELLV 415
Query: 431 RRQISRLLDPSLQCARFIYDELMKISHRC---MFTELQRFPFLRKHMDEVIGNFLRDGLE 487
++Q+ RL +PSL+C +++E+ +I C ELQRFP L + + EV+ + LR L
Sbjct: 416 KKQVKRLEEPSLRCVELVHEEMQRIIQHCSNYSTQELQRFPKLHEAIVEVVTSLLRKRLP 475
Query: 488 PSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSR---VSPPVSLDALESDKG 544
+ M+ +++ +E+ YINT HP+F L +++ + +R + V D ++ G
Sbjct: 476 VTNEMVHNLVAIELAYINTKHPDFADACGVLNNNIEEQRRNRMRELPAAVPRDKVKEAAG 535
Query: 545 LA-------------SERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWR 591
LA ++ +G G A A G+ A G +G D G +WR
Sbjct: 536 LAAPTVVSGDPTASGADMDGAKVGTYA--ACGLRAGAGPQGEQD----------GAGTWR 583
Query: 592 ISSIFGGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEIT 651
+ G D P + P ++ + + L P PV R S + +
Sbjct: 584 -GMLKKGED-----------APGSGPGSPLKGAINL-LDVPVPVARKLSSREQR---DCE 627
Query: 652 VTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAE 711
V + L++SY+ I RKNI+D VPKA+MHFLVN+ K L + + +LY+ L ++L E +
Sbjct: 628 VIERLIKSYFLIVRKNIQDSVPKAVMHFLVNHVKDSLQSELVGQLYKSGLLNDLLTESED 687
Query: 712 VAAKRKRCRELLR 724
+A +RK ++L+
Sbjct: 688 MAQRRKESADMLQ 700
>UniRef100_Q7ZWZ9 Dnm1l-prov protein [Xenopus laevis]
Length = 698
Score = 535 bits (1378), Expect = e-150
Identities = 307/731 (41%), Positives = 440/731 (59%), Gaps = 57/731 (7%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I ++N+LQD+F+ VG S +I LPQ+ VVG+QSSGKSSVLESLVGRD LPRGT + TR
Sbjct: 3 ALIPVINKLQDVFNTVG--SDVIQLPQIVVVGTQSSGKSSVLESLVGRDLLPRGTGVVTR 60
Query: 82 RPLVLQLVHTKPSH-------------PEFGEFLHLPGRKFHDFTQIRHEIQAETDREAG 128
RPL+LQLVH E+G+FLH + + DF +IR EI+ ET+R +G
Sbjct: 61 RPLILQLVHVSSDDRRKTSGDENGVEADEWGKFLHTKNKIYTDFDEIRQEIENETERISG 120
Query: 129 ENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCL 188
NKG+S + I LKIFSPNV+++TLVDLPG+TKVPVGDQP DIE +IR +I+ YI P +
Sbjct: 121 NNKGISSEPIHLKIFSPNVVNLTLVDLPGMTKVPVGDQPKDIEIQIRELILRYISNPNSI 180
Query: 189 ILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLG 248
ILAVT AN+D+A S+AL++A+ +DPDG RT+ VITKLD+MD GTDA ++LLG+VIP++LG
Sbjct: 181 ILAVTAANTDMATSEALKIARESDPDGRRTLAVITKLDLMDAGTDAMDVLLGRVIPVKLG 240
Query: 249 YVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIK 308
+GVVNRSQ DI + + D++ E F + + Y LA G LA+ LN++L HI+
Sbjct: 241 IIGVVNRSQLDINNKKIVADSIRDEYGFLQKK--YPSLATRNGTKYLARTLNRLLMHHIR 298
Query: 309 AVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEM 368
LP LK RI+ SYGE E K+ A +L +++K+ + + +EG K +
Sbjct: 299 DCLPELKTRINVLAAQYQTLLNSYGEPVEDKS---ATLLQLITKFATEYCNTIEGTAKYI 355
Query: 369 STCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEI 428
T EL GGARI YIF + R+LE+VDP LT D+ TA++NATGP+ ALF+PEV FE+
Sbjct: 356 ETSELCGGARICYIFHETFGRTLESVDPLGGLTTIDVLTAIRNATGPRPALFVPEVSFEL 415
Query: 429 FVRRQISRLLDPSLQCARFIYDELMKISHRC---MFTELQRFPFLRKHMDEVIGNFLRDG 485
V+RQ+ RL +PSL+C +++E+ +I C EL RFP L + EV+ + LR
Sbjct: 416 LVKRQVKRLEEPSLRCVELVHEEMQRIIQHCSNYSTQELLRFPKLHDAIVEVVTSLLRKR 475
Query: 486 LEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGL 545
L + M+ +++ +E+ YINT HP+F + +++ + +R+S + A DK
Sbjct: 476 LPVTNEMVHNLVAIELAYINTKHPDFADACGLINNNIEEQRRNRISRDLPPPAASRDKAA 535
Query: 546 ASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSSWRISSIFGGRDNRVYV 605
G G D AG +WR G + V
Sbjct: 536 PGSSTGD-------------------GLPD---------AGTGNWRGMMKAKGEEASV-- 565
Query: 606 KENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVEITVTKLLLRSYYDIAR 665
E + P A P ++ + L P PV R S E + V + L++SY+ I R
Sbjct: 566 -EEKNKAPPAFPASPLRGHAVNLLDVPVPVAR---KLSAREQRDCEVIERLIKSYFLIVR 621
Query: 666 KNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEPAEVAAKRKRCRELLRA 725
KNI+D VPKA+MHFLVN+ K L + + +LY+ L E++L E ++A +R ++L+A
Sbjct: 622 KNIQDSVPKAVMHFLVNHVKDTLQSELVGQLYKSLLLEDLLTESEDMAQRRNEAADMLKA 681
Query: 726 YQQAFRDLDEL 736
Q+A + + E+
Sbjct: 682 LQRASQIIAEI 692
>UniRef100_UPI00003A9933 UPI00003A9933 UniRef100 entry
Length = 737
Score = 532 bits (1371), Expect = e-149
Identities = 311/747 (41%), Positives = 455/747 (60%), Gaps = 50/747 (6%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I ++N+LQD+F+ VG + II LPQ+ VVG+QSSGKSSVLESLVGRD LPRGT + TR
Sbjct: 3 ALIPVINKLQDVFNTVG--ADIIQLPQIVVVGTQSSGKSSVLESLVGRDLLPRGTGVVTR 60
Query: 82 RPLVLQLVHTKPSH-------------PEFGEFLHLPGRKFHDFTQIRHEIQAETDREAG 128
RPL+LQLVH P E+G+FLH + + DF +IR EI+ ET+R +G
Sbjct: 61 RPLILQLVHVSPEDGRKTAGDENEIDAEEWGKFLHTKNKIYTDFDEIRQEIENETERISG 120
Query: 129 ENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCL 188
NKG+S + I LKIFS NV+++TLVDLPG+TKVPVGDQP DIE +IR +I+ +I P +
Sbjct: 121 NNKGISPEPIHLKIFSSNVVNLTLVDLPGMTKVPVGDQPKDIELQIRELILQFISNPNSI 180
Query: 189 ILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLG 248
ILAVT AN+D+A S+AL++A+ DPDG RT+ VITKLD+MD GTDA ++L+G+VIP++LG
Sbjct: 181 ILAVTAANTDMATSEALKIAREVDPDGRRTLAVITKLDLMDAGTDAMDVLMGRVIPVKLG 240
Query: 249 YVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIK 308
+GVVNRSQ DI +S+ D++ E F + + Y LA+ G LA+ LN++L HI+
Sbjct: 241 IIGVVNRSQLDINNKKSVADSIRDEYGFLQKK--YPSLANRNGTKYLARTLNRLLMHHIR 298
Query: 309 AVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEM 368
LP LK RI+ SYGE E K+ A +L +++K+ + + +EG K +
Sbjct: 299 DCLPELKTRINVLAAQYQSLLNSYGEPVEDKS---ATLLQLITKFATEYCNTIEGTAKYI 355
Query: 369 STCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEI 428
T EL GGARI YIF + R+LE+VDP L DI TA++NATGP+ ALF+PEV FE+
Sbjct: 356 ETSELCGGARICYIFHETFGRTLESVDPLGGLNTIDILTAIRNATGPRPALFVPEVSFEL 415
Query: 429 FVRRQISRLLDPSLQCARFIYDELMKISHRC---MFTELQRFPFLRKHMDEVIGNFLRDG 485
V+RQI RL +PSL+C +++E+ +I C EL RFP L + EV+ LR
Sbjct: 416 LVKRQIKRLEEPSLRCVELVHEEMQRIIQHCSNYSTQELLRFPKLHDAIVEVVTCLLRRR 475
Query: 486 LEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSP--PVSLDALESDK 543
L + M+ +++ +E+ YINT HP+F + +++ + +R++ P S+ +S K
Sbjct: 476 LPVTNEMVHNLVAIELAYINTKHPDFADACGLMNNNIEEQRRNRLARELPSSVPRDKSTK 535
Query: 544 GLASERNGKSRGIIARQA-------------NGVVADHGVRGASDVEKVAPSGNAGGSSW 590
+ + A A + V+ GA DV + + +GN W
Sbjct: 536 APGALPPASQETVTAASAEADGKVFLTLTFFSDVLFVQAAAGAGDVTQESGTGN-----W 590
Query: 591 RISSIFGGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLRE-PPPVLRPSESNSEMEGVE 649
R + + V +E +KP A + Q ++L + P PV R S E +
Sbjct: 591 R-GMLKSSKAEEVSAEE--KSKPAAALPASPQKGHAVNLLDVPVPVAR---KLSAREQRD 644
Query: 650 ITVTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEP 709
V + L++SY+ I RKNI+D VPKA+MHFLVN+ K L + + +LY+ L +++L E
Sbjct: 645 CEVIERLIKSYFLIVRKNIQDSVPKAVMHFLVNHVKDTLQSELVGQLYKSLLLDDLLTES 704
Query: 710 AEVAAKRKRCRELLRAYQQAFRDLDEL 736
++A +RK ++L+A Q+A + + E+
Sbjct: 705 EDMAQRRKEAADMLKALQRASQIIAEI 731
>UniRef100_O14541 Dnm1p/Vps1p-like protein [Homo sapiens]
Length = 736
Score = 532 bits (1370), Expect = e-149
Identities = 311/747 (41%), Positives = 451/747 (59%), Gaps = 51/747 (6%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I ++N+LQD+F+ VG + II LPQ+ VVG+QSSGKSSVLESLVGRD LPRGT I TR
Sbjct: 3 ALIPVINKLQDVFNTVG--ADIIQLPQIVVVGTQSSGKSSVLESLVGRDLLPRGTGIVTR 60
Query: 82 RPLVLQLVHTKPSHP-------------EFGEFLHLPGRKFHDFTQIRHEIQAETDREAG 128
RPL+LQLVH E+G+FLH + + DF +IR EI+ ET+R +G
Sbjct: 61 RPLILQLVHVSQEDKRKTTGEENGVEAEEWGKFLHTKNKLYTDFDEIRQEIENETERISG 120
Query: 129 ENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCL 188
NKGVS + I LKIFSPNV+++TLVDLPG+TKVPVGDQP DIE +IR +I+ +I P +
Sbjct: 121 NNKGVSPEPIHLKIFSPNVVNLTLVDLPGMTKVPVGDQPKDIELQIRELILRFISNPNSI 180
Query: 189 ILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLG 248
ILAVT AN+D+A S+AL++++ DPDG RT+ VITKLD+MD GTDA ++L+G+VIP++LG
Sbjct: 181 ILAVTAANTDMATSEALKISREVDPDGRRTLAVITKLDLMDAGTDAMDVLMGRVIPVKLG 240
Query: 249 YVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIK 308
+GVVNRSQ DI +S+ D++ E F + + Y LA+ G LA+ LN++L HI+
Sbjct: 241 IIGVVNRSQLDINNKKSVTDSIRDEYAFLQKK--YPSLANRNGTKYLARTLNRLLMHHIR 298
Query: 309 AVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEM 368
LP LK RI+ SYGE + K+ A +L +++K+ + + +EG K +
Sbjct: 299 DCLPELKTRINVLAAQYQSLLNSYGEPVDDKS---ATLLQLITKFATEYCNTIEGTAKYI 355
Query: 369 STCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEI 428
T EL GGARI YIF + R+LE+VDP L DI TA++NATGP+ ALF+PEV FE+
Sbjct: 356 ETSELCGGARICYIFHETFGRTLESVDPLGGLNTIDILTAIRNATGPRPALFVPEVSFEL 415
Query: 429 FVRRQISRLLDPSLQCARFIYDELMKISHRC---MFTELQRFPFLRKHMDEVIGNFLRDG 485
V+RQI RL +PSL+C +++E+ +I C EL RFP L + EV+ LR
Sbjct: 416 LVKRQIKRLEEPSLRCVELVHEEMQRIIQHCSNYSTQELLRFPKLHDAIVEVVTCLLRKR 475
Query: 486 LEPSETMITHIIEMEMDYINTSHPNFIGG----------------SKALEAAVQQTKPSR 529
L + M+ +++ +E+ YINT HP+F ++ L +AV + K S+
Sbjct: 476 LPVTNEMVHNLVAIELAYINTKHPDFADACGLMNNNIEEQRRNRLARELPSAVSRDKSSK 535
Query: 530 VSPPVSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSS 589
V ++ + E ++E +GK R+ V + G G D + +GN
Sbjct: 536 VPSALAPASQEPSPAASAEADGKLIQDSRRETKNVASGGG--GVGDGVQEPTTGN----- 588
Query: 590 WRISSIFGGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVE 649
WR + + + +E + P P + + L P PV R S E +
Sbjct: 589 WR-GMLKTSKAEELLAEEKSKPIP-IMPASPQKGHAVNLLDVPVPVAR---KLSAREQRD 643
Query: 650 ITVTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEP 709
V + L++SY+ I RKNI+D VPKA+MHFLVN+ K L + + +LY+ L +++L E
Sbjct: 644 CEVIERLIKSYFLIVRKNIQDSVPKAVMHFLVNHVKDTLQSELVGQLYKSSLLDDLLTES 703
Query: 710 AEVAAKRKRCRELLRAYQQAFRDLDEL 736
++A +RK ++L+A Q A + + E+
Sbjct: 704 EDMAQRRKEAADMLKALQGASQIIAEI 730
>UniRef100_O00429 Dynamin-like protein [Homo sapiens]
Length = 736
Score = 531 bits (1369), Expect = e-149
Identities = 311/747 (41%), Positives = 451/747 (59%), Gaps = 51/747 (6%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I ++N+LQD+F+ VG + II LPQ+ VVG+QSSGKSSVLESLVGRD LPRGT I TR
Sbjct: 3 ALIPVINKLQDVFNTVG--ADIIQLPQIVVVGTQSSGKSSVLESLVGRDLLPRGTGIVTR 60
Query: 82 RPLVLQLVHTKPSHP-------------EFGEFLHLPGRKFHDFTQIRHEIQAETDREAG 128
RPL+LQLVH E+G+FLH + + DF +IR EI+ ET+R +G
Sbjct: 61 RPLILQLVHVTQEDKRKTTGEENGVEAEEWGKFLHTKNKLYTDFDEIRQEIENETERISG 120
Query: 129 ENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCL 188
NKGVS + I LKIFSPNV+++TLVDLPG+TKVPVGDQP DIE +IR +I+ +I P +
Sbjct: 121 NNKGVSPEPIHLKIFSPNVVNLTLVDLPGMTKVPVGDQPKDIELQIRELILRFISNPNSI 180
Query: 189 ILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLG 248
ILAVT AN+D+A S+AL++++ DPDG RT+ VITKLD+MD GTDA ++L+G+VIP++LG
Sbjct: 181 ILAVTAANTDMATSEALKISREVDPDGRRTLAVITKLDLMDAGTDAMDVLMGRVIPVKLG 240
Query: 249 YVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIK 308
+GVVNRSQ DI +S+ D++ E F + + Y LA+ G LA+ LN++L HI+
Sbjct: 241 IIGVVNRSQLDINNKKSVTDSIRDEYAFLQKK--YPSLANRNGTKYLARTLNRLLMHHIR 298
Query: 309 AVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEM 368
LP LK RI+ SYGE + K+ A +L +++K+ + + +EG K +
Sbjct: 299 DCLPELKTRINVLAAQYQSLLNSYGEPVDDKS---ATLLQLITKFATEYCNTIEGTAKYI 355
Query: 369 STCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEI 428
T EL GGARI YIF + R+LE+VDP L DI TA++NATGP+ ALF+PEV FE+
Sbjct: 356 ETSELCGGARICYIFHETFGRTLESVDPLGGLNTIDILTAIRNATGPRPALFVPEVSFEL 415
Query: 429 FVRRQISRLLDPSLQCARFIYDELMKISHRC---MFTELQRFPFLRKHMDEVIGNFLRDG 485
V+RQI RL +PSL+C +++E+ +I C EL RFP L + EV+ LR
Sbjct: 416 LVKRQIKRLEEPSLRCVELVHEEMQRIIQHCSNYSTQELLRFPKLHDAIVEVVTCLLRKR 475
Query: 486 LEPSETMITHIIEMEMDYINTSHPNFIGG----------------SKALEAAVQQTKPSR 529
L + M+ +++ +E+ YINT HP+F ++ L +AV + K S+
Sbjct: 476 LPVTNEMVHNLVAIELAYINTKHPDFADACGLMNNNIEEQRRNRLARELPSAVSRDKSSK 535
Query: 530 VSPPVSLDALESDKGLASERNGKSRGIIARQANGVVADHGVRGASDVEKVAPSGNAGGSS 589
V ++ + E ++E +GK R+ V + G G D + +GN
Sbjct: 536 VPSALAPASQEPSPAASAEADGKLIQDSRRETKNVASGGG--GVGDGVQEPTTGN----- 588
Query: 590 WRISSIFGGRDNRVYVKENTSTKPHAEPVHNVQSSSTIHLREPPPVLRPSESNSEMEGVE 649
WR + + + +E + P P + + L P PV R S E +
Sbjct: 589 WR-GMLKTSKAEELLAEEKSKPIP-IMPASPQKGHAVNLLDVPVPVAR---KLSAREQRD 643
Query: 650 ITVTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDLFEEMLQEP 709
V + L++SY+ I RKNI+D VPKA+MHFLVN+ K L + + +LY+ L +++L E
Sbjct: 644 CEVIERLIKSYFLIVRKNIQDSVPKAVMHFLVNHVKDTLQSELVGQLYKSSLLDDLLTES 703
Query: 710 AEVAAKRKRCRELLRAYQQAFRDLDEL 736
++A +RK ++L+A Q A + + E+
Sbjct: 704 EDMAQRRKEAADMLKALQGASQIIAEI 730
>UniRef100_UPI00003BFD07 UPI00003BFD07 UniRef100 entry
Length = 739
Score = 531 bits (1368), Expect = e-149
Identities = 307/755 (40%), Positives = 463/755 (60%), Gaps = 64/755 (8%)
Query: 22 SVISLVNRLQDIFSRVGNQSTIIDLPQVAVVGSQSSGKSSVLESLVGRDFLPRGTDICTR 81
++I ++N+LQD+F+ VG + I LPQ+ V+G+QSSGKSSV+ESLVGR FLPRGT I TR
Sbjct: 3 ALIPVINKLQDVFNTVGADA--IQLPQIVVLGTQSSGKSSVIESLVGRSFLPRGTGIVTR 60
Query: 82 RPLVLQLVHTKPSH-------------PEFGEFLHLPGRKFHDFTQIRHEIQAETDREAG 128
RPL+LQLV+T E+G FLH + + DF++IR EI++ET+R AG
Sbjct: 61 RPLILQLVYTPKDDREHRSAENGTLDLDEWGTFLHTKNKIYIDFSEIRKEIESETERMAG 120
Query: 129 ENKGVSEKQIRLKIFSPNVLDITLVDLPGITKVPVGDQPSDIEARIRTMIMSYIKIPTCL 188
NKG+ + I LKI+S +V+++TL+DLPGITKVPVGDQP DIE++IR +++ YI P +
Sbjct: 121 SNKGICPEPINLKIYSTSVVNLTLIDLPGITKVPVGDQPEDIESQIRQLVLKYICNPNSI 180
Query: 189 ILAVTPANSDLANSDALQMAQVADPDGNRTIGVITKLDIMDRGTDARNLLLGKVIPLRLG 248
ILAV AN+D+A S++L++++ DPDG RT+ V+TKLD+MD GTDA ++L G+VIP++LG
Sbjct: 181 ILAVVTANTDMATSESLKLSKDVDPDGRRTLAVVTKLDLMDAGTDAIDILCGRVIPVKLG 240
Query: 249 YVGVVNRSQEDIQMNRSIKDALVAEEEFFRSRPVYSGLADSCGVPQLAKKLNKILAQHIK 308
+GVVNRSQ+DI N+SI+DAL E F + + Y LA+ G P LAK LN++L HI+
Sbjct: 241 IIGVVNRSQQDIMNNKSIQDALKDEAAFLQRK--YPTLANRNGTPYLAKTLNRLLMHHIR 298
Query: 309 AVLPGLKARISTSLVALAKEHASYGEITESKAGQGALVLNILSKYCDAFSSMLEGKNKEM 368
LP LK R++ + SYGE K+ +L I++K+ ++ S +EG + +
Sbjct: 299 DCLPELKTRVNVMVSQFQTLLNSYGEDVSDKS---QTLLQIITKFASSYCSTIEGTARNI 355
Query: 369 STCELSGGARIHYIFQSIYVRSLEAVDPCEDLTDDDIRTALQNATGPKSALFIPEVPFEI 428
T EL GGARI YIF + ++L+++ P LT DI TA++NATGP+ ALF+PEV FE+
Sbjct: 356 ETTELCGGARICYIFHETFGKTLDSIHPLAGLTKMDILTAIRNATGPRPALFVPEVSFEL 415
Query: 429 FVRRQISRLLDPSLQCARFIYDELMKISHRC---MFTELQRFPFLRKHMDEVIGNFLRDG 485
V+RQI RL +PSL+C +++E+ +I C + E+ RFP L + + +V+ LR
Sbjct: 416 LVKRQIRRLEEPSLRCVELVHEEMQRIIQHCGTEVQQEMLRFPKLHERIVDVVTQLLRRR 475
Query: 486 LEPSETMITHIIEMEMDYINTSHPNFIGGSKALEAAVQQTKPSRVSPPVSLDALESDKGL 545
L P+ M+ +++ +E+ YINT HP+F + + + ++ + ++D ++ ++ +
Sbjct: 476 LPPTNQMVENLVAIELAYINTKHPDFHKDAALVSSLLKNS---------NVDHIKHNRKV 526
Query: 546 ASERNGKSRGIIARQANGVVADHGVRGASDVEKVA--PSGNAGG------SSWRISSIF- 596
S N S I + +AN V+ ++ E+V P N S+W ++++
Sbjct: 527 PSTTNTTSSNITSNEAN-------VKSINNQEEVTSLPEQNKDQGHQMVLSNWLLNNLIP 579
Query: 597 GGR-------DNRVYVKENTSTKPHAEPVHN----VQSSST----IHLREPPPVLRPSES 641
GR DN +ST PH P N VQ +S+ ++L PV + S
Sbjct: 580 QGRTDSTSSIDNSPVRNRESSTSPHPNPSSNPIIEVQKASSQQKPVNLLPEVPV-QTSRK 638
Query: 642 NSEMEGVEITVTKLLLRSYYDIARKNIEDLVPKAIMHFLVNNTKRELHNVFIEKLYREDL 701
SE E + V + L++SY+ I RK+I+D VPKA+MHFLVN K L + + LY+
Sbjct: 639 LSEREQRDCDVIERLIKSYFYIVRKSIQDSVPKAVMHFLVNYVKDNLQSELVTHLYKSHH 698
Query: 702 FEEMLQEPAEVAAKRKRCRELLRAYQQAFRDLDEL 736
E +L E VA +RK ++L+A QA + E+
Sbjct: 699 AEVLLNESEHVAVRRKEAADMLKALTQAGHIISEI 733
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,221,070,447
Number of Sequences: 2790947
Number of extensions: 51615141
Number of successful extensions: 146585
Number of sequences better than 10.0: 496
Number of HSP's better than 10.0 without gapping: 455
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 144568
Number of HSP's gapped (non-prelim): 800
length of query: 773
length of database: 848,049,833
effective HSP length: 135
effective length of query: 638
effective length of database: 471,271,988
effective search space: 300671528344
effective search space used: 300671528344
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Lotus: description of TM0100c.1


